Abstract
Influenza neuraminidase (NA) plays an important role in viral replication, and characterization of viruses resistant to NA inhibitors will help elucidate the role of active-site residues. This information will assist in designing better inhibitors targeted to essential active-site residues that cannot generate drug-resistant mutations. In the present study we used the benzoic acid-based inhibitor BCX-140 to select and characterize resistant viruses. BCX-140 binds to the NA active site in an orientation that is opposite that of a sialic acid-based compound, 4-guanidino-2,4-dideoxy-2,3-dehydro-N-acetylneuraminic acid (GANA). Thus, the guanidino group of BCX-140 binds to Glu-276, whereas in GANA the guanidino group binds to Glu-119. We passaged influenza A/Singapore/1/57 (H2N2) in Madin-Darby canine kidney cells in the presence of BCX-140, and virus resistant to this inhibitor was selected after six passages. The NA of this mutant was still sensitive to inhibition by BCX-140. However, the mutant virus was resistant to BCX-140 in plaque and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays. Sequence analysis of hemagglutinin (HA) and NA genes revealed changes in both, although none were in the active site of the NA. Depending on the method of selection of the resistant virus, two types of changes associated with the sialic acid binding site were seen in the HA. One is a change in HA1 of Ala-133 to Thr, a residue close to the binding site, while the other change was Arg-132 of HA1 to Gln, which in HA1 of serotype H3 is a sialic acid contact (Asn-137). Binding studies revealed that both types of resistant viruses had reduced receptor binding affinity compared to that of the wild type. Thus, resistance to BCX-140 was generated by modifying the HA. NA active-site residue 276 may be essential for activity, and thus, it cannot be changed to generate resistance. However, drug-induced changes in the HA can result in a virus that is less dependent on NA activity for growth in cells and, hence, resistant to NA inhibitors.
Influenza is an acute respiratory illness that has afflicted humans since ancient times. Protection can be afforded by annual immunization with a vaccine consisting of inactivated virus. However, limitations in the efficacy of vaccines, the need to reformulate the vaccine every year in response to the antigenic drift of circulating influenza viruses, and the possibility of an antigenic shift have stimulated the search for antiviral drugs (15). The currently licensed drugs, amantadine and its analog rimantadine, provide only modest benefit to patients because of their limited efficacies (they are active only against influenza A virus strains) and adverse side effects and because of the rapid development of drug-resistant strains (2, 5, 7, 13).
Influenza viruses possess two major surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). The three-dimensional X-ray structure of each has been elucidated for representative strains (3, 24). The function of HA is to recognize and bind to the cell receptor (4). The main function of NA is the promotion of virus release (12, 17). NA is responsible for removing sialic acid from newly synthesized HAs and NAs which are sialylated by cellular enzymes. In the absence of functional NA, virus release is inhibited and virions are formed but remain attached to the cell surface and to each other, forming aggregates on the surfaces of infected cells (17).
NA has been considered a suitable target for antiviral drugs, since it possesses an active site whose amino acid sequence is conserved among all types and subtypes of influenza virus (23). 4-Guanidino-2,4-dideoxy-2,3-dehydro-N-acetylneuraminic acid (GANA) has been described as a highly potent and selective inhibitor of influenza virus NA (22, 25). Recently, it has been shown that the virus develops resistance to GANA in cell culture. The mutations responsible for the resistance are seen in both the HA and the NA genes (6, 14, 18, 21).
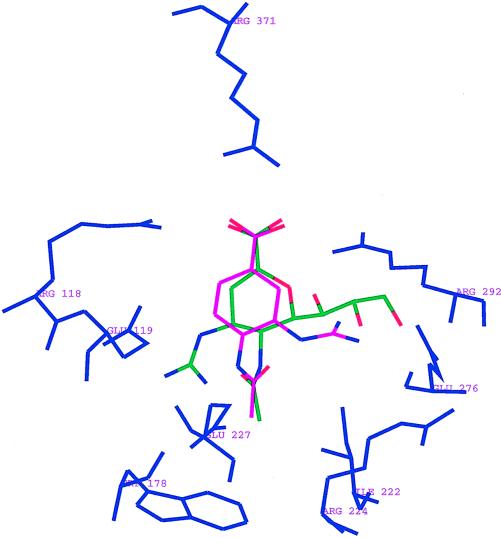
The aromatic compound BCX-140, 4-(acetylamino)-3-guanidinobenzoic acid (designated BANA-113 by Singh et al. [20]), has been shown to inhibit the NA of influenza virus. The two NA inhibitors GANA and BCX-140 bind very differently to the active site of NA N9 (A/Tern/Aus/G70c/75) shown in Fig. 1 (20, 23). In the case of GANA, the terminal two hydroxyl groups of the glycerol moiety interact with Glu-276 and the guanidinyl group interacts with carboxylate groups of Glu-227 and Glu-119. However, in the case of BCX-140 the inhibitor is rotated 180° and the guanidinyl group binds to the glycerol binding pocket of GANA and interacts with Glu-276 (11, 20). Since these two compounds bind differently to the active site, we have investigated the potential for the development of resistance to BCX-140.
FIG. 1.
BCX-140 (pink) and GANA (green) in the active site of NA N9. N2 numbering is used to designate the active-site residues.
