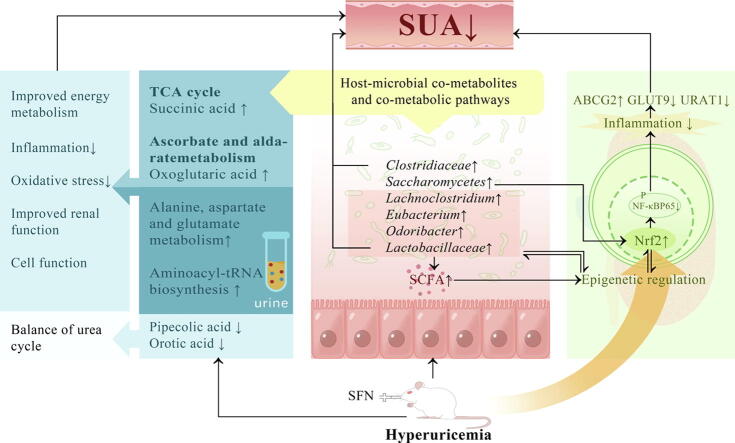
Graphical abstract
Keywords: Hyperuricemia, Sulforaphane, Gut microbiota, Host-gut microbiome co-metabolite
Highlights
-
•
Hyperuricemia has become a public health problem that needs to be solved urgently.
-
•
Sulforaphane may have great potential in reducing uric acid.
-
•
Sulforaphane provides benefits in epigenetic modification of Nrf2, intestinal homeostasis, and host metabolism.
-
•
Succinic acid and oxoglutaric acid were the critical host-gut microbiome co-metabolites of hyperuricaemic rats treated with sulforaphane.
-
•
The findings may provide a good means for efficiently preventing and treating hyperuricemia.
Abstract
Introduction
Currently, revealing how to prevent and control hyperuricemia has become an essential public health issue. Sulforaphane has a wide range of applications in the management of hyperuricemia.
Objective
The study objective was to verify the uric acid-lowering effects and the regulation of the gut-kidney axis mediated by sulforaphane and identify host-microbial co-metabolites in hyperuricemia.
Methods
A hyperuricemia model was established by administering feedstuffs with 4% potassium oxonate and 20% yeast. Forty male Sprague–Dawley rats were randomly divided into the normal control, hyperuricemia, allopurinol, and sulforaphane groups. Animals were treated by oral gavage for six consecutive weeks, and then phenotypic parameters, metabolomic profiling, and metagenomic sequencing were performed.
Results
Sulforaphane could lower uric acid by decreasing urate synthesis and increasing renal urate excretion in hyperuricemic rats (P<0.05). We identified succinic acid and oxoglutaric acid as critical host-gut microbiome co-metabolites. Moreover, sulforaphane improved the diversity of microbial ecosystems and functions, as well as metabolic control of the kidney. Notably, sulforaphane exerted its renoprotective effect through epigenetic modification of Nrf2 and interaction between gut microbiota and epigenetic modification in hyperuricemic rats.
Conclusion
We revealed that sulforaphane could ameliorate the progression of hyperuricemia by reprogramming the gut microbiome and metabolome. Our findings may provide a good means for efficiently preventing and treating hyperuricemia.
Introduction
Hyperuricemia is a chronic disease characterized by abnormally elevated serum uric acid levels. An increasingly aging population and changes in lifestyle have resulted in the yearly increase in the incidence of hyperuricemia. The prevalence of hyperuricemia was approximately 20% in America [1] and even higher in Korea, up to 26.6% [2]. As the largest developing country, the prevalence of hyperuricemia in China was estimated at 16.4% according to the latest meta-analysis of 177 studies and showed a trend of affecting younger individuals in recent years [3]. Hyperuricemia even in asymptomatic patients increases the risk of chronic kidney disease [4], cardiovascular disease [5], metabolic syndrome, and even mortality [6]. These problems are so substantial to families and society that it is essential to identify effective prevention and treatment approaches.
Currently, urate-lowering drug targets are downstream enzymes, and the therapeutic effect is often unsatisfactory and accompanied by liver and kidney damage and myelosuppression. Limited effects and poor adherence to a low-purine diet are also barriers to successful treatment [7]. Notably, increased serum uric acid levels in patients may be a compensatory mechanism for excessive oxidative stress due to its strong antioxidant capacity [8]. Current management strategies, which only target uric acid rather than upstream mechanisms, may not effectively control hyperuricemia.
Hyperuricemia is caused by the excessive production and/or decreased excretion of uric acid. Accumulating studies have indicated that the gut microbiota evidently influences the development of hyperuricemia [9]. Due to the gut-kidney axis [10], dysbiosis of gut microbiota decreases intestinal mucosal barrier function, inducing both low-grade systemic inflammation and local inflammation in the kidney, further leading to elevated serum urate levels and exacerbating gut dysbiosis. Recent research has showed that the effect of the gut microbiota on hyperuricemic hosts partly derives from host and/or microbiome metabolism or co-metabolism [11]. Furthermore, the gut microbiota is reported to communicate with the host through epigenetic mechanisms [12]. Aberrant epigenetic modification in the kidney [13] and intestine [14] may contribute to abnormalities in uric acid excretion. Hence, epigenetic control and the improvement of the intestinal microbiota may become important targets for treating hyperuricemia.
Optimization of the dietary structure has been shown to regulate epigenetics and gut microbiota, representing an important emerging target for lowering uric acid levels. In addition to probiotics directly acting on the gut microbiota, phytochemicals [15] can influence the intestinal microflora through anti-inflammatory and epigenetic modifications [16]. A growing body of evidence suggests that plant-derived substances have been commonly used to combat various diseases. However, the effects of multiple phytochemicals have been greatly limited due to their bioavailability of only 1%-8% [17]. Sulforaphane (SFN), an isothiocyanate derived from cruciferous vegetables, yields a bioavailability of 80% due to its small-size and lipophilic nature [18]. Studies have revealed that SFN is a potent nuclear factor erythroid 2-related Factor 2 (Nrf2) activator [19], which plays an important role in the management of cancers [20] and neurodegeneration [21] by modulating epigenetic mechanisms. Interestingly, SFN can be used to treat acute gout [22] and improve intestinal permeability and intestinal function by decreasing Helicobacter pylori colonization [23] and regulating the imbalance of the intestinal flora [24]. Based on the above research, we hypothesize that SFN can improve hyperuricemia by regulating gut microbiota and activating the Nrf2 pathway through epigenetic modulation.
To validate this hypothesis, we integrated multi-omics data, including metagenomics and metabolomics to verify uric acid-lowering effects and the regulation of the gut-kidney axis of sulforaphane and identify host-microbial co-metabolites.
Material and methods
Materials and reagents
Hyperuricemia feedstuffs made from common feedstuffs and 20% yeast with 4% potassium oxonate were purchased from Beijing Botaihongda Biotechnology Co., Ltd. [SCXK (Jun) 2012-0003]. Glucoraphanin was purchased from Chengdu Geshun Technology Co., Ltd., and myrosinase was provided by Beijing Chemical University. The details regarding the remaining reagents are provided in Appendix A.
Ethics statement
All experiments were conducted according to the Institutional Animal Care and Use Committee of Peking University (2019PHE017) and followed the Guide for the Care and Use of Laboratory Animals (NIH publication no. 85-23, revised 1996).
Animal treatment
Forty male Sprague–Dawley rats (weighing 180–220 g) were purchased from Beijing Vital River Experimental Animal Technology Co., Ltd., and after seven days of acclimatization, forty rats were randomized into 4 groups (n = 10): the normal control group (NC group), the hyperuricemia group (HUA group), the allopurinol group (10 mg/kg, ALL group), and the sulforaphane group (10 mg/kg glucoraphanin and myrosinase, SFN group). Glucoraphanin (10 mg/kg) was metabolized to SFN by myrosinase as described in previous studies [25]. Rats were gavaged for 6 consecutive weeks. The experimental procedures are described in Appendix A.
Uric acid, liver function, and kidney function assays
The levels of serum uric acid (SUA), blood urea nitrogen (BUN), serum creatinine (SCr), cystatin C (CysC), alanine aminotransferase (ALT), aspartate aminotransferase (AST), urinary uric acid (UUA), and urinary creatinine (UCr) were measured with an automatic biochemical device. Uric acid clearance (CUA) and creatinine clearance (CCr) were calculated by the following formulas: CUA = UUA/SUA × Urine volume (mL/min); CCr = UCr/SCr × Urine volume (mL/min).
