Abstract
Disulfidptosis is a novel form of metabolic-related regulated cell death (RCD) that is caused by disulfide stress caused by the accumulation of excess cystine in the cell. Targeting disulfide metabolism imbalance is an emerging strategy for the treatment of cancer. However, it is undetermined how disulfidptosis-related genes (DRGs) influence hepatocellular carcinoma (HCC). Unsupervised clustering analysis was performed on the TCGA-LIHC cohort to identify various phenotypes of disulfidptosis. GSVA was used to measure the activation of characteristic gene sets, while CIBERSORT was employed to estimate the infiltration of immune cells. Disulfidptosis-related signature was generated to quantify the phenotype of disulfidptosis in HCC patients. Next, we examined the disparities among the high and low disulfidptosis score categories by considering clinical characteristics, infiltration of immune cells, functions related to the immune system, sensitivity to chemotherapeutic drugs, and effectiveness of immunotherapy. Two different disulfidptosis phenotypes with different prognoses, clinical traits, biological pathways, and immune cell infiltration were identified. Based on differently expressed genes (DEGs) among 2 disulfidptosis phenotypes, a disulfidptosis-related signature was built. The prognostic value of this signature was then evaluated in the TCGA and GEO datasets. Low disulfidptosis score indicated favorable clinical outcomes, higher levels of immune cell infiltration, lower tumor purity, and enhanced immune responses. Furthermore, we noticed a clear disparity in tumor mutation load and drug responsiveness when comparing the high and low disulfidptosis score categories. Finally, a quantitative nomogram was built with disulfidptosis score and several clinical characteristics. The disulfidptosis-related signature provides new insights into the tumor immune microenvironment and complexity in HCC. The disulfidptosis score can serve as a promising tool for personalized prognostic prediction of HCC patients and for customizing more effective immunotherapeutic strategies.
Keywords: disulfidptosis, hepatocellular carcinoma, immunotherapy, prognosis, tumor microenvironment
1. Introduction
Hepatocellular carcinoma (HCC) is the most common primary malignancy of the liver and represents a major global healthcare challenge.[1] HCC patients have a wide range of treatment options, including surgical excision, liver transplantation, thermal ablation, intra-arterial, radiation, and systemic therapy. Advances in surgical and treatment options have improved treatment response and survival.[2] The outlook for patients with advanced HCC has improved with the introduction of immune checkpoint inhibitors. However, the optimal sequencing of drugs remains to be determined.[1] Hence, there is an urgent need to discover new biomarkers and develop precise prognostic signatures that can effectively anticipate the patient status and assist in personalized treatment for HCC.
As a novel type of metabolic-related regulated cell death (RCD), disulfidptosis has recently become a hot research topic. This recently discovered variant of RCD is caused by disulfide stress resulting from an excessive buildup of intracellular cystine. It is distinct from other known forms of RCD, including apoptosis, ferroptosis, necroptosis, and cuproptosis. These findings were recently published in Nature Cell Biology by Liu et al[3] In their study, the inability of NADPH supply to meet the cell reduction of cystine to cysteine causes disulfide stress, induces actin cytoskeletal protein disulfide bond crosslinking and cytoskeletal contraction, stripping from the plasma membrane, and ultimately cell death known as disulfidptosis. Disulfidptosis may be induced by insufficient glucose intake and excessive cystine intake. The identification and characterization of this cell death mechanism not only advances the basic understanding of cell homeostasis but also suggests that GLUT inhibitor-induced disulfide death may be an effective strategy for tumor treatment. For example, the anti-neoplastic characteristics of specific medications such as cisplatin and paclitaxel are demonstrated by their engagement with intracellular disulfides.[4,5] Furthermore, disulfide isomerases have been observed to be linked to tumorigenicity in various tumors and are regarded as potential targets for cancer treatment.[6,7] Importantly, protein disulfide isomerase family A member 3 plays a role in immunogenic cell death and is implicated in liver damage and hepatocellular carcinoma cell proliferation.[8,9] Nevertheless, the specific role of disulfidptosis in HCC is still unclear. Our objective in this study was to investigate the impact of disulfidptosis on various TME of HCC and develop a unique scoring system to assess patient survival and immunotherapeutic efficacy. This will contribute to the advancement of personalized treatment strategies.
2. Materials and Methods
2.1. Data collection
Data on the transcriptome and comprehensive clinical details of HCC were gathered from the Cancer Genome Atlas (TCGA) repository. After excluding samples that had incomplete or unavailable clinicopathological information, we ended up with a final group of 371 TCGA-LIHC patients. To verify the data, we conducted a search in the Gene Expression Omnibus (GEO) database, available at https://www.ncbi.nlm.nih.gov/gds/. Our search criteria included datasets with over 221 samples and comprehensive clinical information. Additionally, we collected 15 disulfidptosis-related genes (DRGs) from earlier investigation (Table S1, http://links.lww.com/MD/K134).[10] Approval by the Ethics Committee was not required because research data are publicly available.
2.2. Consensus clustering analysis
Cluster analysis was utilized to detect distinctive disulfidptosis phenotypes in HCC based on the expression of 15 DRGs. Each individual cluster size and stability were determined using a consensus clustering technique, and further assurance came from the “ConsensusClusterPlus” package. The ideal number of clusters was identified by analyzing the plot of the empirical cumulative distribution function. The optimum number of subgroups was simultaneously evaluated using cumulative distribution function curves and the proportion of ambiguous clustering.[11] T-distributed stochastic neighbor embedding, principal component analysis (PCA), and uniform manifold approximation and projection methods were applied to verify the subtype assignments. We integrated the patients’ clinicopathological traits such as age, gender, Child-Pugh classification, surgical procedure, tumor status, grade, recurrence, and AJCC stage. The relationship between the different phenotypes and these clinical traits was evaluated. Additionally, OS disparities among various phenotypes were evaluated using Kaplan–Meier analyses. Finally, the CIBERSORT algorithm was also applied to detect the fraction of 22 immune infiltrating cells.
2.3. Functional enrichment analysis and identification of disulfidptosis phenotype-related differentially expressed genes
In order to gain a deeper comprehension of the biological mechanisms and operational cellular routes, we performed gene set variation analysis (GSVA). The R packages “GSVA” and “GSEABase” were used with the reference dataset “c2.cp.kegg.symbols.gmt.” Differentially expressed genes (DEGs) between distinct disulfidptosis phenotypes were identified using the “limma” R package with the following settings: |logFC| ≥ 1 and adjust P value < .001. In order to gain a deeper comprehension of the biological traits and operational cellular pathways, we generated the GO and KEGG pathways that were enriched with distinct genes by utilizing the “clusterprofiler” R package.
