Abstract
It has been suggested that the antibiotic-induced release of lipopolysaccharide (LPS) is an important cause of the development of septic shock in patients treated for severe infections caused by gram-negative bacteria. β-Lactam antibiotics change the integrity of the bacterial cell envelope by binding to penicillin-binding proteins (PBP) in the membrane and thus may affect the amount of LPS that is released and the kinetics of that release. In this respect, ceftazidime at intermediate concentrations binds with a high affinity to PBP 3 and PBP 1a and thus can induce filament formation in addition to killing, whereas imipenem preferentially binds to PBP 2 and PBP 1b, leading to spheroplast formation and rapid cell lysis. We investigated the effects of these antibiotics on the killing and the release of the radioactively labelled LPS of Salmonella typhi Ty 21A. A mathematical model was developed to calculate the delay between bacterial killing and LPS release, designated the lag time. At antibiotic concentrations inducing equal killing, the amount of LPS released was the same for both antibiotics. Only after 6 h of incubation at antibiotic concentrations above 0.5 μg/ml, the amount of 3H-LPS released was slightly higher (∼1.2-fold) in incubations with ceftazidime than in those with imipenem, and the maximum releases of the total label were 33.2% ± 0.89% and 27.1% ± 0.45%, respectively. Despite the clear concentration-dependent effect on the bacterial killing and subsequent LPS release, the lag time was independent of the antibiotic concentration. For ceftazidime as well as imipenem the lag time amounted to approximately 60 min. In conclusion, our findings imply that the mechanism of antibiotic-induced LPS release is independent of the PBP affinities for these β-lactam antibiotics. Furthermore, once the organism is killed by either imipenem or ceftazidime, the rate of LPS release from S. typhi does not differ according to the antibiotic with which the organism is killed, and there is little difference in the relative amount of LPS released.
In early studies of the treatment of patients with typhoid fever, circulatory collapse was observed in some patients shortly after the administration of a loading dose of chloramphenicol (18). Recently, it was suggested that this phenomenon may have been caused by the rapid release of lipopolysaccharide (LPS) from the bacteria as a result of the antibiotic therapy (8). This led to the paradox that antibiotic treatment of bacteremic patients in some cases may actually elicit circulatory shock.
It is generally accepted that LPS from the outer membrane of gram-negative bacteria is responsible for many of the clinical symptoms of sepsis, because it stimulates monocytes and macrophages to produce large amounts of proinflammatory mediators like tumor necrosis factor alpha and interleukins 1β and 6 (2, 13, 21). Although LPS molecules are anchored in the bacterial outer membrane, they are released spontaneously during bacterial growth or after exposure of the bacteria to antibiotics (4, 5, 11).
In the last decade, the antibiotic-induced release of LPS from pathogens like Escherichia coli, Haemophilus influenzae, and Pseudomonas aeruginosa has been studied extensively in vitro as well as in vivo (9). Attention has focused especially on the β-lactam antibiotics ceftazidime and imipenem. The reason for this interest lies in the difference in their binding to penicillin-binding proteins (PBPs). Ceftazidime binds to several PBPs, but at low (14) to intermediate concentrations it binds with a high affinity to PBP 3 and thus may lead to filamentation of the bacteria (4, 23), whereas imipenem preferentially binds to PBP 2 and PBP 1b, resulting in spheroplast formation and a rapid bactericidal effect, respectively (15). It has been suggested that in the case of ceftazidime, the formation of filaments leads to an increase in bacterial biomass and subsequently to a larger pool of LPS to be set free after cell lysis (11). In addition to influencing the amount of LPS that is released, structural changes induced by ceftazidime and imipenem may also affect the kinetics of this process.
The aim of the present study was to gain insight into the kinetics of LPS release as well as the amount of LPS that is released from the outer membrane of a gram-negative organism after incubation with ceftazidime and imipenem. LPS molecules of a galE mutant of Salmonella typhi were labelled by incorporation of 3H-galactose. The release of 3H-LPS after antibiotic-induced killing was determined at various time points and with different antibiotic concentrations based on the MIC. A mathematical model was developed to calculate the delay between bacterial killing and the release of LPS, designated the lag time (Tlag).
MATERIALS AND METHODS
Bacteria.
