Abstract
Genomic tests expand diagnostic and screening opportunities but also identify genetic variants of uncertain clinical significance (VUS). Only a minority of VUS are likely to prove to be pathogenic when later reassessed, but resolution of the uncertainty is rarely timely. That uncertainty adds complexity to clinical decision-making and can result in harms and costs to patients and the healthcare system, including the time-consuming analysis required to interpret a VUS, and the potential for unnecessary treatment and adverse psychological effects. Current efforts to improve variant interpretation will help to reduce the scope of the problem, but the high prevalence of rare and novel variants in the human genome points to VUS as an ongoing challenge. Additional strategies can help to mitigate the potential harms of VUS, including testing protocols that limit identification or reporting of VUS, subclassifying VUS according to the likelihood of pathogenicity, routine family-based variant evaluation, and enhanced counseling efforts. All involve tradeoffs, and the appropriate balance of measures is likely to vary for different test uses and clinical settings. Cross-specialty deliberation and public input could contribute to systematic and broadly supported policies for managing VUS.
Introduction
Genomic tests, including multi-gene panels and exome/genome sequencing, can identify inherited susceptibilities to cancer, cardiovascular diseases, and a wide range of other disorders (1–3). The benefits include improved diagnoses, prognostic information, targeted disease screening, and in some cases, essential information for effective disease management. Variants identified by genomic testing are classified in terms of disease causation as benign; likely benign; VUS; likely pathogenic; and pathogenic (8), based on a wide range of evidence, including variant prevalence, associations between variants and disease, functional and computational predictive data, and similarity between the patient’s clinical features and those associated with alteration of the gene in which a variant is found (Table 1). Because judgment is required in evaluating the evidence (4), laboratories may differ in the classification of a given variant (5,6).
Table 1:
Categories of Evidence Used in Genetic Variant Interpretation*
| Category | Examples |
|---|---|
|
| |
| Population and patient data | • Variant prevalence higher than disease prevalence provides strong evidence for benign classification • Statistical increase in prevalence of variant among affected individuals provides strong evidence of pathogenicity; this criterion is challenging for rare variants. • Match of the patient’s clinical features with those of the condition associated with the gene supports pathogenicity. |
| Segregation data | • Lack of segregation of variant with disease provides strong evidence for benign classification • Segregation of variant with disease provides evidence of pathogenicity, with strength of evidence increasing with number of families studied |
| De novo data | • De novo variant (not present in either parent) in a relevant gene is more likely to be pathogenic; strength of evidence is increased if maternity and paternity are confirmed |
| Functional data | • Studies indicating no deleterious effect on gene function provide strong evidence for benign classification • Studies indicating a deleterious effect on gene function provide strong evidence for pathogenicity |
| Computational and predictive data | • Predictions of functional effects are compared across multiple algorithms that consider cross species conservation of protein sequence, protein folding, critical protein domains, size and change of amino acid substitutions, and predictions of splicing. |
| Other | • A variant observed in combination with another known pathogenic variant may provide evidence for benign classification (if disease is inherited as a dominant condition) or for pathogenicity (if disease is inherited as an autosomal recessive condition) • Categorization of a variant by a reputable source as benign or pathogenic provides supporting evidence for classification |
Based on Richards et al. (8).
Today, VUS substantially outnumber pathogenic findings (7–9); for example, a VUS to pathogenic variant ratio of 2.5 was seen in a metanalysis of studies on genetic testing for breast cancer predisposition (7). Similarly, an 80-gene panel used with 2984 unselected cancer patients identified 1415 patients (47.4%) with a VUS, compared to 397 patients (13.3%) with a pathogenic/likely pathogenic finding (9) In general, the frequency of detection of VUS increases in proportion to the amount of DNA sequenced (10). A VUS result is also more likely to occur for patients who are not of European ancestry (9,11–13) – a consequence of the limited population diversity in genomic datasets (14).
