Abstract
European ash, Fraxinus excelsior is facing the double threat of ongoing devastation by the invasive fungal pathogen, Hymenoscyphus fraxineus and the imminent arrival of the non-native emerald ash borer (EAB), Agrilus planipennis. The spread of EAB which is currently moving westwards from European Russia and Ukraine into central Europe, poses an additional substantial threat to European ash, F. excelsior. While the molecular basis for resistance or variation in resistance among European ash genotypes is heavily investigated, comparatively little is known about the molecular ash traits involved in resistance against EAB. In this study we have gathered transcriptomic data from EAB inoculated genotypes of F. excelsior that have previously shown different levels of susceptibility to EAB. Resultant datasets show differential gene expression in susceptible and resistant genotypes in response to EAB infestation. This data will provide important information on the molecular basis of resistance to the EAB and allow the development of management plans to combat a pending threat of a culturally and ecologically important European tree species.
Subject terms: Forest ecology, Transcriptomics, Biotic
Background & Summary
Fraxinus excelsior, known as common or European ash, is a frequent broad-leaf tree species native to Europe. Despite its ubiquity in the area, the species has come under increasing pressure from a devastating fungal disease, known as ash dieback (ADB), which is decimating F. excelsior across the European continent since the early 1990s1,2. Emerald ash borer (EAB), an invasive alien beetle species in the family Buprestidae, is a new threat for European ash. Like ADB, EAB is native to north-east Asia, where it feeds but poses limited impact on native Asian ash species3. EAB was first reported outside its native range in North America in 20024. Currently, six common North American Fraxinus species are now either endangered or critically endangered on the IUCN Red list5, including Fraxinus pennsylvanica (green ash) and Fraxinus americana (white ash)6,7. This invasive insect has caused substantial ecological and environmental impacts as it killed tens of millions of ash trees in North American countries4,7. To date, there is no report of EAB in Central Europe although the beetle has been reported in European Russia and Ukraine8,9. In Russia, the beetle has caused a serious decline of F. pennsylvanica, which was introduced from North America and local outbreaks of EAB on F. excelsior have also been observed recently10. With the current known location of EAB at Europe’s eastern border, EAB is predicted to spread into central Europe in the next few years, threatening Europe’s already endangered ash populations3. Due to a co-evolution with EAB, the Asian species Fraxinus mandshurica generally exhibits greater resistance to EAB than Eastern North American species, including F. pennsylvanica, F. americana, Fraxinus quadrangulata and Fraxinus nigra10–12. Nevertheless, intraspecific variation in EAB resistance was observed among North American species. For instance, a small number of green ash trees withstood EAB infestations better than others13. Recent evidence, mainly from F. mandshurica and F. pennsylvanica, suggests that resistance to EAB is likely a polygenic trait and several potential genes/proteins in various pathways have been proposed that mediate resistance14–18. Interestingly, a set of F. excelsior saplings deemed to exhibit high resistance to EAB comparable with that of F. mandshurica despite the lack of co-evolution with EAB19. In this study, we present transcriptome analysis of four European ash genotypes subjected to EAB infestation under controlled conditions. Two genotypes (B9 and B20) exhibited increased resistance to EAB in a previous experiment, whereas the other two (B3 and B8) were EAB-susceptible (Gossner et al.20). This study deployed whole-transcriptome sequencing (RNA-Seq) to evaluate the impact of EAB resistance on transcriptional changes in F. excelsior genotypes during infestation with EAB. As EAB will inevitably reach Europe, our datasets are valuable resources for research on the biology of EAB resistance in European ash and for guiding breeding for EABresistance.