To determine whether virus exposed to BCX-140 may generate resistant variants, virus A/Singapore/1/57 was passaged in Madin-Darby canine kidney (MDCK) cells in the presence of BCX-140. The inhibitor binds entirely within the conserved catalytic site of the enzyme and makes no contacts with residues outside that site (11, 20). Any viral variant with a reduced dependency on NA for release from infected cells or an NA variant that can bind and process substrate but is no longer inhibited by the compound could emerge in the presence of the inhibitor. The aim of the present study was to determine if resistant mutants of influenza virus can be generated in the presence of BCX-140 in vitro and determine the phenotypic and molecular characteristics of such mutants.
MATERIALS AND METHODS
Viruses.
Influenza viruses A/PR/8/34, A/FM/1/47, A/New Jersey/8/76, and B/Allen/45 were obtained from the American Type Culture Collection. A/Singapore/1/57 was obtained from Robert Webster (St. Jude Children’s Hospital, Memphis, Tenn.). It had been adapted to MDCK cells and was used as parent virus for the selection of BCX-140-resistant virus. A/Turkey/Mass/76A/Beijing/32/92[R] and CA/Aichi/2/68-PR8/34 were obtained from Edwin Kilbourne (Department of Microbiology and Immunology, New York Medical College, Valhalla, N.Y.). NWS/G70c was obtained from Graeme Laver (Australian National University, Canberra, Australia).
Cells and NAs (bacterial and mammalian).
MDCK cells were obtained from the American Type Culture Collection and were grown in Eagle’s minimal essential medium (Bio-Whitaker) containing 10% fetal bovine serum (FBS; Hyclone Laboratories, Inc.) supplemented with glutamine and antibiotics. NAs from Vibrio cholerae and Salmonella typhimurium were obtained from Sigma. Sheep liver sialidase was partially purified (10) and was used in the NA assay.
Inhibitor.
BCX-140 was synthesized at BioCryst Pharmaceuticals, Inc.
NA assay.
A fluorimetric assay was used to measure influenza virus NA activity (19). The substrate 2′-(4-methylumbelliferyl)-alpha-d-acetylneuraminic acid is cleaved by NA to yield a fluorescent product that can be quantified. The assay mixture contained inhibitor at various concentrations and NA enzyme in 32.5 mM MES [2-(N-morpholino)ethanesulfonic acid] buffer–4 mM calcium chloride (pH 6.5) (total volume, 80 μl). The reaction was started by the addition of 20 μl of the substrate to a final concentration of 75 μM. After 10 min at 37°C, 2.4 ml of 0.1 M glycine–NaOH (pH 10.2) was added to 0.1 ml of the reaction mixture to terminate the reaction. A blank was run with the same substrate solution but with no enzyme. Fluorescence was recorded with an Aminco-Bowman fluorescence spectrophotometer (excitation, 360 nm; emission, 450 nm), and readings from the substrate blanks were subtracted from the sample readings. The 50% inhibitory concentration (IC50) was calculated by plotting percent inhibition of NA activity versus the inhibitor concentration.
In vitro anti-influenza virus activity. (i) MTT assay.
Inhibition of growth of influenza virus in MDCK cells by BCX-140 was performed as described by Nagai et al. (16). MDCK cells were grown in Eagle’s minimal essential medium containing 10% FBS, HEPES, penicillin-streptomycin, and l-glutamine (maintenance medium). Confluent monolayers of MDCK cells in 96-well plates were infected with influenza virus A/Singapore/1/57 (H2N2; 16 PFU) in 0.1 ml of infection medium (maintenance medium without FBS) containing tolylsulfonyl phenylalanyl chloromethyl ketone (TPCK) trypsin. The cells were maintained for 30 min at room temperature to allow the virus to adsorb. The virus inoculum was then removed, and 200 μl of infection medium containing inhibitor at various concentrations was added. A control incubation was performed without virus infection. The plate was incubated at 37°C for 48 h under 5% carbon dioxide. The medium was then removed and the monolayer was washed with phosphate-buffered saline to remove dead cells resulting from infection with the influenza virus. The number of viable cells was determined by a colorimetric method which is based on the in situ metabolic reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) by viable cells. The IC50 of BCX-140 was determined as follows: percent survival = [(Ad − A0d)/(Ac − A0d)] × 100, where Ad is the absorbance at a certain drug concentration, A0d is the absorbance with no drug, and Ac is the absorbance with no virus (control sample). The IC50 was calculated by plotting percent survival versus the inhibitor concentration.
(ii) Plaque inhibition assay.
Plaque inhibition assays were performed as described by Hayden et al. (8). Confluent monolayers of MDCK cells in six-well plates were washed free of maintenance medium before use. The cells were infected with influenza virus A (H2N2; 30 to 50 PFU) in 0.6 ml of infection medium containing 2 μg of TPCK trypsin per ml. The cells were maintained for 30 min at room temperature to allow the virus to adsorb, and the virus inoculum was removed. A 0.5% agar overlay (3 ml) in medium containing trypsin (2 μg/ml) and various concentrations of drug were added to each plate. A control was performed with no drug. The plates were incubated at 37°C under a 5% carbon dioxide atmosphere. After 48 to 72 h the agar was removed and the plates were stained with crystal violet. The IC50 was calculated by plotting plaque numbers as a percentage of that of the control versus the inhibitor concentration.