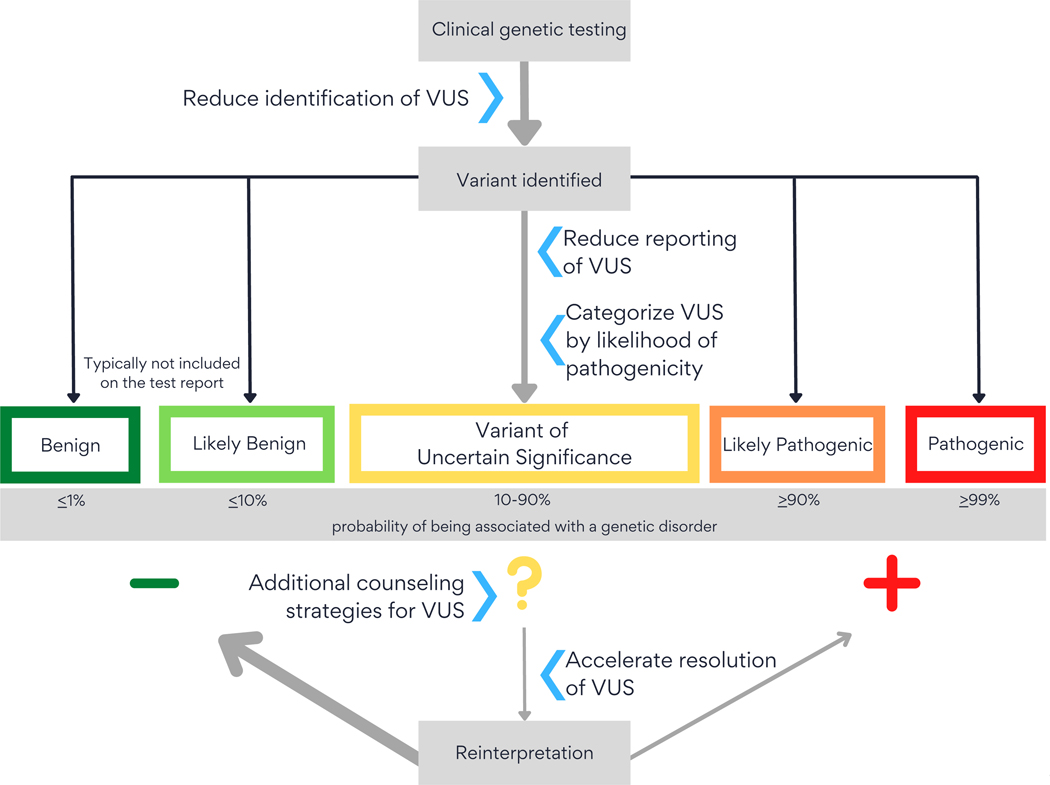
As new evidence becomes available, VUS may be re-classified. Current data suggest that 10 to 15% of re-classified VUS will be upgraded to likely pathogenic/pathogenic, with the reminder downgraded to likely benign/benign (10)(Figure). This article reviews the problems associated with VUS results, current and potential mitigation strategies, and concerns that must be addressed in applying these strategies.
Figure.
Strategies for managing VUSs identified in genomic testing.
VUS = variant of uncertain clinical significance.
Search Strategy
We searched PubMed from 1 January 2018 to 31 December 2021 using the search terms (Variants of uncertain clinical significance) AND (outcome or communication). We also searched PubMed from 1 January 2015 to 31 December 2021 for reviews, structured review, metanalysises and clinical practice guidelines using the search term “variants of uncertain clinical significance.” We included articles that described clinical and psychosocial outcomes related to VUS and reviewed strategies to improve variant interpretation. We excluded clinical case series that lacked a focus on VUS and articles limited to obstetric or pediatric settings.
Practical consequences of a VUS result
When genetic testing is done, both positive and negative results can be helpful, but a VUS result can complicate clinical decision-making, expose patients to potential adverse consequences, and place demands on health care resources (Table 2). For example, identification of a pathogenic variant in a women diagnosed with breast cancer can provide important guidance for clinical management, including potential indications for the use of PARP inhibitors (15) and consideration of both oophorectomy and contralateral mastectomy (1). In addition, family members can be screened for the pathogenic genetic variant and those who test positive can be offered aggressive cancer screening regimens or prophylactic surgery (1). A negative result reduces the likelihood that the cancer has a genetic cause. However, a VUS result leaves the patient and family members without clear guidance.
Table 2:
Problems Associated with Variants of Uncertain Clinical Significance (VUS)
| Clinical Decision-Making |
| • VUS results fail to resolve the clinical question for which testing was done |
| • Resolution of uncertainty is unlikely to occur quickly |
| Potential Adverse Outcomes |
| • VUS results may cause worry and other adverse psychological outcomes |
| • Unnecessary surgery and screening have been reported after VUS results |
| Health System Resources |
| • Variant interpretation requires significant analytic time |
| • VUS incur an obligation to invest in re-interpretation efforts |
Although the majority of VUS are predicted to be benign/likely benign(10), re-classification occurs too slowly to benefit most patients. For example, only 7.7% of unique VUS were resolved over a 10-year period in cancer-related testing done in a major laboratory (16).