Methods
Ash selection, EAB infestation and sample preparation for NGS
In 2018, four European ash genotypes (B3, B8, B9 and B20) were selected from two plots in Switzerland. Replicates of these genotypes (graftings) were previously exposed to EAB-bioassays and showed contrasting levels of resistance to EAB infestation20. To generate replicates of each genotype for the present study, one-year-old, healthy, similar-sized scions were collected from the mother trees in the field in the winter of 2020. Scions were kept on ice in the field and were subsequently stored in a cool room (4 °C) until further processing. All scions were grafted onto common European ash rootstocks (60–80 cm tall) 1–2 weeks after collection. All grafted trees were planted in 4L pots filled with humus-rich soil (Potting soil, Ökohum, Germany) containing 2g long-term fertilizer (18%N, 12%K2O, 6%P2O5 Tardit-Top, Hauert, Switzerland). Trees were kept in outdoor foil tunnels until the beginning of the EAB bioassays. The EAB bioassays are described in detail in Gossner et al.20. Briefly, EAB eggs for the bioassay were obtained from two insect colonies maintained at the Great Lakes Forestry Centre, Sault Ste. Marie (Glfc:IPQL:AplaPPP01 and Glfc:IPQL:AplaPPP02)21. These two families of EAB were initiated from adult insects flushed from green ash log bolts (F. pennsylvanica) collected in Presqu’ile Provincial Park, Brighton, Ontario, Canada and reared according to Roe et al.19.
The import permit was issued by the Federal Office for Agriculture FOAG, Switzerland in 2019 (Letter of authority No. 01 /19) and renewed in 2020 (Letter of authority No. 24/20) and 2021 (Letter of authority No. 36/21). Upon arrival in Switzerland all eggs were transferred to a level 3 biosecurity laboratory in the Plant Protection Lab at WSL (Ecogen nr: A182420) and kept at 25 °C, 55% Relative humidity, 16h Light: 8 h Dark. The eggs were stored for 4–6 days until inoculation for EAB resistance screening. Two weeks prior to EAB infestation the grafted trees were moved from the foil tunnels to biosafety level 3 climate chambers (24 °C, 70% RH, 16 h L: 8h D) at WSL in Switzerland. Trees were infested with EAB during four runs (dates: 12 June–27 July, 14 June-29 July, 19 June-3 August and 29 June- 13 August 2022). In each run 1-2 replicates of each genotype were infested with EAB while a same-sized set of trees remained uninfested (control trees). In total three trees per genotype were inoculated with EAB at two positions on the main stem. EAB eggs were only placed on stem sections that were at least one year old and that were at least 10 cm apart and 10 cm above the graft union. At each inoculation site, four EAB eggs attached to coffee filter strips were directly placed onto the bark and secured with parafilm. Coffee filter strips without EAB-eggs were attached to control trees in a similar fashion. To determine the date of egg-hatching 20–30 EAB eggs were placed in a ventilated Petri dish located next to the experimental trees. Dishes were observed twice a day between 0900–1100 h and 1600–1800 h and the number of freshly hatched EAB larvae were counted. The starting date of a run was defined as the first date on which 50% of all EAB eggs in the dish had hatched. After 45 days all trees were debarked, the EAB larvae were recovered, and the phloem was harvested on dry ice using cold razor blades. All recovered larvae per tree were counted and the dry weight was quantified (Supplementary table 1).Phloem tissue was collected next to beetle galleries in EAB infested trees. Using comparable stem sections phloem tissue was collected from non-infested trees. Harvested phloem was flash-frozen in liquid nitrogen, pulverized with pestle and mortar, and stored at −80 °C.
RNA isolation and sequencing
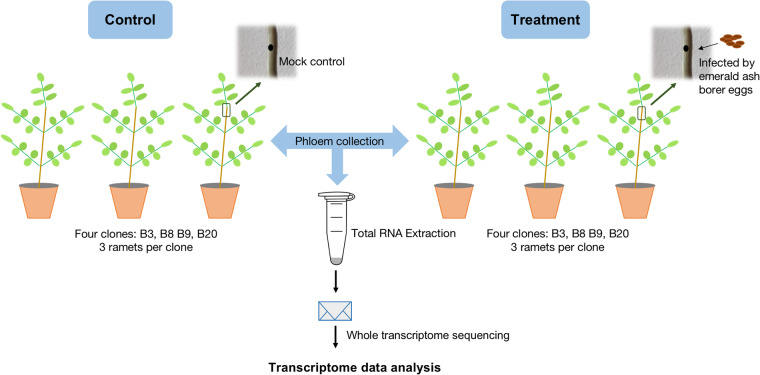
Total RNA was extracted from phloem samples using the E.Z.N.A Plant RNA Kit (Omega BIO-TEK, USA) according to the manufacturer’s instructions. During the extraction, removal of genomic DNA was carried out simultaneously using the RNase-Free DNase I Set (Omega BIO-TEK, USA). RNA quality and quantity were primarily assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA) before sending to Macrogen Europe (The Netherlands) for secondary QC, library preparation with poly-A selection and TruSeq stranded mRNA kit (Illumina, USA) and sequencing with Illumina NovaSeq platform with 100 bp paired-end reads. In total, 24 cDNA libraries from two treatments (control and infested with EAB) were constructed for transcriptome sequencing (Fig. 1; Supplementary table 1).