A galE mutant of S. typhi Ty2 (Ty 21A; Vivotif Berna, Bern, Switzerland) was used. This strain has a UDP-galactose-4-epimerase deficiency and lacks the ability to metabolize normally endogenous galactose, which is necessary for the completion of the outer core oligosaccharide and the production of O side chain pentamers (24). When cultured in the presence of d-galactose, either 3H labelled (Amersham International plc, Buckinghamshire, England) or unlabelled (Sigma Chemical Co., St. Louis, Mo.), synthesis of LPS side chains does occur; this was verified before each experiment by agglutination tests with two specific rabbit anti-LPS serum samples (polyvalent O A-G and 9-0 Salmonella Somatic agglutinating serum; Murex Diagnostics Ltd., Dartford, England). In all experiments nutrient broth supplemented with 0.025% d-glucose (NB-glc) was used. The addition of glucose to the medium was essential for preventing the intracellular accumulation of intermediate products and subsequent osmotic lysis of the bacteria (7). Stock solutions of S. typhi Ty 21A in NB-glc containing 10% glycerol were stored as aliquots at −70°C until use.
Radioactive labelling of S. typhi Ty 21A.
Before each experiment S. typhi Ty 21A was cultured aerobically in NB-glc containing 0.006% 3H-galactose (final concentration 10 μCi/ml; Amersham International plc, Buckinghamshire, England) at 37°C. To remove nonincorporated label the bacteria were washed four times (by centrifugation at 6,000 × g for 10 min) with warm (37°C) medium. Of the total amount of added label, 22.1% ± 1.8% (mean ± standard deviation; n = 6) was incorporated. In preliminary experiments an assessment of whether the radioactively labelled galactose was incorporated specifically into the bacterial membrane and not into the cytosol was performed. To this end, S. typhi Ty 21A was labelled overnight with 3H-galactose, as described above. After washing the bacteria four times in normal medium and twice in 50 mM Tris-HCl containing 2 mM EDTA, the bacteria were resuspended in Tris-HCl. Five freezing-thawing cycles were performed to disintegrate the bacteria, and cell debris was removed by centrifugation (at 1,200 × g for 20 min). The LPS molecules from the cell membrane were separated from the cytosolic fraction by ultracentrifugation at 100,000 × g for 3 h, and the levels of radioactivity in both the pellet and the supernatant fractions were measured. Of the total amount of radioactive label, the pellet contained 95.4%, whereas the cytosolic fraction contained only 4.6%, indicating that LPS molecules in the cell membrane were labelled preferentially.
Antibiotics.
Ceftazidime was obtained from Glaxo Wellcome B.V. (Zeist, The Netherlands), and imipenem was obtained from Merck Sharp & Dome (Haarlem, The Netherlands). The ceftazidime and imipenem MICs for S. typhi Ty 21A, as determined by standard agar sensitivity tests (1), were 0.25 μg/ml. At the start of each experiment stock solutions of the antibiotics were prepared in phosphate-buffered saline (pH 7.4) and were diluted to the appropriate concentrations in NB-glc with 0.05% galactose. The antibiotics were added to the bacterial cultures at final concentrations of 0.0625, 0.125, 0.25, 0.5, 1, 2, 4, and 16 μg/ml (i.e., 0.25 to 64 times the MIC).
Bacterial killing and endotoxin release assay.
An inoculum of 3.23 × 108 ± 1.25 × 108 radioactively labelled S. typhi Ty 21A per ml of NB-glc supplemented with 0.05% unlabelled galactose was used. This high inoculum was necessary in order to obtain enough radioactivity for measurement of the amount of radioactivity in the samples. The bacteria were cultured aerobically at 37°C; after 1 h the antibiotic was added. At various time points thereafter aliquots were collected and the number of viable microorganisms was determined microbiologically by plating serial 10-fold dilutions on blood agar plates. The amount of viable bacteria was expressed as the numbers of CFU per milliliter. For quantification of antibiotic-induced LPS release, the samples were divided into two equal aliquots. The total amount of radioactivity in one aliquot was measured directly. The other aliquot was first centrifuged (at 6,000 × g for 15 min), after which the amount of radioactivity in the supernatant was determined as a measure for the amount of LPS released. All measurements were performed in duplicate and by liquid scintillation counting over 5 min. The amount of LPS released was expressed as a percentage of the total radioactivity.