In the meantime, patients and clinicians may find it difficult to ignore the VUS result (17–21). A recent systematic review and metanalysis of studies of clinical management after breast cancer genetic testing found, reassuringly, that rates of prophylactic and therapeutic surgery did not differ between women with VUS and those with benign findings (22). Nevertheless, instances of unnecessary procedures or clinical surveillance following a VUS result have been reported (17, 21,23,24), and a physician survey suggested that VUS may prompt unneccesary family testing (20).
A VUS finding may also be worrisome and confusing or cause other negative responses, including disappointment, sadness, frustration and decisional regret (21,25–28). Some responses may be based on misunderstanding, such as an over-estimation of the likelihood that the VUS is pathogenic, but others reflect the discomfort of uncertainty. In one study, a patient receiving a cancer-related VUS noted that she “spent two years after getting this test result anxious, upset and a bit paralyzed, not sure what to do” (25).
Despite these outcomes, many patients express a preference for receiving all results from genomic testing, including VUS (12,29). VUS may produce positive as well as negative responses, including relief, hope, empowerment, and satisfaction from participating in the generation of knowledge that may have value to one’s family or the public in the future (27). As with negative emotions, these responses are sometimes based on misunderstanding; for example, hope may derive from the expectation that the VUS can inform clinical care or that definitive re-classification will occur quickly (27,29).
In addition to these patient outcomes, VUS have implications for health resources. Variant interpretation is time-consuming (4), and VUS incur an obligation to pursue re-interpretation to resolve the uncertainty (30).
Addressing the VUS Challenge
Efforts to launch genomic medicine have included significant investments to improve variant interpretation and reduce VUS. These include standardization of interpretive methods (4), the creation of a national database incorporating available population, clinical, structural and functional data (31), and collaborative efforts to review variant data and refine interpretation of both specific variants and protocols for variant interpretation (31–36). New evidence for variant interpretation is being generated in clinical studies that emphasize inclusion of diverse populations(37,38) and through systematic study of the functional significance of variation in different genes and gene regions (39–41). As evidence accumulates, machine learning and artificial intelligence offer the potential to scale predictions of pathogenicity for novel variants (42–44).
Additional strategies to address the challenge of VUS
Both research about VUS and current practice trends suggest additional strategies that, if standardized, could help to mitigate the adverse consequences of VUS.
Reduced identification of VUS in clinical testing.
Multi-gene panels have increased in size due to decreasing sequencing costs. Increasingly, virtual panels are used; that is, exome/genome sequence data are generated, with reporting limited to the genes comprising the panel. However, increasing gene panel size can lead to inclusion of genes with doubtful claims to disease association and clinical utility. For example, a recent review of evidence for genes included on a Long QT Syndrome panel found that 9 of the 17 genes had limited or disputed evidence and only 3 of 17 had definitive evidence for the syndrome (45). Similarly, in a panel assessing hereditary colon cancer, only 20 of 42 genes had definitive (14/42) or strong (6/42) evidence of association with a relevant syndrome, and 3 of the included genes had either no reported evidence or were disputed or refuted (46). As noted in technical guidance from the American College of Medical Genetics and Genomics (47), rigorous standards for the construction of multi-gene panels, including only those genes for which there is strong evidence of a clinical association, would reduce identification of VUS without appreciable loss of clinical utility (Figure). This approach would also address the concerns of payers who have denied coverage of large multi-gene panels on the grounds that they include experimental tests (48). However, it requires on-going evaluation of an evolving evidence base and consensus about the evidence threshold for gene inclusion.
Reduced reporting of VUS.
When a multi-gene panel test is performed for diagnostic purposes, most laboratory reports include all VUS identified by the test, but some laboratories now offer a choice. For example, one laboratory does not routinely report VUS from a multi-gene panel for the diagnosis of inherited seizure disorders (49); instead, a clinician may opt in to VUS reporting at the time the test is ordered.
Current practice also indicates that de facto limits are being set on VUS reported from exome/genome sequencing. Because this testing approach identifies thousands of rare variants in each individual, most of them VUS (50), providing comprehensive VUS information would be overwhelming to laboratories, patients and clinicians. Therefore, laboratories seek to limit reporting to VUS most likely to prove pathogenic (51–54) (Figure). Because variant interpretation is based on a constellation of different types of evidence (Table 1), VUS may sometimes be categorized as more or less likely to be pathogenic, even when the evidence is not definitive. Some experts have suggested the use of a reporting system that subdivides VUS into qualitative or quantitative categories that more transparently communicate the likelihood of pathogenicity (55,56).