2.4. Establishment and verification of disulfidptosis-related signature
Prognostic DEGs were selected using a univariate Cox analysis. To avoid overfitting, we performed a regression analysis using the Least absolute shrinkage and selection operator (LASSO) method on the prognostic DEGs. This analysis was carried out using the “glmnet” R package. Subsequently, we created a signature called disulfidptosis-related signature (DRS) based on the results of the analysis. The disulfidptosis score was calculated as follows:
where n denotes the number of remaining genes; Coefi denotes the risk coefficient from the final Cox model, and Expi denotes the mRNA expression level of each gene. Next, the TCGA-LIHC dataset was utilized to determine the cutoff value for dividing the HCC samples into groups based on their disulfidptosis score, using the median score as a reference. Scatter plots were used to show the correlation between survival status and disulfidptosis score across the TCGA-LIHC and GSE14520 datasets. Subsequently, PCA was performed to separate low- and high-risk HCC using data from both TCGA-LIHC and GSE14520 datasets. Survival curves were generated using the Kaplan-Meier technique, and the significance of differences was evaluated using log-rank tests. We utilized the concordance index (C-index) to further compare the prediction efficacy of the disulfidptosis score and clinical traits. We also employed the receiver operating characteristic curve (ROC) method to validate the prediction efficacy of the disulfidptosis score in the TCGA-LIHC and GSE14520 cohorts, respectively.
2.5. Clinical value of the disulfidptosis signature
To examine the correlation between disulfidptosis scores and different clinical characteristics, the chi-square test was utilized. We further stratified the predictive power of disulfidptosis scores for different clinical features of HCC patients. Independent predictive abilities were identified by conducting univariate and multivariate Cox regression analyses on disulfidptosis scores and clinical traits in the TCGA-LIHC and GSE14520 datasets. To evaluate the accuracy of the disulfidptosis scores in predicting clinical outcomes of HCC, the “rms” R package was employed to generate a nomogram comprising of disulfidptosis score and clinical traits. To validate the effectiveness of the nomogram, ROC curves and calibration plots were employed.
2.6. Analysis of immune microenvironment and immunotherapeutic response
We first analyzed the differences in the immune microenvironment between the high and low disulfidptosis score groups and mined for differences in infiltrated immune cells, immune function, and TME score by the ssGSEA and ESTIMATE algorithms. We employed the Immunophenoscore (IPS) algorithm to calculate IPS scores, which are based on unbiased gene expression of representative cell types using machine learning methods, in order to assess immunogenicity through the analysis of immunomodulators, immunosuppressive cells, MHC molecules, and effector cells. Immunotherapy yields a more favorable outcome with an increased IPS score. Obtained from The Cancer Immunome Atlas (TCIA) database, the IPS scores of TCGA-LIHC patient samples were acquired. The Tumor Immune Dysfunction and Exclusion (TIDE) score was used to predict the antitumor immune escape possibility of high and low disulfidptosis score groups (http://tide.dfci.harvard.edu/).
2.7. Mutation analysis and drug sensitivity
Using the “oncoPredict” package, the half-maximal inhibitory concentration (IC50) of each patient was determined in order to predict the sensitivity of these chemotherapeutic agents. IC50 indicated the effectiveness of an agent in suppressing specific biological or biochemical functions. We used the “maftools” R package to generate waterfall plots, which show the mutation patterns of HCC patients with high and low disulfidptosis scores, in order to examine the somatic mutations linked to disulfidptosis.
2.8. Statistical analysis
R (v4.3.0; http://www.Rproject.org) was used for statistical comparisons. Wilcoxon tests were utilized to compare the difference between the 2 groups. P value < .05 was considered statistically significant.
3. Results
3.1. Identification of 2 distinct disulfidptosis phenotypes in HCC
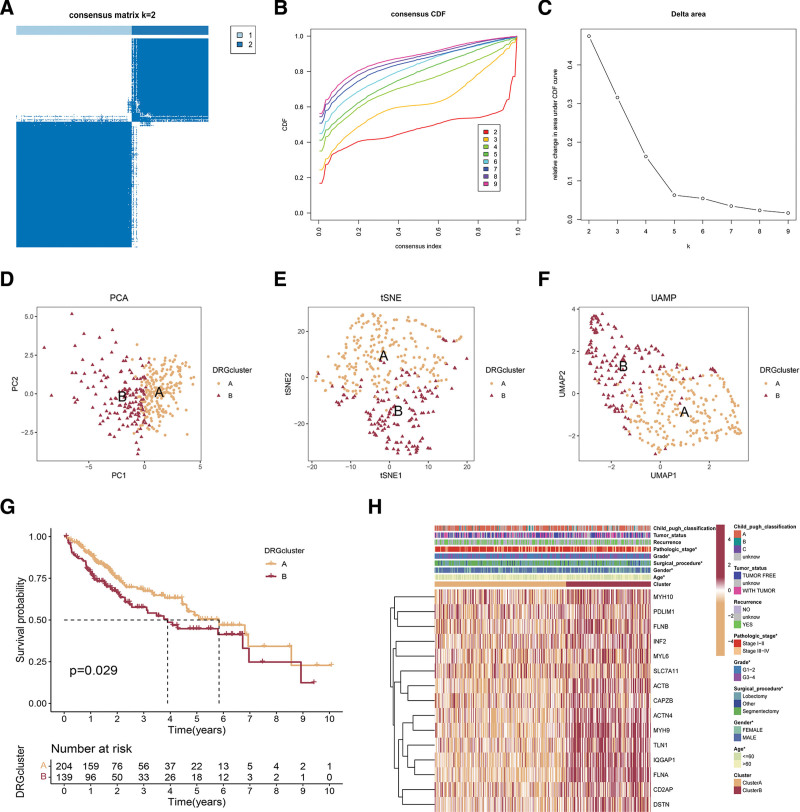
To acquire HCC subtypes related to disulfidptosis, we conducted consistency clustering with 15 DRGs. The clustering demonstrated optimal stability (Fig. 1A–C) at k = 2. Finally, 2 clusters were identified: cluster A (n = 223) and B (n = 147). The transcriptional profiles of the DRGs exhibited significant variations between the 2 distinct disulfidptosis phenotypes (Fig. 1D–F). In order to demonstrate the dominance of clustering, a K-M analysis was employed to showcase the predictive ability, revealing the unfavorable result of cluster B in TCGA-LIHC (P = .028; Fig. 1G). In Figure 1H, the heatmap demonstrated a clear distinction in the expression of DRGs among the 2 clusters, with cluster B exhibiting higher expression levels of DRGs. Moreover, we observed a significant correlation between disulfidptosis phenotype, age, gender, surgical procedure, grade, and pathologic stage (P < .05) (Fig. 1H).