Mathematical model for Tlag analysis.
A mathematical model was developed to calculate the time relation between the antibiotic-induced killing of S. typhi Ty 21A and the release of LPS from its membrane. The time course of the numbers of CFU at a given concentration was regarded as a reflection of the distribution of survival times of individual bacteria in the inoculum (12). It was assumed that at the start of the experiment the radioactively labelled LPS content is not different between bacteria. Correction for the ongoing cell division in the surviving fraction of bacteria is necessary because of twofold dilution of the initial label at each cell division. Because the bacteria in the inoculum were supposed to multiply at the same rate as the controls in the absence of antibiotics, the actual number of CFU was corrected as follows: N′ = N · e−rt, in which N′ is the corrected number of CFU, N is the actual number of CFU, t is the time, and r is the exponential rate of growth of S. typhi Ty 21A in the absence of antibiotics. For the corrected number of CFU, the mean survival time was calculated as the first statistical moment (19) by using an algorithm described by Brockmeier (3). This algorithm is based on the area under the curve (AUC) of the numbers of CFU extrapolated to infinity. Since the number of CFU at 6 h was generally less than 1% of the inoculum, the extrapolated AUC was regarded as an insignificant contribution to the total AUC. N′ is proportional to the amount of LPS bound in the membranes of viable bacteria. Thus, the mean survival time of each bacterium is the same as the mean time during which LPS is bound in the membrane of a viable bacterium (TLPSV).
The LPS release was supposed to occur some time after a bacterial cell had become nonviable. Furthermore, the total amount of LPS set free at the highest antibiotic concentration was regarded as a reflection of the total amount of labelled LPS in the inoculum that eventually is available to be set free. From this maximum the amount of LPS released at various time points during the experiment was subtracted, resulting in the value for the amount of LPS still bound in bacteria, either viable or nonviable. By the same algorithm mentioned above, the first statistical moment of this value was calculated. This was regarded as the mean time that LPS is bound in viable as well as nonviable bacteria (TLPSV/NV). The difference between TLPSV and TLPSV/NV represents the mean time that LPS is bound to the membranes of nonviable bacteria and therefore is a measure of the delay between the death of a bacterium and the release of LPS. This delay was designated Tlag. Calculations were performed in Mathcad (version 3.0; MathSoft Inc., Cambridge, Mass.).
Analysis of data.
Two separate sets of experiments were performed for each β-lactam antibiotic, and a wide concentration range was used. The numbers of viable bacteria, the amount of LPS released, TLPSV, and TLPSV/NV were expressed as means ± standard errors of the means (SEMs) and were analyzed for time, concentration, and antibiotic dependency by analysis of variance. Tlag calculations performed with the two sets of data from the separate experiments were performed for each antibiotic concentration and were statistically examined by analysis of variance and regression analysis. Because the results of the two experiments were not significantly different and Tlags were found to be independent of antibiotic concentrations, all Tlags for each antibiotic were combined. Mean Tlags were expressed as means ± SEMs (n = 15 or 16) and were tested by a Student t test. P values of ≤0.05 were considered significant.
RESULTS
Antibiotic-induced killing of S. typhi Ty 21A.
The growth of S. typhi Ty 21A in antibiotic-free medium was determined in control experiments (n = 8), and the log10 numbers of viable bacteria increased from 8.69 ± 0.06 to 9.10 ± 0.08 during 6 h, a small increase probably due to the high inoculum at the start of the experiment. The growth rate of the bacteria was determined from 0 to 2, 2 to 4, and 4 to 6 h and amounted overall to 0.15 ± 0.07 log10 h−1.