In keeping with this approach, there is an emerging consensus that VUS should not be reported when genomic testing is undertaken for population screening (57,58), as for example in the proposal to routinely screen women for pathogenic variants in the BRCA1 and BRCA2 genes (58). This consensus acknowledges that in a screening setting the prior probability for a pathogenic finding is low, and in consequence VUS have an increased likelihood of being benign compared to testing patients with disease (10). A related strategy is to report VUS only from genes with a well-established gene-disease association, when a gene panel includes some genes with less definitive evidence (56).
Although these approaches suggest a benefit from reducing VUS reporting, consistent policies have not yet been fully debated or established.
Accelerate resolution of VUS through family-based or variant-based evaluation.
Many VUS are family-specific (10), and family-based analysis can often clarify interpretation. Family members can be tested to determine whether the variant is de novo or, if inherited, whether the VUS and the disease consistently occur together in the family (“co-segregation”). De novo variants have a higher likelihood of being pathogenic (Table 1) (4), a factor that can be taken into account in determining appropriate clinical actions. If the VUS is inherited and the condition in question is dominant (meaning that a single variant is sufficient to cause the disease, as is the case for most inherited cancer and cardiovascular syndromes), the VUS can often be effectively evaluated by family testing that extends beyond first- and second-degree relatives (59–61). In a pilot study, family co-segregation data were obtained by a process in which patients were coached to identify and reach out to appropriate family members for testing. This study led to timely resolution of 60% of VUS, 84% of which were re-classified as benign or likely benign (59). Similarly, family co-segregation analysis clarified the clinical implications of 9 of 13 VUS (69%) identified in clinical breast cancer testing; of these, 7 were benign and 2 had evidence of pathogenicity (61). Variant interpretation can also be enhanced for rare conditions through systematic identification of patients with VUS. In one instance, electronic health records were used to identify three patients with similar clinical findings and VUS in the same gene, leading to further study that established that suggested the gene was associated with a new genetic disorder (62). Routine use of these approaches could result in more timely re-classification for many patients, but would require reimbursement of the associated counseling, testing, and variant interpretation costs.
Additional counseling strategies.
Principles for genetic counseling highlight the importance of a patient-centered approach (63), including pre-test counseling that prepares patients for potential VUS findings, and post-test counseling that offers additional discussion and education if a VUS is found (27). Preparing patients for a potential VUS findings appears to reduce adverse psychological outcomes (21). However, counseling patients about a VUS result is challenging (27,64–66). On-going research seeks to identify optimal strategies to explain the uncertainties inherent in VUS results and to support patients receiving them (21,54,67,68) In addition to research effort, enhanced counseling would require additional clinical resources.
Discussion
A patchwork of strategies is emerging to minimize the adverse effects of VUS. These include improved construction of multi-gene panels, reduced reporting of VUS, family-based studies to accelerate VUS re-classification, and research to improve counseling and education. Moving to consistent policies will require consensus building.
As an example, efforts to improve multi-gene panels will require consideration of whether to include genes with suggestive but non-definitive evidence for pathogenicity (47). Inclusion would allow for the identification of rare variants highly likely to be pathogenic, such as variants predicted to result in loss of gene function (Table 1), and could contribute to increasing knowledge about gene-disease associations. But the potential uncertainty of such results would need to be conveyed in test reports (47). A corollary issue would be the need to ensure periodic updating, to remove or add genes as evidence about gene-disease associations accumulates. A related question is the degree to which panels should be focused narrowly on specific clinical presentations (such as arrhythmia disorders or inherited breast cancer risk) or more broadly, for example, with the construction of cardiac or cancer panels covering a large range of inherited conditions. Smaller panels would reduce discovery of VUS, but overly narrow panels could reduce testing efficiency and sensitivity.
Although clinical expertise is needed to address these questions, patients also have a stake in VUS policies, particularly those that involve decisions about return of VUS results. When a VUS is not reported, clinicians and patients are likely to assume the test result was negative. Thus, justifications for returning some VUS and not others require careful consideration and patient input. A systematic approach to promote timely re-interpretation could also be considered, involving iterative reevaluation of all VUS generated by clinical testing. In this approach, the patient and ordering physician could be notified if a VUS was reclassified as likely pathogenic or pathogenic, and patients could be informed at the time of testing about the possibility that additional results might be forthcoming due to ongoing evaluation.