Morphological observation
Half of the right kidney samples were fixed in 4% paraformaldehyde. Then, the fixed kidney was dehydrated, made transparent, dipped in wax, embedded, sliced, and then encased in paraffin as previously described [26]. Then, the sections were stained by haematoxylin-eosin (HE), dehydrated, cleared and sealed. An examination of tissues under a light microscope was completed as a final step. Five fields per sample at 400 × magnification were analyzed. A tubulointerstitial damage score was assigned as described previously [27].
The other half of the right kidney samples were fixed in 4% glutaraldehyde. Dehydration, transparency preparation, wax dipping, and embedding were used to prepare paraffin sections as previously described [28]. Sections were then processed for electron microscopic examination. Five fields per sample at 3000 × magnification were analyzed. The foot process fusion rate was estimated by dividing the total length of the glomerular basement membrane by the total length of the foot process fusion.
Xanthine oxidase (XOD) and adenosine deaminase (ADA) activity assay
A 10% homogenate of liver samples was prepared with saline. Subsequently, the liver homogenate supernatant was used for the measurement of XOD activity by a commercial kit (A002-1–1; Nanjing Jiancheng Bioengineering Institute). The coefficient of variation (CV) was 1.6%. Serum supernatant was used for the measurement of XOD and ADA activity by commercial kits (A048-1–1; Nanjing Jiancheng Bioengineering Institute). The CV of the ADA kit was 1.7%. Samples were measured in duplicate.
Western blotting
Western blotting was used to measure Nrf2, toll-like receptor 4 (TLR4), nuclear factor-κB (NF-κB), myeloid differentiation Factor 88 (MyD88), NOD-like receptor protein 3 (NLRP3), tissue inhibitor of metalloproteinase 1 (TIMP-1), tissue injury markers metalloproteinase‐2 (MMP‐2) and metalloproteinase-9 (MMP-9), and urate transporters urate ATP-binding cassette, subfamily G, member 2 (ABCG2), transporter 1 (URAT1), and glucose transporter 9 (GLUT9) expression in the kidney tissue obtained from rats. Protein concentration was quantified by a BCA assay. SDS–PAGE and transmembrane electrophoresis were performed on 10% separation gels and 5% stacking gels. The samples were electrophoresed and transferred to 5% skim milk-blocked polyvinylidene fluoride membranes. Then, the primary antibodies were incubated overnight followed by incubation with the secondary antibodies. A quantitative analysis of the protein bands was conducted using ImageJ software after the image was captured after development.
Metabolomics study based on GC–MS/MS
Urine samples were thawed at 4 ℃ before analysis and centrifuged at 10,000 rpm at 4 ℃ for 10 min. The supernatant was subsequently injected into the GC–MS/MS system for analysis. GC–MS analysis was carried out using the Hybrid Quadrupole-Orbitrap GC–MS/MS System (Q Exactive GC, Thermo Fisher). The raw data files generated by GC–MS/MS were processed using Compound Discoverer 3.1 (CD3.1, Thermo Fisher) to perform peak alignment, peak selection, and quantitation for each metabolite. The normalized data were used to predict the molecular formula based on additive ions, molecular ion peaks, and fragment ions. Then, peaks were matched with the mzCloud (https://www.mzcloud.org/), mzVault, and MassList databases to obtain accurate qualitative and relative quantitative results.
Metagenomic sequencing analysis of gut microbiota
Fecal contents were collected, and microbial DNA was extracted. The amplified DNA was subjected to library preparation (Agilent 2100/Q-PCR) and sequenced on the Illumina NovaSeq platform according to the manufacturer’s instructions (PE 150). Sequencing data preprocessing was performed to obtain clean data, including quality control, quality trimming, and adapter clipping. To study the species composition and diversity information in the samples, we annotated and classified all valid sequences of all samples with Kraken2. To functionally annotate genes, we used HUMAnN2 software to compare the sequences after quality control (based on DIAMOND) and host removal with the protein database (UniRef90), further filtering out reads that failed to be compared. According to the results of mapping between the ID obtained from UniRef90 and the ID obtained from the KEGG databases, the relative abundance of the corresponding functions of each functional database was calculated.
Statistical analysis
Non-omics data analysis
Data were analysed with SPSS V.27 and expressed as the mean ± standard deviation (X̅±SD). Normality and homogeneity of variance were determined using the Shapiro–Wilk normality test and Levene’s test. Analysis of variance (ANOVA) was used to compare significant differences between multiple groups. Pairwise comparisons were tested by the least significant difference (LSD) test. The criterion of significance was set as P < 0.05.
Metabolomics analysis
The metabolite databases (KEGG and HMDB) were used for metabolite annotation. The normalization function in MetabolAnalystR was used to make the distribution of the data closer to a normal distribution. Orthogonal partial least squares discriminant analysis (OPLS-DA) analyses were conducted, and metabolites were separated with value importance in projection (VIP)>1. ANOVA and Kruskal–Wallis tests were used in this metabolite analysis (FC ≥ 2 or FC ≤ 0.5, and FDR P<0.05). Random forest (RF) was also performed to determine differentially abundant metabolites. Overrepresentation analysis (ORA) enrichment analysis and topology analysis were conducted, and the key metabolic pathways were identified based on P<0.05 (ORA) and the pathway impact score.
Metagenomics analysis
Principle coordinates analysis (PCoA) plots of Bray–Curtis distance were used to determine beta diversity. LDA effect size (LEfSe) analysis was applied to discover the differential microbiota (LDA scores>4 and FDR P<0.05). KEGG and Enzyme nomenclature databases were used to analyze gut microbiota functions. Heatmap analysis and LEfSe analysis (LDA scores>2 and FDR P<0.05) were performed to compare functional relative abundances.
Association analysis
We used a three‐pronged approach to determine the associations between the metabolome, gut microbiome, and phenotype. Spearman correlation coefficients were calculated, a heatmap was plotted by the Pheatmap package in R language, and a network association graph was generated using Cytoscape 3.7.2.
Results
SFN reduced the uric acid level and improved urate excretion in rats
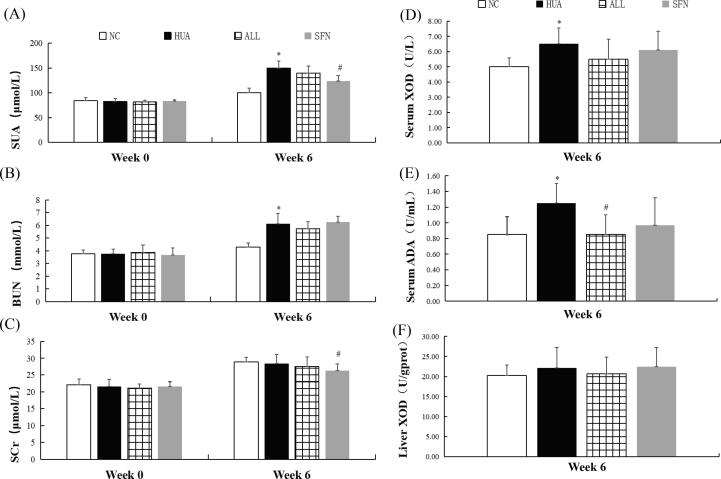
At week 0, no statistically significant differences in the SUA, BUN, and SCr levels were found among all groups (P = 0.548, 0.191, and 0.429, respectively). At week 6, the SUA level in the HUA group significantly higher than that in the NC group (P = 0.007). The SUA level in HUA rats was decreased by SFN, while allopurinol treatment was not as effective (P = 0.045). The BUN level was significantly higher in the HUA group than in the NC group (P = 0.034). The BUN level was not significantly different between the SFN and ALL groups and the HUA group (P = 0.183 and 0.924, respectively), while the SCr level was lower in the SFN group (P = 0.042) (Fig. 1A-C).
Fig. 1.
Serum uric acid levels, blood urea nitrogen levels, serum creatinine levels, serum XOD activity, serum ADA activity, and liver XOD activity in hyperuricemic rats treated for 6 weeks with SFN (n = 10). Serum uric acid (A), blood urea nitrogen (B), serum creatinine (C), serum XOD activity (D), serum ADA activity (E), and liver XOD activity (F). * indicates that the difference is statistically significant compared with the NC group (P<0.05); # indicates that the difference is statistically significant compared to the HUA group (P<0.05); ANOVA, followed by LSD for post hoc comparisons.
