Abstract
Aureobasidin A (AbA) has strong antifungal effects arising from an unusual mechanism. We show that AbA interacts with ATP-binding cassette (ABC) transporters in yeast and mammalian cells. We isolated a gene of Saccharomyces cerevisiae that conferred resistance to AbA when the gene was present in multiple copies. The gene was identical to YOR1/YRS1, which confers resistance to oligomycin, reveromycin, and organic anions, none of which have structures similar to that of AbA. We also isolated an aur3R recessive mutant of S. cerevisiae with increased resistance to AbA. Northern hybridization showed that the aur3R mutant expressed not only YOR1 but also the ABC transporter-encoding gene PDR5 at high levels. Genetic studies showed that the aur3R mutant had a mutation in the PDR1 gene, which encodes a transcriptional regulator of PDR5 and YOR1. Analysis of a yor1 disruptant of the aur3/pdr1 mutant showed that both the functional YOR1 gene and the mutation in PDR1 were necessary for AbA resistance. These results suggest that YOR1 is more important than PDR5 for AbA resistance. We found in Candida albicans a novel gene whose sequence was similar to the sequence of YOR1 in S. cerevisiae. The amino acid sequence of the C. albicans YOR1 homolog showed no significant similarity to the sequences of CDR1 and CDR2, which are ABC transporters of C. albicans. Furthermore, AbA inhibited the efflux of the anticancer agent vincristine through P glycoproteins in cancer cells with multidrug resistance.
Aureobasidin A (AbA) is an antifungal antibiotic produced by Aureobasidium pullulans R106. It is a cyclic depsipeptide with a molecular weight of 1,100, and it contains eight amino acids and a hydroxy acid (15, 33). AbA is active against a variety of fungi including the budding yeast Saccharomyces cerevisiae, killing it by inhibiting bud growth due to abnormal deposition of actin and chitin (9). To identify the mechanism of action of AbA against yeasts, we and another group isolated an AbA resistance gene from the budding yeast (12, 13). The dominant resistance gene isolated from S. cerevisiae, AUR1, encodes a putative integral membrane protein and is essential for growth. The protein encoded by the AUR1 gene is associated with the activity of inositol phosphatidylceramide synthase, which is involved in sphingolipid synthesis (20), and seems to be an intracellular target in the resistance (12).
The resistance of tumor cells and pathogenic fungi to some chemotherapeutic drugs is a problem in the treatment of cancer and fungal infections. The multidrug resistance of tumors is caused by overexpression of a 170-kDa plasma membrane protein, the P glycoprotein belonging to the superfamily of ATP-binding cassette (ABC) transporters (8, 11, 14). In recent years, the frequent use of the antifungal agent fluconazole for the treatment of oropharyngeal candidiasis in AIDS patients has caused the appearance of C. albicans strains resistant to this and other azoles. Resistance to antifungal agents in C. albicans can be mediated by multidrug efflux transporters (26). Multidrug transporters are divided into two classes: the ABC multidrug transporters (22, 27) and the major facilitated transporter (10). Furthermore, ABC transporter proteins are classified into two subgroups according to their structures (14). One is the MDR subgroup represented by the mammalian multidrug resistance P glycoprotein (encoded by the MDR1 gene), and the other is the CFTR subgroup represented by human cystic fibrosis transmembrane conductance regulator (CFTR) (23) and multidrug resistance-associated protein (MRP1) (5). In human tumor cells, two ABC transporter genes, MDR1 and MRP1, elicit multidrug resistance when the genes are overexpressed. The budding yeast S. cerevisiae also has several ABC transporters from each subgroup. The mating factor transporter gene STE6 (18), the pleiotropic drug resistance gene PDR5/STS1 (1, 4), and the 4-nitroquinoline-N-oxide resistance gene SNQ2 (29) have sequences similar to the sequence of MDR1. The PDR5 and SNQ2 genes are involved in the cross-resistance of S. cerevisiae to antifungal azole drugs (19, 26). Pdr5p contributes to the resistance by pumping azoles out of the cell. A gene with a sequence similar to that of PDR5 in C. albicans, CDR1 (22), is overexpressed in azole-resistant clinical isolates (26). In contrast, a cadmium resistance gene (YCF1) and a gene involved in resistance to oligomycin and organic anions, YOR1/YRS1, encode proteins of the CFTR subgroup in S. cerevisiae (6, 16, 32).
In this paper, we report the role of ABC transporters in the AbA resistance of S. cerevisiae. AbA was a substrate of ABC transporters in both S. cerevisiae and human tumor cells.
MATERIALS AND METHODS
Yeast strains, media, and genetic methods.
