Abstract
The objective of this study was to reveal the effect of rumen degradable starch (RDS) on bile acid metabolism and liver transcription in dairy goats using metabolomics and transcriptomics. Eighteen Guanzhong dairy goats of a similar weight and production level (body weight = 45.8 ± 1.54 kg, milk yield = 1.75 ± 0.08 kg, and second parity) were randomly assigned to 3 treatment groups where they were fed a low RDS (LRDS, RDS = 20.52% DM) diet, medium RDS (MRDS, RDS = 22.15% DM) diet, or high RDS (HRDS, RDS = 24.88% DM) diet, respectively. The goats were fed with the experimental diets for 5 weeks. On the last day of the experiment, all goats were anesthetized, and peripheral blood and liver tissue samples were collected. The peripheral blood samples were used in metabolomic analysis and white blood cell (WBC) count, whereas the liver tissue samples were used in transcriptomic analysis. Based on the metabolomics results, the relative abundances of primary bile acids in the peripheral blood were significantly reduced in the group that was fed the HRDS diet (P < 0.05). The WBC count was significantly increased in the HRDS group compared with that in the LRDS and MRDS groups (P < 0.01), indicating that there was inflammation in the HRDS group. Transcriptomic analysis showed that 4 genes related to bile acid secretion (genes: MDR1, RXRα, AE2, SULT2A1) were significantly downregulated in the HRDS group. In addition, genes related to the immune response were upregulated in the HRDS group, suggesting the HRDS diet induced a hepatic inflammatory response mediated by lipopolysaccharides (LPS) (gene: LBP), activated the Toll-like receptor 4 binding (genes: S100A8, S100A9) and the NF-kappa B signaling pathway (genes: LOC106503980, LOC108638497, CD40, LOC102180880, LOC102170970, LOC102175177, LBP, LOC102168903, LOC102185461, LY96 and CXCL8), triggered inflammation and complement responses (genes: C1QB, C1QC, and CFD). The HRDS diet induced a hepatic inflammatory response may be mediated by activating the Toll-like receptor 4 binding and NF-kappa B signaling pathway after free LPS entered the liver. The changes of bile acids profile in blood and the down-regulation of 4 key genes (MDR1, RXRα, AE2, SULT2A1) involved in bile secretion in liver are probably related to liver inflammation.
Keywords: Rumen-degradable starch, Bile acid metabolism, Liver inflammation, Dairy goat
1. Introduction
Starch is an important carbohydrate in the ruminant diet, and can provide energy for high-yield dairy ruminants. Based on the digestion site of starch in ruminants, dietary starch can be divided into rumen-degradable starch (RDS) and rumen-undegradable starch. The amount of RDS in the diet depends on the sources and processing methods of grains (Zebeli et al., 2010). A high amount of RDS in the diet poses a huge risk to the health of the rumen in dairy ruminants, which can lead to subacute ruminal acidosis and inflammatory response (Minuti et al., 2014; Kim et al., 2018).
In previous studies, we reported that a high RDS (HRDS) diet significantly reduced rumen pH and acetate-to-propionate ratio in dairy goats, and increased free lipopolysaccharides (LPS) in rumen fluid and plasma and induced inflammatory response in liver (Shen et al., 2020a, 2020b). In this way, the de novo fatty acid synthesis and milk fat production was reduced, causing milk fat depression (MFD) in lactating goats (Zheng et al., 2020). Moreover, it was found that feeding goats a HRDS diet can significantly reduce the relative abundances of primary bile acids in mammary vein plasma (Zheng et al., 2020). Thus, we performed a metabolomic analysis of the peripheral blood of dairy goats to investigate metabolic mechanisms behind observed differences in the abundances of primary bile acids.
The liver is an important hub for lipid metabolism and is responsible for the synthesis of fatty acids, cholesterol, and primary bile acids (Chiang, 2009). Bile acids are important physiological agents for intestinal nutrient absorption and can regulate hepatic lipid metabolism mediated by nuclear receptors and G protein-coupled receptor (Chiang, 2013). Therefore, in this study, we focused on the effect of the HRDS diet on bile acid metabolism in the liver of dairy goats in an attempt to reveal the underlying mechanism by which an HRDS diet induces abnormal bile acid metabolism.
Previous studies have used dairy cows as a model to reveal the effect of high-concentration feed on liver tissue (Guo et al., 2017; Ohtaki et al., 2020; Abaker et al., 2017). However, there are limited data on the potential effects of the same total starch content, but different amounts of RDS, on the liver tissue and bile acid metabolism in dairy goats. In this study, we adopted metabolomic and transcriptomic methods to explore the underlying mechanism by which an HRDS diet induces abnormal bile acid metabolism in the liver of dairy goats. We hypothesized that, under the same total dietary starch content, high RDS included in the diet may interfere with bile acid metabolism through inducing liver inflammation in dairy goats.
2. Materials and methods
2.1. Animal ethics
The experimental animals and protocols (protocol number 100403) were approved by the Institutional Animal Care and Use Committee of Northwest A & F University (Yangling, Shaanxi, China).
