Abstract
PURPOSE
Allogeneic hematopoietic cell transplantation (HCT) in patients with myelodysplastic syndrome (MDS) improves overall survival (OS). We evaluated the impact of MDS genetics on the benefit of HCT in a biological assignment (donor v no donor) study.
METHODS
We performed targeted sequencing in 309 patients age 50-75 years with International Prognostic Scoring System (IPSS) intermediate-2 or high-risk MDS, enrolled in the Blood and Marrow Transplant Clinical Trials Network 1102 study and assessed the association of gene mutations with OS. Patients with TP53 mutations were classified as TP53multihit if two alleles were altered (via point mutation, deletion, or copy-neutral loss of heterozygosity).
RESULTS
The distribution of gene mutations was similar in the donor and no donor arms, with TP53 (28% v 29%; P = .89), ASXL1 (23% v 29%; P = .37), and SRSF2 (16% v 16%; P = .99) being most common. OS in patients with a TP53 mutation was worse compared with patients without TP53 mutation (21% ± 5% [SE] v 52% ± 4% at 3 years; P < .001). Among those with a TP53 mutation, OS was similar between TP53single versus TP53multihit (22% ± 8% v 20% ± 6% at 3 years; P = .31). Considering HCT as a time-dependent covariate, patients with a TP53 mutation who underwent HCT had improved OS compared with non-HCT treatment (OS at 3 years: 23% ± 7% v 11% ± 7%; P = .04), associated with a hazard ratio of 3.89; 95% CI, 1.87 to 8.12; P < .001 after adjustment for covariates. OS among patients with molecular IPSS (IPSS-M) very high risk without a TP53 mutation was significantly improved if they had a donor (68% ± 10% v 0% ± 12% at 3 years; P = .001).
CONCLUSION
HCT improved OS compared with non-HCT treatment in patients with TP53 mutations irrespective of TP53 allelic status. Patients with IPSS-M very high risk without a TP53 mutation had favorable outcomes when a donor was available.
INTRODUCTION
Allogeneic hematopoietic cell transplantation (HCT) is the only curative treatment for patients with myelodysplastic syndrome (MDS). Two biological assignment studies demonstrated improved overall survival (OS) for older patients with high-risk MDS and an available donor compared with those without a donor.1,2 Although the survival benefit of HCT was observed across different clinical parameters, these studies did not assess somatic or germline gene mutations, which have been shown to predict outcomes after allogeneic HCT in retrospective cohorts.3-7
CONTEXT
Key Objective
To determine whether the improved survival of allogeneic hematopoietic cell transplantation (HCT) in a high-risk myelodysplastic syndrome (MDS) biological assignment trial of HCT was independent of gene mutations.
Knowledge Generated
Overall survival was significantly improved by HCT in high-risk genetic subgroups including TP53-mutated MDS and International Prognostic Scoring System-Molecular very high risk. The improvement by HCT was independent of baseline clinical or genetic characteristics including TP53 mutational clearance.
Relevance (C.F. Craddock)
-
This pivotal prospective study demonstrates that allogeneic HCT represents an important, potentially curative, therapeutic strategy in patients with high-risk genetic sub-groups of MDS, including patients with TP53 mutated MDS.*
*Relevance section written by JCO Associate Editor Charles F. Craddock, MD.
Mutations in TP53 are unequivocally associated with dismal outcomes after HCT because of high rates of disease relapse or progression to AML.3-5 Consequently, the role of HCT for patients with TP53-mutated MDS or AML is debated.8 Retrospective analyses have evaluated the potential impact of disease-, patient-, and transplant-related variables, but results are conflicting, and conclusions are fundamentally limited by the lack of a comparative non-HCT group.3,7,9,10
We performed a genetic analysis of the Blood and Marrow Transplant Clinical Trials Network (BMT CTN) 1102 study of older patients with advanced MDS to identify whether the survival benefit observed in patients biologically assigned to HCT compared with non-HCT approaches was independent of gene mutations. We specifically focused on mutations associated with outcome in this high-risk MDS cohort, including TP53.
METHODS
Clinical Cohort
Samples were obtained from the BMT CTN 1102 study (ClinicalTrials.gov identifier: NCT02016781), a multicenter trial comparing reduced intensity conditioning (RIC) HCT with hypomethylating therapy or best supportive care in patients age 50-75 years with International Prognostic Scoring System (IPSS) intermediate-2 or high-risk de novo MDS.1 Frozen whole blood collected at the time of enrollment was available from 309 of 384 enrolled patients in the Center for International Blood and Marrow Transplant Research (CIBMTR) Research Sample Repository or the NMDP Repository (Fig 1). Sample availability was higher in patients assigned to the donor arm compared with the no donor arm (n = 229, 88.1% v n = 80, 64.5%; P < .001). Patient characteristics and clinical outcomes were aligned with those previously reported for this trial. Baseline characteristics were not significantly different between patients with an available sample compared with those without (Data Supplement [Table S1], online only) and, among patients with samples, were similar in the donor and no donor group. The median follow-up in survivors was 32 months (range, 6-38). All patients provided written informed consent to participate in both the BMT CTN 1102 trial and the CIBMTR research database. This study was approved by the BMT CTN and CIBMTR and conducted with approval of the Dana-Farber Cancer Institute institutional review board.
FIG 1.
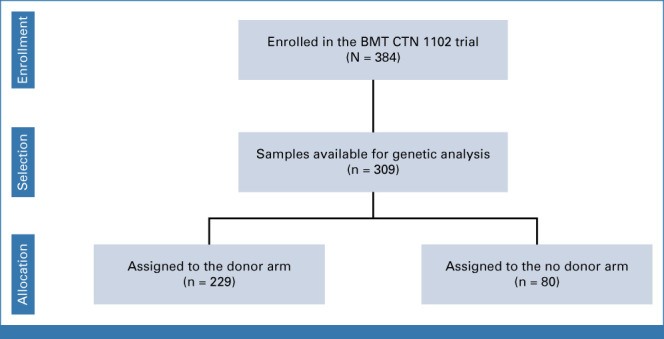
Patients included in this study. BMT CTN, Blood and Marrow Transplant Clinical Trials Network.
Genetic Analysis
Targeted DNA sequencing on samples at the time of enrollment was performed on 113 genes known to be recurrently mutated in myeloid malignancies or associated with germline predisposition to develop myeloid malignancies (Data Supplement [Table S2]), using a variant allele fraction (VAF) cutoff of 0.02 (Custom SureSelect, Agilent Technologies, Santa Clara, CA). TP53 mutation allele abundance was quantified just before transplantation in DNA extracted from preconditioning blood samples using a custom targeted error-corrected DNA sequencing panel covering the entire coding sequence of the TP53 gene (VariantPlex, ArcherDx, Boulder, CO).11 Detailed sequencing information is provided in the Data Supplement (Appendix). MDS with TP53 mutations was further categorized on the basis of the number of TP53 mutations (single-nucleotide variants or small indels) and the presence of TP53 deletion or copy-neutral loss of heterozygosity (CN-LOH). Those with ≥2 TP53 mutations or ≥1 point mutation in combination with TP53 CN-LOH, TP53 deletion, or chromosome 17/17p deletion by karyotype were classified as TP53multihit, whereas those with a single TP53 point mutation without LOH were classified as TP53single.12,13 FLT3 internal tandem duplications and KMT2A partial tandem duplications were identified as described.14,15 The genetic analysis was locked before merging with clinical data.