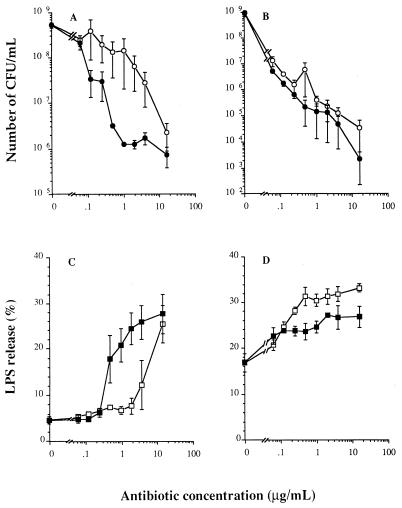
Ceftazidime and imipenem had clear bactericidal effects (Fig. 1A and B; data for 0.5, 1, and 4 h not shown). The killing depended on the antibiotic concentration and the incubation time. Incubation with various concentrations (0.25 to 64 times the MICs) of imipenem for 2 h led to the killing of a larger proportion of the bacteria than incubation with ceftazidime at similar concentrations. From Fig. 1A it can be deduced that approximately 10 times more ceftazidime than imipenem is needed to obtain the same extent of killing obtained at 2 h. This was observed for almost all antibiotic concentrations, especially those higher than 0.25 μg/ml (Fig. 1A). After 6 h of incubation, the number of viable bacteria remaining was similar for bacteria incubated with imipenem and ceftazidime, i.e., <0.1% of the inoculum at the start of the experiment for most concentrations (Fig. 1B). Maximal killing at 6 h was obtained with 16 μg of ceftazidime or imipenem per ml, resulting in a decrease in the viable numbers of 4.42 ± 0.50 or 6.04 ± 0.45 log10, respectively, compared with the numbers of bacteria cultured for 6 h in the absence of an antibiotic.
FIG. 1.
Numbers of viable bacteria (A and B) and amount of 3H-labelled LPS (C and D) released from S. typhi Ty 21A incubated for 2 h (A and C) and 6 h (B and D) with ceftazidime (○, □) or imipenem (•, ▪) at different concentrations. Values are the means ± SEMs of two experiments.
Antibiotic-induced release of 3H-LPS from S. typhi Ty 21A.
Bacteria cultured in antibiotic-free medium showed a basal release of 3H-labelled LPS of 2,739 ± 494 and 9,771 ± 1,255 cpm (mean ± SEM; n = 4) at 2 and 6 h, respectively, which represented 4.6% ± 0.3% and 16.8% ± 0.8% of the total amount of incorporated label. Incubation with ceftazidime or imipenem resulted in time- and concentration-dependent effects on the release of 3H-LPS (Fig. 1C and D; data for 0.5, 1, and 4 h not shown). After 2 h (Fig. 1C), the ceftazidime-induced release of LPS was increased only for concentrations of 4 μg/ml and higher, whereas imipenem had already induced a significantly higher level of release at much lower concentrations (≥0.25 μg/ml).
Compared to incubations with identical concentrations of ceftazidime, imipenem induced an approximately 10-fold higher level of release of LPS. A maximum amount of LPS was already released after 2 h of incubation with the highest concentrations of imipenem. After 6 h the amount of radioactively labelled LPS released was somewhat higher (∼1.2-fold) in the incubations with ceftazidime than in those with imipenem. This small difference was significant at ceftazidime concentrations above 0.5 μg/ml. The maximal 3H-LPS release induced by ceftazidime was 33.2% ± 0.89% of the total amount of labelled LPS, whereas with imipenem the maximal release amounted to 27.10% ± 0.45% (Fig. 1D).
Effects of the antibiotics on the binding of LPS in the bacterial membrane.
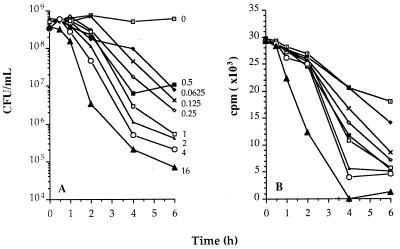
The results of a representative experiment with ceftazidime are presented in Fig. 2. The numbers of CFU, after correction for ongoing multiplication (Fig. 2A), were used to calculate the mean survival time, i.e., the time that LPS was bound in the membranes of viable bacteria (TLPSV), for each microorganism incubated with this given concentration. For example, the TLPSV at the highest concentration (16 μg/ml) was 0.58 ± 0.09 h. The corresponding values of TLPSV are presented in Fig. 3A; these values were significantly dependent on the antibiotic concentration (P ≤ 0.01). For both antibiotics the concentration range allowed the establishment of the maximal effect, i.e., the minimal values of TLPSV, which were similar for ceftazidime and imipenem and which amounted to approximately 0.5 h. However, at similar concentrations the killing activity of imipenem is about 10-fold greater than that of ceftazidime.