Figure 1.
Consensus clustering of DRGs in hepatocellular carcinoma. (A) The heatmap of the consensus matrix defines 2 clusters (k = 2) and indicates their correlation region. (B, C) Consensus clustering cumulative distribution function (CDF), relative change in area under CDF curve, and tracking plot for k = 2 to 9. (D–F) The PCA, t-SNE, and UMAP reveal a clear distinction in transcriptomes among the 2 clusters. (G) Survival analysis of cluster A and cluster B in TCGA-LIHC cohort. (H) The heatmap showed the expression of DRGs and clinical characteristics in 2 clusters. DRGs = disulfidptosis-related genes, PCA = principal component analysis, t-SNE = T-distributed stochastic neighbor embedding, UMAP = uniform manifold approximation and projection.
3.2. TME and functional enrichment analysis in 2 distinct disulfidptosis phenotypes
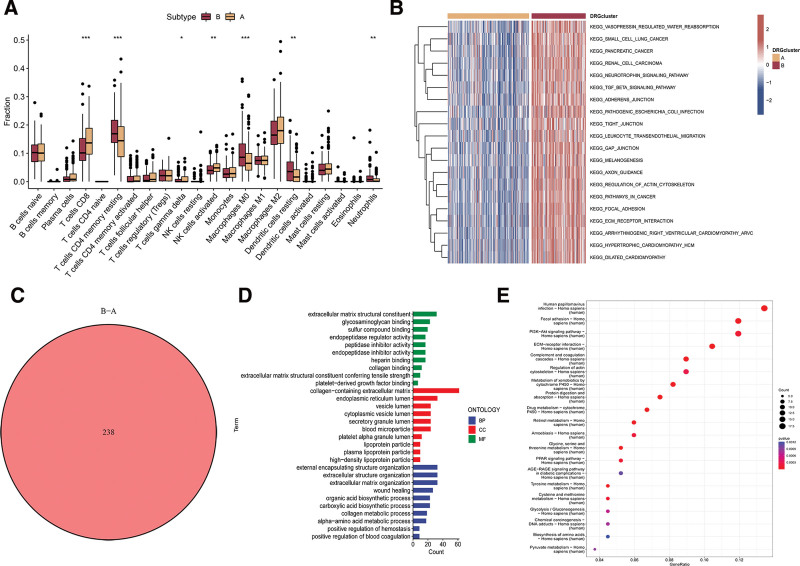
To understand the role of TME in HCC, we examined the infiltration of immune cells in 2 clusters within TCGA-LIHC dataset. Figure 2A demonstrated the presence of fewer CD8 + T cells, activated NK cells, and gamma delta T cells in cluster B, while cluster B exhibited higher levels of resting memory CD4 + T cells, M0 Macrophages, resting dendritic cells, and neutrophil infiltrations. To reveal possible biomolecular changes in 2 distinct disulfidptosis phenotypes, this study conducted GSVA enrichment analyses. In our investigation, we found that cluster B of disulfidptosis showed significant enrichment in pathways associated with the activation of carcinogenesis, including the TGF-β signaling pathway, pathways related to cancer, and specifically pancreatic cancer (Fig. 2B). To further explore the potential biological activity of disulfidptosis phenotypes, a total of 238 DEGs were identified from the overlap across different disulfidptosis phenotypes (Fig. 2C). GO enrichment analysis reveals that DEGs are highly enriched in extracellular matrix structural constituent (Fig. 2D). KEGG of the above DEGs suggested that tumor- and metabolism-related biological processes and pathways were markedly enriched, such as PI3K-Akt signaling pathway and regulation of actin cytoskeleton (Fig. 2E).
Figure 2.
TME and functional enrichment analysis in 2 distinct disulfidptosis phenotypes. (A) Comparison of immune infiltrating cells between 2 clusters across. (B) GSVA uncovering potential biomolecular alterations in 2 different disulfidptosis phenotypes. (C) Identification of differentially expressed genes across 2 disulfidptosis phenotypes. (D) Gene ontology analysis of the genes differentially expressed between the 2 disulfidptosis phenotypes. (E) KEGG analysis of the DEGs between 2 disulfidptosis phenotypes. DEGs = differently expressed genes, GSVA = gene set variation analysis.
3.3. Establishment and verification of disulfidptosis-related signature
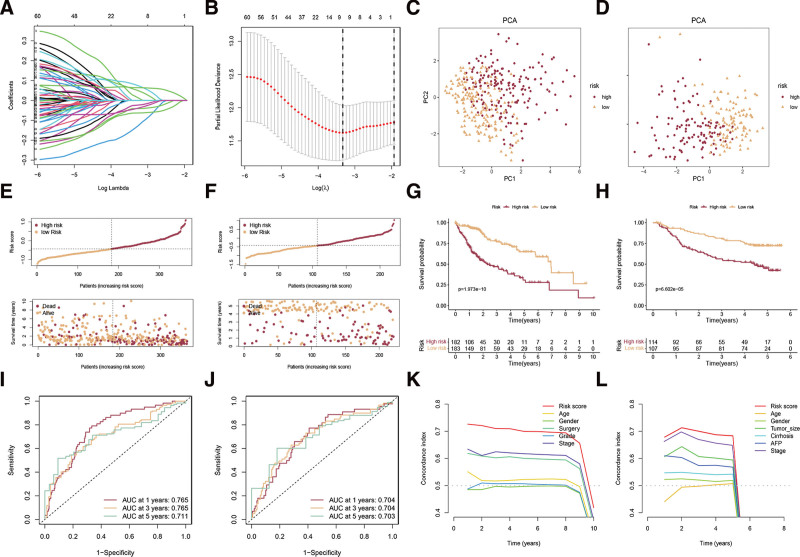
To obtain prognostic DEGs, we integrated the expression data of the above DEGs with corresponding survival data and identified 69 DEGs that are associated with HCC prognosis, based on univariate Cox regression analysis (Table S2, http://links.lww.com/MD/K135). Then, LASSO Cox regression was employed to build a disulfidptosis-related signature (DRS) named the disulfidptosis score (Fig. 3A and B). Afterward, we computed the disulfidptosis score for every individual, enabling us to categorize all individuals into either the high-risk or low-risk category depending on the median disulfidptosis score of the TCGA-LIHC dataset. The effective separation of low- and high-risk samples was demonstrated through PCA analysis using the TCGA-LIHC and GSE14520 datasets (Fig. 3C and D). The scores and survival status of each HCC patient in the TCGA-LIHC and GSE14520 datasets are shown in Figure 3E and 3F. Patients of high risk in the 2 cohorts showed dismal prognoses of OS (Fig. 3G and H). To verify the efficacy of the DRS, we plotted an ROC curve to demonstrate its accuracy in determining the survival of HCC patients. The results in the TCGA-LIHC cohort showed that the AUCs for the 1-, 3-, and 5-year overall survival ROC curves based on the DRS were 0.765, 0.765, and 0.711, respectively (Fig. 3I). The results in the GSE14520 cohort also showed the signature had excellent sensitivity for predicting survival (Fig. 3J). Moreover, the C-index values of the disulfidptosis score in the 2 cohorts were higher than those of other clinical traits (Fig. 3K and L).