Statistical Analysis
To identify mutations associated with OS in the whole study cohort, we evaluated 17 genes that were mutated in ≥10 patients in the study cohort (Data Supplement [Fig S1 and Table S3]). Genes that were mutated less frequently were subject to descriptive analysis. OS was considered as the time from consent until death from any cause or until censoring at the date of last contact being alive. OS curves were estimated using the Kaplan-Meier method, and cumulative incidences of relapse or progression to leukemia were estimated with the Aalen-Johansen method, with death without relapse or disease progression being treated as competing events. Outcomes were compared in univariate analysis of survival and competing risks using log-rank and Gray's test, respectively. Comparisons between the two groups were performed using the Mann-Whitney U test for continuous variables, whereas the Fisher's exact test was used for categorical variables.
The impact of allogeneic HCT was assessed using two methods: (1) a time-dependent analysis allowing the HCT covariate to change at the time of HCT, where OS curves were shown using Simon-Makuch plots16 and (2) a dynamic landmarking analysis at 3, 6, and 9 months from consent by treatment arm in which patients were assigned to no HCT group if they were not transplanted at the landmark time.17,18
Multivariable analysis was performed using a Cox proportional hazards model with adjustment for prespecified variables, which included age at enrollment (older than 65 v 65 years and younger), performance status (Karnofsky <90 v 90-100 or Eastern Cooperative Oncology Group 1 v 0), IPSS risk status (high v intermediate-2), and MDS disease duration (≥3 v <3 months). Stepwise selection of variables with an univariate of P < .2 for OS was used to generate a multivariable model integrating the remaining clinical and genetic features, with P < .1 as the threshold for variable inclusion in the model.
RESULTS
Genetic Characteristics
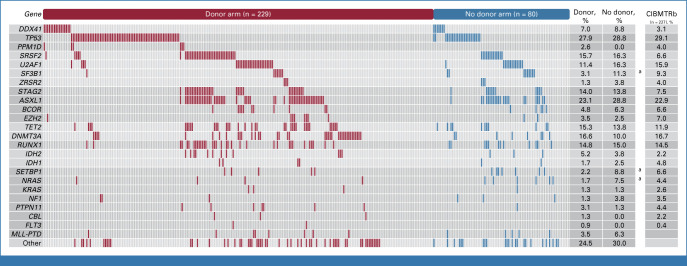
We identified ≥1 mutation in 272 of 309 (88%) patients. The overall distribution of somatic gene mutations was similar in the donor and no donor arms, with TP53 (28% v 29%; P = .89), ASXL1 (23% v 29%; P = .37), SRSF2 (16% v 16%; P = .99), and DNMT3A (17% v 10%; P = .20) being the most common (Fig 2). Inferred germline mutations in DDX41 were found in 7% (n = 23) of patients, and rare variants affecting core telomerase components TERT (n = 9) or TERC (n = 1) were observed in 3% of patients, consistent with a recent report.6 The frequency and distribution of gene mutations in the BMT CTN 1102 cohort were similar to those from a retrospective registry-level MDS transplant cohort (n = 227) matched for age, IPSS risk group, and primary versus therapy-related MDS status (Fig 2).3
FIG 2.
Spectrum of myeloid driver mutations in the BMT CTN 1102 study. A comutation plot shows mutations in individual genes per study arm as labeled on the top. Mutations are depicted by colored bars and each column represents one of the 309 patients. aSignificant with P < .05 (Fisher's exact test). bSelection of patients with de novo MDS with IPSS intermediate-2 or high risk age 50-75 years from a matched retrospective cohort.3 BMT CTN, Blood and Marrow Transplant Clinical Trials Network; IPSS, International Prognostic Scoring System; MDS, myelodysplastic syndrome.
Clinical and Genetic Characteristics of TP53 Mutations
Among 87 patients with a TP53 mutation, 48 (55%) were classified as TP53multihit, including 27 with ≥2 TP53 mutations, 15 with one TP53 mutation and TP53 LOH identified by NGS, and six with one TP53 mutation and deletion of chromosome 17/17p by karyotype (Data Supplement [Fig S2]). The presence of a TP53 mutation, but not TP53 allelic state, was significantly associated with clinical and molecular characteristics, including a higher frequency of complex karyotype (TP53multihit = 67% and TP53single = 62% v TP53WT = 10%; P < .001 and P < .001, respectively) and lower platelet count at enrollment (TP53multihit = 42 × 109/L and TP53single = 62 × 109/L v TP53WT = 90 × 109/L; P = .002 and P = .032, respectively; Data Supplement [Table S4]). Consistent with these differences, patients with a TP53 mutation were significantly more likely to have IPSS high-risk disease than those without a TP53 mutation (52% v 26%; P < .001). Other clinical and transplant characteristics were not different between patients with and without a TP53 mutation (Data Supplement [Table S4]).
Genetic Determinants of Outcomes
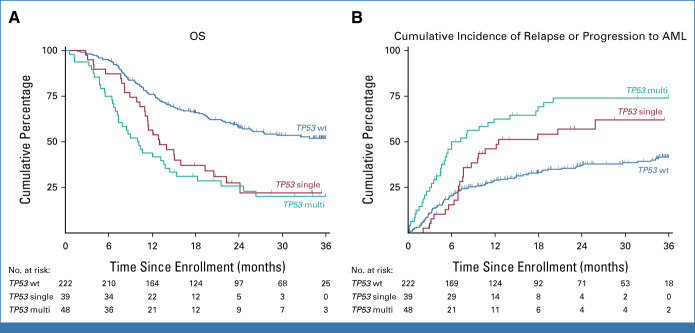
In univariate analysis, the presences of a TP53 mutation and KMT2APTD were significantly associated with shorter OS compared with patients without those mutations (TP53: 21% ± 5% [SE] v 52% ± 4% at 3 years; hazard ratio [HR], 2.55; 95% CI, 1.86 to 3.50; P < .001; KMT2APTD: 8% ± 7% v 45% ± 3% at 3 years; HR, 2.21; 95% CI, 1.22 to 3.99; P = .009), whereas the presence of a germline DDX41 mutation (74% ± 9% v 41% ± 3% at 3 years; HR, 0.39; 95% CI, 0.17 to 0.87; P = .022) and somatic mutations in STAG2 (HR, 0.57; 95% CI, 0.34 to 0.96; P = .034) was associated with favorable OS (Data Supplement [Table S3]). OS at 3 years was similar in patients with TP53single and TP53multihit allelic states (22% ± 8% v 20% ± 6%; HR, 1.29; 95% CI, 0.79 to 2.11; P = .31; Fig 3A). The cumulative incidence of MDS relapse or progression to AML was significantly higher in patients with a TP53 mutation compared with those without a TP53 mutation (68% ± 5% v 42% ± 4% at 3 years; P < .001), and among patients with a TP53 mutation, the incidence was significantly higher in those with TP53multihit compared with TP53single (74% ± 6% v 62% ± 8% at 3 years; P = .03; Fig 3B). Similarly, OS and cumulative incidence of MDS relapse or progression to AML were not different comparing TP53 with or without complex karyotype or deletion 17/17p (Data Supplement [Fig S3]).