Fig. 1.
Schematic representation of experimental setup. Ash saplings were divided into control and EAB infested. RNA was extracted from phloem and sequenced.
Data processing and differential gene expression analyses
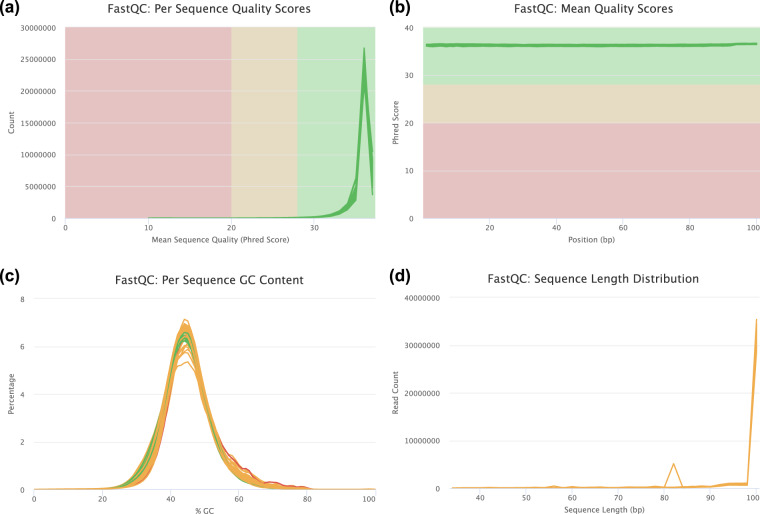
The raw reads were quality controlled by removing adapter sequences, low-quality bases (≤Q20) and reads shorter than 35 bases using the BBDuk program from the BBTools suite version 38.90 (Fig. 2)22. Ambiguous bases were removed. High quality reads were mapped to Fraxinus excelsior BATG-0.5 reference genome23 using HISAT v2-2.2.124 with default parameters. We used featureCounts v 2.0.225 from the Subread package and the F. excelsior annotation version 4 (TGAC v2) of the BATG-0.5 genome assembly to summarize counts at gene level.
Fig. 2.
Quality control of sequenced transcripts showing (a) sequence quality (Phred score), (b) quality score per sequence base position, (c) percentage GC content across sequences, (d) sequence length distribution per number of transcripts (reads).