Selection of an influenza virus (H2N2) mutant resistant to BCX-140.
Confluent monolayers of MDCK cells in six-well plates were washed free of maintenance medium before use. Two sets of cells were infected with influenza virus A (H2N2; 30 to 50 PFU) in 0.6 ml of infection medium containing 2 μg of TPCK trypsin per ml. The cells were maintained for 30 min at room temperature to allow the virus to adsorb, and then the medium containing unadsorbed virus was removed. The first set of cells was incubated in the presence of BCX-140 (500 μM), and the second set was used as an untreated control. The plates were incubated at 37°C under a 5% carbon dioxide atmosphere for 48 h. The cells were centrifuged out, and the supernatant was used to infect the next batch of cells (second passage). Again, the first set of cells was incubated in the presence of BCX-140 (500 μM) and the second set was used as an untreated control. After each passage both MTT and plaque assays were performed to look for the development of resistant strains. The resistant stock obtained after six passages in the presence of BCX-140 was designated BCX-140RM, and virus passaged six times in the absence of the drug is designated C6A. BCX-140RM was grown in MDCK cells in the absence of the drug before its use in the enzyme assays. Both the plaque and the MTT assays showed that BCX-140RM remains resistant to the drug even after it is grown in the absence of the drug. A second selection was done in the same way but with 250 μM BCX-140, and the resulting resistant stock was passaged twice at limiting dilution in the presence of the inhibitor.
Sequencing of HA and NA genes.
The resistant viruses BCX-140RM and the wild-type virus were grown in MDCK cells and were purified with sucrose gradients. The purified viruses were disrupted with sodium dodecyl sulfate and were digested with proteinase K at 56°C for 20 min. The RNA was extracted with hot phenol, followed by phenol-chloroform extraction and ethanol precipitation (1). Full-length NA and HA cDNAs were synthesized from virion RNA with avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim). These were then amplified by PCR (94°C for 1 min, 34°C for 1 min, and 72°C for 2 min, for 34 cycles and then 72°C for 8 min). The PCR fragments were gel purified and extracted with the Wizard kit (Promega). The sequences of these PCR fragments were determined with the ABI PRISM dye terminator cycle sequencing kit (Perkin-Elmer, Applied Biosystems Inc.).
Hemagglutination and RBC elution assays.
Hemagglutination assays were performed in microtiter U-bottom plates with 50 μl of virus and 50 μl of washed 1% chicken or human erythrocytes (RBCs) suspended in 0.9% saline. The plates were incubated at 4°C for approximately 1 h. To assay elution by the viral NA, 8 hemagglutination units of virus was preincubated for 30 min at room temperature either without inhibitor or with twofold serial dilutions of BCX-140 starting with a concentration of 12 μM in the first well. The virus was allowed to agglutinate chicken or human RBCs at 4°C for 1 h. Elution of the virus was monitored at 37°C by looking for the appearance of the RBC button.
RESULTS
Inhibition of the NAs of various influenza viruses by BCX-140.
The results of the NA inhibition assays are presented in Table 1. BCX-140 inhibited the enzyme activities of influenza virus type A subtypes N1, N2, and N9 and effectively inhibited influenza virus type B. The IC50s ranged from 2 to 55 μM.
TABLE 1.
Inhibition of the NA of influenza A and B viruses by BCX-140
| Virus | IC50 (μM)a |
|---|---|
| A/PR/8/34 (H1N1) | 55 ± 8 |
| A1/FM/1/47 (H1N1) | 15 ± 1 |
| A/New Jersey/8/76 (H1N1) | 22.0 ± 0.3 |
| A/Singapore/1/57 (H2N2) | 5.3 ± 0.3 |
| CA/Aichi/68(HA)PR8/34(NA)(H3N2) | 5.5b |
| A/Turkey/Mass/76(HA)/Beijing/32/92(NA) (H6N2) | 5.1 ± 0.6 |
| A/NWS/G70C (H1N9) | 2.5 ± 0.5 |
| B/Allen/45 | 24 ± 1 |
Values are means ± standard errors of the means for three determinations except where indicated.
Determined only once.
Inhibition of other NAs.
The selectivity of BCX-140 for influenza virus NA was investigated. BCX-140 showed no significant inhibition (10 to 20%) of bacterial NA (V. cholerae and S. typhimurium NAs) up to 1 mM. Also, BCX-140 did not inhibit mammalian NA (partially purified sheep liver NA) when it was used at a concentration of up to 1 mM.
Inhibition of virus replication by BCX-140.
BCX-140 inhibited the growth of influenza virus in MDCK cells. The results of the MTT assay are presented in Table 2. BCX-140 inhibited the replication of most of the strains studied. Two H1N1 strains were not inhibited by BCX-140 when it was used at a concentration of up to 500 μM. BCX-140 also inhibited virus growth in the plaque assay, with an IC50 of approximately 30 μM (by plaque numbers) except for those for the H1N1 strains, for which the IC50s were greater than 100 μM (data not shown).
TABLE 2.