FIG 3.
Clinical outcomes by TP53 allelic state. (A) OS by TP53 allelic state. (B) Cumulative incidence of MDS relapse or progression to AML by TP53 allelic state, with death considered as a competing event. Time is measured from consent. MDS, myelodysplastic syndrome; OS, overall survival.
Outcome Analysis Adjusted for Clinical and Genetic Variables
To identify the impact of (1) donor availability and (2) the actual application of HCT in this high-risk MDS cohort, we constructed two multivariable models adjusted for prespecified clinical variables including age at enrollment, performance status, IPSS risk status, MDS disease duration, and clinical and genetic variables identified with stepwise selection. The first model is based on the random assignment on the basis of donor availability, whereas the second model addresses the HCT versus no HCT comparison, where HCT was treated as a time-dependent covariate.
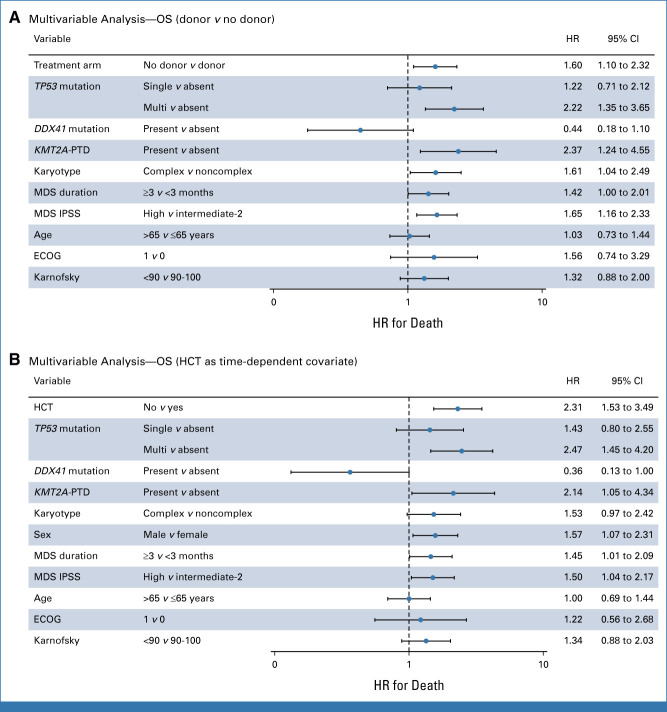
In the donor versus no donor analysis, TP53, KMT2APTD, and DDX41 mutations were associated with OS after adjustment for covariates (Fig 4A). Patients who were assigned to the donor arm had improved OS compared with patients in the no donor arm (HR, 1.60; 95% CI, 1.10 to 2.32; P = .013; Fig 4A). TP53 allelic state was associated with worse outcome, particularly patients with TP53multihit compared with those without TP53 mutations (HR, 2.22; 95% CI, 1.35 to 3.65; P = .002; Fig 4A). Applying that multivariable model to patients with a TP53 mutation, we found that patients in the donor arm had a nonsignificant improved OS compared with those in the no donor arm (HR, 1.76; 95% CI, 0.95 to 3.26; P = .073; Data Supplement [Fig S4]). No interactions were found between treatment arms and mutations in both multivariable models.
FIG 4.
Forest plots of the multivariable analysis. Forest plot of subgroup analyses showing the HR for death and 95% CI in (A) the multivariable analysis of the donor versus no donor comparison and (B) the multivariable time-dependent analysis where HCT was considered as a time-dependent variable. Multivariable Cox regression analysis was used, with adjustment for treatment arm (A) or HCT (B), TP53 allelic state, DDX41 mutation, KMT2APTD, complex karyotype, duration of disease, IPSS score, sex, age, and performance score. ECOG, Eastern Cooperative Oncology Group; HCT, hematopoietic cell transplantation; HR, hazard ratio; IPSS, International Prognostic Scoring System; MDS, myelodysplastic syndrome; OS, overall survival.
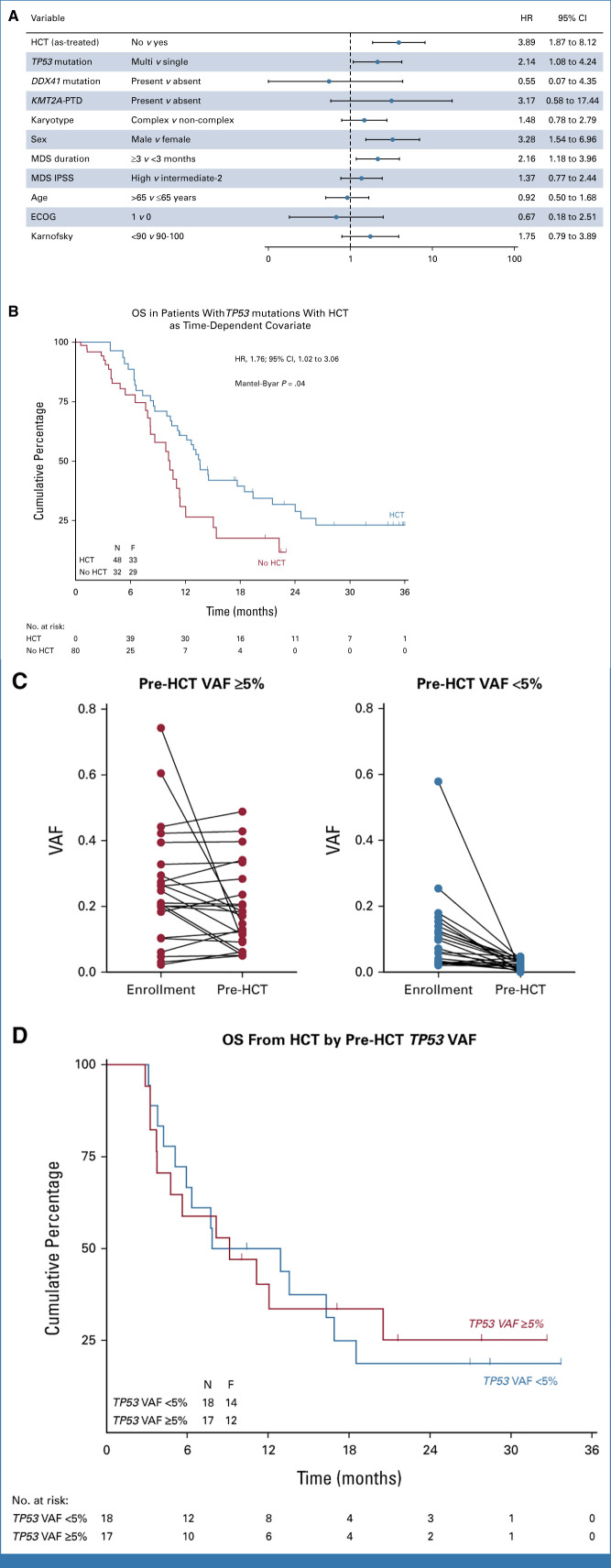
In the time-dependent analysis of HCT, we included 197 patients who actually received HCT after RIC and 78 patients who did not receive HCT (Data Supplement [Fig S5]). Using a multivariable model in which HCT was considered as time-dependent covariate, adjusted for age, sex, performance status, IPSS, MDS duration, and complex karyotype, TP53 and KMT2APTD were associated with differential survival (Fig 4B). HCT was associated with a 2-fold lower instantaneous risk of death compared with patients not receiving HCT after adjustment for covariates (HR, 2.31; 95% CI, 1.53 to 3.49; P < .001). To assess the impact of HCT in the subset of patients with the highest risk of death, we specifically applied this multivariable time-dependent model to patients with a TP53 mutation (Fig 5A). We found that patients with a TP53 mutation who were not transplanted had reduced OS compared with patients who received HCT (HR, 3.89; 95% CI, 1.87 to 8.12; P < .001; Fig 5A), indicating that HCT might improve long-term survival in patients with mutated TP53, independent of other risk factors. No interactions were found between HCT and mutations in both multivariable models. OS in patients with TP53 mutations who actually received RIC HCT estimated 23% ± 7% at 3 years which was significantly improved compared with no HCT in a time-dependent survival analysis (Fig 5B). These observations were also consistently found at multiple landmark time points, although only significant at the 9-month landmark (Data Supplement [Fig S6]).