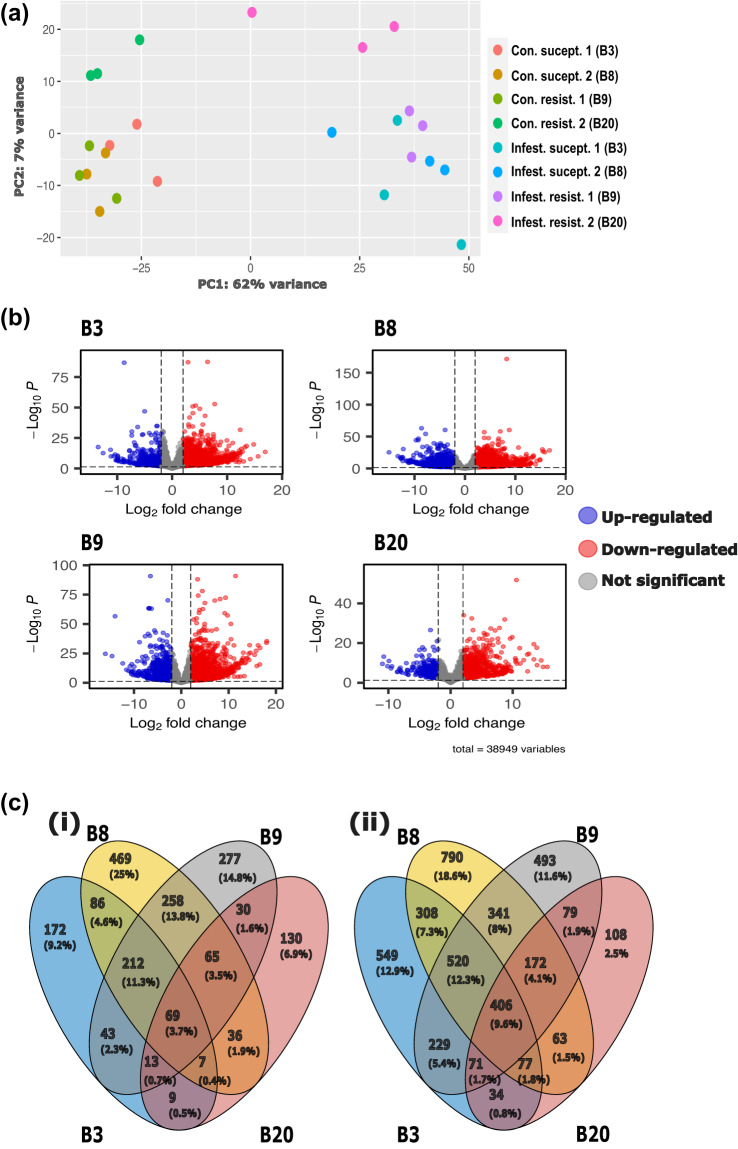
Gene level read count data was compared using the DESeq2 v1.38.3 software package26. Read counts from each of the four F. excelsior genotypes (B3, B8, B9 and B20) were compared using three control ramets against three EAB inoculated ramets. Therefore, all comparisons were made using six ramets of each genotype, with a 3 × 3 design. Differentially expressed genes (DEGs) were designated as those with a P adjusted value <0.05 and fold change (FC) > |2|. P-value distribution is shown in Supplementary figure 1. To reduce noise but preserve large effects within fold change estimates the ‘apeglm’v1.20.0 shrinkage estimator27 was applied. The number of differentially expressed genes and direction of regulation (i.e., up or down) for each genotype is presented in Supplementary table 2. To visualise variation within the transcriptome datasets, principal components from normalized read counts were plotted using the ‘plotPCA’ option within DESeq2. Resultant differentially expressed genes were visualised using the EnhancedVolcano software package v1.1628. Combined sets of differentially expressed genes were visualised in a Venn diagram using ggvenn v0.1.929 and ggVennDiagram v1.2.230.
Data Records
Raw transcriptome data of F. excelsior genotypes B3, B8, B9 and B20 infested with EAB were deposited at the DDBJ Sequence Read Archive (DRA) under the BioProject PRJDB1533631. Count data of F. excelsior genotypes B3, B8, B9 and B20 genotypes were deposited in Figshare under a CC-By licence32.
Technical Validation
Quality control
Low quality and ambiguous bases were trimmed. The quality score of the bases per position were greater than the Phred score of 20 and each read contained a minimum of 35 bases. The GC content of all samples fell within a normal distribution range. These measurements validate the high quality and lack of contaminants in the data. Alignment of the validated reads to the BATG-0.5 F. excelsior reference genome gave high mapping rates (minimum 76.31%, maximum 84.58%; Supplementary table 1), ensuring the high quality of data generated in this study. Sequencing output quality was visualized using FastQC33 and MultiQC34 (Fig. 2). Differentially expressed genes produced by DESeq2 were compared to those produced using EdgeR v3.40.235. The congruence between the programs in each of the four clones was between 60–80%. The code and resultant data for comparative analysis is presented in Figshare32.