FIG. 2.
Killing of S. typhi by ceftazidime and subsequent release of LPS at various concentrations of the antibiotic. (A) The number of viable bacteria was corrected for ongoing multiplication of the inoculum. For each curve the mean survival time of the bacteria was calculated as the first statistical moment. This calculation is based on the AUC extrapolated to infinity (see Materials and Methods section). The mean survival time is a measure of TLPSV. For example, the effect at 16 μg/ml corresponds to a TLPSV value of 0.58 ± 0.09 h. (B) The amount of labelled LPS bound in the membrane of viable as well as nonviable bacteria was calculated by subtracting the actual amount of LPS released at each time point from the maximum amount of label released at the highest concentration. The first statistical moment of these curves was calculated, and this resulted in a value for TLPSV/NV. For example, the effect at 16 μg/ml corresponds to a TLPSV/NV value of 1.77 ± 0.23 h.
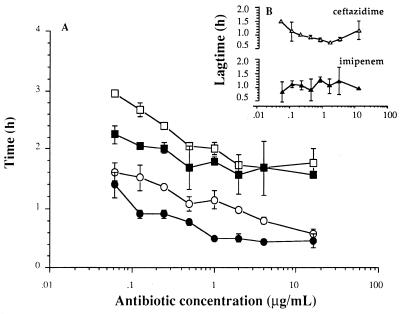
FIG. 3.
(A) TLPSV (circles) and TLPSV/NV (squares) for S. typhi incubated with different concentrations of ceftazidime (○, □) or imipenem (•, ▪). (B) The delay in the time between killing of the bacteria and the release of 3H-labelled LPS was calculated from the difference between TLPSV and TLPSV/NV, and was called Tlag. Values are means ± SEMs for one to two experiments.
The amount of LPS not released, calculated as the difference between the maximal amount released and the actual amount released, is shown for the same experiment (Fig. 2B). These values were used to calculate the mean time that LPS was bound in viable and nonviable bacteria, TLPSV/NV. For example, the TLPSV/NV at the highest concentration (16 μg/ml) was 1.77 ± 0.23 h. The corresponding TLPSV/NVs were also clearly concentration dependent (Fig. 3A), and minimal values were approximately 1.65 h for both ceftazidime and imipenem.
Tlag of LPS release after incubation with antibiotics.
Tlag, the delay in time between the time of killing of the bacteria and the time of the subsequent release of LPS from the bacterial membrane, represents the time that LPS molecules are still contained in the membranes of nonviable bacteria. Tlags were calculated from the difference between the mean TLPSV and the mean TLPSV/NV at each antibiotic concentration (Fig. 3B). Tlags ranged from 0.75 ± 0.01 to 1.49 h for ceftazidime and 0.85 ± 0.37 to 1.29 ± 0.09 h for imipenem. Although both TLPSV and TLPSV/NV were concentration dependent, the Tlags for ceftazidime and imipenem were not found to be significantly dependent on the antibiotic concentration (P values, 0.52 and 0.98, respectively). The mean Tlags for ceftazidime and imipenem did not vary significantly and were approximately 1 h (1.01 ± 0.07 h for ceftazidime and 1.08 ± 0.09 h for imipenem [values are means ± SEMs for 15 to 16 separately calculated Tlags]). In additional experiments the Tlags for two other antibiotics, i.e., aztreonam and gentamicin, were determined as described above for ceftazidime and imipenem. For aztreonam, an antibiotic that binds to PBP 3 with a high affinity, Tlag was also independent of antibiotic concentration and amounted to 1.05 ± 0.03 h (n = 16), which is the same as those found for imipenem and ceftazidime. Tlags for the aminoglycoside gentamicin could not be calculated because the release of 3H-labelled LPS was so low that it did not exceed control levels.