The relative cost, resource requirements, feasibility and priority of this and other approaches to VUS would need to be considered as part of consensus building. The key stakeholders include genetics professionals, healthcare payers and administrators, patients, and specialists in areas in which genetic testing plays an important role, such as oncology, neurology, cardiology, pediatrics, obstetrics, and primary care. Professional organizations already engaged in policymaking on genetic testing include the American College of Medical Genetics and Genomics, the Association of Molecular Pathology, and the National Society of Genetic Counselors. Other professional societies engaged in guideline development, including the American College of Physicians and other organizations representing major specialties, represent important potential partners for deliberation on these issues.
Conclusion
Although a number of initiatives to improve genetic variant interpretation are underway, the VUS problem will continue to grow as genomic testing expands. There is a pressing need for deliberation among the full range of stakeholders to determine an optimal mix of test design, reporting and counseling approaches to manage this problem.
Take Home Points.
Gene variants of uncertain clinical significance (VUS) are a frequent outcome of genomic testing.
VUS complicate clinical decision-making, and can result in costs to the health care system, worry, and unnecessary clinical procedures.
More widespread adoption of strategies to mitigate the potential harms of VUS should be considered, including testing protocols that limit identification or reporting of VUS, routine family-based variant evaluation, and enhanced educational efforts.
Because all strategies involve tradeoffs, cross-specialty deliberation and public input could contribute to systematic and broadly supported policies for managing VUS.
Acknowledgements
This work was supported by grant no. R01-HG010365 from the National Institutes of Health. Drs. Appelbaum, Chung and Parens were also supported by NIH grant no. RM1-HG007257. The content is solely the responsibility of the authors and does not reflect the official views of the National Institutes of Health or the authors’ institutions.
References
- 1.Valencia OM, Samuel SE, Viscusi RK, Riall TS, Neumayer LA, Aziz H. The role of genetic testing in patients with breast cancer: a review. JAMA Surg. 2017. Jun 1;152(6):589–594. [DOI] [PubMed] [Google Scholar]
- 2.Sinicrope FA. Lynch Syndrome-associated colorectal cancer. N Engl J Med. 2018. Aug 23;379(8):764–773. [DOI] [PubMed] [Google Scholar]
- 3.O’Donnell CJ, Nabel EG. Genomics of cardiovascular disease. N Engl J Med. 2011. Dec 1;365(22):2098–109. [DOI] [PubMed] [Google Scholar]
- 4.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and the Association for Molecular Pathology/ Genet Med 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.SoRelle JA, Gemmell AP, Ross TS. Different Interpretations of the same genetic data. Ann Intern Med. 2020. Aug 4;173(3):239–240. [DOI] [PubMed] [Google Scholar]
- 6.Amendola LM, Jarvik GP, Leo MC et al. Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016. Jun 2;98(6):1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Marcke C, Collard A, Vikkula M, Duhoux FP. Prevalence of pathogenic variants and variants of unknown significance in patients at high risk of breast cancer: A systematic review and meta-analysis of gene-panel data. Crit Rev Oncol Hematol. 2018. Dec;132:138–144. [DOI] [PubMed] [Google Scholar]
- 8.Kim J et al. Prevalence of pathogenic/likely pathogenic variants in the 24 cancer genes of the ACMG Secondary Findings v2.o list in a large cancer cohort and ethnicity-matched controls. Genome Med 2018;10:99 doi. 10.1186/s13073-018-0607-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samadder NJ, Riegert-Johnson D, Boardman L et al. Comparison of Universal Genetic Testing vs Guideline-Directed Targeted Testing for Patients With Hereditary Cancer Syndrome. JAMA Oncol. 2021. Feb 1;7(2):230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shirts BH, Pritchard CC, Walsh T. Family-specific variants and the limits of human genetics. Trends Mol Med. 2016. Nov;22(11):925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurian AW. BRCA1 and BRCA2 mutations across race and ethnicity: distribution and clinical implications. Curr Opinion Obst Gynecol 2010; 22:72–78. [DOI] [PubMed] [Google Scholar]