FIG 5.
Forest plot of the multivariable analysis in patients with mutated TP53. (A) Forest plot of subgroup analyses in patients with mutated TP53 showing the HR for death and 95% CI in the multivariable time-dependent analysis where HCT was considered as a time-dependent variable. Multivariable Cox regression analysis was used, with adjustment for HCT, TP53 allelic state, DDX41 mutation, KMT2APTD, complex karyotype, duration of disease, IPSS score, sex, age, and performance score. (B) OS in patients with TP53 mutations in which HCT is considered as a time-dependent covariate according to a Simon-Makuch plot. Time is measured from consent and patients switch from the no HCT to the HCT at the time of HCT if they received HCT. (C) Serial analysis of enrollment and pre-HCT TP53 samples (n = 35). Patients with pre-HCT TP53 VAF ≥5% (left) and pre-HCT TP53 VAF <5% (right) are shown. (D) OS in patients with TP53 mutations by pre-HCT TP53 VAF cutoff of 5%. Time is measured from transplantation. ECOG, Eastern Cooperative Oncology Group; HCT, hematopoietic cell transplantation; HR, hazard ratio; IPSS, International Prognostic Scoring System; MDS, myelodysplastic syndrome; OS, overall survival; VAF, variant allele fraction.
Molecular Clearance of TP53 Mutation Before HCT Does Not Predict Long-Term Survival
It has been proposed that long-term survival after HCT of patients with TP53-mutated MDS is limited to those whose mutation burden can be reduced below a VAF of 5%.19 Therefore, we obtained samples from all patients who received HCT and had an available sample in the CIBMTR Research Sample Repository (n = 35 of n = 55 total) and then performed targeted, error-corrected sequencing of the TP53 coding sequence. We determined whether TP53 variants present at the time of enrollment were persistent in the preconditioning blood sample, quantified the allele abundance, and analyzed the association between quantitative molecular responses and clinical outcome. Using a high-sensitivity analysis, we found that 33 of 35 patients (94%) had persistent TP53 mutations (median VAF, 0.045; range, 0.0049-0.489; Fig 5C). Using a 5% VAF cutoff per previous published analyses, 17 of 35 (48.6%) had persistent mutations. To test the hypothesis that pre-HCT molecular clearance explained the observed long-term survival among patients with TP53-mutated MDS, we analyzed the association between pre-HCT molecular positivity (at either 2% or 5% VAF cutoffs) with OS after transplantation. OS at 3 years was similar for patients with a pre-HCT TP53 VAF cutoff of ≥5% versus <5% (22% ± 12% v 18% ± 10%; P = .95; Fig 5D) and also not different for a cutoff of ≥2% versus <2% (24% ± 9% v 11% ± 11%; P = .26; Data Supplement [Fig S7]).
Outcome on the Basis of Molecular IPSS Risk Classification
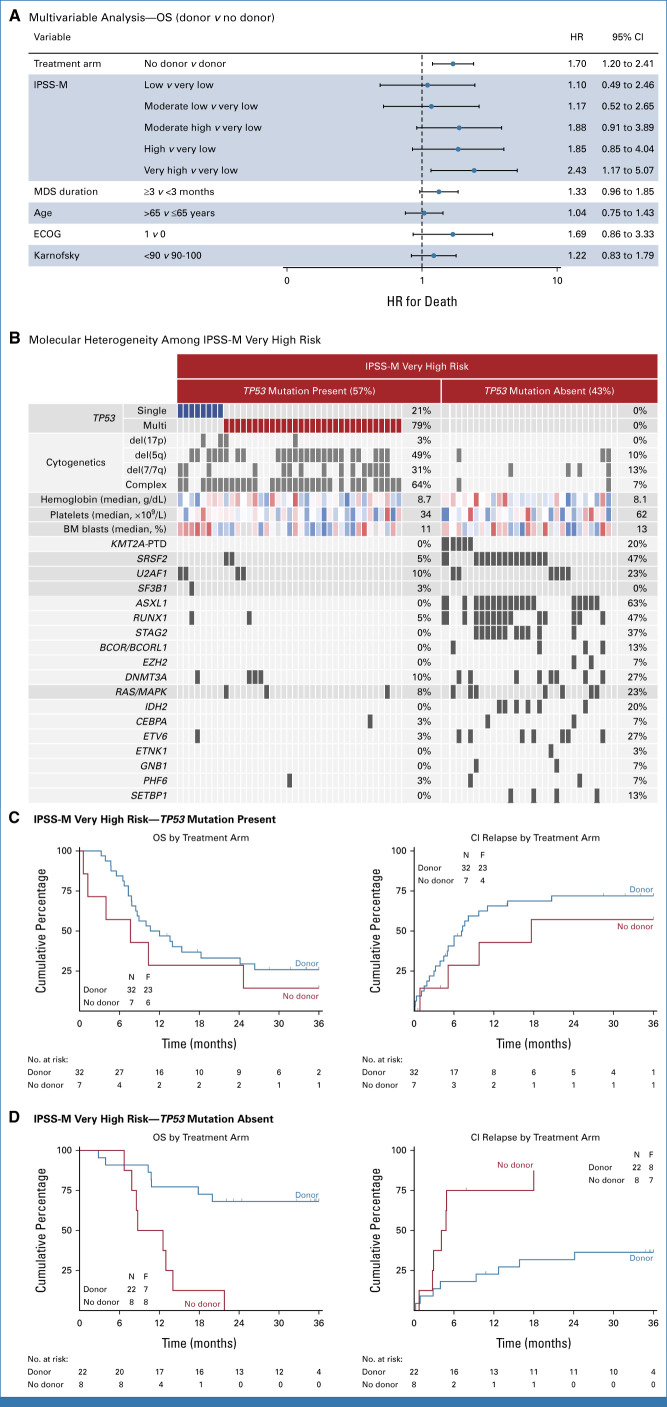
The molecular IPSS (IPSS-M) is a six-tiered MDS risk classification that was developed by combining hematologic parameters, cytogenetic risk, and somatic mutations in 31 genes.13 Since the relative weights of selected variables in the IPSS-M were determined in an unselected cohort that spanned IPSS low- and high-risk disease and in which <10% received allogeneic transplantation, we sought to determine whether IPSS-M risk model was predictive of transplantation outcomes. In the donor versus no donor analysis, only the IPSS-M very high-risk subgroup was associated with inferior survival compared with very low-risk patients after adjustment for clinical- and transplant-related covariates (HR, 2.43; 95% CI, 1.17 to 5.07; P = .018; Fig 6A). Patients in the IPSS-M very high-risk subgroup had a heterogeneous molecular profile, with 57% having a TP53 mutation (of which 79% were TP53multihit), and the remaining 43% with TP53 wild-type disease most commonly had ASXL1, RUNX1, and SRSF2 mutations (Fig 6B). Although patients with IPSS-M very high risk with a TP53 mutation had poor outcome irrespective of donor availability (26% ± 8% v 14% ± 13% at 3 years; P = .28; Fig 6C), IPSS-M very high risk without a TP53 mutation had significantly improved survival in those with a donor compared with those in the no donor arm (68% ± 10% v 0% ± 12% at 3 years; P = .001; Fig 6D). No interactions were found between treatment arms and IPSS-M risk groups.