Figure 3.
Establishment and verification of disulfidptosis-related signature. (A, B) The LASSO was used to identify the best parameter (lambda) and determine the coefficient profiles of the disulfidptosis-related DEGs. (C, D) PCA analyses of low and high disulfidptosis score subgroups in TCGA-LIHC cohort (C) and GSE14520 cohort (D). (E, F) The scores and survival status of each HCC patient in the TCGA-LIHC cohort (E) and GSE14520 cohort (F). (G, H) Survival analysis of low and high disulfidptosis score subgroups in the TCGA-LIHC cohort (G) and GSE14520 cohort (H). (I, J) ROC curve analysis was conducted on DRS in the TCGA-LIHC cohort (I) and GSE14520 cohort (J). The C-index values of the disulfidptosis score and clinical traits in the TCGA-LIHC cohort (K) and GSE14520 cohort (L). DEGs = differently expressed genes. DRS = disulfidptosis-related signature, HCC = hepatocellular carcinoma, PCA = principal component analysis, ROC = receiver operating characteristic.
3.4. Clinical significance of the disulfidptosis signature
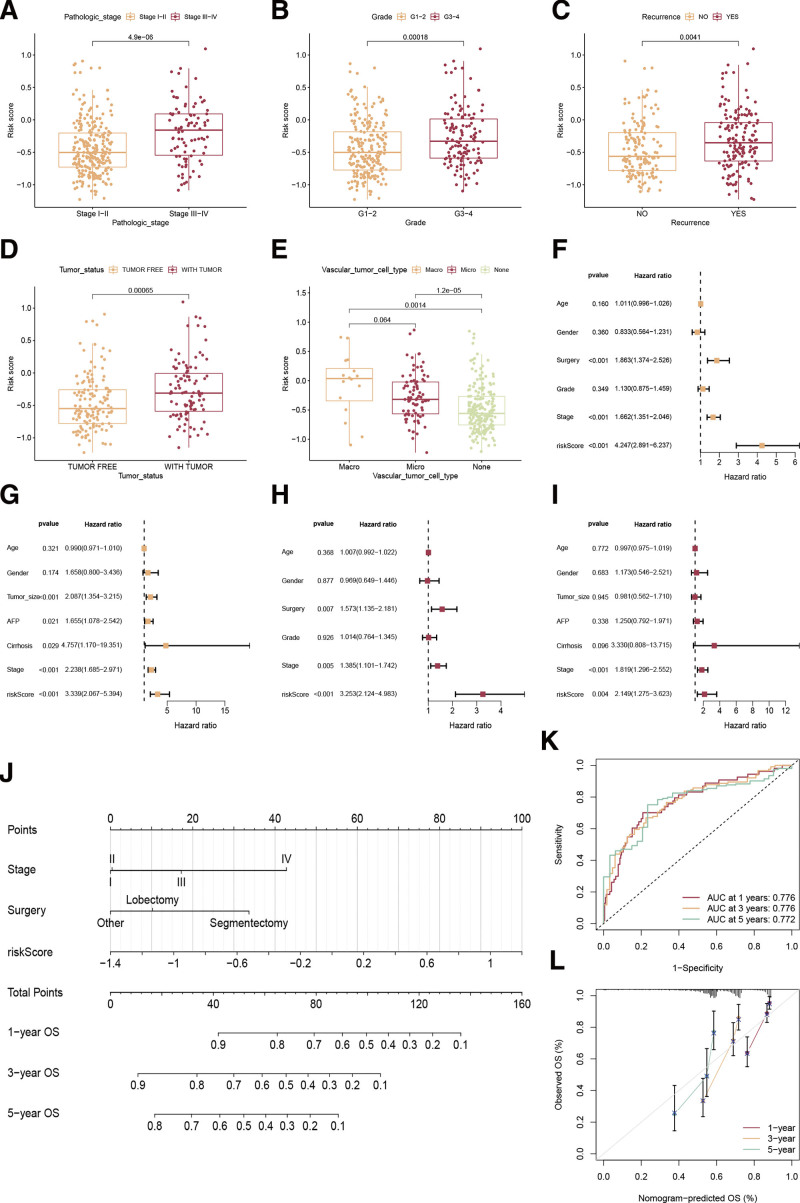
HCC patients were stratified into groups according to age, grade, recurrence, surgical procedure tumor status, Child-Pugh classification, TNM stage, and vascular tumor cell type. Patients with stage III-IV, G3-G4, recurrence, tumor status, and macro/microvascular invasion may have higher disulfidptosis scores (Fig. 4A–E). Univariate Cox regression demonstrated disulfidptosis score, surgery, and TNM stage were prognostic factors to HCC (Fig. 4F and G), and the disulfidptosis score independently indicated patients’ OS according to multivariate analysis in both TCGA-LIHC and GSE14520 datasets (Fig. 4H and I). To determine the disulfidptosis scores in the subgroups, we also conducted a subgroup survival analysis. As shown in Figure S1A-P, http://links.lww.com/MD/K133, observations of low-risk outlived in age groups (≤60 or >60 years), gender (Female or male), grade (G1-G2 or G3-G4), TNM stage (I-II or III-IV), recurrence (no or yes), surgery (segmentectomy, lobectomy, or other), tumor status (tumor-free or with tumor), and Child-Pugh classification A group compared to the high-risk.