DISCUSSION
The main findings of the present study are that when ceftazidime and imipenem are examined at concentrations that induce an equivalent killing of an inoculum, the rates of release of radioactively labelled LPS from S. typhi do not differ, and there is little difference in the relative amount of LPS released. Furthermore, the delay between the time of antibiotic-induced killing and the time of the subsequent release of LPS from the bacterial membrane was concentration independent and was approximately the same, i.e., 1 h, for ceftazidime and imipenem. These conclusions are based on the observation that in incubations with S. typhi for up to 6 h, imipenem not only induces a more rapid killing of bacteria than ceftazidime but also induces a more rapid release of LPS. At equivalent levels of bacterial killing, however, the amount of LPS released is similar for both antibiotics. Only after 6 h of incubation at the highest concentrations did ceftazidime release somewhat higher (i.e., 1.2-fold) amounts of LPS. Tlags were calculated by microbiological determination of the numbers of viable bacteria, together with measurements of the amount of radioactively labelled LPS. Labelling of LPS can easily be accomplished in galactose-4-epimerase-deficient microorganisms, such as the vaccine strain S. typhi Ty 21A used in the present study. A drawback of this technique is that LPS molecules synthesized during the experiment, and therefore not labelled, are not detected. However, the number of filaments formed during our experiments must be very low due to the rapid killing (mean survival time, about 30 min), and the relatively slow growth, as evidenced by a generation time of approximately 1 h in the absence of antibiotics. Thus, under the present experimental conditions, the amount of unlabelled, newly formed LPS is probably very low. This indicates that our data give a fair estimate of the total amount of LPS that is released. Although by our method of radioactive labelling the actual molar amount of LPS molecules could not be measured, it did enable us to measure LPS in a direct way and to identify Tlag as a new parameter in the kinetics of LPS release.
When the release of LPS was analyzed, we found that bacterial killing, antibiotic concentration, and incubation period had a significant effect on the amount of LPS that was released. When examined at equivalent levels of bacterial killing, however, this amount of LPS was independent of the type of antibiotic used in the incubation. Although our results indicated a small difference (∼1.2-fold) in the amount of released LPS induced by ceftazidime compared to the amount induced by imipenem for bacteria incubated with the highest antibiotic concentrations for 6 h, they do not support earlier studies in which incubation of E. coli with ceftazidime resulted in a much higher (2.5- to 5-fold) release of LPS than incubation with imipenem, as determined by bioreactivity measurements in a Limulus amoebocyte lysate assay (4, 11). This discrepancy in the amount of LPS released could be related to differences in bacterial species, experimental conditions, bacterial inocula, or antibiotic concentrations. For instance, in some studies very high concentrations of antibiotics were examined (4), whereas we used concentrations in the range up to the peak levels in patient serum during antibiotic treatment. Moreover, experimental conditions in which comparisons between antibiotics are based only on MICs ignore the pharmacodynamic characteristics of the antibiotics: for instance, although two antibiotics may have identical MICs when they are evaluated after 24 h of incubation, their rates of bacterial cell turnover may differ strikingly during this period (1).
In the past, the release of LPS was thought to be a direct effect of bacteriolysis, which would imply that the bacterial killing is instantly followed by the release of LPS. This is probably the case for complement-mediated lysis: when E. coli is incubated with whole serum, LPS release occurs instantly and is complete within 10 to 12 min of exposure to serum (6, 22). In contrast to complement-mediated lysis, the present study shows that there is a delay between the antibiotic-induced killing and the release of LPS. The mean Tlag for S. typhi incubated with ceftazidime or imipenem is approximately 1 h and is independent of the antibiotic concentration. Studies with P. aeruginosa and E. coli bacteria showed that ceftazidime at intermediate to low concentrations binds to PBP 3 with a high affinity, whereas at high concentrations there is additional binding to PBP 1, probably leading to cell lysis (17). Although the PBP affinities of S. typhi are unknown, we assumed that they are highly similar to those of other members of the family Enterobacteriaceae. Thus, the finding that Tlag is the same for imipenem and ceftazidime suggests that the mechanism of LPS release is independent of PBP binding. This view is supported by the finding that the Tlag for ceftazidime as well as for aztreonam, an antibiotic with PBP binding characteristics similar to those of ceftazidime, was independent of the antibiotic concentration. Thus, once a microorganism is killed by a β-lactam antibiotic, neither the PBP affinity nor the concentration of the antibiotic influences the delay until the release of its LPS.