At week 6, the UUA, UCr, and CCr levels increased and the CUA level significantly decreased in rats after feeding with hyperuricemia-inducing feedstuff (P = 0.001, 0.033, 0.191, and 0.429, respectively). SFN increased the CUA level and significantly decreased the UCr level (P<0.001). The level of CUA was higher and the levels of UCr and CCr were lower in the ALL group than in the HUA group (P = 0.021, 0.047, and 0.026, respectively). Among all groups, there were no statistically significant differences in CysC levels (P = 0.755) (Supplementary Table S1).
SFN inhibited uric acid production in rats
The activity of serum XOD and ADA was significantly higher in hyperuricemic rats than in rats in the NC group, and there was no significant difference in the liver XOD activity of hyperuricemic rats at week 6 (P = 0.010, 0.004, and 0.308, respectively). The serum ADA activity was lower in rats given SFN treatment than in rats in the HUA group (P = 0.041). Similarly, allopurinol also reduced serum ADA activity (P = 0.004) (Fig. 1D-F).
SFN improved liver function in rats
At week 6, both serum ALT and AST levels were significantly higher in the HUA group than in the NC group (P = 0.013 and 0.045, respectively). SFN had certain protective effects on the liver in hyperuricemic rats (P = 0.012 and 0.020, respectively). However, allopurinol poorly ameliorated liver function in hyperuricemic rats (Supplementary Table S1).
SFN ameliorated kidney pathology in hyperuricemic rats
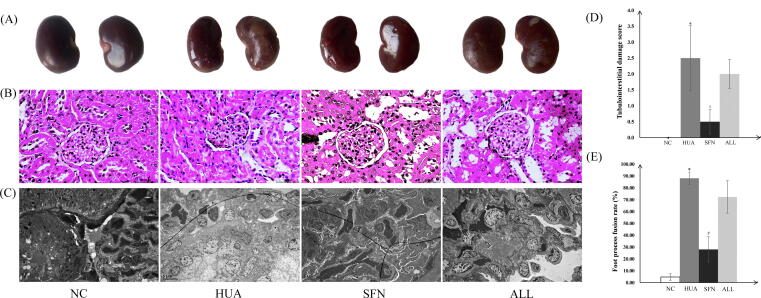
At week 6, observation of gross pathology showed normal kidney morphology with a smooth surface in the NC group. The HUA kidneys had developed a rough surface with whitish vacuoles. The surface of the kidneys following SFN intervention was indistinguishable from that of normal kidneys. The ALL group showed some improvement in the rough surface of the kidneys, but enlargement and vacuoles were still observed (Fig. 2A).
Fig. 2.
Gross pathology (A), light microscopy (B), electron microscopy observation (C), tubulointerstitial damage scores (D), and foot process fusion rates (E) of the kidney of hyperuricemic rats treated for 6 weeks with SFN. Scale bar: 100 μm; original magnification 400× (B); and scale bar: 5 μm; original magnification 3000× (C). Five fields of view per section and three rats per group were tested. * indicates that the difference is statistically significant compared with the NC group (P<0.05); # indicates that the difference is statistically significant compared to the HUA group (P<0.05); ANOVA, followed by LSD for post-hoc comparisons.
Under a light microscope, rats with hyperuricemia had dilated renal tubules, disorderly renal tubular epithelial cells, and scattered infiltrations of inflammatory cells in their renal interstitial tissue unlike in the NC group. The structures of glomeruli, renal tubules, and epithelial cells in the SFN group were similar to those in the NC group. The extent of tubular dilation was lower in the ALL group than in the HUA group, and neatly arranged renal tubular epithelial cells were observed. Specific details about the tubulointerstitial damage scores are provided in Fig. 2D.
Under an electron microscope, renal tissues obtained from hyperuricemic rats showed swollen glomeruli and extensive fusion of foot processes unlike in the NC group. In the SFN group, swelling to the glomerular epithelial cells was notably ameliorated, with the foot process arranging clearly visible, which was similar to the NC group. The ALL group showed less glomerular epithelial swelling than the HUA group, while the foot processes of the podocytes fused. Specific details about the foot process fusion rates are provided in Fig. 2E.
SFN regulated urate transporter expression
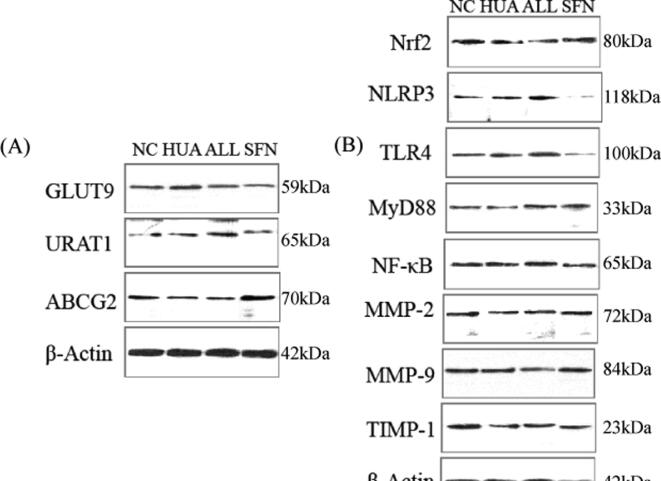
The kidneys of hyperuricemic rats displayed increased expression of the uric acid reabsorption transporters GLUT9 and URAT1 (P = 0.030 and 0.034, respectively). After 6 weeks of SFN intervention, GLUT9 and URAT1 expression was significantly decreased. The expression of the uric acid excretion transporter ABCG2 was significantly lower in the HUA group than in the NC group. ABCG2 expression was significantly increased in the rats after SFN treatment. However, allopurinol had no effect on transport-related proteins (Fig. 3A).
Fig. 3.
GLUT9, URAT1, ABCG2, Nrf2, NLRP3, TLR4, MyD88, NF-κB, MMP-2, MMP-9, and TIMP-1 levels in hyperuricemic rats treated for 6 weeks with SFN. GLUT9, URAT1, and ABCG2 (A) and Nrf2, NLRP3, TLR4, MyD88, NF-κB, MMP-2, MMP-9, and TIMP-1 (B) (n = 5). The samples derived from the same experiment and that blots were processed in parallel.
SFN diminished renal inflammatory responses by regulating Nrf2 activity
Hyperuricemic rats displayed lower expression of Nrf2 in the kidneys than rats in the NC group. SFN intervention enhanced the expression of Nrf2 (P = 0.001), but allopurinol treatment failed. The NLRP3 level was increased in the HUA group, and SFN treatment significantly inhibited the levels of NLRP3 (P = 0.026). TLR4, MyD88, and NF-κB expression in the HUA group was significantly higher than that in the NC group (P = 0.043, 0.005, and 0.003, respectively). In contrast, SFN treatment significantly downregulated the expression of these signalling molecules (P = 0.014, 0.001, and 0.030, respectively). Furthermore, SFN had better efficacy than allopurinol. MMP‐2 and MMP-9 expression showed no significant changes among all the groups. Hyperuricemic rats displayed a reduced level of TIMP-1, which was significantly attenuated by SFN treatment (Fig. 3B).
SFN improved metabolic function in hyperuricemic rats
Screening of differential metabolites
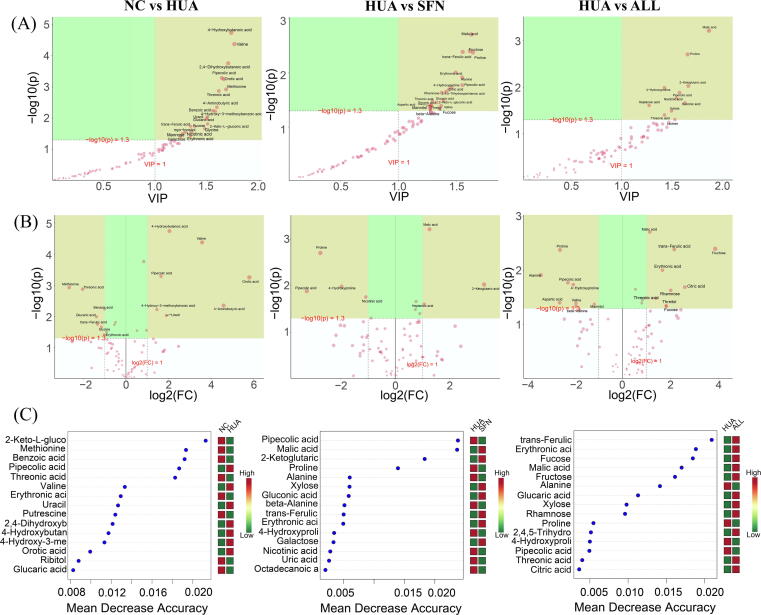
The metabolic profile of rats in the SFN group differed from that of the HUA group by OPLS-DA (R2X = 0.686, R2Y = 0.954, Q2 = 0.731, P<0.05). To identify important metabolites that may be related to how SFN influenced hyperuricemia, we used three approaches, including OPLS-DA, Kruskal–Wallis tests, and random forest (Supplementary Fig. 1). The details for significantly differential metabolites are shown in Supplementary Table S2.