Figure 4.
Clinical value of the disulfidptosis signature. (A–E) The relationships between disulfidptosis scores and TNM stage (A), cancer grade (B), recurrence (C), tumor status (D), and vascular tumor cell type (E). (F, G) Univariate Cox regression analyses of clinical factors and disulfidptosis score in the TCGA-LIHC cohort (F) and GSE14520 cohort (G). (H, I) The independent prognostic efficacy of clinical factors and disulfidptosis score by multivariate Cox regression analyses in the TCGA-LIHC cohort (H) and GSE14520 cohort (I). (J) A nomogram plot to develop a personalized prognosis score for each patient. (K) Receiver operating characteristic curve of the nomogram at 1-, 3-, and 5-yr. (L) Calibration plots are used for internally validating the nomogram.
To quantify the disulfidptosis score better in combination with other clinical factors for the OS of HCC patients, we developed a nomogram plot to develop a personalized prognosis score for each patient (Fig. 4J). Figure 4K shows that the nomogram had a high predictive accuracy with AUC values of 0.776, 0.776, and 0.772 at 1-, 3-, and 5-year intervals. The calibration curve (Fig. 4L) confirmed the excellent performance of the nomogram.
3.5. Analysis of immune microenvironment and immunotherapeutic response
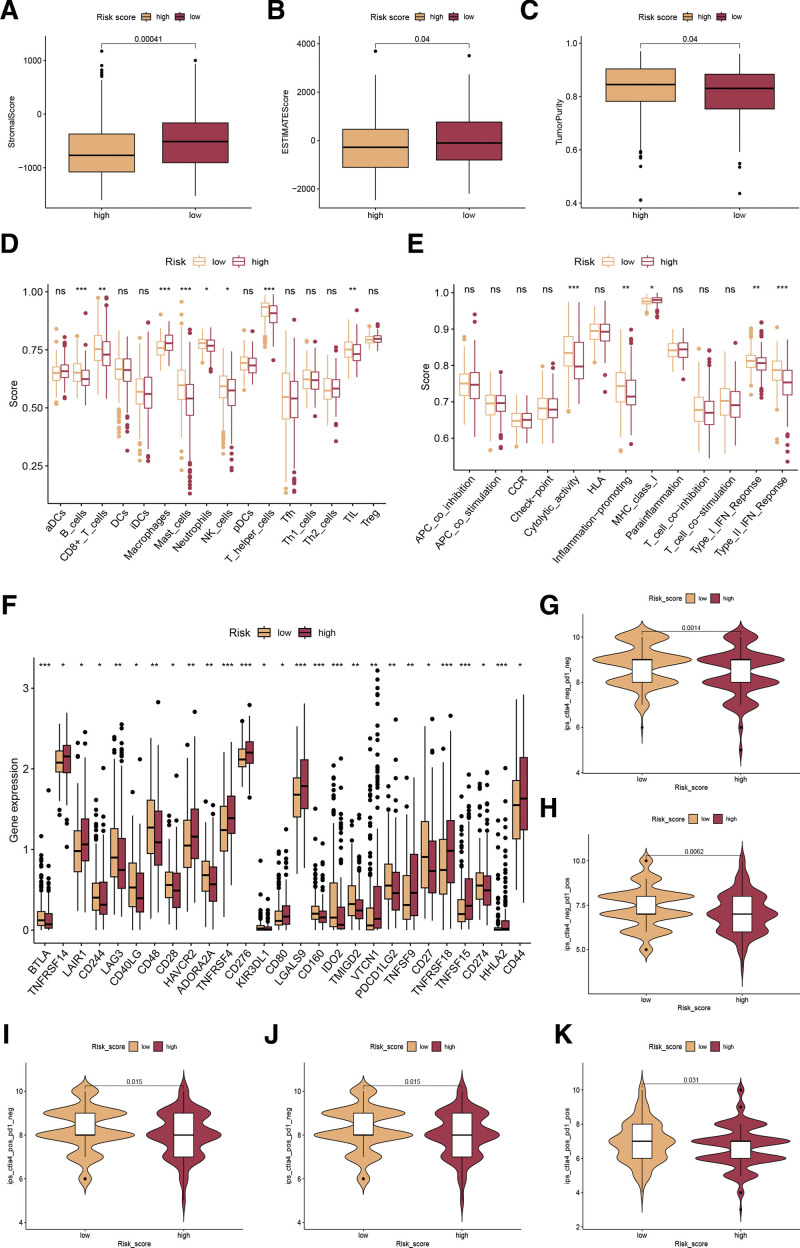
We conducted a thorough analysis to compare the TME in groups with low and high disulfidptosis scores. In the initial step, we measured the overall presence of immune, stromal cells, and tumor purity in the 2 disulfidptosis score categories. Our findings revealed a decreased stromal score and ESTIMATE score, along with an increased tumor purity in the high disulfidptosis score category (Fig. 5A–C). Furthermore, we investigated the differences in immune cell infiltration and immune-related functions among the 2 disulfidptosis score groups. Our results showed that macrophages exhibited elevated levels in the high disulfidptosis score group and low levels of B cells, CD8 + T cells, mast cells, neutrophils, NK cells, T helper cells, and TIL were found in the high disulfidptosis score group (Fig. 5D). To find out the potential outcomes of ICI therapy, we compared the immune function of the 2 disulfidptosis score groups. As a result, the high-risk patients showed lower immune functions, such as cytolytic_activity, Inflammation-promoting, Type_I_IFN_Reponse, and Type_II_IFN_Reponse (Fig. 5E). We chose a few significant immune checkpoints and assessed the expression of each in groups with high and low disulfidptosis scores. According to the results (Fig. 5F), significant differences were observed in most of the immune-related genes, including PD-L1 (CD274) (Fig. 5F). Additionally, IPS is suggested as an indicator of the immune reaction. In the TCGA-LIHC cohort, our examination showed a significant increase in the IPS score of anti-CTLA-4/PD-1 treatment among individuals with a low disulfidptosis score (Fig. 5G–J). Furthermore, it was observed that individuals with elevated disulfidptosis scores exhibited increased TIDE scores (Fig. 5K), suggesting that the group at higher risk had a greater likelihood of immune evasion, thereby reducing the efficacy of immunotherapy in such instances.