The remaining question is whether and to what extent antibiotic-induced LPS release and subsequent induction of cytokine production aggravate the clinical symptoms of septic shock. A number of clinical studies have addressed this issue but have yielded contradictory results (4, 10, 16, 20). In this respect, it is important to realize that the endotoxin concentration in blood is not the only factor determining the severity of septic shock caused by gram-negative bacteria. The biological effects of LPS in blood not only depend on the amount and the kinetics of LPS release but are also regulated by a complex system of serum proteins like LPS-binding protein, high-density lipoprotein, albumin, endogenous anticore antibodies, and bactericidal-permeability increasing protein, which can either enhance or neutralize the reactivity of LPS. Therefore, the clinical symptoms of endotoxemia will in the end be determined by the interplay between all of these factors.
In conclusion, this study shows that the Tlag in LPS release amounted to 1 h for ceftazidime as well as imipenem and was independent of the antibiotic concentration, despite a clear concentration-dependent effect of both antibiotics on the killing of S. typhi and the subsequent release of LPS. This argues for a mechanism of LPS release from the bacterial membrane that is independent of the PBP affinity of the antibiotic. Furthermore, the results indicate that the LPS release is intricately correlated with the bacterial killing and that at concentrations inducing equivalent killing of the inoculum there is little difference in the amount of LPS that is released and no difference in its rate of release.
ACKNOWLEDGMENTS
This study was financially supported by the Praeventiefonds (project 28-2293) and an educational grant from Glaxo Wellcome B.V. (Zeist, The Netherlands).
REFERENCES
- 1.Acar J F, Goldstein F W. Dilution in agar. In: Lorian V, editor. Antibiotics in laboratory medicine. 4th ed. Baltimore, Md: The Williams & Wilkins Co.; 1996. pp. 28–32. [Google Scholar]
- 2.Bone R C. The pathogenesis of sepsis. Ann Intern Med. 1991;115:457–469. doi: 10.7326/0003-4819-115-6-457. [DOI] [PubMed] [Google Scholar]
- 3.Brockmeier D. In vitro-in vivo correlation, a time scaling problem? Arzneim Forsch/Drug Res. 1984;34:1604–1607. [PubMed] [Google Scholar]
- 4.Dofferhoff A S M, Nijland J H, de Vries-Hospers H G, Mulder P O M, Weits J, Bom V J J. Effects of different types and combinations of antimicrobial agents on endotoxin release from gram-negative bacteria: an in-vitro and in-vivo study. Scand J Infect Dis. 1991;23:745–754. doi: 10.3109/00365549109024303. [DOI] [PubMed] [Google Scholar]
- 5.Evans M E, Pollack M. Effects of antibiotic class and concentration on the release of lipopolysaccharide from Escherichia coli. J Infect Dis. 1993;167:1336–1343. doi: 10.1093/infdis/167.6.1336. [DOI] [PubMed] [Google Scholar]
- 6.Fierer J, Finley F, Braude A I. Release of 51Cr-endotoxin from bacteria as an assay of serum bactericidal activity. J Immunol. 1974;112:2184–2192. [PubMed] [Google Scholar]
- 7.Germanier R, Furer E. Isolation and characterization of galE mutant Ty 21A of Salmonella typhi: a candidate strain for a live, oral typhoid vaccine. J Infect Dis. 1975;131:553. doi: 10.1093/infdis/131.5.553. [DOI] [PubMed] [Google Scholar]
- 8.Hopkin B D A. Frapper fort ou doucement: a gram-negative dilemma. Lancet. 1978;ii:1193–1194. [PubMed] [Google Scholar]
- 9.Hurley J C. Antibiotic-induced release of endotoxin: a reappraisal. Clin Infect Dis. 1992;15:840–854. doi: 10.1093/clind/15.5.840. [DOI] [PubMed] [Google Scholar]