The S. cerevisiae strains used in this study are listed in Table 1. Candida albicans TIMM 0136 was also used. Yeast cells were grown aerobically in YPD (1% yeast extract, 2% Bacto Peptone, 2% dextrose) at 30°C. Synthetic minimal medium (SD; 2% glucose, 0.7% yeast nitrogen base without amino acids, appropriate amino acid supplements) and sporulation medium (1% potassium acetate) were used. Standard genetic techniques of crossing, sporulation, and tetrad analysis were performed as described by Sherman et al. (30).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SH3328 | MATα his4 leu2-3,112 thr4 ura3-52 | S. Harashima |
| DKD-5D | MATa his3 leu2-3,112 trp1 | S. Harashima |
| AL22-4A | MATa his3 leu2-3,112 trp1 pdr1R-A1 | This study |
| AL49-3A | MATa his3 leu2-3,112 trp1 pdr1R-A3 | This study |
| AL33-9C | MATα his4 leu2-3,112 thr4 pdr1R-A2 | This study |
| AL33-18C | MATa his4 leu2-3,112 trp1 ura3-52 pdr1R-A2 | This study |
| A141-9A | MATα his4 leu2-3,112 thr4 ura3-52 yor1::URA3 | This study |
| AR9-4A | MATa his3 leu2-3,112 trp1 AURR1-1 | Hashida-Okado et al. (12) |
| AOD1 | MATa/MATα trp1/+ +/thr4 +/his4 his3/+ ura3-52/ura3-52 leu2-3,112/leu2-3,112 | Hashida-Okado et al. (12) |
| AOD3 | MATa/MATα trp1/+ +/thr4 +/his4 his3/+ ura3-52/ura3-52 leu2-3,112/leu2-3,112 yor1::URA3/+ | This study |
| A07 | MATα his4 leu2-3,112 thr4 ura3-52 (pWL7) | This study |
| T73-1 | MATa his3 leu2-3,112 trp1 pdr1R::LEU2 | This study |
Bioassay of drug sensitivity.
The sensitivities of yeast cells to various drugs and toxic compounds were assayed by measuring the MIC as follows. Yeast cells suspended in water were streaked with sterile toothpicks onto YPD agar plates containing various concentrations of drugs or other compounds. The plates were incubated at 30°C for 2 days.
Isolation and sequencing of YOR1.
Standard molecular cloning techniques were performed as described by Sambrook et al. (25). For construction of a genomic DNA library, chromosomal DNA was isolated from AbA-resistant mutant AR9-4A and wild-type strain DKD-5D of S. cerevisiae, as reported by Philipsen et al. (21). Each DNA was partially digested with HindIII, and fragments of 3 to 15 kb were obtained by agarose gel electrophoresis. The fragments were ligated to a HindIII-digested pWH5 vector and were then introduced into Escherichia coli HB101. The mutant genomic library was introduced by the modified lithium acetate procedure (28) into the wild-type strain SH3328, for which the MIC of AbA was 0.4 μg/ml. Colonies of Leu+ transformants were replicated on YPD agar plates with AbA at 1.5 μg/ml. From one transformant for which the MIC was 5 μg/ml, plasmid DNA was recovered and was designated pWL7. The ability of pWL7 to confer AbA resistance was checked by reintroduction of pWL7 into the wild-type strain.
Isolation of YOR1 homolog of C. albicans.
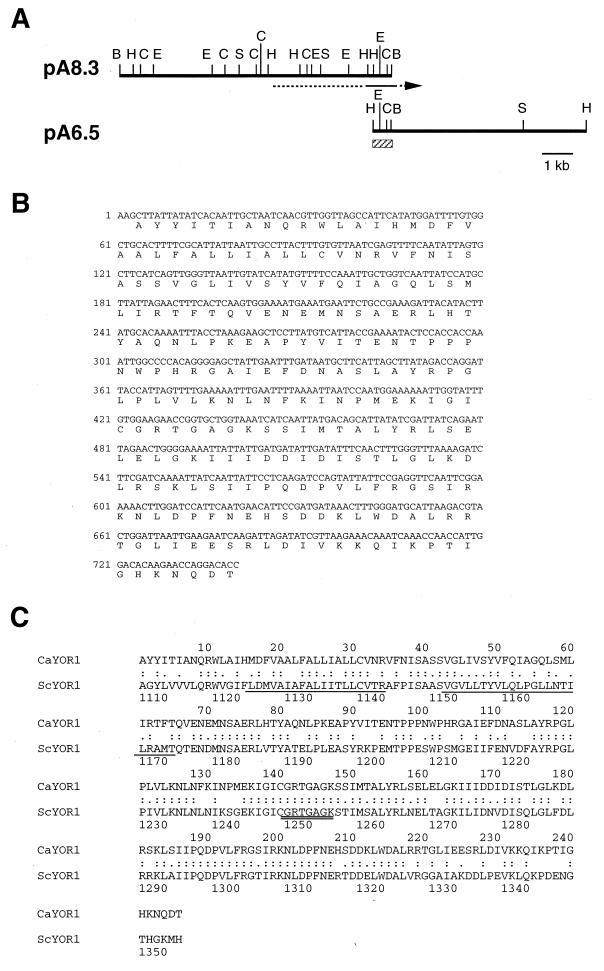
Chromosomal DNA was isolated from C. albicans TIMM 0136 as described by Philippsen et al. (21). The DNA was partially digested with either HindIII or BamHI and was ligated to pTV119 for construction of genomic DNA libraries. Plasmid pA8.3 containing the YOR1 homolog of C. albicans was isolated from the library by colony hybridization with a 1.2-kb HindIII-PstI fragment of S. cerevisiae YOR1 (see Fig. 1) as the probe. Plasmid pA6.5 was isolated by colony hybridization with a PCR product containing the carboxyl-terminal region of pA8.3 as the probe.