Figure 5.
Analysis of immune microenvironment and immunotherapeutic response. (A, B) Comparison of the stromal score (A), ESTIMATE score (B), and tumor purity (C) among the 2 disulfidptosis score groups. (D) Variances in the immune cell populations among the 2 disulfidptosis score groups, quantified by the ssGSEA algorithm. (E) Variances in immune-related pathways’ activity between the 2 disulfidptosis score groups. (F) Differences in the manifestations of immune checkpoints among the low- and high-risk categories. (G-J) Differences in IPS score of CTLA-4/PD-1 among the low- and high-risk categories. (K) Differences in TIDE score among the low- and high-risk categories.
3.6. Mutation analysis and drug sensitivity
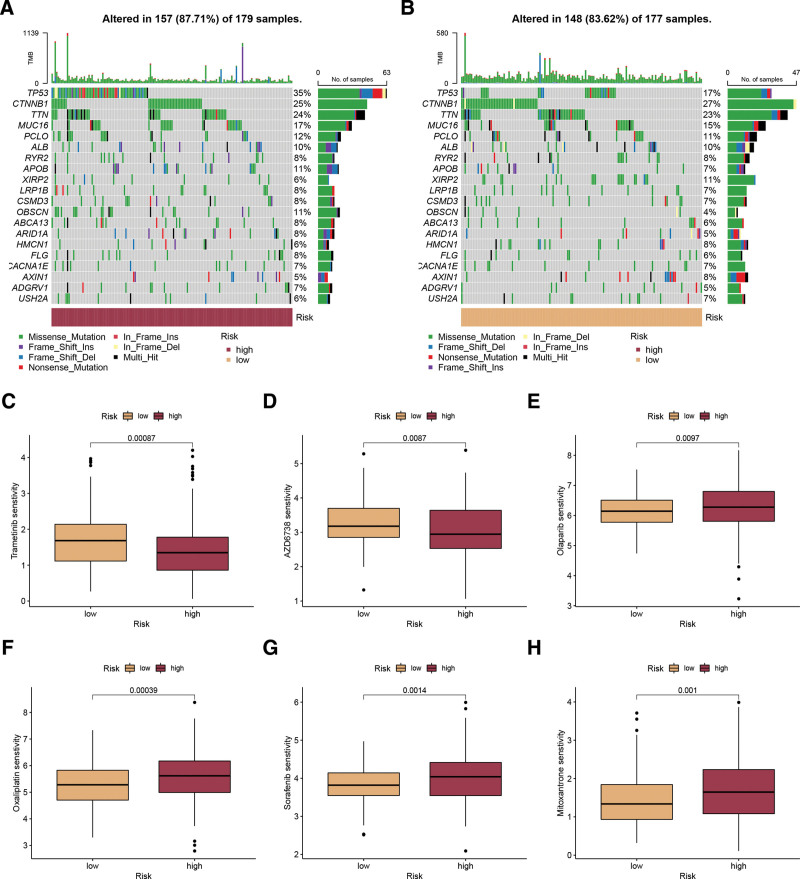
In order to examine the disparities in genomic mutations among 2 subgroups categorized by disulfidptosis score, we illustrated the mutation landscape comparing the high and low disulfidptosis score groups (Fig. 6A and B). A unique mutation profile was identified in the 2 groups. As an example, TP53, a significant gene that suppresses tumors, exhibited a mutation rate of 35% in the group with a high disulfidptosis score, which was considerably greater than the 17% observed in the group with a low disulfidptosis score (Fig. 6A and B). Moreover, the responsiveness to typical chemotherapy drugs was assessed in every sample obtained from the TCGA-LIHC dataset. In Figure 6C–H, it was noticed that a high disulfidptosis score resulted in considerably lower IC50 values for Trametinib and AZD6738, whereas a low disulfidptosis score led to significant decreases in IC50 values for Olaparib, Oxaliplatin, Sorafenib, and Mitoxantrone. Consequently, individuals exhibiting elevated disulfidptosis scores may display heightened responsiveness to Trametinib and AZD6738, whereas those with a low disulfidptosis score are more inclined to experience advantages from Olaparib, Oxaliplatin, and Sorafenib.
Figure 6.
Mutation analysis and drug sensitivity. (A, B) The mutation landscape between high (A) and low (B) disulfidptosis score groups. (C-H) The estimated IC50 values of Trametinib, AZD6738, Olaparib, Oxaliplatin, Sorafenib, and Mitoxantrone in patients with high and low disulfidptosis scores.
4. Discussion
HCC remains a significant global healthcare problem.[12] Although multiple treatment modalities are available for advanced HCC, including liver transplantation, percutaneous ablation, radiation, immunotherapy, and targeted therapy, the overall prognosis of HCC patients remains unfavorable.[13,14] These calls for the identification of novel treatment targets to improve survival rates as well as new biomarkers for assessing HCC progression.
Recently, Professor Gan Boyi and his team members discovered and identified a new type of cell death called disulfidptosis.[3] They found that high expression of SLC7A11 restricts NADPH production during glucose starvation, leading to the large accumulation of small molecules of disulfide, a series of redox defects, and cell death. Many cancer therapies use apoptosis to kill cancer cells.[3] However, many cancer cells can find ways to evade treatment-induced apoptosis, eventually leading to treatment resistance and disease recurrence. This suggests that targeted disulfide death as a cancer therapy deserves further investigation. The SLC7A11 transporter, belonging to the SLC family, has a crucial function in preserving cellular glutathione levels and safeguarding cells against cell death caused by oxidative stress.[15] Numerous studies have indicated that SLC7A11 is responsible for promoting resistance to ferroptosis and has a regulatory function in the context of cancer. SLC7A11 is not only an effective target for ferroptosis, but also has a significant impact on the treatment of drug-resistant tumors, including lung adenocarcinoma, bladder cancer, and colorectal cancer.[16–19] While certain research has indicated that drug-induced disulfidptosis in tumor cells may involve multiple DRGs, the complexity of the molecular level of HCC makes it inefficient to rely solely on a single gene or factor for constructing a predictive model. On the other hand, a strong and more precise model can be created by merging several genes. Hence, a dependable predictive gene pattern is necessary to support personalized prognosis and precision medical intervention, assisting in the anticipation of patient survival in HCC cases.