Metabolic pathway analysis
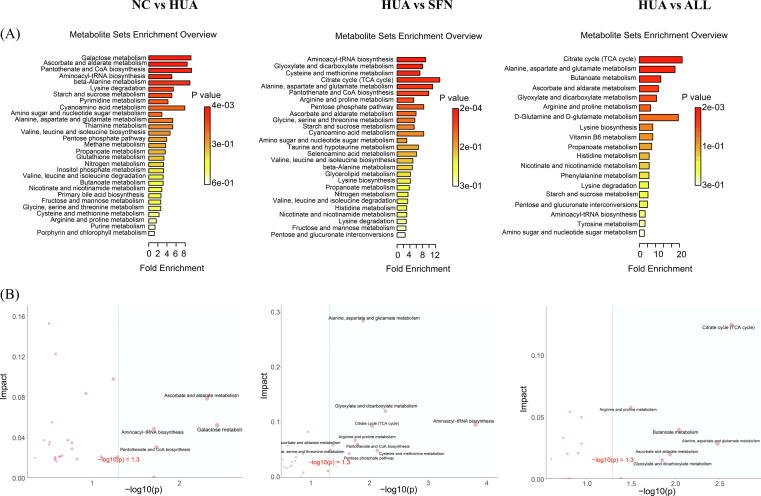
To identify the key metabolic pathways, we applied pathway enrichment analysis and topology analysis (Fig. 4; Supplementary Table S3). In the HUA group, glycine, threonic acid, D-mannose, sucrose, and L-methionine levels were significantly lower, while uracil and pipecolic acid levels were higher than those in the NC group. SFN treatment significantly elevated the levels of threonic acid, oxoglutaric acid, and succinic acid, while SFN reduced the contents of L-proline, pipecolic acid, and 4-hydroxyproline. The pathways involved in the response to SFN treatment of hyperuricemia were ascorbate and aldarate metabolism; glyoxylate and dicarboxylate metabolism; alanine, aspartate and glutamate metabolism; the citrate cycle (TCA cycle); and arginine and proline metabolism. Among them, the ascorbate and aldarate metabolism pathway was obviously disturbed by hyperuricemia and could act as a target for SFN treatment. Interestingly, although both SFN and allopurinol functioned in hyperuricemia progression by modulating these pathways, they acted by correcting and reversing the levels of different metabolic biomarkers.
Fig. 4.
Metabolic alterations of hyperuricemic rats treated for 6 weeks with SFN (n = 10). Pathway enrichment analysis of ORA (the darker the color, the smaller the p value) (A) and the scatter plots for a summary of the joint evidence from enrichment analysis (p value) and topology analysis (pathway impact) (B).
SFN intervention altered gut microbiota in hyperuricemic rats
SFN altered the gut microbiota composition in hyperuricemic rats
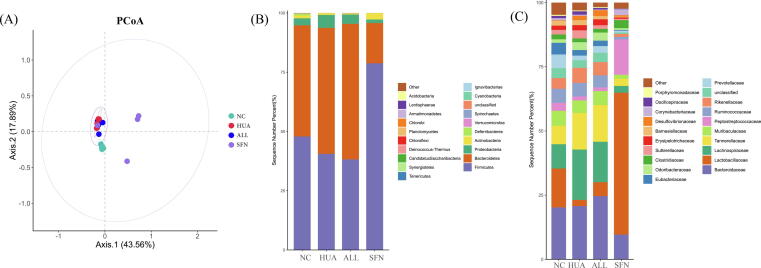
SFN treatment significantly increased the beta diversity of hyperuricemic rats, while allopurinol intervention did not achieve comparable effects (Supplementary Fig. 2A). At the phylum level, the rats in the HUA group showed a lower abundance of Firmicutes and an increased abundance of Bacteroidetes than rats in the NC group. The SFN group displayed a significantly higher Firmicutes/Bacteroidetes ratio than the HUA group (Supplementary Fig. 2B). At the family level, the abundance of Lactobacillaceae and Clostridiaceae was decreased in hyperuricemic rats. Of interest, SFN greatly increased the relative abundance of both (Supplementary Fig. 2C).
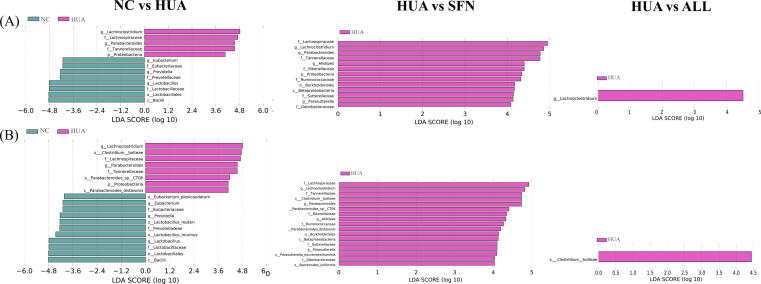
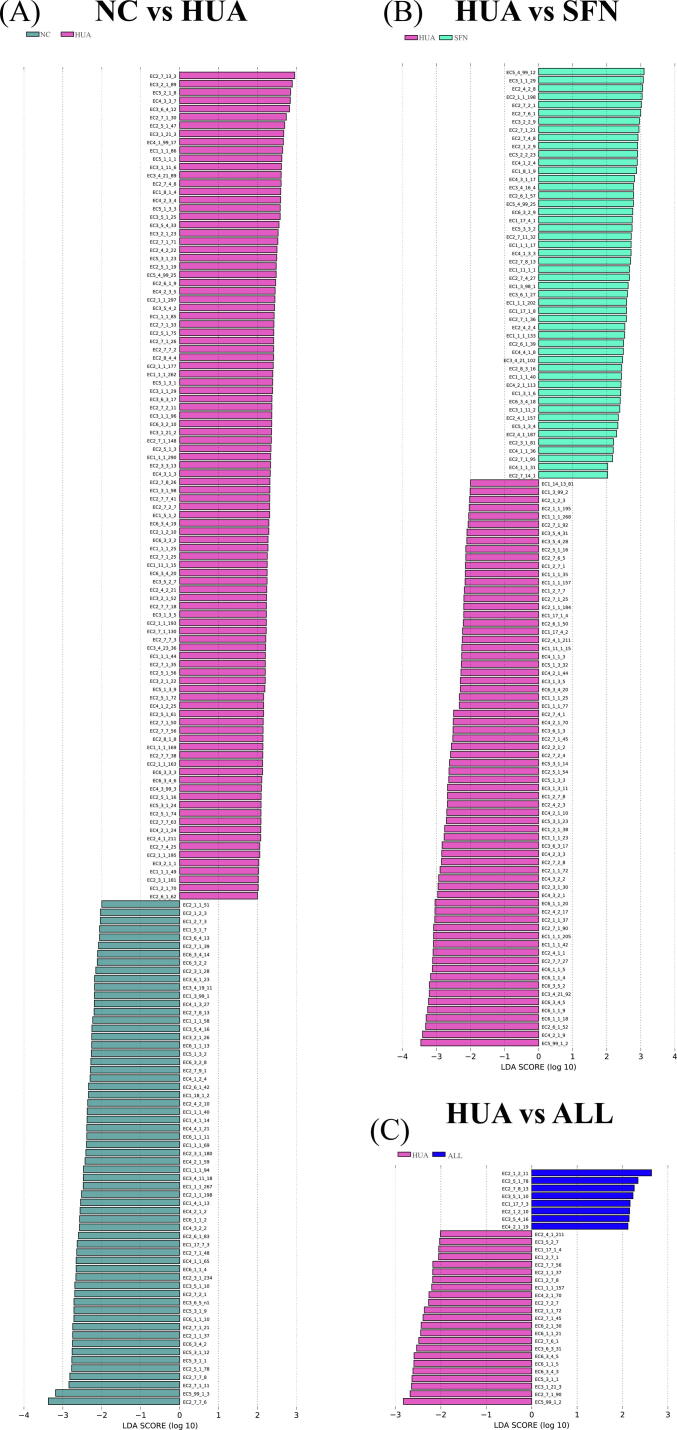
The dominant microorganisms in the groups were explored using LefSe analysis (Fig. 5). Lactobacillus, Parabacteroides, Eubacterium, and Lachnoclostridium were depleted in the HUA group but enriched with SFN treatment (P<0.05). Moreover, SFN treatment significantly increased the abundance of Saccharomyces and decreased the abundances of Bacteroides, Parasutterella, and Alistipes in hyperuricemic rats (LDA>2, P<0.05). At the species level, the hyperuricemic rats exhibited a lower abundance of Lactobacillus murinus, Lactobacillus reuteri, Eubacterium plexicaudatum, and Odoribacter splanchnicus than normal rats (P<0.05). Moreover, the hyperuricemic rats had significantly higher abundances of Bacteroides cellulosilyticus and Clostridium bolteae than rats in the NC group, which was reversed after SFN intervention (P<0.05). SFN can also increase the abundance of Odoribacter splanchnicus and Actinomyces ruminicola. The ALL group had significantly lower abundance of Clostridium bolteae than the HUA group.