Inhibition of viral growth in MDCK cells by BCX-140 determined by the MTT assay
| Virus | IC50 (μM)a |
|---|---|
| A/PR/8/34 (H1N1) | No inhibition up to 500 μM |
| A1/FM/1/47 (H1N1) | 181 ± 50 |
| A/New Jersey/8/76 (H1N1) | No inhibition up to 500 μM |
| A/Singapore/1/57 (H2N2) | 29 ± 5b |
| CA/Aichi/68HAPR8/34NA(H3N2) | 261 ± 133b |
| A/Turkey/Mass/76HA/Beijing/32/92NA[R] (H6N2) | 59 ± 25 |
| B/Allen/45 | 109 ± 93c |
Values are means ± standard errors of the means for three determinations except where indicated.
Values are means ± standard errors of the means for five determinations.
Values are means ± standard errors of the means for four determinations.
Selection and characteristics of resistant mutant.
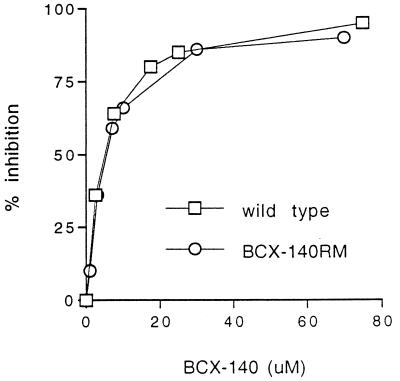
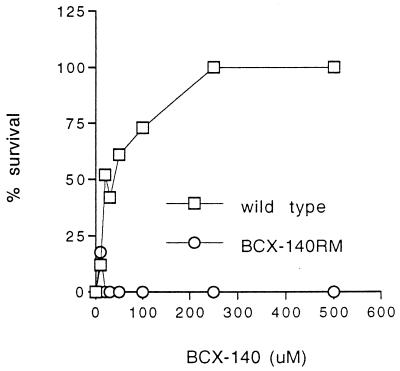
After six passages of the A/Singapore/1/57 virus in the presence of the drug a resistant stock was obtained. The NA activity of the BCX-140-resistant mutant (BCX-140RM) was sensitive to the drug (Fig. 2); however, its growth in MDCK cells was unaffected by the drug (up to 500 μM), as seen both in the plaque assay (Fig. 3) and in the MTT assay (Fig. 4). The degree of resistance of a cloned mutant (BCX-140RM2) to the inhibitor was comparable to that of the uncloned mixed population tested with 500 μM BCX-140. The levels of NA activity were comparable for BCX-140RM and wild-type virus when the activity was standardized to the amount of protein.
FIG. 2.
Effect of BCX-140 on the NA activity of the wild-type strain and a BCX-140-resistant strain of influenza virus A/Singapore/1/57.
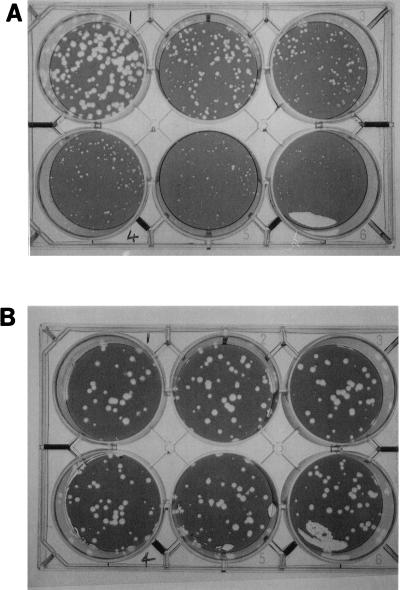
FIG. 3.
Effect of BCX-140 on plaque formation of the wild-type strain (A) and a BCX-140-resistant strain (B) of influenza virus A/Singapore/1/57. The concentrations of the inhibitor were 0, 30, and 50 μM (left to right in the upper row, respectively, and 100, 250, and 500 μM left to right in the lower row, respectively).
FIG. 4.
Effect of BCX-140 on viral growth in MDCK cells followed by the MTT assay.
To ensure that we were working with a single virus population, BCX-140RM (D6A), a stock of the resistant virus that had been passaged once without drug, was subjected to four limiting dilution passages to give BCX-140RM2 or D6A*, and this virus was used for sequence analysis. In another experiment the selection procedure was repeated in the presence of 250 μM BCX-140. After six passages the hemagglutination titer of the resistant stock (termed BCX-140R) was the same as that of the control without drug, and this stock was passaged two more times at the limiting dilution with BCX-140.
Sequences of the NA and HA genes.
The HA and NA genes of the resistant viruses were sequenced. The changes are presented in Table 3. There were some ambiguities in the D6A sequence, but these may have been the result of the presence of a mixed virus population. When D6A was further passaged at limiting dilution (BCX-140RM2) one of the ambiguities (Gly-348 to Arg) disappeared. A few other changes seen in the NA of BCX-140RM2 were not in the area of the active site (Table 3). There were no changes in the active site of the NA in any of the resistant viruses. Interestingly, two sites of change were seen in the HAs of these viruses. Arg-132 of HA1 was changed to Gln in BCX-140RM and BCX-140R, while Ala-133 was changed to Thr in BCX-140RM2, as noted in Table 3. Some differences were found in the sequence of the control virus HA (C6A) compared with the sequence of the A/Singapore/1/57 HA reported previously (GenBank accession no. L11142). Arg-132 in HA1 of influenza A virus serotype H2 corresponds to residue 137 in HA1 (a sialic acid contact residue) of serotype H3, while Ala-133 corresponds to the adjacent residue (residue 138) in HA1 of serotype H3.