Analysis of transcriptome data
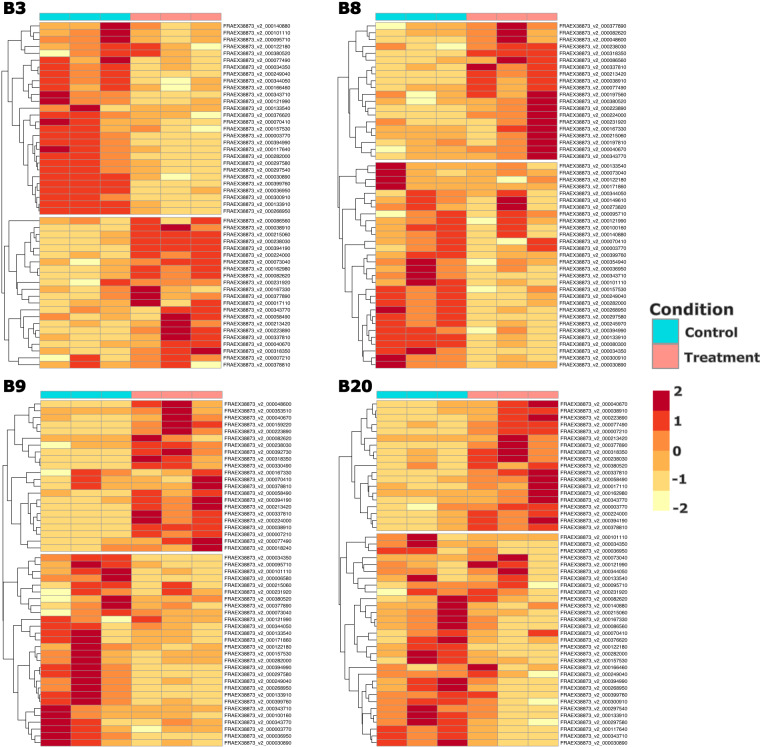
The read counts of all samples were similar after normalization (Supplementary table 1), which was performed using the internal library size correction methods within DeSeq2. A principal component analysis (PCA) showed clear separation and control trees, with samples from each genotype × infestation treatment combination clustered together (Fig. 3a). Greater dispersion of infested samples compared to control in Fig. 3a may be due to movement of larvae underneath the bark and subsequent difficulty in assigning larvae to infestation sites 45 days post infestation. By comparing the EAB-infested trees with controls, we identified genes that were differentially expressed (DEGs) between each control treatment and EAB infested treatment for each genotype (Fig. 3b-c). Heatmaps show the gene expression pattern of the 50 top ranked DEGs (Fig. 4), where notable DEGs include ethylene responsive transcription factor (FRAEX38873_v2_000164410.1), pathogenesis related protein (FRAEX38873_v2_000394990.1) in clone B3, a plant disease resistance response protein (FRAEX38873_v2_000312240.1) in clone B8, auxin response factor (FRAEX38873_v2_000165740.1) in clone B9 and ethylene responsive transcription factor (FRAEX38873_v2_000007640.1), two pathogen related proteins (FRAEX38873_v2_000146470.1 and FRAEX38873_v2_000308360.1), and a auxin efflux carrier protein (FRAEX38873_v2_000273780.1) in clone B20. The unique responses to EAB infestation in each genotype attest that our data is highly valuable for understanding the biology of resistance to EAB in European ash.
Fig. 3.
Differential gene expression in EAB susceptible and EAB resistant ash trees. (a) Principal component analysis of all biological replicates showing susceptible and resistant control genotypes, and susceptible and resistant EAB infested genotypes. (b) Volcano plots showing differentially expressed genes where red indicates significant differential expression in genotype (i) B3, (ii) B8, (iii) B9, and (iv) B20. (c) Venn diagrams showing independent and overlapping (i) upregulated genes, and (ii) downregulated genes.
Fig. 4.
Heatmaps showing the top 50 differentially expressed genes across ramets of each genotype, (i) B3, (ii) B8, (iii) B9, and (iv) B20. N.B. y-axes vary between heat maps.