FIG 6.
Outcome based on IPSS-M risk classification. (A) Forest plot of subgroup analyses showing the hazard ratio for death and 95% CI in the intention-to-treat analysis of donor versus no donor. Multivariable Cox regression analysis was used, with adjustment for treatment arm, IPSS-M score, duration of disease, age, and performance score. (B) A comutation plot shows mutations in individual genes in patients with IPSS-M very high risk per TP53 mutation status as labeled on the top. Mutations, cytogenetic abnormalities, hemoglobin, platelet count, and bone marrow blasts are depicted by colored bars and each column represents one of the 69 patients. For hemoglobin, platelet count, and bone marrow blast, the color scheme ranges from red (high-risk value) to blue (low-risk value). (C) OS (left) and cumulative incidence of MDS relapse or progression to AML (right) by donor versus no donor comparison in patients with IPSS-M very high risk with a TP53 mutation. (D) OS (left) and cumulative incidence of MDS relapse or progression to AML (right) by donor versus no donor comparison in patients with IPSS-M very high risk without a TP53 mutation. Time is measured from consent. ECOG, Eastern Cooperative Oncology Group; HR, hazard ratio; IPSS-M, molecular International Prognostic Scoring System; MDS, myelodysplastic syndrome; OS, overall survival.
Outcomes in Patients With Germline DDX41 Mutations
Inferred germline DDX41 mutations were present in 7% (n = 23) of patients with MDS in this study. Patients with mutated DDX41 had higher hemoglobin levels at enrollment (11.6 v 9.1 g/dL; P < .001) and higher bone marrow blast count (12% v 8%; P = .040) compared with patients without DDX41 mutations (Data Supplement [Table S5]). Other clinical and transplantation characteristics were not significantly different among patients with a germline DDX41 mutation versus those without.
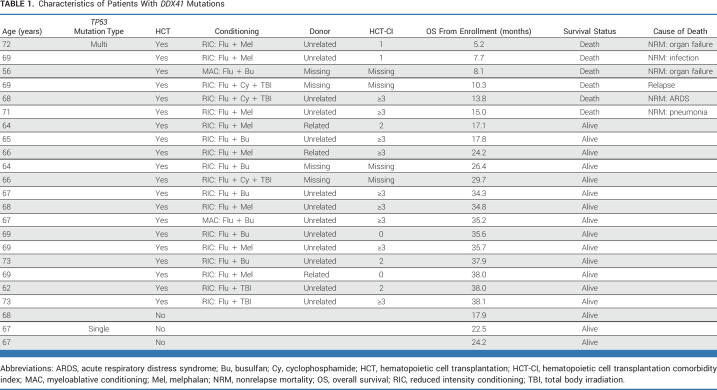
The presence of a germline DDX41 mutation was associated with favorable outcomes, consistent with previous studies.13,20 Twenty of 23 patients proceeded to HCT (Table 1). Only one patient, who also had somatic biallelic TP53 mutations, experienced MDS relapse after HCT. Non-relapse mortality (NRM) was observed in five patients, including one patient who received myeloablative conditioning, three of seven who received melphalan, and one patient who received fludarabine, cycloposphamide, and total body irradiation who had HCT-CI score ≥3. There was no NRM among DDX41 patients receiving fludarabine with 2 days of busulfan or fludarabine and total body irradiation.
TABLE 1.
Characteristics of Patients With DDX41 Mutations
DISCUSSION
Allogeneic HCT confers superior survival in transplant eligible patients with high-risk MDS and an available donor.1,2,21 Analyses of retrospective, registry-level transplant cohorts have suggested that the benefit of transplantation may not extend across MDS molecular subtypes,3-5,13,22 but these studies all lacked direct comparison with non-HCT treatment. To determine directly whether HCT improves MDS outcomes independent of gene mutations, we performed genetic analysis of the prospective BMT CTN 1102 biologic assignment trial. Even after adjustment for genetic variables, survival after allogeneic HCT remained superior compared with non-HCT treatment with no interaction between genetic subtype and treatment effect.
Previous retrospective studies have found that the presence of a TP53 mutation is the most powerful predictor of poor survival of patients with MDS after transplantation, with long-term survival varying from 0% to 25% across studies.3-5,7,23 The absence of a non-HCT control group in such retrospective analyses has thus called into question whether the long-term survival observed in these studies was reasonably attributable to the transplantation intervention. In this study, we directly addressed this question and now conclude definitively that reduced intensity transplantation mediates long-term survival for patients with TP53-mutated MDS compared with non-HCT treatment. Moreover, we show that the benefit of HCT over non-HCT treatment was independent of TP53 allelic state and not restricted to specific subgroups of TP53 mutated MDS, including VAF, complex karyotype, or mutation clearance after pre-HCT hypomethylating agent treatment.