- 12.Idos GE, Kurian AW, Ricker C et al. Multicenter Prospective Cohort Study of the Diagnostic Yield and Patient Experience of Multiplex Gene Panel Testing For Hereditary Cancer Risk. JCO Precis Oncol. 2019. Mar 28;3:PO.18.00217. doi: 10.1200/PO.18.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouse SL, Florentine MM, Taketa E, Chan DK. Racial and ethnic disparities in genetic testing for hearing loss: a systematic review and synthesis. Hum Genet. 2021. Sep 7. doi: 10.1007/s00439-021-02335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popejoy A, Fullerton SM. Genomics is failing on diversity. Nature 2015;538(7624):161–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slade D. PARP and PARG inhibitors in cancer treatment. Genes Dev. 2020. Mar 1;34(5–6):360–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mersch J, Brown N, Pirzadeh-Miller S, Mundt E, Cox HC, Brown K, Aston M et al. Prevalence of Variant Reclassification Following Hereditary Cancer Genetic Testing. JAMA. 2018. Sep 25;320(12):1266–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ackerman MJ. Genetic purgatory and the cardiac channelopathies: Exposing the variants of uncertain/unknown significance issue. Heart Rhythm. 2015. Nov;12(11):2325–31. [DOI] [PubMed] [Google Scholar]
- 18.Korngiebel DM, Zech JM, Chappelle A, Burke W, Carline JD, Gallagher TH, Fullerton SM. Practice Implications of Expanded Genetic Testing in Oncology. Cancer Invest. 2019;37(1):39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scherr CL, Ross Arguedas AA, Getachew-Smith H, Marshall-Fricker C, Shrestha N, Brooks K, Fischhoff B, Vadaparampil ST. A modern dilemma: how experts grapple with ambiguous genetic test results. Med Decis Making 2020;40(5):655–668. [DOI] [PubMed] [Google Scholar]
- 20.Plon SE, Cooper HP, Parks B et al. Genetic testing and cancer risk management recommendations by physicians for at-risk relatives. Genet Med. 2011. Feb;13(2):148–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mighton C, Shickh S, Uleryk E, Pechlivanoglou P, Bombard Y. Clinical and psychological outcomes of receiving a variant of uncertain significance from multigene panel testing or genomic sequencing: a systematic review and meta-analysis. Genet Med. 2021. Jan;23(1):22–33. [DOI] [PubMed] [Google Scholar]
- 22.Makhnoon S, Bednar EM, Krause KJ, Peterson SK, Lopez-Olivo MA. Clinical management among individuals with variant of uncertain significance in hereditary cancer: A systematic review and meta-analysis. Clin Genet. 2021. Aug;100(2):119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray ML, Cerrato F, Bennett RL, Jarvik GP. Follow-up of carriers of BRCA1 and BRCA2 variants of unknown significance: variant reclassification and surgical decisions. Genet Med. 2011. Dec;13(12):998–1005. [DOI] [PubMed] [Google Scholar]
- 24.Ray T. Patients having ovaries removed without clear genetic, medical rationale, study suggests. Precision Oncology News. May 29, 2020. https://www.precisiononcologynews.com/cancer/patients-having-ovaries-removed-without-clear-genetic-medical-rationale-study-suggests#.Yhe5zi1h1qs. Accessed February 24, 2022. [Google Scholar]
- 25.Makhnoon S, Shirts BH, Bowen DJ. Patients’ perspectives of variants of uncertain significance and strategies for uncertainty management. J Genet Couns. 2019. Apr;28(2):313–325. [DOI] [PubMed] [Google Scholar]
- 26.Tsai GJ, Garrett LT, Makhnoon S, Bowen DJ, Burke W, Shirts BH. Patient goals, motivations, and attitudes in a patient-driven variant reclassification study. J Genet Couns. 2019. Jun;28(3):558–569. [DOI] [PubMed] [Google Scholar]
- 27.Clift K, Macklin S, Halverson C, McCormick JB, Dabrh AMA, Hines S. Patients’ views on variants of uncertain significance across indication. J Comm Genet 2020; 11:139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dijk S, Timmermans DR, Meijers-Heijboer H et al. Clinical characteristics affect the impact of an uninformative DNA test result: the course of worry and distress experienced by women who apply for genetic testing for breast cancer. J Clin Oncol. 2006. Aug 1;24(22):3672–7. [DOI] [PubMed] [Google Scholar]