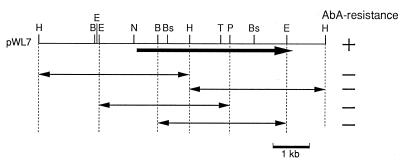
FIG. 1.
Restriction map and subcloning of the YOR1 locus. The restriction map of an 8.5-kb genomic insert of pWL7 is shown at the top. The thick arrow indicates the location and direction of an ORF. DNAs subcloned on the pWH5 vector were examined for their ability to confer AbA resistance on wild-type cells. B, BamHI; Bs, BstXI; E, EcoRI; H, HindIII; N, NheI; P, PstI; T, Tth111I. +, resistance conferred; −, resistance not conferred.
Isolation of AbA-resistant mutants.
The S. cerevisiae wild-type strain DKD-5D formed no colonies on YPD agar plates containing AbA at 0.4 μg/ml. Cells grown in YPD liquid medium were suspended in 0.2 M phosphate buffer (pH 8.0) containing 0.2% glucose and were treated with 3% ethyl methanesulfonate for 90 min, leaving about 40% of the cells viable. Mutagenized cells were cultured overnight in YPD medium containing 1 μg of AbA per ml and were plated onto YPD agar plates containing 1 μg of AbA per ml. The MICs of the antifungal agents cycloheximide, miconazole, amphotericin B, and AbA for the mutants were tested.
Gene disruption.
Gene disruption was done by a one-step method (24). For disruption of the YOR1 gene, an 8.5-kb HindIII fragment of pWL7 was subcloned into pUC119, and the plasmid obtained was cleaved with BstXI and blunted with T4 DNA polymerase. A 1.1-kb HindIII-EcoRI fragment of URA3 was blunted and inserted into the blunted BstXI sites of pWL7. By ligation of the blunted BstXI and HindIII, a HindIII recognition sequence was created. A linear 4-kb fragment containing yor1::URA3 was obtained by EcoRI digestion. This linear fragment was introduced into diploid strain AOD1, which was spread onto SD plates without uracil. The stable Ura+ transformants obtained were sporulated, and the tetrads that were obtained were dissected on YPD agar plates. All four spores from the tetrads formed colonies, indicating that YOR1 is not essential for growth. The segregation of Ura+:Ura− strains was 2:2. Disruption of YOR1 was confirmed by Southern hybridization of genomic DNAs from each of the four segregants. The pattern of hybridizing bands was identical to that expected from the restriction map, with the results showing disruption of the YOR1 gene of the Ura+ spores.
For the disruption of PDR1, a DNA fragment containing this gene was obtained by PCR. The primers used for PCR were 5′-ATCTTCGATATCATCTGCAGGG-3′ (positions +1032 to +1053) as the 5′ primer and 5′-TGCTGAGCGACCATTGAATGGC-3′ (positions +2820 to +2799) as the 3′ primer; the primers were based on the DNA sequence of PDR1 (1). Amplification by PCR was done with S. cerevisiae genomic DNA as the template. An amplified fragment was cloned into the HincII site of modified pUC19 lacking the HindIII site, generating pUCPDR1. The 0.8-kb HindIII fragment in PDR1 was replaced with a 2.2-kb HindIII fragment containing LEU2. The resulting plasmid, pUCPDR1::LEU2, was digested with SphI and BamHI to generate a linear 3.1-kb fragment, which was introduced into the aur3 mutant AL22-4A.
DNA and RNA analysis.
Southern hybridization was done as described by Sambrook et al. (25). S. cerevisiae RNA for Northern hybridization was prepared as described previously (12). The quantitation of the RNA was done by measuring the autoradiograph by densitometry. The PDR5 probe was obtained by PCR with oligonucleotides 5′-ACGTTACTAGCTACTCCTCCG-3′ (positions +35 to +55 of PDR5) as the 5′ primer, 5′-TTATTGAACAAGTCGTACGC-3′ (positions +1136 to +1117) as the 3′ primer, and S. cerevisiae genomic DNA as the template. The resulting 1.1-kb fragment was labeled and used as the probe.
Intracellular accumulation of [3H]vincristine.
AbA and verapamil were dissolved in dimethyl sulfoxide and diluted with phosphate-buffered saline (pH 7.2). An adriamycin-resistant cell line, A2780AD, of a human ovarian tumor was used as described previously (34). In brief, A2780AD cells (106/ml) in RPMI 1640 medium containing 5% fetal calf serum and 100 μg of kanamycin per ml were plated in wells of 24-well tissue culture dishes. After incubation of the cells at 37°C for 24 h, [3H]vincristine (222 GBq/mmol; Amersham) was added to a final concentration of 20 nM. Then various concentrations of drugs in a volume of 5 μl or the same volume of saline was added. After incubation of the cells at 37°C for 2 h, the intracellular concentration of vincristine was assayed. Means for triplicate samples were calculated.