Together, these data indicate that no patient with TP53-mutated MDS should be excluded from consideration for HCT a priori on the basis of TP53 status alone. Despite the relative benefit of HCT over non-HCT treatment, however, the absolute survival benefit remains modest, meriting value-based discussions between physicians and patients on the appropriateness of transplantation. Recent data have indicated that TP53-mutated disease consists of an immune-privileged, evasive phenotype in the bone marrow microenvironment, which might result in reduced sensitivity to alloreactive T cells.24 Strategies to exploit alloreactivity and restore the microenvironment might improve outcomes after HCT. Several other approaches could be considered to improve long-term outcomes, including early allogeneic HCT in patients with TP53-mutated MDS25 or pre-, peri-, and post-HCT interventions aimed at mitigating the risk of relapse. Pretransplant treatment with hypomethylating agents has been associated with clinical responses in patients with mutated TP53, which has also been shown feasible to bridge time to HCT during a donor search period.19,26,27 Combination of hypomethylating treatment with novel agents, for example eprenetapopt or magrolimab, yielded promising results with high rates of complete remission, mutational clearance, and prolonged survival in patients with TP53-mutated myeloid disease.28-31 Data on efficacy and especially tolerability of these combinations for disease reduction before HCT are needed.
With the development of the IPSS-M, prognostic modeling in MDS now integrates clinical variables, cytogenetic risk, and molecular genetic profile to define six risk categories based on leukemia-free survival and OS outcomes.13 However, in the IPSS-M cohort, fewer than 10% of patients received allogeneic HCT, raising the possibility that the relative weights of selected variables could differ in the context of transplantation. In the BMT CTN 1102 cohort, half of patients fell within the IPSS-M high-risk and very high-risk groups (28% and 22%, respectively), consistent with the clinical practice to prioritize higher-risk patients for transplantation. Although we found that patients in the IPSS-M very high-risk group had inferior transplant outcomes, we noted that this group was heterogeneous, including patients with biallelic TP53 mutations and patients without TP53 mutations who had a rather different clinical and genetic profile, including ASXL1, RUNX1, and splicing factor mutations in the context of relatively high blast counts. In the relatively small subgroup of IPSS-M very high-risk patients without TP53 mutations who had no available donor, we observed poor outcomes, consistent with the non-HCT IPSS-M model. However, when a donor was available, outcomes of these IPSS-M very high-risk patients were favorable. These findings indicate that IPSS-M very high risk MDS without a TP53 mutation may be very sensitive to allogeneic HCT and could be ideal candidates for early transplantation as a path to long-term survival.32
We found germline DDX41 mutations in 7% of patients, and these were associated with favorable outcomes with a low risk of relapse, consistent with previous reports.33,34 The only patient with germline DDX41 mutation who experienced disease relapse had somatic biallelic TP53 alterations. These data indicate that MDS with a DDX41 mutation is highly curable with RIC-HCT, and treatment strategies should focus on minimizing toxicity to reduce the risk of NRM.
In conclusion, our data indicate that the benefit of HCT in patients with IPSS intermediate-2 and high-risk MDS extends to high-risk genetic subgroups. Moreover, patients with TP53-mutated MDS, irrespective of additional clinical or genetic variables, including allelic state, VAF, and pre-HCT mutation clearance, have superior survival with RIC allogeneic HCT compared with non-HCT treatment approaches, indicating that these patients should not be excluded for HCT on the basis of genetic findings alone, further reinforcing the conclusion that such patients should be offered transplantation when a donor is available. Patients with IPSS-M very high-risk MDS without a TP53 mutation had favorable outcomes when a donor was available, suggesting that such patients see particular benefit from early transplantation.
ACKNOWLEDGMENT
The authors thank the transplantation center teams for enrolling patients in this trial and the National Marrow Donor Program and Center for International Blood and Marrow Transplant Research for collecting samples and patient data.
Jurjen Versluis
This author is a member of the Journal of Clinical Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Honoraria: AbbVie
Consulting or Advisory Role: ExCellThera
Harrison K. Tsai
Consulting or Advisory Role: Vertex
Asmita Mishra
Research Funding: Novartis
Joseph McGuirk
Honoraria: Kite, a Gilead company, AlloVir, Magenta Therapeutics, Nektar, Sana Biotechnology
Consulting or Advisory Role: Kite, a Gilead company, Juno Therapeutics, Allovir, Magenta Therapeutics, EcoR1 Capital, CRISPR Therapeutics
Speakers' Bureau: Kite/Gilead
Research Funding: Novartis (Inst), Fresenius Biotech (Inst), Astellas Pharma (Inst), Bellicum Pharmaceuticals (Inst), Novartis (Inst), Gamida Cell (Inst), Pluristem Therapeutics (Inst), Kite, a Gilead company (Inst), AlloVir (Inst)
Travel, Accommodations, Expenses: Kite, a Gilead company, Syncopation Life Sciences, SITC/ACCC
Richard T. Maziarz
Leadership: Artiva
Honoraria: Novartis
Consulting or Advisory Role: Novartis, Incyte, Kite/Gilead
Research Funding: Bristol Myers Squibb/Celgene/Juno, ORCA Therapeutics, Gamida Cell, Allovir, Novartis
Patents, Royalties, Other Intellectual Property: Athersys, Inc shared patent re: use of mesenchymal stromal cells for treatment of GVHD
Peter Westervelt
Travel, Accommodations, Expenses: Magenta Therapeutics
Mary Horowitz
Consulting or Advisory Role: Medac (Inst)
Research Funding: Jazz Pharmaceuticals (Inst), Novartis (Inst), Sanofi (Inst), Astellas Pharma (Inst), Xenikos (Inst), Gamida Cell (Inst)
Christopher S. Hourigan
Open Payments Link: https://openpaymentsdata.cms.gov/physician/1021742
Ryotaro Nakamura
Consulting or Advisory Role: Viracor Eurofins, Magenta Therapeutics, Kadmon, Napajen Pharma, Omeros, Bluebird Bio, Ono Pharmaceutical
Research Funding: Helocyte (Inst), Miyarisan pharmaceutical (Inst)
Travel, Accommodations, Expenses: Kyowa Hakko Kirin, Alexion Pharmaceuticals
Corey Cutler
Stock and Other Ownership Interests: Bluebird Bio, Verastem, Northwest Biotherapeutics, Actinium Pharmaceuticals, 2Seventy Bio, Alimera Sciences
Honoraria: Omeros, Janssen, Cimeio, Deciphera, Jazz Pharmaceuticals, Incyte, Sanofi, Bristol Myers Squibb, Mallinckrodt, CTI BioPharma Corp, Jasper Therapeutics, CSL Behring, InhibRx, Astellas Pharma, Rigel, Oxford Immune Algorithmics
Consulting or Advisory Role: Incyte, Jazz Pharmaceuticals, CareDX, Mallinckrodt/Therakos, Sanofi, CTI BioPharma Corp, Equillium, Bristol Myers Squibb, Cimeio, Editas Medicine
Uncompensated Relationships: Kadmon
R. Coleman Lindsley
Consulting or Advisory Role: Bluebird Bio, Takeda, Sarepta Therapeutics, Vertex, Verve Therapeutics, Qiagen, Jazz Pharmaceuticals
Patents, Royalties, Other Intellectual Property: International Patent Application No. PCT/US2020/049257. Title: CRISPR effector system based multiplex cancer diagnostics. International Filing Date: September 3, 2020. Inventors: Jonathan Gootenberg, Omar Abudayyeh, Jeremy Koob, Rahul Vedula, Coleman Lindsley, Feng Zhang Publication No/Date: WO 2021/046257, March 11, 2021. Applicants: The Broad Institute, Inc, Massachusetts Institute of Technology, and Dana-Farber Cancer Institute, Inc. Broad Ref: BI-10578 MIT Ref: 21822JR DFCI Ref.: DFCI 2775.010 JMIN Ref: BROD-4630WP
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in abstract form at the 50th Tandem Meetings, Transplantation & Cellular Therapy Meetings of ASTCT and CIBMTR, Salt Lake City, UT, April 23-26, 2022.