TABLE 3.
Nucleotide and predicted amino acid changes in HAs and NAs of viruses resistant to inhibitor BCX-140 (strains BCX-140RM, BCX-140RM2, and BCX-140R) compared to parental virus A/Singapore/1/57 passaged without inhibitor (strain C6A)
| Virus stock | NA changes
|
HA changesa
|
||
|---|---|---|---|---|
| Nucleotide | Amino acid | Nucleotide | Amino acid | |
| BCX-140RM (D6A) | C-484→T/C | G-483→A | Arg-132→Gln (HA1) | |
| T-964→C | ||||
| G-1061→G/C | Gly-348→Gly/Arg | |||
| A-1099→T | ||||
| BCX-140RM2 (D6A*) | T-964→C | G-485→A | Ala-133→Thr (HA1) | |
| G-630→C | Ser-204→Thr | |||
| G-951→G/C | Ser-311→Ser/Thr | |||
| BCX-140R | T-964→C | G-483→A | Arg-132→Gln (HA1) | |
The HA sequence contained the following changes compared to the HA sequence of A/Singapore/1/57 HA sequence in GenBank: G-539 to A (Glu-151 to Lys), A-639 to C (Lys-189 to Thr), C-877 to T, A-996 to T (Lys-303 to Ile), A-853 to A/T, A-1115 to G (HA Ile-218 to Val), T-1201 to C, G-1720 to A.
HA properties.
Influenza virus agglutinates RBCs at 4°C, and the viral NA activity normally disaggregates them at 37°C. All viruses grew to similar hemagglutination titers when the titers were measured with chicken or human RBCs. The RBC binding affinities of the resistant viruses were characterized by elution assays with chicken and human RBCs. The NA activities of the control virus (C6A) and the two mutant viruses (BCX-140RM2 and BCX-140R) with different HA receptor binding site changes were monitored by studying virus elution from chicken and human RBCs. The results are presented in Table 4. C6A did not elute from either type of RBC on incubation at 37°C overnight with or without the presence of BCX-140. BCX-140R was completely eluted from chicken RBCs in 1 h with or without inhibitor, but elution of BCX-140R from human cells was inhibited by BCX-140. In the case of BCX-140RM2, the chicken RBCs eluted the virus in 2 to 3 h. In the presence of the inhibitor, inhibition of elution occurred with high inhibitor concentrations, even with overnight incubation at 37°C.
TABLE 4.
Elution from RBCsa
| Virus strain (amino acid change) | Elution time (h) from the following:
|
|||
|---|---|---|---|---|
| Chicken RBCs
|
Human RBCs
|
|||
| BCX-140 − | BCX-140 + | BCX-140 − | BCX-140 + | |
| C6A | >18 | >18 | >18 | >18 |
| BCX-140RM2 (Ala-133 to Thr) | 2–3 | Partial | >18 | >18 |
| BCX-140R (Arg-132 to Gln) | 1 | 1 | 1 | >18 |
Virus strains C6A, BCX-140RM2, and BCX-140R (8 hemagglutination units) were allowed to agglutinate either chicken or human RBCs at 4°C, and then the elution times observed on warming to 37°C in the presence (+) or absence (−) of BCX-140 were determined.
Thus, in C6A the HA does not elute as a result of either NA activity or thermal motion at 37°C. In BCX-140RM2, since there is no change in the NA active site, the rapid elution must result from a decreased affinity to chicken cells (but not human cells) such that the NA activity elutes the virus, but incubation at 37°C when NA is inhibited gives very slow or partial elution. In BCX-140R the change in the HA site (R137Q) allows the virus to rapidly elute from human cells as a result of NA activity or from chicken cells simply as a result of incubation at 37°C.
Thus, in BCX-140R the mutation in the sialic acid contact residue (Arg-132 to Gln) in HA reduces the affinity of its HA for sialic acids compared to that of the HA of the control virus (C6A). The change in the receptor binding site of HA1 (Ala-133 to Thr) of BCX-140RM2 also appears to reduce the affinity of its HA for the binding of sialic acid, but the affinity is less than that of the HA of BCX-140R. The difference in elution patterns of the resistant viruses from chicken or human RBCs compared to that of control virus also suggests a change in the specificities of the HAs of these resistant viruses.
DISCUSSION
Recent studies have generated influenza viruses resistant to the NA inhibitor GANA (6, 14, 18, 21). In vitro, the emergence of virus resistant to GANA required multiple passages under the pressure of increasing concentrations of the drug; for the influenza virus inhibitor amantadine, however, only one to two passages are required to generate resistant virus (2, 7). Two types of mutants emerged with GANA; the phenotype of one type of mutant is reduced dependence on NA function, and that of the other type of mutant is resistance of its NA to inhibition by GANA as a result of a mutation in the NA gene (14). The molecular basis of the first phenotype is proposed to be related to changes in the HA molecule that cause a reduction in the affinity of HA for sialic acid receptors on cells or virion particles, making it easier for progeny virus to be released from the cells or from each other. In the second type, a mutation in the NA gene was observed, in which Glu-119 was replaced by alanine or glycine (6, 20). The glutamate at position 119 is one of two conserved residues that interact with the guanidino group of GANA to provide a tight and specific interaction between the enzyme and the active-site inhibitor (23). This mutation resulted in a resistant strain in which there was a significant reduction in the affinity of the enzyme for the inhibitor. It has been suggested that when the virus is passaged in the presence of GANA, initially changes in HA appear which make the virus less sensitive to the drug. Further passaging of this virus results in changes in the Glu-119 residue of the NA (6); however, in one case, the Glu-119 residue of the NA was modified and no changes in the HA sequence were observed (21).