Nucleotide sequence accession number.
The C. albicans YOR1 gene sequence has been deposited in the GenBank database under accession no. AF034608.
RESULTS
Cloning of a gene conferring AbA resistance.
While isolating an AUR1R mutant gene from a genomic library of resistant mutants constructed with the multicopy vector pWH5, we obtained a transformant with somewhat more AbA resistance than wild-type cells; the transformant grew on YPD agar plates containing 1.5 μg of AbA per ml but not on plates containing 5 μg of AbA per ml. Most of the other transformants obtained had higher levels of resistance, even growing in the presence of 25 μg of AbA per ml, so resistance was high; the AUR1R gene has been recovered from such transformants (12). Plasmid pWL7, which contained an 8.5-kb DNA fragment, was recovered from the transformant with the lowest level of resistance. Digested DNA fragments derived from the 8.5-kb insert shown in Fig. 1 did not confer AbA resistance on wild-type cells, so the insert contained an AbA resistance gene other than AUR1R. The ability of pWL7 to confer AbA resistance was checked by reintroduction of pWL7 into wild-type strain SH3328. Nucleotide sequencing showed that the insert DNA had a large open reading frame (ORF) of 4,431 bp encoding a protein of 1,477 amino acid residues. Searches for sequences similar to the predicted polypeptide sequence have shown that the sequence of the ORF is identical to that of YOR1/YRS1, which confers resistance to oligomycin, reveromycin, and organic anions (6, 16). Yor1p/Yrs1p is a member of the ABC transporter superfamily, like MDR1 and CFTR, and is most closely related to human MRP1 (5) and the cadmium resistance factor Ycf1p in S. cerevisiae (32). We constructed by one-step gene disruption a strain depleted of the YOR1 gene. The disrupted cells (Ura+; clones b and c, Fig. 2) did not grow on YPD agar plates containing 0.2 μg of AbA per ml, but the YOR1+ cells (clones a and b, Fig. 2) grew well. This result showed that the yor1-null cells were hypersensitive to AbA. Next, we examined the sensitivities of wild-type (DKD-5D, SH3328), yor1-null (A141-9A), multicopy YOR1-containing (A07), and aur3R (AL22-3A; see below) cells to various drugs and compounds on YPD agar plates (Table 2). The MICs of these drugs and compounds on YPD agar plates were the same for these strains and the wild-type strain. This result indicated that the YOR1 gene is specifically involved in resistance to AbA.
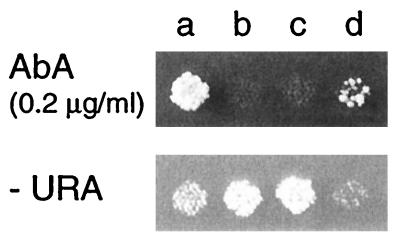
FIG. 2.
Sensitivity of yor1-disrupted cells to AbA. Tetrads derived from heterozygous diploid strain AOD3 (YOR1/yor1::URA3) were incubated on YPD agar plates and replicated onto YPD agar plates with 0.2 μg of AbA per ml or SD plates without uracil (−URA). Drug supersensitivity and Ura+ also segregated together in other tetrads (data not shown).
TABLE 2.
Sensitivities of the yor1 gene mutants
| Compound (MIC units) | MICa
|
||||
|---|---|---|---|---|---|
| DKD-5D (wild type) | SH3328 (wild type) | AL22-4A (aur3R) | A141-9A (yor1::URA3) | A07 (pWL7) | |
| AbA (μg/ml) | 0.4 | 0.4 | >25 | 0.2 | 5.0 |
| Cycloheximide (μg/ml) | 0.2 | 0.4 | 0.2 | 0.2 | 0.2 |
| Miconazole (μg/ml) | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Amphotericin B (μg/ml) | 12.5 | 6.3 | 6.3 | 6.3 | 3.2 |
| CdCl2 (μg/ml) | 50 | 50 | 50 | 50 | 50 |
| LiCl (μg/ml) | 8.4 | 4.2 | 8.4 | 4.2 | 4.2 |
| CaCl2 (mg/ml) | 55.5 | 11.1 | 11.1 | 27.8 | 27.8 |
| NaCl (mg/ml) | 87.6 | 87.6 | 87.6 | 87.6 | 87.6 |
MICs were determined on YPD (pH 6.4) agar plates containing each compound. The plates were incubated at 30°C for 2 days.
Overexpression of the YOR1 gene by aur3 mutation.