- 29.Jamal L, Robinson JO, Christensen KD, Blumenthal-Barby J, Slashinski MJ, Perry DL, Vassy JL et al. When bins blur: patient perspectives on categories of results from clinical whole genome sequencing. AJOB Emp Bioethics 2017:8(2):82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Appelbaum PS, Parens E, Berger SM, Chung WK, Burke W. Is there a duty to reinterpret genetic data? The ethical dimensions. Genet Med. 2020. Mar;22(3):633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ClinGen. Clinical Genome Resource:ClinVar and ClinGen partnership, Available at https://clinicalgenome.org/about/clingen-clinvar-collaboration/, Accessed August 4, 2021. [Google Scholar]
- 32.Thompson BA, Spurdle AB, Plazzer JP, Greenblatt MS, Akagi K, Al-Mulla F, Bapat B et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet. 2014. Feb;46(2):107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davieson CD, Joyce KE, Sharma L, Shovlin CL. DNA variant classification-reconsidering “allele rarity” and “phenotype” criteria in ACMG/AMP guidelines. Eur J Med Genet. 2021. Oct;64(10):104312. [DOI] [PubMed] [Google Scholar]
- 34.Patel MJ, DiStefano MT, Oza AM et al. ; ClinGen Hearing Loss Clinical Domain Working Group. Disease-specific ACMG/AMP guidelines improve sequence variant interpretation for hearing loss. Genet Med. 2021. Nov;23(11):2208–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morales A, Kinnamon DD, Jordan E, Platt J et al. ; DCM Precision Medicine study of the DCM Consortium. Variant Interpretation for Dilated Cardiomyopathy: Refinement of the American College of Medical Genetics and Genomics/ClinGen Guidelines for the DCM Precision Medicine Study. Circ Genom Precis Med. 2020. Apr;13(2):e002480. doi: 10.1161/CIRCGEN.119.002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JS, Oh S, Park SK, Lee MH et al. Reclassification of BRCA1 and BRCA2 variants of uncertain significance: a multifactorial analysis of multicentre prospective cohort. J Med Genet. 2018;55(12):794–802. [DOI] [PubMed] [Google Scholar]
- 37.Electronic Medical Records and Genomics (eMERGE) Network. Available at https://www.genome.gov/Funded-Programs-Projects/Electronic-Medical-Records-and-Genomics-Network-eMERGE, Accessed February 24, 2022. [Google Scholar]
- 38.H3Africa. Human heredity and health in Africa. Available at https://h3africa.org, Accessed February 24, 2022. [Google Scholar]
- 39.Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, Janizek JD et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018. Oct;562(7726):217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amr SS, Al Turki SH, Lebo M, Sarmady M, Rehm HL, Abou Tayoun AN. Using large sequencing data sets to refine intragenic disease regions and prioritize clinical variant interpretation. Genet Med. 2017. May;19(5):496–504. [DOI] [PubMed] [Google Scholar]
- 41.Monteiro AN, Bouwman P, Kousholt AN et al. Variants of uncertain clinical significance in hereditary breast and ovarian cancer genes: best practices in functional analysis for clinical annotation. J Med Genet. 2020;57(8):509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014. Mar;46(3):310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet. 2016. Oct 6;99(4):877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi H, Chen C, Zhang H, et al. MVP: predicting pathogenicity of missense variants by deep learning. bioRxiv. Published online April 2, 2018:259390. doi: 10.1101/259390 [DOI] [Google Scholar]
- 45.Adler A, Novelli V, Amin AS, Abiusi E, Care M, Nannenberg EA, Feilotter H et al. An International, Multicentered, Evidence-Based Reappraisal of Genes Reported to Cause Congenital Long QT Syndrome. Circulation. 2020. Feb 11;141(6):418–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seifert BA, McGlaughon JL, Jackson SA, Ritter DI, Roberts ME, Schmidt RJ, Thompson BA et al. Determining the clinical validity of hereditary colorectal cancer and polyposis susceptibility genes using the Clinical Genome Resource Clinical Validity Framework. Genet Med. 2019. Jul;21(7):1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bean LJH, Funke B, Carlston CM et al. ; ACMG Laboratory Quality Assurance Committee. Diagnostic gene sequencing panels: from design to report-a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2020. Mar;22(3):453–461. [DOI] [PubMed] [Google Scholar]
- 48.Trosman JR. Weldon CB, Douglas MP, Kurian AW, Kelley RK, Deverka PA, Phillips KA. Payer coverage for hereditary cancer panels: barriers, opportunities, and implications for precision medicine. J Natl Compr Canc Netw 2017:15(2):219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ambry Genetics. Epilepsy Next. Available at: https://www.ambrygen.com/providers/genetic-testing/8/neurology/epilepsynext, Accessed October 15, 2021.