BCX-140 selectively inhibits the NA activities of the various viruses, with IC50s ranging from 2 to 55 μM. X-ray crystallographic studies have shown that the guanidino groups in BCX-140 and GANA interact with different parts of the NA active site (Fig. 1). The guanidinyl group in GANA replaces a water molecule at the active site and interacts with Glu-227 and Glu-119. The terminal two hydroxyls of the glycerol moiety in GANA interact with Glu-276. However, in the case of BCX-140 the molecule rotates 180° and the guanidino group occupies the glycerol binding subsite of GANA, forming a salt bridge interaction with residue Glu-276, and the Glu-119 pocket remains vacant. In comparison to Glu-119, Glu-276, being a sialic acid contact residue, may be less likely to be mutated.
BCX-140 inhibits the growth of several influenza viruses in MDCK cells. The IC50 ranges from 29 to 261 μM (Table 2). Two H1N1 viruses, A/PR/8/34 and A/New Jersey/8/76, were not inhibited by BCX-140 when it was used at concentrations of up to 500 μM. Even though BCX-140 inhibits the NA enzymes of all the viruses tested, the compound does not inhibit the growth of some of the viruses in MDCK cells. Different strains of influenza virus show nearly 1,000-fold differences in their susceptibilities to GANA in vitro (plaque assay), even though the IC50s for NA enzyme inhibition are comparable (25). This suggests that some viruses are less dependent than others on NA activity for growth in MDCK cells.
Even though BCX-140 is a relatively weak inhibitor of virus growth in cells, it is possible to select a resistant virus by passage in the presence of the drug. When the A/Singapore/1/57 virus was passaged in MDCK cells in the presence of BCX-140, a strain of virus which was significantly less sensitive to the inhibitor was selected. NA enzyme inhibition assays showed no significant differences in inhibition between the mutant and the wild type. Furthermore, sequence analysis of the NA gene showed no changes in the substrate or inhibitor binding site. In the case of some of the GANA-resistant mutants, Ala or Gly replaces Glu-119 in the NA active site. This replacement is a direct result of the critical interaction between Glu-119 and the guanidino group of GANA. However, in the case of BCX-140 this interaction is absent and the guanidino group points 180° away from the Glu-119 pocket. The guanidino group in its position in BCX-140 interacts with residues that are critical for substrate binding.
The NA changes observed in the mutant were of no significance to the observed decrease in sensitivity of mutant virus to BCX-140 in the cell assays. There was no significant detectable difference between the wild type and the mutant virus in the BCX-140 NA enzyme inhibition assay, and the amino acid residues involved in the binding of substrate or inhibitor were identical for the viruses.
The release of virus from infected cells depends on the equilibrium between the binding of HA to sialic acid residues on viral and cellular glycoconjugates and the cleaving of the same residues by NA, resulting in the release of infectious virus. However, a mutation in the HA gene causing a decrease in the affinity of HA for sialic acid could result in the release of virus without the need for significant NA activity. Therefore, the HA gene of the mutant virus was also sequenced.
A study of the mutations in the HA genes of GANA-resistant mutants (Table 5) revealed that there is no common mutation as in the case of the NA gene, in which Glu-119 is preferentially mutated. The majority of the changes in the HA are clustered around the sialic acid binding site, but none of these changes are known to interact directly with sialic acid in the HA receptor site. However, it has been speculated that these changes could alter the steric and electronic properties of the receptor site to affect the binding affinity of HA for its substrate.
TABLE 5.
Deduced amino acid sequence changes in the NAs and HAs of GANA-resistant mutants
| Virus | HA mutation | NA mutation | Reference no. |
|---|---|---|---|
| NWS/G70c (H1N9) | Thr-155→Ala | Asn-346→Ser | 14 |
| NWS/G70c (H1N9) | No change | Glu-119→Gly | 21 |
| B/HK/8/73 (HG) | Asn-145→Ser | Glu-119→Gly | 21 |
| Asn-150→Ser | |||
| A/Singapore/1/57 (H2N2) | Gly-135→Asp | No change | 18 |
| A/Turkey/Minnesota 833/80 (H4N2) | Gly-75→Glu (HA2) | Glu-119→Gly | 6 |
| Arg-249→Lys |
Sequence analysis of the HA gene of strain BCX-140RM revealed a change in a residue associated with the receptor binding site. Arg-132 was changed to Gln. X-ray crystallographic studies have shown that the carboxylate group of sialic acid is stabilized by a hydrogen bond to the main-chain NH of Asn-137 (H3HA1). Preliminary modeling studies suggest that the side chain of Arg-132 in H2HA1 (which corresponds to residue 137 in H3HA1) could be positioned for direct interaction with the carboxylate and the C-1 hydroxyl group of the sialic acid (Fig. 5). A change in this basic residue to a neutral residue could lead to a decrease in the binding affinity to the receptors. A weaker affinity of HA for the MDCK cellular receptor or for the receptor on the surface of another virion particle might allow the release of virus from infected cells with reduced dependence on NA activity. This is supported by the result of our RBC binding and elution assays. A change in the receptor binding region (Arg-132 to Gln) of the resistant virus causes rapid elution of the virus from either chicken or human RBCs compared to the rate of elution from control virus. Thus, changing the HA for the generation of resistance appears to be the mechanism of escape of H2N2 viruses because the active-site residues of the NA gene cannot be mutated without losing activity. The resistant viruses have a lower affinity than the control virus for chicken RBCs. The change of Arg-132 to Gln allows elution from chicken RBCs, even when NA is inhibited, and allows the NA to elute virus from human RBCs. The effect of the change of Ala-133 to Thr is less marked, as might be expected if it is not a direct contact with sialic acid. Unfortunately, although differences in the binding of virus to human or chicken RBCs have long been recognized, the chemical basis for this difference remains unknown. This mutation in an HA residue that directly interacts with sialic acid has not been reported previously.