Fig. 5.
Gut microbial alterations in hyperuricemic rats treated for 6 weeks with SFN (n = 10). LEfSe significant differences in abundance at the genus level between groups (LDA score>4) (A) and LEfSe significant differences in abundance at the species level between groups (LDA score>4) (B).
SFN altered microbial function in hyperuricemic rats
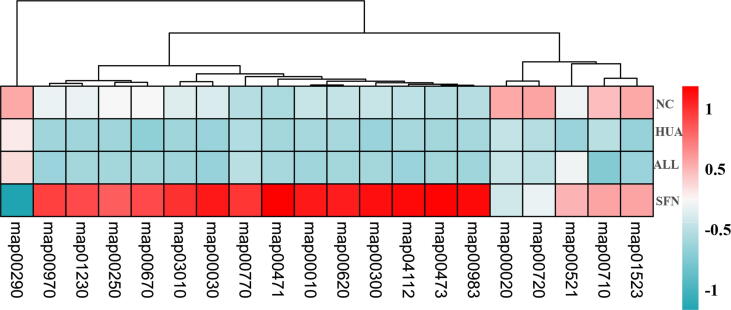
A change in bacterial taxonomy could affect the metabolic functions of the gut microbiota (Supplementary Fig. 3). Activation of the aminoacyl-tRNA biosynthesis and pantothenate and CoA biosynthesis pathways was significantly lower in the HUA group than in the NC group, and rats given SFN treatment exhibited significantly higher activation of the TCA cycle and alanine, aspartate and glutamate metabolism pathways than rats in the HUA group, which was consistent with the findings in metabolomic analysis. Moreover, SFN treatment also downregulated valine, leucine and isoleucine biosynthesis and upregulated D-glutamine and D-glutamate metabolism, D-alanine metabolism, lysine biosynthesis, pyruvate metabolism, the pentose phosphate pathway, biosynthesis of amino acids, glycolysis/gluconeogenesis, one carbon pool by folate, antifolate resistance, and drug metabolism.
In addition, the functional enzymes of the gut microbiota are shown in Supplementary Fig. 4. The activities of malate dehydrogenase, acetate kinase, and thymidine kinase, which are involved in the TCA cycle, and adenylosuccinate lyase, which is involved in purine metabolism, significantly decreased in the HUA group. The expression of shikimate dehydrogenase and peroxiredoxin, which exert antioxidant and anti-inflammatory effects, was significantly increased in the HUA group. All these changes were significantly reversed after SFN treatment. Intriguingly, there was a significant difference in many epigenetic enzymes for pairwise comparisons between groups. The activity of 16S rRNA (cytidine(1402)-2′-O)-methyltransferase, leucine-tRNA ligase, and DNA (cytosine-5-)-methyltransferase) was lower in the HUA group than in the NC group and higher in the SFN group than in the HUA group. The activity of cobalt-precorrin-5B (C(1))-methyltransferase was higher in the HUA group than in the NC group and lower in the SFN group than in the HUA group. Moreover, SFN significantly increased the expression of aminoacyl-tRNA hydrolase and tRNA pseudouridine synthase (P<0.05).
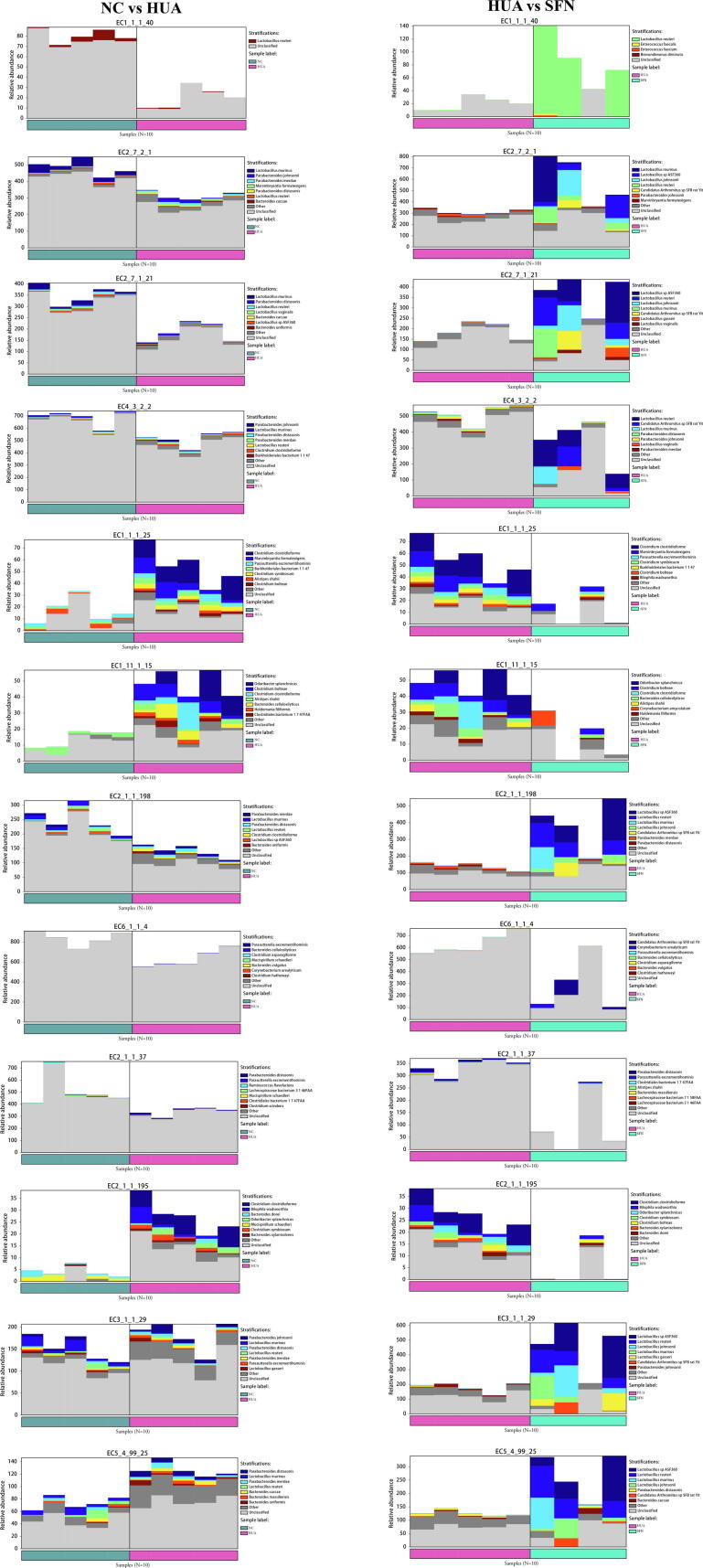
We used HUMAnN2 to identify the species source of function and drew bar charts (Supplementary Fig. 5). The energy metabolism, purine metabolism, nucleotide methylation, and tRNA modification functions were derived from Lactobacillus reuteri and Lactobacillus murinus. The antioxidant and anti-inflammatory functions and the amino acid methylation function were derived from Clostridium clostridioforme, Clostridium symbiosum, Clostridium bolteae, and Odoribacter splanchnicus.