SUPPORT
The Center for International Blood and Marrow Transplant Research is supported primarily by U24CA076518; the Health Resources and Services Administration, Department of Health and Human Services contract HHSH250201200016C; and the Office of Naval Research grants N00014-17-1-2388 and N00014-16-1-2020. Support for this study was provided by grants U10HL069294 U24HL138660 to the Blood and Marrow Transplant Clinical Trials Network from the National Heart, Lung, and Blood Institute and the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH). This work was supported by a Scholar award from The Leukemia & Lymphoma Society (R.C.L.), the Edward P. Evans Center for myelodysplastic syndrome at Dana-Farber Cancer Institute (R.C.L.), the NIH P01CA229092 (R.C.L.), Rubicon fellowship from the Netherlands Organization for Scientific Research (J.V.), Erasmus Medical Center Foundation-Daniel den Hoed (J.V.), Rene Vogels stipend (J.V.), and the Intramural Research Program of the National Heart, Lung, and Blood Institute (C.S.H).
CLINICAL TRAIL INFORMATION
NCT02016781 (BMTCTN1102)
DATA SHARING STATEMENT
The authors attest that all genetic data required for replication are contained in the article and Data Supplement.
AUTHOR CONTRIBUTIONS
Conception and design: Jurjen Versluis, Wael Saber, Brent Logan, Ryotaro Nakamura, Corey Cutler, R. Coleman Lindsley
Financial support: Ryotaro Nakamura, R. Coleman Lindsley
Administrative support: R. Coleman Lindsley
Provision of study materials or patients: Joseph McGuirk, Richard T. Maziarz, Peter Westervelt, Mary Horowitz, Corey Cutler
Collection and assembly of data: Jurjen Versluis, Wael Saber, Laura W. Dillon, Asmita Mishra, Pranay Hegde, Devdeep Mukherjee, Mary Horowitz, Ryotaro Nakamura, Corey Cutler, R. Coleman Lindsley
Data analysis and interpretation: Jurjen Versluis, Wael Saber, Harrison K. Tsai, Christopher J. Gibson, Laura W. Dillon, Asmita Mishra, Joseph McGuirk, Richard T. Maziarz, Peter Westervelt, Devdeep Mukherjee, Michael J. Martens, Brent Logan, Mary Horowitz, Christopher S. Hourigan, Ryotaro Nakamura, R. Coleman Lindsley
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Allogeneic Hematopoietic Cell Transplantation Improves Outcome in Myelodysplastic Syndrome Across High-Risk Genetic Subgroups: Genetic Analysis of the Blood and Marrow Transplant Clinical Trials Network 1102 Study
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jurjen Versluis
This author is a member of the Journal of Clinical Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Honoraria: AbbVie
Consulting or Advisory Role: ExCellThera
Harrison K. Tsai
Consulting or Advisory Role: Vertex
Asmita Mishra
Research Funding: Novartis
Joseph McGuirk
Honoraria: Kite, a Gilead company, AlloVir, Magenta Therapeutics, Nektar, Sana Biotechnology
Consulting or Advisory Role: Kite, a Gilead company, Juno Therapeutics, Allovir, Magenta Therapeutics, EcoR1 Capital, CRISPR Therapeutics
Speakers' Bureau: Kite/Gilead
Research Funding: Novartis (Inst), Fresenius Biotech (Inst), Astellas Pharma (Inst), Bellicum Pharmaceuticals (Inst), Novartis (Inst), Gamida Cell (Inst), Pluristem Therapeutics (Inst), Kite, a Gilead company (Inst), AlloVir (Inst)
Travel, Accommodations, Expenses: Kite, a Gilead company, Syncopation Life Sciences, SITC/ACCC
Richard T. Maziarz
Leadership: Artiva
Honoraria: Novartis
Consulting or Advisory Role: Novartis, Incyte, Kite/Gilead
Research Funding: Bristol Myers Squibb/Celgene/Juno, ORCA Therapeutics, Gamida Cell, Allovir, Novartis
Patents, Royalties, Other Intellectual Property: Athersys, Inc shared patent re: use of mesenchymal stromal cells for treatment of GVHD
Peter Westervelt
Travel, Accommodations, Expenses: Magenta Therapeutics
Mary Horowitz
Consulting or Advisory Role: Medac (Inst)
Research Funding: Jazz Pharmaceuticals (Inst), Novartis (Inst), Sanofi (Inst), Astellas Pharma (Inst), Xenikos (Inst), Gamida Cell (Inst)
Christopher S. Hourigan
Open Payments Link: https://openpaymentsdata.cms.gov/physician/1021742
Ryotaro Nakamura
Consulting or Advisory Role: Viracor Eurofins, Magenta Therapeutics, Kadmon, Napajen Pharma, Omeros, Bluebird Bio, Ono Pharmaceutical
Research Funding: Helocyte (Inst), Miyarisan pharmaceutical (Inst)
Travel, Accommodations, Expenses: Kyowa Hakko Kirin, Alexion Pharmaceuticals
Corey Cutler
Stock and Other Ownership Interests: Bluebird Bio, Verastem, Northwest Biotherapeutics, Actinium Pharmaceuticals, 2Seventy Bio, Alimera Sciences
Honoraria: Omeros, Janssen, Cimeio, Deciphera, Jazz Pharmaceuticals, Incyte, Sanofi, Bristol Myers Squibb, Mallinckrodt, CTI BioPharma Corp, Jasper Therapeutics, CSL Behring, InhibRx, Astellas Pharma, Rigel, Oxford Immune Algorithmics
Consulting or Advisory Role: Incyte, Jazz Pharmaceuticals, CareDX, Mallinckrodt/Therakos, Sanofi, CTI BioPharma Corp, Equillium, Bristol Myers Squibb, Cimeio, Editas Medicine
Uncompensated Relationships: Kadmon
R. Coleman Lindsley
Consulting or Advisory Role: Bluebird Bio, Takeda, Sarepta Therapeutics, Vertex, Verve Therapeutics, Qiagen, Jazz Pharmaceuticals
Patents, Royalties, Other Intellectual Property: International Patent Application No. PCT/US2020/049257. Title: CRISPR effector system based multiplex cancer diagnostics. International Filing Date: September 3, 2020. Inventors: Jonathan Gootenberg, Omar Abudayyeh, Jeremy Koob, Rahul Vedula, Coleman Lindsley, Feng Zhang Publication No/Date: WO 2021/046257, March 11, 2021. Applicants: The Broad Institute, Inc, Massachusetts Institute of Technology, and Dana-Farber Cancer Institute, Inc. Broad Ref: BI-10578 MIT Ref: 21822JR DFCI Ref.: DFCI 2775.010 JMIN Ref: BROD-4630WP
No other potential conflicts of interest were reported.