- 50.Dewey FE, Grove ME, Pan C, Goldstein BA, Bernstein JA, Chaib H, Merker JD et al. Clinical interpretation and implications of whole-genome sequencing. JAMA. 2014. Mar 12;311(10):1035–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vears DF, Senecal K, Borry P. Reporting practices for variants of uncertain significance from next generation sequencing technologies. Eur J Med Genet 2017;60:553–8. [DOI] [PubMed] [Google Scholar]
- 52.Vears DF, Senecal K, Clarke AJ, Jackson L, Laberge AM, Lovrecic L, Piton A et al. Points to consider for laboratories reporting results from diagnostic genome sequencing. Eur J Hum Genet 2018;26:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Timmermans S, Tietbohl, Skaperdas E. Narrating uncertainty: variants of uncertain significance (VUS) in clinical exome sequencing. BioSocieties 2017;12:439–58. [Google Scholar]
- 54.McLaughlin HM, Ceyhan-Birsoy O, Christensen KD et al. ; MedSeq Project. A systematic approach to the reporting of medically relevant findings from whole genome sequencing. BMC Med Genet. 2014. Dec 14;15:134. doi: 10.1186/s12881-014-0134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tavtigian SV, Harrison SM, Boucher KM, Biesecker LG. Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines. Hum Mutat. 2020; 41(10):1734–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ellard. et al. https://www.acgs.uk.com/media/11631/uk-practice-guidelines-for-variant-classification-v4-01-2020.pdf. [Google Scholar]
- 57.Prince AE, Berg JS, Evans JP, Jonas DE, Henderson G. Genomic screening of the general adult population: key concepts for assessing net benefit with systematic evidence reviews. Genet Med. 2015. Jun;17(6):441–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.King MC, Levy-Lahad E, Lahad A. Population-based screening for BRCA1 and BRCA2: 2014 Lasker Award. JAMA. 2014;312(11):1091–2. [DOI] [PubMed] [Google Scholar]
- 59.Tsai GJ, Rañola JMO, Smith C, Garrett LT, Bergquist T, Casadei S, Bowen DJ, Shirts BH. Outcomes of 92 patient-driven family studies for reclassification of variants of uncertain significance. Genet Med. 2019. Jun;21(6):1435–1442. [DOI] [PubMed] [Google Scholar]
- 60.Caputo SM, Golmard L, Léone M et al. Classification of 101 BRCA1 and BRCA2 variants of uncertain significance by cosegregation study: A powerful approach. Am J Hum Genet. 2021;108(10):1907–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuntini R, Ferrari S, Bonora E, Buscherini F, Bertonazzi B, Grippa M, Godino L, Miccoli S, Turchetti D. Dealing With BRCA1/2 Unclassified Variants in a Cancer Genetics Clinic: Does Cosegregation Analysis Help? Front Genet. 2018. Sep 11;9:378. doi: 10.3389/fgene.2018.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brokamp E, Koziura ME, Phillips JA, Tang LA, Cogan JD, Rives LC, Robertson AK, Duncan L, Bican A, Peterson JF, Newman JH, Hamid R, Bastarache L, Undiagnosed Diseases Network. Genet Med. Doi.org/10/1038/s41436-021-01179-w [Google Scholar]
- 63.Biesecker BB, Lillie SE, Amendola LM, Donohue KE, East KM, Foreman AKM, Gilmore MJ, Greve V, Liangolou B, O’Daniel JM, Odgis JA, Rego S, Rolf B, Scollon S, Suckiel SA, Zepp J, Joseph G. A review and definition of ‘usual care’ in genetic counseling trials to standardize use in research. J Genet Couns. 2021. Feb;30(1):42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slavin TP, Niell-Swiller M, Solomon I, Nehoray B, Rybak C, Blazer KR, Weitzel JN. Clinical Application of Multigene Panels: Challenges of Next-Generation Counseling and Cancer Risk Management. Front Oncol. 2015. Sep 29;5:208. doi: 10.3389/fonc.2015.00208. Erratum in: Front Oncol. 2015;5:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Makhnoon S, Mork M, Arun B, Volk RJ, Peterson SK. Perceptions of provider’s epistemic authority in response to variant of uncertain significance-related recommendations. J Genet Couns. 2020. Oct 8:10.1002/jgc4.1337. doi: 10.1002/jgc4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vears DF, Borry P, Savulescu J, Koplin JJ. Old Challenges or New Issues? Genetic Health Professionals’ Experiences Obtaining Informed Consent in Diagnostic Genomic Sequencing. AJOB Empir Bioeth. 2021. Jan-Mar;12(1):12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suckiel SA, O’Daniel JM, Donohue KE et al. Genomic Sequencing Results Disclosure in Diverse and Medically Underserved Populations: Themes, Challenges, and Strategies from the CSER Consortium. J Pers Med. 2021. Mar 13;11(3):202. doi: 10.3390/jpm11030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhong L, Donovan EE, Vangelisti AL. Examining the Effectiveness of Genetic Counselors’ Communication of Variant of Uncertain Significance Results of Breast Cancer Genes. Health Commun. 2021. May;36(5):606–615. [DOI] [PubMed] [Google Scholar]