In this study, we identified 2 distinct disulfidptosis phenotypes characterized by varying clinical manifestations, survival rates, and infiltration of immune cells. Cluster B was highly expressed DRGs and exhibited a better prognosis compared with cluster A. Disulfidptosis cluster A presented the infiltrations of CD8 + T cells, activated NK cells, and gamma delta T cells, while disulfidptosis cluster B exhibited the infiltrations of M0 macrophages, resting dendritic cells, and neutrophils. We employed GSVA enrichment analysis to explore the differences in biological behavior among these 2 subtypes. The enrichment of pathways related to carcinogenic activation pathways such as TGF-β signaling pathway, pathways in cancer, and pancreatic cancer, was observed in disulfidptosis cluster B. According to suggestions, the initiation of TGF-β-associated routes hinders the infiltration of lymphocytes into the tumor parenchyma.[20] Modulating TGF-β has the potential to alter the TME and boost the body immune response against cancer.[21,22] Therefore, we deduced that individuals with HCC in the disulfidptosis cluster B might experience advantages from the amalgamation of immunotherapy and TGF-β inhibitors. To further explore the potential biological activity of disulfidptosis phenotypes, a total of 238 DEGs were identified from the overlap across different disulfidptosis phenotypes. KEGG enrichment analysis reveals that DEGs are highly enriched in ECM and tumor invasion, particularly ECM-receptor interaction and Focal adhesion, and PI3K-Akt signaling pathway. Certain studies have demonstrated that the ECM receptor plays a role in the advancement of HCC and is associated with unfavorable clinical outcomes. HCC patients with poor clinical prognosis were independently predicted by proteins associated with focal adhesion.
Next, we discovered 238 shared DEGs associated with 2 disulfidptosis phenotypes, which exhibited significant connections to pathways related to metabolism and tumors. This finding confirms that the aforementioned genes serve as gene signatures for disulfidptosis phenotypes. Afterward, in order to build a DRS for forecasting HCC prognosis, a total of 9 DEGs were utilized, namely LAMB1, FBLN2, FTCD, RDH16, MSC, SERPINE1, SPP1, CYP2C9, and IGHG1. Some of the DEGs have been reported to be associated with HCC.[23–25] The DRS can classify HCC patients into low and high disulfidptosis score groups in the training set (TCGA-LIHC) and external validation set (GSE14520). As expected, patients in the high disulfidptosis score group exhibited a worse prognosis and advanced clinical features. Furthermore, disulfidptosis score showed a strong independent prognostic ability. To enhance the precision of prognostic forecasting, we developed and verified a nomogram through the evaluation of different factors. The findings demonstrated a significant correlation between the prognosis of HCC and factors such as DRS, surgery, and pathological stage. Furthermore, we created a numerical chart that enhanced effectiveness and simplified the utilization of the Drs Additionally, this DRS reflects immunocyte infiltration, immune-related functions, immunotherapy, and drug sensitivity of risk groups. The utilization of this prognostic DRS may help to enable personalized prognostic prediction and expedite the advancement of individualized cancer treatment.
The tumor microenvironment (TME) is a complex environment for the survival and development of tumor cells, and immune cells in the microenvironment and their regulatory modes have a crucial impact on the onset and advancement of cancer.[26] In our study, there was less infiltration of stromal cells in the high disulfidptosis score group, indicating higher tumor purity in the high disulfidptosis score group. Tumor purity was remarkably associated with the clinicopathological traits, genomic expression, and biological characteristics of patients with tumors. Using the ssGSEA algorithms, patients of high risk had lower levels of immune infiltrating cells, particularly B cells and CD8 + T cells. T cell-mediated antitumor immunity is complemented by tumor-infiltrating B cells.[27] The secretion of immunoglobulins by B cells, along with promoting T-cell response and directly killing cancer cells, can inhibit tumor progression.[28] Furthermore, the risk score was negatively correlated with cytolytic activity and IFN response. High cytolytic activity in TME from NK cells or T cells against tumor cells leads to better immunotherapy and prognosis.[29] IFN family regulates both innate and adaptive immune responses that mediate the anti-tumor effect.[30] Immunotherapies, which focus on immune checkpoints, are regarded as highly effective therapeutic approaches for enhancing cancer outcomes.[31,32] Herein, we observed that PD-L1 expression was higher in the low-risk group than in the high-risk group. The IPS and TIDE scores further demonstrated that the low-risk group had a better response to immunotherapy. Taken together, this research validated the dependable prognostic effectiveness of the disulfidptosis score in the immune system reaction, indicating that disulfidptosis could potentially be utilized in medical settings to recognize the immunophenotypes and direct treatment approaches.
Nevertheless, several limitations should be pointed out. First, we acquired gene expression and clinical HCC data from publicly accessible websites, and it is imperative to confirm our findings through supplementary experimental assays. Furthermore, the phenotypes of disulfidptosis and the disulfidptosis score were assessed using retrospective cohorts. Hence, it is necessary to employ potential groups to validate our findings. Furthermore, it is imperative to conduct functional and mechanistic investigations to elucidate the precise role and mechanisms of DRGs in the advancement of HCC.
5. Conclusions
This study comprehensively assessed the disulfidptosis phenotypes and developed a disulfidptosis-related score that provides new insights into the tumor immune microenvironment and complexity in HCC. The disulfidptosis score can serve as a reliable tool for estimating the prognosis of HCC patients and customizing more effective immunotherapeutic strategies.
Author contributions
Conceptualization: Xi Chen, Yongan Zhou.
Data curation: Xi Chen, Qun Liang.
Formal analysis: Xi Chen, Qun Liang.
Investigation: Xi Chen, Qun Liang.
Methodology: Xi Chen, Qun Liang.
Project administration: Xi Chen, Qun Liang.
Resources: Xi Chen, Qun Liang.
Software: Xi Chen, Qun Liang.
Supervision: Yongan Zhou.
Validation: Qun Liang, Yongan Zhou.
Visualization: Xi Chen, Qun Liang.
Writing – original draft: Xi Chen, Qun Liang, Yongan Zhou.
Writing – review & editing: Xi Chen, Qun Liang, Yongan Zhou.