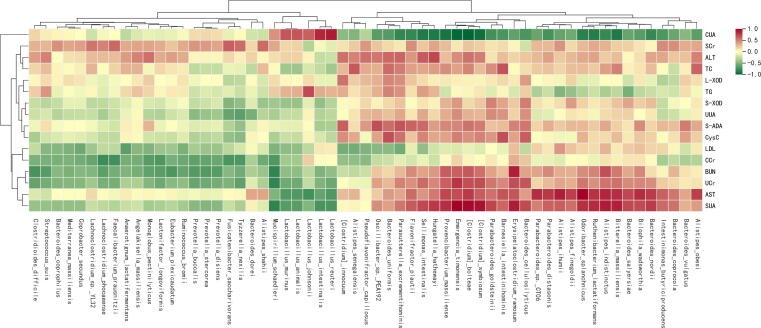
Relationships between hyperuricemic gut microbiota and phenotypes
As shown in Supplementary Fig. 6, the SUA, UCr, and AST levels were negatively related to the abundances of two probiotic bacteria, Lactobacillus murinus and Lactobacillus reuteri, and positively related to the abundance of Clostridium bolteae (P<0.05, r>0.4 or r<-0.4). The BUN level was negatively related to Eubacterium plexicaudatum abundance and positively related to Clostridium bolteae abundance (P<0.05, r>0.4 or r<-0.4). The CUA level was negatively related to Clostridium bolteae abundance and positively related to Lactobacillus murinus and Lactobacillus reuteri abundance (P<0.05, r>0.4 or r<-0.4). The CCr level was negatively related to Eubacterium plexicaudatum abundance (P<0.05, r<-0.4).
Correlation of metagenomics and metabolome
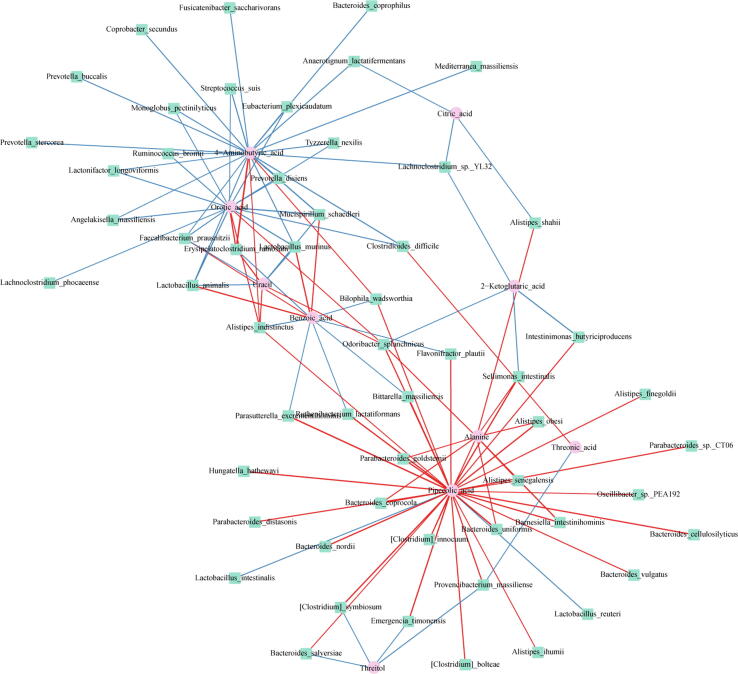
We identified four host-gut microbiome co-metabolic pathways from metabolomics and metagenomics analysis, including pantothenate and CoA biosynthesis, aminoacyl-tRNA biosynthesis, alanine, aspartate and glutamate metabolism, and the TCA cycle. Among the key metabolites in these pathways, succinic acid and oxoglutaric acid are critical host-gut microbiome co-metabolites. Furthermore, we analyzed the correlation between the levels of sixty differentially abundant bacteria at the species level and the levels of twelve key metabolites by Spearman's correlation analysis. In this association network plot (Fig. 6), pipecolic acid, orotic acid, and 4-aminobutyric acid were crucial nodes closely connected with the gut microbiota. Notably, pipecolic acid showed the largest number of associations (thirty-two associations) with microbial species, and its levels were strongly associated with the metabolism of lysine biosynthesis in microorganisms. Pipecolic acid levels were significantly positively correlated with Clostridium bolteae, Clostridium symbiosum, and Clostridium innocuum abundance. Orotic acid levels were significantly negatively correlated with Eubacterium plexicaudatum abundance (P<0.05, r>0.4 or r<-0.4). 4-Aminobutyric acid levels were significantly negatively correlated with Lactobacillus animalis, Eubacterium plexicaudatum, Bacteroides coprophilus, Ruminococcus bromii, and Faecalibacterium prausnitzii abundance (P<0.05, r>0.4 or r<-0.4).
Fig. 6.
Network plot of network analysis for the correlation between metabolite types and the gut microbiome. Each node in the network represents a metabolite or microbe, and each edge represents the relationship between any two nodes, with the line color determined by the direction of association (red: positive; blue: negative). Correlations with differences in correlation strengths >0.6 were selected for visualization. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
In this study, we demonstrated the excellent curative potential of SFN in lowering uric acid levels by decreasing urate synthesis and increasing renal urate excretion. For the first time, we revealed that SFN could ameliorate the progression of hyperuricemia by reprogramming the gut microbiome and metabolome.
We first explored the effects of SFN on uric acid production and revealed that SFN can decrease XOD and ADA activity and partly reduce liver damage to decrease urate synthesis. Promisingly, SFN can achieve a similar performance to allopurinol. We next focused on the influence of SFN on the excretory function of the kidney by assessing the following four dimensions. We discovered that SFN could improve renal morphological changes. Moreover, SFN not only increased uric acid excretion by upregulating renal ABCG2 protein expression but also inhibited uric acid reabsorption by downregulating renal URAT1 and GLUT9 expression. We further revealed that SFN could suppress NF-κB-mediated NLRP3 inflammasome activation by inhibiting TLR4/MyD88 signaling, which is in partial agreement with previous literature [29]. We also found that SFN activated TIMP-1, which was different from previous studies [30].
We integrated metagenomic and metabolomic analyses to identify two host-microbial co-metabolites, oxoglutaric acid and succinic acid. SFN treatment increased the levels of oxoglutaric acid and then improved ascorbate and aldarate metabolism. Succinic acid can be affected by short-chain fatty acids (SCFAs) and modulate DNA methylation through ten-eleven translocation methylcytosine dioxygenases [31]. Coincidentally, succinic acid can enter the host-gut microbiome co-metabolic pathway TCA cycle. The metabolic fluxes in the TCA cycle were suppressed when rats were administered SFN, suggesting that SFN might hamper hyperuricemia in rats by regulating this important metabolite. In our study, the decreased levels of the host-gut microbiome co-metabolic pathway alanine, aspartate and glutamate metabolism in the HUA group were possibly related to abnormal renal function. Under hyperuricemic conditions, the kidney maintains the acid balance by taking up more glutamine and glutamate [32], causing the levels of both to decline. SFN treatment affected alanine, aspartate and glutamate metabolism to alleviate oxidative stress in rats, resulting in further improvements in intestinal barrier function [33]. Alterations in another host-gut microbiome co-metabolic pathway, aminoacyl-tRNA biosynthesis, probably affect cell function, leading to the development of hyperuricemia. Recent research suggests that tRNA metabolism contributes to metabolic diseases by affecting insulin sensitivity [34]. By altering the activity of tRNA-modifying enzymes, oxidative stress may influence the tRNA epigenome [35]. SFN could upregulate the activity of aminoacyl-tRNA hydrolase. Notably, we found that pipecolic acid is the metabolite most strongly associated with gut microflora. It is important to regulate immunity with pipecolic acid, a non-proteinacious product of lysine catabolism. Our findings suggest that pipecolic acid levels were elevated in hyperuricemic rats and positively related to the abundance of intestinal pathogens, including Clostridium bolteae, Clostridium innocuum, and Clostridium symbiosum. Fortunately, SFN treatment reversed the increases in the abundance of the three species in this genus. Our results also showed that Clostridium bolteae abundance was positively related to the indices of kidney and liver injury and urate excretion. Furthermore, we found that orotic acid was the second most connected with the gut microbiota and negatively correlated with Eubacterium plexicaudatum abundance and that the latter was negatively correlated with the indices of kidney injury. Research has shown that the urea cycle can be disrupted by excessive orotic acid [36]. As a result of SFN, intestinal flora are regulated, and orotic acid levels are reduced to maintain the urea cycle balance. Therefore, interactions between the host gut microbiome and metabolome are perturbed in hyperuricemic individuals, and fortunately SFN can treat or ameliorate the interactions.