- 10.Hurley J C, Louis W J, Tosolini F A, Carlin J B. Antibiotic-induced release of endotoxin in chronically bacteriuric patients. Antimicrob Agents Chemother. 1991;35:2388–2394. doi: 10.1128/aac.35.11.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson J J, Kropp H. β-Lactam antibiotic-induced release of free endotoxin: in vitro comparison of penicillin-binding protein (PBP)2-specific imipenem and PBP3-specific ceftazidime. J Infect Dis. 1992;165:1033–1041. doi: 10.1093/infdis/165.6.1033. [DOI] [PubMed] [Google Scholar]
- 12.Mattie H, Zhang L-C, van Strijen E, Sekh B R, Douwes-Idema A E A. Pharmacokinetic and pharmacodynamic models of the antistaphylococcal effects of meropenem and cloxacillin in vitro and in experimental infection. Antimicrob Agents Chemother. 1997;41:2083–2088. doi: 10.1128/aac.41.10.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michie H R, Manogue K R, Spriggs D R, et al. Detection of circulating tumor necrosis factor after endotoxin administration. N Engl J Med. 1988;318:1481–1486. doi: 10.1056/NEJM198806093182301. [DOI] [PubMed] [Google Scholar]
- 14.Milatovic D. Influence of subinhibitory concentrations of antibiotics on opsonization and phagocytosis of P. aeruginosa by human polymorphonuclear leucocytes. Eur J Clin Microbiol. 1984;3:288–293. doi: 10.1007/BF01977475. [DOI] [PubMed] [Google Scholar]
- 15.Neu, H. C. 1985. Relation of structural properties of beta-lactam antibiotics to antibacterial activity. Am. J. Med. 79(Suppl. 2A):2–13. [DOI] [PubMed]
- 16.Prins J M, van Agtmael M A, Kuijper E J, van Deventer S J H, Speelman P. Antibiotic-induced endotoxin release in patients with gram-negative urosepsis: a double-blind study comparing imipenem and ceftazidime. J Infect Dis. 1995;172:886–891. doi: 10.1093/infdis/172.3.886. [DOI] [PubMed] [Google Scholar]
- 17.Pucci M J, Boice-Sowek J, Kessler R E, Dougherty T J. Comparison of cefepime, cefpirome, and cefaclidine binding affinities for penicillin-binding proteins in Escherichia coli K-12 and Pseudomonas aeruginosa SC8329. Antimicrob Agents Chemother. 1991;35:2312–2317. doi: 10.1128/aac.35.11.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reilly J, Compagnon A, Tournier P, Bastin R, Du Buit H. Les accidents du traitement des fièvres typhoides par la chloromycétine. Ann Med. 1950;51:598. , 602, 607, 627–629, 644. [PubMed] [Google Scholar]
- 19.Sachs L. Additional statistics for the characterization of a one dimensional frequency distribution. In: Sachs J, editor. Applied statistics, a handbook of techniques. New York, N.Y: Springer-Verlag; 1982. pp. 103–106. [Google Scholar]
- 20.Shenep J L, Flynn P M, Barrett F F, Stidham G L, Westenkirchner D F. Serial quantitation of endotoxemia and bacteraemia during therapy for gram-negative bacterial sepsis. J Infect Dis. 1988;157:565–568. doi: 10.1093/infdis/157.3.565. [DOI] [PubMed] [Google Scholar]
- 21.Suffredini A F, Fromm R E, Parker M M, et al. The cardiovascular response of normal humans to the administration of endotoxin. N Engl J Med. 1989;321:280–287. doi: 10.1056/NEJM198908033210503. [DOI] [PubMed] [Google Scholar]
- 22.Tesh V L, Duncan R L, Jr, Morrison D C. The interaction of Escherichia coli with normal human serum: the kinetics of serum-mediated lipopolysaccharide release and its dissociation from bacterial killing. J Immunol. 1986;137:1329–1335. [PubMed] [Google Scholar]
- 23.Tuomanen E, Gilbert K, Tomasz A. Modulation of bacteriolysis by cooperative effects of penicillin-binding proteins 1a and 3 in Escherichia coli. Antimicrob Agents Chemother. 1986;30:659–663. doi: 10.1128/aac.30.5.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Dissel J T, Kwappenberg K M C, van Furth R. S. typhi vaccine strain Ty21A can cause a generalized infection in whole body-irradiated but not in hydrocortisone-treated mice. Scand J Immunol. 1995;41:457–461. doi: 10.1111/j.1365-3083.1995.tb03592.x. [DOI] [PubMed] [Google Scholar]