Supplementary Material
Abbreviations:
- DEGs
- differently expressed genes
- DRGs
- disulfidptosis-related genes
- DRS
- disulfidptosis-related signature
- GSVA
- gene set variation analysis
- HCC
- hepatocellular carcinoma
- PCA
- principal component analysis
- RCD
- regulated cell death
- ROC
- receiver operating characteristic
XC and QL contributed equally to this work.
Supplemental Digital Content is available for this article.
The authors have no funding and conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
How to cite this article: Chen X, Liang Q, Zhou Y. Construction of a novel disulfidptosis-related signature for improving outcomes in hepatocellular carcinoma: Observational study. Medicine 2023;102:40(e35423).
Contributor Information
Xi Chen, Email: chenxi202305@sina.com.
Qun Liang, Email: liangqun2023@sina.com.
References
- [1].Vogel A, Meyer T, Sapisochin G, et al. Hepatocellular carcinoma. Lancet. 2022;400:1345–62. [DOI] [PubMed] [Google Scholar]
- [2].Gilles H, Garbutt T, Landrum J. Hepatocellular carcinoma. Crit Care Nurs Clin North Am. 2022;34:289–301. [DOI] [PubMed] [Google Scholar]
- [3].Liu X, Nie L, Zhang Y, et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol. 2023;25:404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sideris S, Aoun F, Zanaty M, et al. Efficacy of weekly paclitaxel treatment as a single agent chemotherapy following first-line cisplatin treatment in urothelial bladder cancer. Mol Clin Oncol. 2016;4:1063–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mitin T, Hunt D, Shipley WU, et al. Transurethral surgery and twice-daily radiation plus paclitaxel-cisplatin or fluorouracil-cisplatin with selective bladder preservation and adjuvant chemotherapy for patients with muscle invasive bladder cancer (RTOG 0233): a randomised multicentre phase 2 trial. Lancet Oncol. 2013;14:863–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen TY, Yang CY, Yang MT, et al. Protein disulfide isomerase a4 promotes lung cancer development via the Stat3 pathway in stromal cells. Clin Transl Med. 2022;12:e606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang S, Jackson C, Karapetyan E, et al. Roles of protein disulfide isomerase in breast cancer. Cancers (Basel). 2022;14:745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Clement CC, Osan J, Buque A, et al. PDIA3 epitope-driven immune autoreactivity contributes to hepatic damage in type 2 diabetes. Sci Immunol. 2022;7:eabl3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kondo R, Ishino K, Wada R, et al. Downregulation of protein disulfide-isomerase A3 expression inhibits cell proliferation and induces apoptosis through STAT3 signaling in hepatocellular carcinoma. Int J Oncol. 2019;54:1409–21. [DOI] [PubMed] [Google Scholar]
- [10].Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Șenbabaoğlu Y, Michailidis G, Li JZ. Critical limitations of consensus clustering in class discovery. Sci Rep. 2014;4:6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nagaraju GP, Dariya B, Kasa P, et al. Epigenetics in hepatocellular carcinoma. Semin Cancer Biol. 2022;86(Pt 3):622–32. [DOI] [PubMed] [Google Scholar]
- [13].Su GL, Altayar O, O’Shea R, et al. AGA clinical practice guideline on systemic therapy for hepatocellular carcinoma. Gastroenterology. 2022;162:920–34. [DOI] [PubMed] [Google Scholar]
- [14].Galle PR, Dufour JF, Peck-Radosavljevic M, et al. Systemic therapy of advanced hepatocellular carcinoma. Future Oncol. 2021;17:1237–51. [DOI] [PubMed] [Google Scholar]
- [15].Goji T, Takahara K, Negishi M, et al. Cystine uptake through the cystine/glutamate antiporter xCT triggers glioblastoma cell death under glucose deprivation. J Biol Chem. 2017;292:19721–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12:599–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang L, Liu W, Liu F, et al. IMCA induces ferroptosis mediated by SLC7A11 through the AMPK/mTOR pathway in colorectal cancer. Oxid Med Cell Longev. 2020;2020:1675613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shen L, Zhang J, Zheng Z, et al. PHGDH inhibits ferroptosis and promotes malignant progression by upregulating SLC7A11 in bladder cancer. Int J Biol Sci. 2022;18:5459–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang W, Sun Y, Bai L, et al. RBMS1 regulates lung cancer ferroptosis through translational control of SLC7A11. J Clin Invest. 2021;131:e152067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tauriello DVF, Palomo-Ponce S, Stork D, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554:538–43. [DOI] [PubMed] [Google Scholar]
- [21].Peng D, Fu M, Wang M, et al. Targeting TGF-β signal transduction for fibrosis and cancer therapy. Mol Cancer. 2022;21:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50:924–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu T, Gan H, He S, et al. RNA helicase DDX24 stabilizes LAMB1 to promote hepatocellular carcinoma progression. Cancer Res. 2022;82:3074–87. [DOI] [PubMed] [Google Scholar]
- [24].Zhu YH, Li JB, Wu RY, et al. Clinical significance and function of RDH16 as a tumor-suppressing gene in hepatocellular carcinoma. Hepatol Res. 2020;50:110–20. [DOI] [PubMed] [Google Scholar]
- [25].Yu D, Green B, Marrone A, et al. Suppression of CYP2C9 by microRNA hsa-miR-128-3p in human liver cells and association with hepatocellular carcinoma. Sci Rep. 2015;5:8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xiao Y, Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther. 2021;221:107753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Engelhard V, Conejo-Garcia JR, Ahmed R, et al. B cells and cancer. Cancer Cell. 2021;39:1293–6. [DOI] [PubMed] [Google Scholar]
- [28].Tokunaga R, Naseem M, Lo JH, et al. B cell and B cell-related pathways for novel cancer treatments. Cancer Treat Rev. 2019;73:10–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sivori S, Pende D, Quatrini L, et al. NK cells and ILCs in tumor immunotherapy. Mol Aspects Med. 2021;80:100870. [DOI] [PubMed] [Google Scholar]
- [30].Alspach E, Lussier DM, Schreiber RD. Interferon γ and its important roles in promoting and inhibiting spontaneous and therapeutic cancer immunity. Cold Spring Harb Perspect Biol. 2019;11:a028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Riley RS, June CH, Langer R, et al. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discovery. 2019;18:175–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 2020;17:807–21. [DOI] [PMC free article] [PubMed] [Google Scholar]