The balance in host microflora is closely related to health. Our results showed that feeding mice hyperuricemia-inducing feedstuff decreased the abundance of Lactobacillaceae and Clostridiaceae, while SFN was effective in reversing these changes. Lactobacillaceae is well known for its beneficial effects in modulating intestinal microbiota and its contribution to reducing uric acid levels [37]. It has been reported that Clostridiaceae can degrade uric acid [38]. The change in probiotic abundances, such as the abundances of Lactobacillus, Parabacteroides, and Saccharomyces, was consistent with that in previous studies, which showed that this genus was depleted in hyperuricemia [39]. The gene function analysis of the predicted metagenomes showed high agreement with the metabolomic profiling results, mainly reflected in pathways involved in energy metabolism, purine metabolism, and antioxidant and anti-inflammatory effects. Interestingly, we showed that these functions are largely due to Lactobacillus spp. This finding was also confirmed by the results of correlation analysis, which suggested that Lactobacillus spp. abundance was negatively associated with SUA, UCr, and AST levels and positively associated with CUA levels. Fortunately, SFN treatment contributed to a higher abundance of Saccharomyces. A cohort study demonstrated that Saccharomyces spp. can improve insulin resistance by influencing bacterial diversity [40]. Saccharomyces was also revealed to promote Nrf2 in ulcerative colitis mice [41]. Therefore, the modulation of Saccharomyces and activation of Nrf2-modulated antioxidant defence and immunomodulatory effects may partially explain how SFN ameliorates hyperuricemia. Studies have identified that the abundance of Bacteroides was higher in the gut microbiota of hyperuricemic patients than in healthy subjects [42], which is consistent with the results reported here. Fortunately, we found SFN reduced the relative abundance of Bacteroides. Our results, therefore, indicated that SFN may reduce antigen load to the host and help alleviate inflammation and immune reactions by decreasing the levels of opportunistic pathogens.
SFN treatment increased the abundance of the SCFA producers Lachnoclostridium, Eubacterium, and Odoribacter. SCFAs promote uric acid excretion by providing energy to cells in the intestinal wall [43], enhancing epithelial barrier function [44], suppressing pro-inflammatory cytokine release, and leading to amelioration of kidney inflammation [45]. We also studied gut microbiota at the species level, which is distinct from the family or genus level observations performed in other studies; this approach made our pooled estimate more exact and credible. Hyperuricemic rats showed a significant increase in the abundances of pathogenic and potentially harmful enterobacteria. Interestingly, SFN could also enhance Actinomyces ruminicola abundance, which produces formic, acetic, and lactic acids as end products from glucose [46]. Taken together, these data suggest that the gain of protective bacteria and loss of pathobionts mediated by SFN treatment at least in part contribute to improvements in hyperuricemia.
Epigenetic modifications could be regulated by SFN-mediated changes in the gut microbiome. SCFAs, as mentioned above, represent an important group of epigenetically relevant molecules [47]. We revealed that SFN treatment may increase the levels of the one carbon pool by folate and antifolate resistance through functional analyses of intestinal microbiota. Folate produced by Lactobacillus species is known to contribute to DNA methylation or histone methylation by donating methyl groups [48]. We also found that SFN-mediated alterations in the gut microbiome could regulate enzyme expression related to epigenetic modifications. SFN-mediated activation of Nrf2 was partially responsible for the many anti-inflammatory effects. Intriguingly, Nrf2 status was reported to affect HDAC gene promoter associations [49]. In our opinion, this was a crucial strategy for lowering uric acid levels.
The limitations of this study include three points. We used guinea glucoraphanin bioactivated with myrosinase for our experiments, but future experiments may directly involve sulforaphane. Bioinformatics analysis resulted in speculations that require further experimental testing. In addition, further investigation of the interaction between the microbiota and the host epigenome is still needed.
Conclusion
In summary, we were the first to identify the role of SFN in lowering uric acid levels. Notably, SFN provides benefits in epigenetic modification of Nrf2, intestinal homeostasis, and host metabolism. Our findings may provide a good means for efficiently preventing and treating hyperuricemia.
CRediT authorship contribution statement
Ruoyu Wang: Conceptualization, Methodology, Software, Data curation, Visualization, Writing – original draft. Mairepaiti Halimulati: Software, Validation. Xiaojie Huang: Software, Validation. Yuxin Ma: Methodology, Supervision. Lutong Li: Visualization, Investigation. Zhaofeng Zhang: Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Compliance with Ethics Requirements
All Institutional and National Guidelines for the care and use of animals (fisheries) were followed.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.11.003.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Supplementary figure 1.
Supplementary figure 2.
Supplementary figure 3.
Supplementary figure 4.
Supplementary figure 5.
Supplementary figure 6.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov; PRJNA 847951.
References
- 1.Singh G., Lingala B., Mithal A. Gout and hyperuricaemia in the USA: prevalence and trends. Rheumatology (Oxford) 2019;58(12):2177–2180. doi: 10.1093/rheumatology/kez196. [DOI] [PubMed] [Google Scholar]
- 2.Koo B.S., et al. Distribution of serum uric acid levels and prevalence of hyper- and hypouricemia in a Korean general population of 172,970. Korean J Intern Med. 2021;36(Suppl 1):S264–S272. doi: 10.3904/kjim.2020.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J., et al. Sonneratia apetala seed oil attenuates potassium oxonate/hypoxanthine-induced hyperuricemia and renal injury in mice. Food Funct. 2021;12(19):9416–9431. doi: 10.1039/d1fo01830b. [DOI] [PubMed] [Google Scholar]
- 4.Roughley M.J., Belcher J., Mallen C.D., Roddy E. Gout and risk of chronic kidney disease and nephrolithiasis: meta-analysis of observational studies. Arthritis Res Ther. 2015;17(1):90. doi: 10.1186/s13075-015-0610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao T.F., et al. Hyperuricemia and the risk of ischemic stroke in patients with atrial fibrillation–could it refine clinical risk stratification in AF? Int J Cardiol. 2014;170(3):344–349. doi: 10.1016/j.ijcard.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Browne L.D., et al. Serum uric acid and mortality thresholds among men and women in the Irish health system: a cohort study. Eur J Intern Med. 2021;84:46–55. doi: 10.1016/j.ejim.2020.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen S.M., Zobbe K., Kristensen L.E., Christensen R. Nutritional recommendations for gout: an update from clinical epidemiology. Autoimmun Rev. 2018;17(11):1090–1096. doi: 10.1016/j.autrev.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Nieto F.J., Iribarren C., Gross M.D., Comstock G.W., Cutler R.G. Uric acid and serum antioxidant capacity: a reaction to atherosclerosis? Atherosclerosis. 2000;148(1):131–139. doi: 10.1016/s0021-9150(99)00214-2. [DOI] [PubMed] [Google Scholar]
- 9.Dalbeth N., Merriman T.R., Stamp L.K. Gout. Lancet. 2016;388(10055):2039–2052. doi: 10.1016/s0140-6736(16)00346-9. [DOI] [PubMed] [Google Scholar]
- 10.Yang T., Richards E.M., Pepine C.J., Raizada M.K. The gut microbiota and the brain-gut-kidney axis in hypertension and chronic kidney disease. Nat Rev Nephrol. 2018;14(7):442–456. doi: 10.1038/s41581-018-0018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan L., et al. Abnormal metabolism of gut microbiota reveals the possible molecular mechanism of nephropathy induced by hyperuricemia. Acta Pharm Sin B. 2020;10(2):249–261. doi: 10.1016/j.apsb.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuevas-Sierra A., Ramos-Lopez O., Riezu-Boj J.I., Milagro F.I., Martinez J.A.D. Gut microbiota, and obesity: links with host genetics and epigenetics and potential applications. Adv Nutr. 2019;10(suppl_1):S17–S30. doi: 10.1093/advances/nmy078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yip K., Cohen R.E., Pillinger M.H. Asymptomatic hyperuricemia: is it really asymptomatic? Curr Opin Rheumatol. 2020;32(1):71–79. doi: 10.1097/bor.0000000000000679. [DOI] [PubMed] [Google Scholar]