We searched for mutants that were specifically resistant to AbA and that grew in the presence of 1 μg of AbA per ml after mutagenization of wild-type DKD-5D cells. Several haploid AbA-resistant mutants were crossed with wild-type strain SH3328, and heterozygous diploids were obtained. The diploids derived from three mutants, AL22-4A, AL33-9C, and AL49-3A, were as sensitive to AbA as the wild-type strain, indicating that in these mutants the mutations are recessive. In contrast, other mutants had higher levels of resistance (to more than 25 μg of AbA per ml) than those of the three AL mutants, indicating that the mutations in these mutants are dominant, and the mutations were in the aur1+ gene. Analysis of tetrads from the diploids of the AL mutants indicated that they all carried a single chromosomal mutation, and complementation tests showed that the mutations were all in one locus, designated aur3. Analysis of tetrads from the diploids obtained by crosses between aur3R mutant AL33-9C and AUR1R mutant AR9-4A indicated that aur3 is not an allele of AUR1 (data not shown). Tetrads from diploids obtained by crosses between the yor1::URA3 mutant A141-9A and aur3R mutant AL33-18C were tested for resistance to AbA at 1.5 μg/ml and for the Ura+ phenotype. Of the 22 four-spored tetrads analyzed, 4 tetrads had two resistant spores, 15 tetrads had one resistant spore, and 3 tetrads had no resistant spores. This ratio, 4:15:3, is close to 1:4:1, showing that aur3 is not linked genetically with YOR1. All Ura+ spores having the yor1-null allele, some of which had the aur3R mutation, were sensitive to AbA (Fig. 3), suggesting that a functional YOR1 gene is needed for the aur3R mutant to be resistant to AbA. These results also suggest the possibility that the aur3R mutation causes overexpression of the YOR1 gene, leading to resistance to AbA, although the possibility that the function of AUR3 may be downstream of the YOR1 gene function remains.
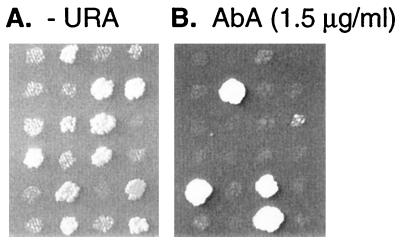
FIG. 3.
Correlation between the YOR1 gene and the aur3R mutation in AbA resistance. yor1-disrupted haploid A141-9A cells were crossed with aur3R mutant AL33-18C, and the diploids obtained were allowed to sporulate. Tetrad segregants from the diploids were streaked onto YPD agar plates, the plates were incubated at 30°C for 2 days, and the segregants were replicated on a YPD agar plate with 1.5 μg of AbA per ml (B) or an SD plate without uracil (−URA) (A).
To find whether expression of the YOR1 gene was controlled by AUR3, the YOR1 mRNA in aur3R mutants was examined by Northern hybridization (Fig. 4A). The AL22-4A mutant contained 20-fold as much YOR1 mRNA as the parental strain. The mutant also overexpressed mRNA of another ABC transporter gene, PDR5. Southern blot analysis (Fig. 4B) showed that mutant AL22-4A had the same number of copies of YOR1 as the parental strain. These results suggest that the transcription of both YOR1 and PDR5 is regulated by the AUR3 gene product and that overexpression of the YOR1 gene occurs because its regulation is abnormal as a result of a mutation in the AUR3+ gene. Genetic mapping by tetrad analysis showed that the aur3 locus is linked loosely to the centromere (for aur3-trp1, parental ditype:nonparental ditype:tetrad [PD:NPD:T] = 18:17:5; for aur3-met14, PD:NPD:T = 7:6:0) and is linked tightly to LEU1 (PD:NPD:T = 12:0:0) on chromosome VII. LEU1 has been mapped to a position near PDR1 (1), a transcriptional regulator gene of PDR5 and YOR1, suggesting that the mutation in the aur3R mutants was in the PDR1 locus. To examine this suggestion, we introduced pdr1::LEU2 DNA into aur3 mutant AL22-4A to disrupt the PDR1 gene and selected Leu+ transformants. All transformants had lost their resistance to AbA and had the same sensitivity to AbA as wild-type cells (Fig. 5). Furthermore, one of these transformants was crossed with wild-type strain SH3328, and the diploid that was obtained was sporulated. Segregation of the Leu+ and AbA resistance phenotype among the resulting spores was examined by random spore analysis (Fig. 5). No AbA-resistant clone (aur3R PDR1) appeared among ∼104 viable segregants. These results indicate that AUR3 is identical to PDR1. Therefore, the aur3R mutations in strains AL22-4A, AL33-9C, and AL49-3A were designated pdr1R-A1, pdr1R-A2, and pdr1R-A3, respectively.
FIG. 4.
Northern blot analysis of the YOR1 gene in aur3R mutants. (A) Total RNA from wild-type DKD-5D cells (lanes 1, 3, and 5) or aur3R mutant AL22-4A cells (lanes 2, 4, and 6) was studied by Northern hybridization. The probes used were a 1.2-kb HindIII-PstI fragment of YOR1 (lanes 1 and 2), a 1.1-kb PDR5 fragment (lanes 3 and 4), and a 1.5-kb actin DNA fragment (lanes 5 and 6). (B) Genomic DNAs from DKD-5D cells (lane 7) and AL22-4A cells (lane 8) were digested with HindIII and studied by Southern hybridization with the 1.0-kb NheI-BstXI fragment of YOR1 as the probe (see Fig. 1).