Supplementary information
Acknowledgements
This research was supported by the Independent Research Fund Denmark (DFF|Technology and Production Sciences) under the grant no. 8022-00355B and the Swiss National Science Foundation SNF (grant number: 310030_189075). We thank Feng Long for help with Fig. 1 illustrations.
Author contributions
L.R.N., M.M.G., K.B.B., C.K., M.E. - Conceived the study; T.L., A.D.R. - provided E.A.B. eggs; V.Q. – provided genotypes and its replicates; M.E. - responsible for infestation experiments. J.M.D., C.K. - analysed the data; J.M.D. - wrote the first draft of the manuscript and all authors contributed to revisions; M.M.G., L.R.N. – received grant funding.
Code availability
All code used in this study is freely available at Figshare 10.6084/m9.figshare.2376140132 and GitHub https://github.com/clydeandforth/RNA_seq_EAB.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: James M. Doonan, Chatchai Kosawang.
Contributor Information
James M. Doonan, Email: jd@ign.ku.dk
Lene R. Nielsen, Email: lron@ign.ku.dk
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-023-02588-z.
References
- 1.Gross A, Holdenrieder O, Pautasso M, Queloz V, Sieber TN. Hymenoscyphus pseudoalbidus, the causal agent of European ash dieback. Mol. Plant Pathol. 2014;15:5–21. doi: 10.1111/mpp.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKinney LV, et al. The ash dieback crisis: genetic variation in resistance can prove a long-term solution. Plant Pathol. 2014;63:485–499. doi: 10.1111/ppa.12196. [DOI] [Google Scholar]
- 3.Valenta V, Moser D, Kuttner M, Peterseil J, Essl F. A High-Resolution Map of Emerald Ash Borer Invasion Risk for Southern Central Europe. Forests. 2015;6:3075–3086. doi: 10.3390/f6093075. [DOI] [Google Scholar]
- 4.Herms DA, McCullough DG. Emerald Ash Borer Invasion of North America: History, Biology, Ecology, Impacts, and Management. Annu. Rev. Entomol. 2014;59:13–30. doi: 10.1146/annurev-ento-011613-162051. [DOI] [PubMed] [Google Scholar]
- 5.Barstow, M. et al. The Red List of Fraxinus. (BGCI, 2018).
- 6.Cappaert D, McCullough DG, Poland TM, Siegert NW. Emerald Ash Borer in North America: A Research and Regulatory Challenge. Am. Entomol. 2005;51:152–165. doi: 10.1093/ae/51.3.152. [DOI] [Google Scholar]
- 7.Poland TM, McCullough DG. Emerald Ash Borer: Invasion of the Urban Forest and the Threat to North America’s Ash Resource. J. For. 2006;104:118–124. [Google Scholar]
- 8.Volkovitsh MG, Bieńkowski AO, Orlova-Bienkowskaja MJ. Emerald Ash Borer Approaches the Borders of the European Union and Kazakhstan and Is Confirmed to Infest European Ash. Forests. 2021;12:691. doi: 10.3390/f12060691. [DOI] [Google Scholar]
- 9.Orlova-Bienkowskaja MJ, et al. Current range of Agrilus planipennis Fairmaire, an alien pest of ash trees, in European Russia and Ukraine. Ann. For. Sci. 2020;77:1–14. doi: 10.1007/s13595-020-0930-z. [DOI] [Google Scholar]
- 10.Rebek EJ, Herms DA, Smitley DR. Interspecific variation in resistance to emerald ash borer (Coleoptera: Buprestidae) among North American and Asian ash (Fraxinus spp.) Environ. Entomol. 2008;37:242–246. doi: 10.1603/0046-225X(2008)37[242:IVIRTE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 11.Tanis SR, McCullough DG. Differential persistence of blue ash and white ash following emerald ash borer invasion. Can. J. For. Res. 2012;42:1542–1550. doi: 10.1139/x2012-103. [DOI] [Google Scholar]
- 12.Klooster WS, et al. Ash (Fraxinus spp.) mortality, regeneration, and seed bank dynamics in mixed hardwood forests following invasion by emerald ash borer (Agrilus planipennis) Biol. Invasions. 2014;16:859–873. doi: 10.1007/s10530-013-0543-7. [DOI] [Google Scholar]