REFERENCES
- 1.Nakamura R, Saber W, Martens MJ, et al. : Biologic assignment trial of reduced-intensity hematopoietic cell transplantation based on donor availability in patients 50-75 years of age with advanced myelodysplastic syndrome. J Clin Oncol 39:3328-3339, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kröger N, Sockel K, Wolschke C, et al. : Comparison between 5-azacytidine treatment and allogeneic stem-cell transplantation in elderly patients with advanced MDS according to donor availability (VidazaAllo study). J Clin Oncol 39:3318-3327, 2021 [DOI] [PubMed] [Google Scholar]
- 3.Lindsley RC, Saber W, Mar BG, et al. : Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med 376:536-547, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Della Porta MG, Gallì A, Bacigalupo A, et al. : Clinical effects of driver somatic mutations on the outcomes of patients with myelodysplastic syndromes treated with allogeneic hematopoietic stem-cell transplantation. J Clin Oncol 34:3627-3637, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bejar R, Stevenson KE, Caughey B, et al. : Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J Clin Oncol 32:2691-2698, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reilly CR, Myllymäki M, Redd RA, et al. : The clinical and functional effects of TERT variants in myelodysplastic syndrome. Blood 138:898-911, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshizato T, Nannya Y, Atsuta Y, et al. : Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: Impact on outcome of stem cell transplantation. Blood 129:2347-2358, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Versluis J, Lindsley RC: Transplant for TP53-mutated MDS and AML: Because we can or because we should? Hematol Am Soc Hematol Educ Program 2022:522-527, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciurea SO, Chilkulwar A, Saliba RM, et al. : Prognostic factors influencing survival after allogeneic transplantation for AML/MDS patients with mutations. Blood 131:2989-2992, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loke J, Labopin M, Craddock C, et al. : Additional cytogenetic features determine outcome in patients allografted for TP53 mutant acute myeloid leukemia. Cancer 128:2922-2931, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dillon LW, Gui G, Page KM, et al. : DNA sequencing to detect residual disease in adults with acute myeloid leukemia prior to hematopoietic cell transplant. JAMA 329:745-755, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernard E, Nannya Y, Hasserjian RP, et al. : Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med 26:1549-1556, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernard E, Tuechler H, Greenberg PL, et al. : Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evid 1:1-14, 2022 [DOI] [PubMed] [Google Scholar]
- 14.Tsai HK, Brackett DG, Szeto D, et al. : Targeted informatics for optimal detection, characterization, and quantification of FLT3 internal tandem duplications across multiple next-generation sequencing platforms. J Mol Diagn 22:1162-1178, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai HK, Gibson CJ, Murdock HM, et al. : Allelic complexity of KMT2A partial tandem duplications in acute myeloid leukemia and myelodysplastic syndromes. Blood Adv 6:4236-4240, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon R, Makuch RW: A non-parametric graphical representation of the relationship between survival and the occurrence of an event: Application to responder versus non-responder bias. Stat Med 3:35-44, 1984 [DOI] [PubMed] [Google Scholar]
- 17.Putter H, van Houwelingen HC: Understanding landmarking and its relation with time-dependent Cox regression. Stat Biosci 9:489-503, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mantel N, Byar DP: Evaluation of response-time data involving transient states: An illustration using heart-transplant data. J Am Stat Assoc 69:81-86, 1974 [Google Scholar]
- 19.Hunter AM, Komrokji RS, Yun S, et al. : Baseline and serial molecular profiling predicts outcomes with hypomethylating agents in myelodysplastic syndromes. Blood Adv 5:1017-1028, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sébert M, Passet M, Raimbault A, et al. : Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood 134:1441-1444, 2019 [DOI] [PubMed] [Google Scholar]
- 21.Robin M, Porcher R, Adès L, et al. : HLA-matched allogeneic stem cell transplantation improves outcome of higher risk myelodysplastic syndrome A prospective study on behalf of SFGM-TC and GFM. Leukemia 29:1496-1501, 2015 [DOI] [PubMed] [Google Scholar]
- 22.Bejar R, Stevenson K, Abdel-Wahab O, et al. : Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 364:2496-2506, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grob T, Al Hinai ASA, Sanders MA, et al. : Molecular characterization of mutant TP53 acute myeloid leukemia and high-risk myelodysplastic syndrome. Blood 139:2347-2354, 2022 [DOI] [PubMed] [Google Scholar]
- 24.Sallman DA, McLemore AF, Aldrich AL, et al. : TP53 mutations in myelodysplastic syndromes and secondary AML confer an immunosuppressive phenotype. Blood 136:2812-2823, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koreth J, Pidala J, Perez WS, et al. : Role of reduced-intensity conditioning allogeneic hematopoietic stem-cell transplantation in older patients with de novo myelodysplastic syndromes: An international collaborative decision analysis. J Clin Oncol 31:2662-2670, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voso MT, Leone G, Piciocchi A, et al. : Feasibility of allogeneic stem-cell transplantation after azacitidine bridge in higher-risk myelodysplastic syndromes and low blast count acute myeloid leukemia: Results of the BMT-AZA prospective study. Ann Oncol 28:1547-1553, 2017 [DOI] [PubMed] [Google Scholar]
- 27.Welch JS, Petti AA, Miller CA, et al. : TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med 375:2023-2036, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter AM, Sallman DA: Targeting TP53 mutations in myelodysplastic syndromes. Hematol Oncol Clin North Am 34:421-440, 2020 [DOI] [PubMed] [Google Scholar]
- 29.Sallman DA, DeZern AE, Garcia-Manero G, et al. : Eprenetapopt (APR-246) and azacitidine in -mutant myelodysplastic syndromes. J Clin Oncol 39:1584-1594, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cluzeau T, Sebert M, Rahmé R, et al. : Eprenetapopt plus azacitidine in mutated myelodysplastic syndromes and acute myeloid leukemia: A phase II study by the Groupe Francophone des Myélodysplasies (GFM). J Clin Oncol 39:1575-1583, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra A, Tamari R, DeZern AE, et al. : Eprenetapopt plus azacitidine after allogeneic hematopoietic stem-cell transplantation for mutant acute myeloid leukemia and myelodysplastic syndromes. J Clin Oncol 40:3985-3993, 2022 [DOI] [PubMed] [Google Scholar]
- 32.Sauta E, Robin M, Bersanelli M, et al. : Real-world validation of molecular international prognostic scoring system for myelodysplastic syndromes. J Clin Oncol 41:2827-2842, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polprasert C, Schulze I, Sekeres MA, et al. : Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell 27:658-670, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quesada AE, Routbort MJ, DiNardo CD, et al. : DDX41 mutations in myeloid neoplasms are associated with male gender, TP53 mutations and high-risk disease. Am J Hematol 94:757-766, 2019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors attest that all genetic data required for replication are contained in the article and Data Supplement.