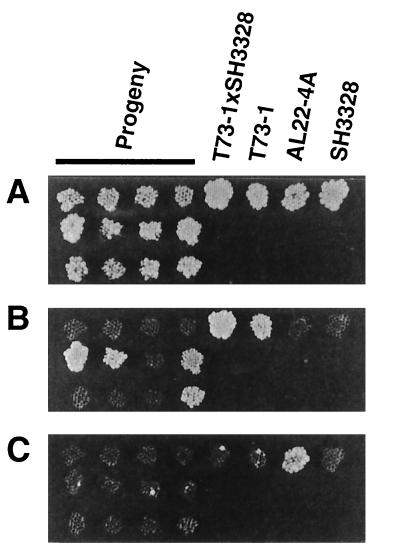
FIG. 5.
Genetic analysis of a linkage between aur3 and PDR1. The wild-type strain (SH3328), aur3R mutant AL22-4A, and aur3R mutant with a disrupted PDR1 gene T73-1, a diploid strain (T73-1 × SH3328), and random spore isolates derived from the diploid cells were streaked onto a YPD agar plate. The plate was incubated at 30°C for 2 days and then replicated onto YPD agar plates (A), an SD plate lacking leucine (B), and a YPD agar plate containing 2 μg of AbA per ml (C). Incubation was carried out for 2 days at 30°C. No progeny showing AbA resistance appeared. Only 12 of the progenies examined are shown.
Isolation of YOR1 homolog from C. albicans.
To search for a C. albicans gene with a sequence similar to that of the S. cerevisiae YOR1 gene, Southern hybridization of genomic DNA of C. albicans was performed with a 1.2-kb HindIII-PstI fragment of S. cerevisiae YOR1 (Fig. 1) as a probe. A single band hybridized with the probe (data not shown), indicating that C. albicans has a gene hybridizable to the YOR1 gene and, therefore, a gene with a high degree of sequence similarity to it. The YOR1 homolog was isolated from the genomic DNA library of C. albicans by hybridization with the same probe described above. A plasmid, pA8.3, that contained an 8.3-kb BamHI fragment was selected. Nucleotide sequencing of the fragment showed that it lacked the region coding for the carboxyl terminus of the protein. A residual coding region, plasmid pA6.5, was isolated by screening of another genomic library with a part of the 8.3-kb fragment as a probe. Figure 6A shows a restriction map of these DNA fragments. Parts of the 8.3-kb BamHI and 6.5-kb HindIII fragments were sequenced. The results indicated that the predicted amino acid sequence had a high degree of similarity (61%) to the sequence at residues 1107 to 1352 of S. cerevisiae Yor1p (Fig. 6B and C), but no similarity to the sequences of CDR1 (22) and CDR2 (27), which have been identified as multidrug resistance genes in C. albicans. Therefore, this gene was designated YOR1 of C. albicans.
FIG. 6.
YOR1 homolog of C. albicans. (A) Restriction map of the cloned DNA. The hatched box indicates the region sequenced. The arrow indicates the direction of transcription. Restriction sites are as follows: B, BamHI; C, ClaI; E, EcoRI; H, HindIII; S, SpeI. (B) Nucleotide sequence and predicted amino acid sequence of the partially sequenced region of the YOR1 homolog. (C) Alignment of the C. albicans YOR1 homolog (CaYOR1) and the S. cerevisiae YOR1 gene (ScYOR1). The Walker A motif in the nucleotide-binding domain and the membrane-spanning domains of S. cerevisiae YOR1p are indicated by double underlines and underlines, respectively. Identical and similar amino acid residues are marked by colons and dots, respectively.
Inhibition of ABC transporter by AbA.
To identify the direct effect of AbA on the ABC transporter, we used multidrug-resistant tumor cells and tested AbA for its ability to inhibit the efflux of vincristine out of the cells. This efflux is caused by the P glycoprotein. AbA had no cytotoxic effects on the cells at the concentrations tested. AbA caused as much of an increase in the amount of vincristine that accumulated in A2780AD cells (Table 3) as verapamil did. This result indicates that AbA inhibits the drug efflux caused by mammalian ABC transporters.
TABLE 3.
Change in vincristine accumulation caused by AbA in tumor cells with multidrug resistance
| Drug | Intracellular concn (pmol/106 cells) (% changea in concn of [3H]vincristine at the following different doses [μg/ml] of AbA or verapamil):
|
|||
|---|---|---|---|---|
| 0 | 0.1 | 1 | 10 | |
| AbA | 0.052 (100) | 0.042 (82) | 0.077 (149) | 0.247 (478) |
| Verapamil | 0.060 (116) | 0.162 (312) | 0.366 (708) | |
The change in concentration is given as a percentage of the concentration without AbA or verapamil.
DISCUSSION
We showed that overexpression of YOR1/YRS1 confers resistance to AbA. Katzmann et al. (16) have shown that the expression of YOR1 is regulated by PDR1 and PDR3, both of which also regulate the expression of the PDR5 gene. We showed by genetic analysis that the aur3R mutation causes overexpression of YOR1 as well as PDR5 and that AUR3 is identical to PDR1.