FIG. 5.
Sialic acid binding site of HA (H3). A model proposing the interaction between Arg-132 (Asn-137 in H3HA1) and the carboxyl and hydroxyl groups of sialic acid.
The emergence of mutants resistant to drugs like amantadine and rimantadine occurs very rapidly both in vitro and in vivo (2, 5, 7, 13). However, resistance to the NA inhibitor does not seem to occur rapidly in vitro (4, 6, 21). Also, no strain resistant to GANA has been isolated in vivo from mice, ferrets, or humans (9, 23).
The resistance of the BCX-140RM strains in vivo has yet to be established. The resistance of the virus in tissue culture to NA inhibition suggests a decreased affinity of HA to the HA receptor. The same phenomenon may not be observed in vivo. In fact, it has been shown that a GANA-resistant variant developed in MDCK cell culture (with a mutation in the HA gene) was sensitive to the drug in vivo in mice and ferrets (18). The viruses may exhibit specificity for a different oligosaccharide linkage in vivo, and the HA gene mutations may have little impact on receptor binding in the natural host. This is consistent with the results of RBC binding and elution assays. Resistant virus (strain BCX-140R) readily elutes from chicken RBCs in the absence of NA activity, whereas no virus elution from human RBCs was observed in the absence of NA activity. This suggests that in vitro resistance studies may not be able to predict the outcome in vivo.
The results of these studies have indicated that BCX-140 inhibits influenza virus infection in cell culture and induces resistance in influenza virus without altering the activity or sensitivity of NA to BCX-140. BCX-140 and GANA bind to the active site very differently, but both produce resistant mutants with an altered HA and apparently altered HA function. These findings suggest that the residues in the active site of NA are relatively resistant to mutation.
ACKNOWLEDGMENTS
This work was supported in part by grant AI-18203 from the National Institutes of Health.
We thank Aleta Dean for expert technical assistance, Naiming Chu for the structure figures, and W. G. Laver for providing virus and protein crystals. We thank Janet Rogers of the Recombinant DNA/Protein Resource Facility at Oklahoma State University and Susan Hollingshead of the UAB Microbiology Sequencing Core Facility for excellent sequencing services.
REFERENCES
- 1.Air G M, Els M C, Brown L E, Laver W G, Webster R G. Location of antigenic sites on the three-dimensional structure of the influenza N2 virus neuraminidase. Virology. 1985;145:237–248. doi: 10.1016/0042-6822(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 2.Bean W J, Threlkeld S C, Webster R G. Biologic potential of amantadine-resistant influenza A virus in an avian model. J Infect Dis. 1989;159:1050–1056. doi: 10.1093/infdis/159.6.1050. [DOI] [PubMed] [Google Scholar]
- 3.Colman P M. Influenza virus neuraminidase. Structure, antibodies, and inhibitors. Protein Sci. 1994;3:1687–1696. doi: 10.1002/pro.5560031007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colman P M, Varghese J N, Laver W G. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature (London) 1983;303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 5.Grambas S, Bennett M S, Hay A J. Influence of amantadine resistance mutations on the pH regulatory function of the M2 protein of influenza A viruses. Virology. 1992;191:541–549. doi: 10.1016/0042-6822(92)90229-i. [DOI] [PubMed] [Google Scholar]
- 6.Gubareva L V, Bethell R, Hart G J, Murti K G, Penn C R, Webster R G. Characterization of mutants of influenza A virus selected with the neuraminidase inhibitor 4-guanidino-Neu5Ac2en. J Virol. 1996;70:1818–1827. doi: 10.1128/jvi.70.3.1818-1827.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayden F G, Sperber S J, Belshe R B, Clover R D, Hay A J, Pyke S. Recovery of drug-resistant influenza A virus during therapeutic use of rimantadine. Antimicrob Agents Chemother. 1991;35:1741–1747. doi: 10.1128/aac.35.9.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayden F G, Cote K M, Douglas R G., Jr Plaque inhibition assay for drug susceptibility testing of influenza viruses. Antimicrob Agents Chemother. 1980;17:865–870. doi: 10.1128/aac.17.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayden F G, Lobo M, Hussey E K, Eason C U. Efficacy of intranasal GG167 in experimental human influenza A and B virus infection. In: Brown L E, Hampson A W, Webster R G, editors. Options for the control of influenza III. Amsterdam, The Netherlands: Elsevier Science B.V.; 1996. pp. 718–725. [Google Scholar]