- 13.Koch JL, Carey DW, Mason ME, Poland TM, Knight KS. Intraspecific variation in Fraxinus pennsylvanica responses to emerald ash borer (Agrilus planipennis) New For. 2015;46:995–1011. doi: 10.1007/s11056-015-9494-4. [DOI] [Google Scholar]
- 14.Bai X, et al. Transcriptomic Signatures of Ash (Fraxinus spp.) Phloem. PLOS ONE. 2011;6:e16368. doi: 10.1371/journal.pone.0016368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitehill JGA, et al. Interspecific Proteomic Comparisons Reveal Ash Phloem Genes Potentially Involved in Constitutive Resistance to the Emerald Ash Borer. PLOS ONE. 2011;6:e24863. doi: 10.1371/journal.pone.0024863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitehill JGA, Rigsby C, Cipollini D, Herms DA, Bonello P. Decreased emergence of emerald ash borer from ash treated with methyl jasmonate is associated with induction of general defense traits and the toxic phenolic compound verbascoside. Oecologia. 2014;176:1047–1059. doi: 10.1007/s00442-014-3082-8. [DOI] [PubMed] [Google Scholar]
- 17.Lane T, et al. The green ash transcriptome and identification of genes responding to abiotic and biotic stresses. BMC Genomics. 2016;17:702. doi: 10.1186/s12864-016-3052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly LJ, et al. Convergent molecular evolution among ash species resistant to the emerald ash borer. Nat. Ecol. Evol. 2020;4:1116–1128. doi: 10.1038/s41559-020-1209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Showalter DN, et al. Resistance of European ash (Fraxinus excelsior) saplings to larval feeding by the emerald ash borer (Agrilus planipennis) PLANTS PEOPLE PLANET. 2020;2:41–46. doi: 10.1002/ppp3.10077. [DOI] [Google Scholar]
- 20.Gossner, M. et al. A glimmer of hope - Ash genotypes with increased resistance to ash dieback pathogen show cross-resistance. New Phytol. (2023). [DOI] [PubMed]
- 21.Roe AD, Demidovich M, Dedes J. Origins and History of Laboratory Insect Stocks in a Multispecies Insect Production Facility, With the Proposal of Standardized Nomenclature and Designation of Formal Standard Names. J. Insect Sci. 2018;18:1. doi: 10.1093/jisesa/iey037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bushnell, B. BBMap. sourceforge.net/projects/bbmap/10.1186/1471-2105-13-238.
- 23.Sollars ESA, et al. Genome sequence and genetic diversity of European ash trees. Nature. 2017;541:212–216. doi: 10.1038/nature20786. [DOI] [PubMed] [Google Scholar]
- 24.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu A, Ibrahim JG, Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019;35:2084–2092. doi: 10.1093/bioinformatics/bty895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blighe, K., Rana, S. & Lewis, M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. (2023).
- 29.Yan, L. ggvenn: Draw Venn Diagram by ‘ggplot2’. (2022).
- 30.Gao, C.-H., Yu, G. & Cai, P. ggVennDiagram: An Intuitive, Easy-to-Use, and Highly Customizable R Package to Generate Venn Diagram. Front. Genet. 12, (2021). [DOI] [PMC free article] [PubMed]
- 31.DNA Data Bank of Japan https://ddbj.nig.ac.jp/resource/sra-submission/DRA004814 (2023).
- 32.Doonan JM, 2023. Transcriptome profiling of Fraxinus excelsior genotypes infested by emerald ash borer. Figshare. [DOI] [PMC free article] [PubMed]
- 33.Andrews, S. A Quality Control Tool for High Throughput Sequence Data. (2010).
- 34.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Doonan JM, 2023. Transcriptome profiling of Fraxinus excelsior genotypes infested by emerald ash borer. Figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All code used in this study is freely available at Figshare 10.6084/m9.figshare.2376140132 and GitHub https://github.com/clydeandforth/RNA_seq_EAB.