The fact that the amount of the YOR1 gene had more of an effect than PDR5 on resistance to AbA suggests that YOR1 is more important for AbA resistance in S. cerevisiae. The Yor1 protein that confers resistance was more similar to the CFTR subgroup (including MRP1) than the MDR1 subgroup. Therefore, AbA may be a better substrate for members of the CFTR subgroup than the MDR1 subgroup in human cells as well. In small-cell lung cancer cells, overexpression of MRP1 causes resistance to several anticancer agents (5). AbA may overcome the multidrug resistance of such cancer cells overexpressing MRP1 better than it overcomes the resistance of the A2780AD cancer cells overexpressing MDR1 examined in this study.
We identified a gene of C. albicans whose sequence was similar to that of YOR1 of S. cerevisiae. A search for a sequence homologous to that of C. albicans Yor1p showed that it is more similar to the CFTR subgroup than to the MDR1 subgroup, to which C. albicans Cdr1p and Cdr2p belong (22, 27). Of the fungal pathogens, clinical isolates of C. albicans resistant to fluconazole overexpress the PDR5 homolog CDR1 gene (22) or the gene for another efflux pump, BENR (10). Whether these strains have proteins in the CFTR subgroup including Yor1p is not known. In S. cerevisiae, several loci are involved in pleiotropic drug resistance (PDR) (2), and they seem to interact, causing the expression of resistance (7, 19). At times, the mutation of a PDR locus such as AUR3/PDR1 or PDR3 causes overexpression of other PDR genes encoding ABC transporters (3). This fact suggests the possible emergence of multidrug resistant strains of pathogenic Candida isolates which have mutations in yet unidentified genes similar to PDR1 and PDR3 of S. cerevisiae, and the resistance may develop particularly readily in haploid species such as Candida glabrata.
It is important that the intrinsic roles and substrates of ABC transporters in cell metabolism be known. Some are already known; for example, mouse mdr2 is essential in the liver for the export of phospholipids from the apical surface of the canalicular membrane into the bile (31), CFTR acts as a chloride ion channel in the lungs (23), and yeast Ste6p is a transporter of the a factor (18). Cells overexpressing YOR1/YRS1 are resistant to oligomycin when the cells are grown with unfermentable carbon sources such as glycerol and ethanol (6). Such cells are also resistant to reveromycin and organic anions when they are grown in medium with a low pH (pH 4.5) (16). However, resistance to AbA is seen at pH 6.4 in ordinary YPD medium, which contains glucose as the carbon source. It seems likely that YOR1 contributes to cellular resistance to AbA and structurally related compounds rather than to resistance to other toxic compounds.
A variety of hydrophobic and ionic compounds can be transported through membranes by ABC transporters, and some also inhibit the transporters. Thus, the inhibitory action of AbA in tumor cells may result from competition because of structural or ionic similarity to the usual substrate of ABC transporters. The antifungal activity of AbA against yeasts could be used to investigate the effects of inhibitors on various ABC transporters, including mammalian ones, as described by Kino et al. (17).
ACKNOWLEDGMENTS
We thank Takashi Takabatake for technical support. We also thank S. Harashima (Osaka University) for generous gifts of yeast strains and C. Shimoda (Osaka City University) for providing plasmids and technical advice.
REFERENCES
- 1.Balzi E, Chen W, Ulaszewski S, Capieaux E, Goffeau A. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J Biol Chem. 1987;262:16871–16879. [PubMed] [Google Scholar]
- 2.Balzi E, Goffeau A. Multiple or pleiotropic drug resistance in yeast. Biochem Biophys Acta. 1991;1073:241–252. doi: 10.1016/0304-4165(91)90128-4. [DOI] [PubMed] [Google Scholar]
- 3.Balzi E, Wang M, Leterme S, Dyck L V, Goffeau A. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J Biol Chem. 1994;269:2206–2214. [PubMed] [Google Scholar]
- 4.Bissinger P H, Kuchler K. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. J Biol Chem. 1994;269:4180–4186. [PubMed] [Google Scholar]
- 5.Cole S P C, Bhardwaj G, Gerlach J H, Mackie J E, Grant C E, Almquist K C, Stewart A J, Kurz E U, Duncan A M V, Deeley R G. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 6.Cui Z, Hirata D, Tsuchiya E, Osada H, Miyakawa T. The multidrug resistance-associated protein (MRP) subfamily (Yrs1/Yor1) of Saccharomyces cerevisiae is important for the tolerance to a broad range of organic anions. J Biol Chem. 1996;271:14712–14716. doi: 10.1074/jbc.271.25.14712. [DOI] [PubMed] [Google Scholar]