- 10.Holzer C T, von Itzstein M, Jin B, Pegg M S, Stewart W P, Wu W-Y. Inhibition of sialidases from viral, bacterial and mammalian sources by analogues of 2-deoxy-2,3-didehydro-N-acetylneuraminic acid modified at the C-4 position. Glycoconjugate J. 1993;10:40–45. doi: 10.1007/BF00731185. [DOI] [PubMed] [Google Scholar]
- 11.Jedrzejas M J, Singh S, Brouillette W J, Air G M, Luo M. A strategy for theoretical binding constants, Ki, calculations for neuraminidase aromatic inhibitors designed on the basis of the active site structure of influenza virus neuraminidase. Proteins Struct Funct Genet. 1995;23:264–277. doi: 10.1002/prot.340230215. [DOI] [PubMed] [Google Scholar]
- 12.Liu C, Eichelberger M C, Compans R W, Air G M. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol. 1995;69:1099–1106. doi: 10.1128/jvi.69.2.1099-1106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mast E E, Davis J P, Harmon M W, Arden N H, Circo R, Tyzka G E. Emergence and possible transmission of amantadine-resistant viruses during nursing home outbreaks of influenza A (H3N2) Am J Epidemiol. 1991;134:988–997. doi: 10.1093/oxfordjournals.aje.a116184. [DOI] [PubMed] [Google Scholar]
- 14.McKimm-Breschkin J L, Blick T J, Sahasrabudhe A, Tiong T, Marshall D, Hart G J, Bethell R C, Penn C R. Generation and characterization of variants of NWS/G70C influenza virus after in vitro passage in 4-amino-Neu5Ac2en and 4-guanidino-Neu5Ac2en. Antimicrob Agents Chemother. 1996;40:40–46. doi: 10.1128/aac.40.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy B R, Webster R G. Orthomyxoviruses. In: Fields B N, Knipe D M, editors. Virology. 2nd ed. Vol. 1. New York, N.Y: Raven Press; 1990. pp. 1091–1152. [Google Scholar]
- 16.Nagai T, Miyaichi Y, Tomimori T, Suzuki Y, Yamada H. In vivo anti-influenza virus activity of plant flavonoids possessing inhibitory activity for influenza virus sialidase. Antivir Res. 1992;19:207–217. doi: 10.1016/0166-3542(92)90080-o. [DOI] [PubMed] [Google Scholar]
- 17.Palese P, Tobita K, Ueda M, Compans R W. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology. 1974;61:397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- 18.Penn C R, Barnett J M, Bethell R C, Fenton R, Gearing K L, Healy N, Jowett A J. Selection of Influenza virus with reduced sensitivity in vitro to the neuraminidase inhibitor GG167 (4-guanidino-Neu5Ac2en): changes in hemagglutinin may compensate for loss of neuraminidase activity. In: Brown L E, Hampson A W, Webster R G, editors. Options for the control of influenza III. Amsterdam, The Netherlands: Elsevier Science B.V.; 1996. pp. 735–740. [Google Scholar]
- 19.Potier M, Mameli L, Belislem M, Dallaire L, Melanxon S B. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-d-N-acetylneuraminate) substrate. Anal Biochem. 1979;94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 20.Singh S, Jedrzejas M J, Air G M, Luo M, Laver W G, Brouillette W J. Structure based inhibitors of influenza virus sialidase. A benzoic acid lead with novel interaction. J Med Chem. 1995;38:3217–3225. doi: 10.1021/jm00017a005. [DOI] [PubMed] [Google Scholar]
- 21.Staschke K A, Colacino J M, Baxter A J, Air G M, Bansal A, Hornback W J, Munroe J E, Laver W G. Molecular basis for the resistance of influenza viruses to 4-guanidino-Neu5Ac2en. Virology. 1995;214:642–646. doi: 10.1006/viro.1995.0078. [DOI] [PubMed] [Google Scholar]
- 22.Thomas G P, Forsyth M, Penn C R, McCauley J W. Inhibition of the growth of influenza viruses in vitro by 4-guanidino-2,4-dideoxy-N-acetylneuraminic acid. Antivir Res. 1994;24:351–356. doi: 10.1016/0166-3542(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 23.von Itzstein M, Wu W-Y, Kok G B, Pegg M S, Dyason J C, Jin B, Van Phan T, Smythe M L, White H F, Oliver S W, Colman P M, Varghese J N, Ryan D M, Woods J M, Bethell R C, Hotham V J, Cameron J M, Penn C R. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature (London) 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 24.Wilson I A, Skehel J J, Wiley D C. Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature (London) 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 25.Woods J M, Bethell R C, Coates J A V, Healy N, Hiscox S A, Pearson B A, Ryan D M, Ticehurst J, Tilling J, Walcott S M, Penn C R. 4-Guanidino-2,4-dideoxy-2,3-dehydro-N-acetylneuraminic acid is a highly effective inhibitor both of the sialidase (neuraminidase) and of growth of a wide range of influenza A and B viruses in vitro. Antimicrob Agents Chemother. 1993;37:1473–1479. doi: 10.1128/aac.37.7.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]