- 14.Wu W., Dnyanmote A.V., Nigam S.K. Remote communication through solute carriers and ATP binding cassette drug transporter pathways: an update on the remote sensing and signaling hypothesis. Mol Pharmacol. 2011;79(5):795–805. doi: 10.1124/mol.110.070607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P., et al. Resveratrol-induced gut microbiota reduces obesity in high-fat diet-fed mice. Int J Obes (Lond) 2020;44(1):213–225. doi: 10.1038/s41366-019-0332-1. [DOI] [PubMed] [Google Scholar]
- 16.Shock T, Badang L, Ferguson B, Martinez-Guryn K. The interplay between diet, gut microbes, and host epigenetics in health and disease. J Nutr Biochem 95 (2021) 108631. https://doi.org/10.1016/j.jnutbio.2021.108631. [DOI] [PMC free article] [PubMed]
- 17.Houghton C.A. Sulforaphane: Its “Coming of Age” as a Clinically relevant nutraceutical in the prevention and treatment of chronic disease. Oxid Med Cell Longev. 2019;2019:2716870. doi: 10.1155/2019/2716870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vermeulen M., Klöpping-Ketelaars I.W., van den Berg R., Vaes W.H. Bioavailability and kinetics of sulforaphane in humans after consumption of cooked versus raw broccoli. J Agric Food Chem. 2008;56(22):10505–10509. doi: 10.1021/jf801989e. [DOI] [PubMed] [Google Scholar]
- 19.Bai Y., et al. Sulforaphane Protects against cardiovascular disease via Nrf2 activation. Oxid Med Cell Longev. 2015;2015 doi: 10.1155/2015/407580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su X., et al. Anticancer activity of sulforaphane: the epigenetic mechanisms and the Nrf2 Signaling pathway. Oxid Med Cell Longev. 2018;2018:5438179. doi: 10.1155/2018/5438179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santín-Márquez R., Alarcón-Aguilar A., López-Diazguerrero N.E., Chondrogianni N., Königsberg M. Sulforaphane – role in aging and neurodegeneration. Geroscience. 2019;41(5):655–670. doi: 10.1007/s11357-019-00061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu K., et al. Association of solid fuel use with risk of cardiovascular and all-cause mortality in rural China. JAMA. 2018;319(13):1351–1361. doi: 10.1001/jama.2018.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang Y.W., Jang J.Y., Kim Y.H., Kim J.W., Shim J.J. The effects of broccoli sprout extract containing sulforaphane on lipid peroxidation and helicobacter pylori infection in the gastric mucosa. Gut Liver. 2015;9(4):486–493. doi: 10.5009/gnl14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He C., et al. Sulforaphane normalizes intestinal flora and enhances gut barrier in mice with BBN-induced bladder cancer. Mol Nutr Food Res. 2018;62(24):e1800427. doi: 10.1002/mnfr.201800427. [DOI] [PubMed] [Google Scholar]
- 25.Giacoppo S., et al. (RS)-glucoraphanin purified from Tuscan black kale and bioactivated with myrosinase enzyme protects against cerebral ischemia/reperfusion injury in rats. Fitoterapia. 2014;99:166–177. doi: 10.1016/j.fitote.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 26.Henderson N.C., et al. Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat Med. 2013;19(12):1617–1624. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka Y., et al. Fenofibrate, a PPARα agonist, has renoprotective effects in mice by enhancing renal lipolysis. Kidney Int. 2011;79(8):871–882. doi: 10.1038/ki.2010.530. [DOI] [PubMed] [Google Scholar]
- 28.Azeloglu E.U., et al. Interconnected network motifs control podocyte morphology and kidney function. Sci Signal. 2014;7(311):ra12. doi: 10.1126/scisignal.2004621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishida K., et al. Sulforaphane ameliorates ethanol plus carbon tetrachloride-induced liver fibrosis in mice through the Nrf2-mediated antioxidant response and acetaldehyde metabolization with inhibition of the LPS/TLR4 signaling pathway. J Nutr Biochem. 2021;89 doi: 10.1016/j.jnutbio.2020.108573. [DOI] [PubMed] [Google Scholar]
- 30.Liu W., et al. Activation in M1 but not M2 macrophages contributes to cardiac remodeling after myocardial infarction in rats: a critical role of the calcium sensing receptor/NRLP3 inflammasome. Cell Physiol Biochem. 2015;35(6):2483–2500. doi: 10.1159/000374048. [DOI] [PubMed] [Google Scholar]
- 31.Etchegaray J.P., Mostoslavsky R. Interplay between Metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol Cell. 2016;62(5):695–711. doi: 10.1016/j.molcel.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dryer S.E. Glutamate receptors in the kidney. Nephrol Dial Transplant. 2015;30(10):1630–1638. doi: 10.1093/ndt/gfv028. [DOI] [PubMed] [Google Scholar]
- 33.Sun L., et al. Cecal gut microbiota and metabolites might contribute to the severity of acute myocardial ischemia by impacting the intestinal permeability, oxidative stress, and energy metabolism. Front Microbiol. 2019;10:1745. doi: 10.3389/fmicb.2019.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med 2014;20(6):306–314. https://doi.org/10.1016/j.molmed.2014.01.008. [DOI] [PubMed]
- 35.Wei F.Y., et al. Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans. Cell Metab. 2015;21(3):428–442. doi: 10.1016/j.cmet.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 36.Wendler P.A., Blanding J.H., Tremblay G.C. Interaction between the urea cycle and the orotate pathway: studies with isolated hepatocytes. Arch Biochem Biophys. 1983;224(1):36–48. doi: 10.1016/0003-9861(83)90188-1. [DOI] [PubMed] [Google Scholar]
- 37.Gerritsen J, Smidt H, Rijkers GT, de Vos WM. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr 2011;6(3):209-240. https://doi.org/10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed]
- 38.Hartwich K., Poehlein A., Daniel R. The purine-utilizing bacterium Clostridium acidurici 9a: a genome-guided metabolic reconsideration. PLoS ONE. 2012;7(12):e51662. doi: 10.1371/journal.pone.0051662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., et al. The gut microbiota as a target to control hyperuricemia pathogenesis: potential mechanisms and therapeutic strategies. Crit Rev Food Sci Nutr. 2021;1–11 doi: 10.1080/10408398.2021.1874287. [DOI] [PubMed] [Google Scholar]
- 40.Shuai M., et al. Mapping the human gut mycobiome in middle-aged and elderly adults: multiomics insights and implications for host metabolic health. Gut. 2022 doi: 10.1136/gutjnl-2021-326298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao H., et al. Saccharomyces boulardii ameliorates dextran sulfate sodium-induced ulcerative colitis in mice by regulating NF-κB and Nrf2 signaling pathways. Oxid Med Cell Longev. 2021;2021:1622375. doi: 10.1155/2021/1622375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xing S.C., et al. Study on the diversity of bacteroides and clostridium in patients with primary gout. Cell Biochem Biophys. 2015;71(2):707–715. doi: 10.1007/s12013-014-0253-5. [DOI] [PubMed] [Google Scholar]
- 43.Nieuwdorp M., Gilijamse P.W., Pai N., Kaplan L.M. Role of the microbiome in energy regulation and metabolism. Gastroenterology. 2014;146(6):1525–1533. doi: 10.1053/j.gastro.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Jin U.H., et al. Short chain fatty acids enhance aryl hydrocarbon (Ah) responsiveness in mouse colonocytes and caco-2 human colon cancer cells. Sci Rep. 2017;7(1):10163. doi: 10.1038/s41598-017-10824-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu TJ, Hsieh YJ, Lu CW, Lee CJ, Hsu BG. Linagliptin protects against endotoxin-induced acute kidney injury in rats by decreasing inflammatory cytokines and reactive oxygen species. Int J Mol Sci 2021;22(20). https://doi.org/10.3390/ijms222011190. [DOI] [PMC free article] [PubMed]
- 46.An D., Cai S., Dong X. Actinomyces ruminicola sp. nov., isolated from cattle rumen. Int J Syst Evol Microbiol. 2006;56(Pt 9):2043–2048. doi: 10.1099/ijs.0.64059-0. [DOI] [PubMed] [Google Scholar]
- 47.Krautkramer K.A., et al. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell. 2016;64(5):982–992. doi: 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossi M., Amaretti A., Raimondi S. Folate production by probiotic bacteria. Nutrients. 2011;3(1):118–134. doi: 10.3390/nu3010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajendran P., et al. Nrf2 status affects tumor growth, HDAC3 gene promoter associations, and the response to sulforaphane in the colon. Clin Epigenetics. 2015;7(1):102. doi: 10.1186/s13148-015-0132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov; PRJNA 847951.