- 7.Dexter D, Moye-Rowley W S, Wu A, Golin J. Mutations in the yeast PDR3, PDR4, PDR7, and PDR9 pleiotropic (multiple) drug resistance loci affect the transcript level of an ATP binding cassette transporter encoding gene, PDR5. Genetics. 1994;136:505–515. doi: 10.1093/genetics/136.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Endicott J A, Ling V. The biochemistry of P-glycoprotein-mediated multidrug resistance. Annu Rev Biochem. 1989;58:137–171. doi: 10.1146/annurev.bi.58.070189.001033. [DOI] [PubMed] [Google Scholar]
- 9.Endo M, Takesako K, Kato I, Yamaguchi H. Fungicidal action of aureobasidin A, a cyclic depsipeptide antifungal antibiotic, against Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1997;41:672–676. doi: 10.1128/aac.41.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fling M E, Kopf J, Tamarkin A, Gorman J A, Smith H A, Koltin Y. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol Gen Genet. 1991;227:318–329. doi: 10.1007/BF00259685. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman M M, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 12.Hashida-Okado T, Ogawa A, Endo M, Yasumoto R, Takesako K, Kato I. AUR1, a novel gene conferring aureobasidin resistance on Saccharomyces cerevisiae: a study of defective morphologies in Aur1p-depleted cells. Mol Gen Genet. 1996;251:236–244. doi: 10.1007/BF02172923. [DOI] [PubMed] [Google Scholar]
- 13.Heidler S A, Radding J A. The AUR1 gene in Saccharomyces cerevisiae encodes dominant resistance to the antifungal agent aureobasidin A ( LY295337) Antimicrob Agents Chemother. 1995;39:2765–2769. doi: 10.1128/aac.39.12.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 15.Ikai K, Takesako K, Shiomi K, Moriguchi M, Umeda Y, Yamamoto J, Kato I, Naganawa H. Structure of aureobasidin A. J Antibiot. 1991;44:925–933. doi: 10.7164/antibiotics.44.925. [DOI] [PubMed] [Google Scholar]
- 16.Katzmann D J, Hallstrom T C, Voet M, Wysock W, Golin J, Volckaert G, Moye-Rowley W S. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6875–6883. doi: 10.1128/mcb.15.12.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kino K, Taguchi Y, Yamada K, Komano T, Ueda K. Aureobasidin A, an antifungal cyclic depsipeptide antibiotic, is a substrate for both human MDR1 and MDR2/P-glycoproteins. FEBS Lett. 1996;399:29–32. doi: 10.1016/s0014-5793(96)01265-3. [DOI] [PubMed] [Google Scholar]
- 18.McGrath J P, Varshavsky A. The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature. 1989;340:400–404. doi: 10.1038/340400a0. [DOI] [PubMed] [Google Scholar]
- 19.Meyers S, Schauer W, Balzi E, Wagner M, Goffeau A, Golin J. Interaction of the yeast pleiotropic drug resistance genes PDR1 and PDR5. Curr Genet. 1992;21:431–436. doi: 10.1007/BF00351651. [DOI] [PubMed] [Google Scholar]
- 20.Nagiec M M, Nagiec E E, Baltisberger J A, Wells G B, Lester R L, Dickson R C. Sphingolipid synthesis as a target for antifungal drugs: complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- 21.Philippsen P, Stotz A, Scherf C. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:169–182. doi: 10.1016/0076-6879(91)94014-4. [DOI] [PubMed] [Google Scholar]
- 22.Prasad R, De Wergifosse P, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr Genet. 1995;27:320–329. doi: 10.1007/BF00352101. [DOI] [PubMed] [Google Scholar]
- 23.Riordan J R, Rommens J M, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J, Drumm M L, Iannuzzi M C, Collins F S, Tsui L. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 24.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J F, Fritsch T, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Sanglard D, Kuchler K, Ischer F, Pagani J-L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 28.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 29.Servos J, Haase E, Brendel M. Gene SNQ2 of Saccharomyces cerevisiae, which confers resistance to 4-nitroquinoline-N-oxide and other chemicals, encodes a 169 kDa protein homologous to ATP-dependent permeases. Mol Gen Genet. 1993;236:214–218. doi: 10.1007/BF00277115. [DOI] [PubMed] [Google Scholar]
- 30.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 31.Smit J J M, Schinkel A H, Oude Elferink R P J, Groen A K, Wagenaar E, van Deemter L, Mol C A A M, Ottenhoff R, van der Lugt N M T, van Roon M A, van der Valk M A, Offerhaus G J A, Berns A J M, Borst P. Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell. 1993;75:451–462. doi: 10.1016/0092-8674(93)90380-9. [DOI] [PubMed] [Google Scholar]
- 32.Szczypka M S, Wemmie J A, Moye-Rowley W S, Thiele D J. A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J Biol Chem. 1994;269:22853–22857. [PubMed] [Google Scholar]
- 33.Takesako K, Ikai K, Haruna F, Endo M, Shimanaka K, Sono E, Nakamura T, Kato I, Yamaguchi H. Aureobasidins, new antifungal antibiotics: taxonomy, fermentation, and properties. J Antibiot. 1991;44:919–924. doi: 10.7164/antibiotics.44.919. [DOI] [PubMed] [Google Scholar]
- 34.Tsuruo T, Iida H, Tsukagoshi S, Sakurai Y. Increased accumulation of vincristine and adriamycin in drug-resistant tumor cells following incubation with calcium antagonists and calmodulin inhibitors. Cancer Res. 1982;42:4730–4733. [PubMed] [Google Scholar]