2.2. Experimental design and sample collection
Eighteen Guanzhong goats in their second lactation were used in this study. The animal management practices used in this study, as well as the data on dry matter intake, milk yield, and milk composition of these goats have been reported previously (Zheng et al., 2020), and are shown in Appendix Table 1. Briefly, animals received a low RDS (LRDS) diet during a 2-week preliminary period to obtain baseline values (Appendix Table 2). Over the 2-week period, they were randomly fed 3 diets with similar total starch contents but different RDS contents: low RDS (LRDS; RDS = 20.52% DM, n = 6 goats, body weight = 45.8 ± 1.99 kg, 90 milk yield = 1.74 ± 0.09 kg), medium RDS (MRDS; RDS = 22.15% DM, n = 6 goats, body weight = 45.8 ± 1.47 kg, milk yield = 1.76 ± 0.09 kg) and high RDS (HRDS; RDS = 24.88% DM, n = 6 goats, body weight = 45.7 ± 1.37 kg, milk yield = 1.74 ± 0.07 kg). The goats were fed the experimental diets for 5 weeks, which consisted of 4 weeks for diet adaptation and 1 week for sample collection. After being fed for 3 h in the morning of the last day of the experiment, all goats were anesthetized according to Ismail et al. (2010). Thereafter, a 5-mL vacuum blood collection tube containing EDTA-K2 anticoagulant was used to collect blood samples from the mammary artery. Subsequently, all goats were euthanized and dissected to collect liver tissue samples. The liver tissue was aseptically sampled from the left rear quarter of the liver and stored in liquid nitrogen until transcriptomic analysis.
2.3. Dietary RDS calculation and diet formulation
The 3 experimental diets have been formulated by partially replacing corn with wheat. Dietary RDS was calculated using the following formula: × ERDi, where Pi represents the proportion of dietary starch in feed i in the diet, ERDi represents the starch effective degradability of feed i, and n is the number of ingredients containing starch in the feed formula (Zebeli et al., 2008). The ingredients and nutrient compositions of the 3 diets are listed in Appendix Table 3. Samples of the 3 diets were dried at 55 °C for 72 h and then ground through a 1-mm screen. They were analyzed for their DM (dried at 105 °C for 3 h) and crude protein (Kjeldahl determination) contents according to the AOAC, NDF, and ADF in Van Soest et al. (1991). Starch content was analyzed using an amylase assay kit (Megazyme, International Ireland Ltd., Bray, Co. Ireland).
2.4. Metabolomic analysis
Blood samples from the mammary artery were centrifuged to prepare plasma and stored at −80 °C prior to metabolomic analysis. The average storage time of the samples is about 20 d. The analysis of metabolites in the samples of mammary artery plasma from the LRDS, MRDS, and HRDS groups was performed using liquid chromatography-tandem mass spectrometry (LC-MS/MS, 1290 Infinity LC, Agilent Technologies) coupled to a triple time-of-flight mass spectrometer (AB Sciex TripleTOF 5600). The data collection and raw data processing were performed according to previously described methods (Zhou et al., 2019a). The database for the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (http://www.genome.jp/kegg/) was used to determine the metabolic pathways of differential metabolites between MRDS and LRDS groups, HRDS and LRDS groups, and HRDS and MRDS groups.
2.5. White blood cells count and differential
The white blood cells (WBC) count of the blood samples from the mammary artery was analyzed using an automated blood cell analyzer (Hitachi 7600, Japan). The detection indicators included total WBC, lymphocyte (LYMPH), monocyte (MONO), neutrophil (NEUT), eosinophil (EO), basophil (BASO), and atypical lymphocyte (ALY) counts.
2.6. Transcriptomic analysis
Transcriptomic analysis was performed only on the LRDS and HRDS groups. The RNA in the liver samples of the LRDS and HRDS treatments was extracted using RNAiso Plus reagent (TaKaRa, Dalian, China) according to the standard procedures of the manufacturer's instructions. High-quality RNA (OD260/280 > 1.8, OD260/230 > 2.0, and RNA integrity number > 8) was used to prepare the mRNA library using a NovaSeq sequencing system (Illumina HiSeq 4000, San Diego, USA). The detailed methods of sequencing and the processing of raw data have already been described (Yu et al., 2018). The sequences were aligned to the Capra hircus genome (https://www.ncbi.nlm.nih.gov/genome/?term=txid9925[orgn]). The expression level of each transcript was calculated as the fragments per kilobase of exon per million mapped reads (FRKM) by using RSEM software (http://deweylab.biostat.wisc.edu/rsem/). Gene Ontology (GO) functional enrichment and KEGG pathway analysis were carried out by Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do).
2.7. Construction of the WGCNA co-expression network
R Software (v.3.3) and the WGCNA package (v.1.67) were used to perform weighted gene co-expression network analysis (WGCNA) of the differentially expressed genes (DEG) in the liver tissue samples. The detailed methods for constructing the WGCNA co-expression network have already been described (Zhang and Horvath, 2005). Genes with similar expression profiles were clustered into the same module using WGCNA software. Subsequently, the WBC count was regarded as a phenotype, and the correlation between the eigengene modules and the WBC count was analyzed using Spearman's correlation tests. The connections of the top 20 nodes in the eigengene modules that were significantly related to the phenotype were visually analyzed using the Cytoscape software (v.3.6.1, https://cytoscape.org/).
2.8. Real-time quantitative PCR
To verify the reliability of the results from transcriptome sequencing of the liver tissue from LRDS and HRDS treatments, quantitative real-time PCR was performed on 6 selected DEG (ABCB1, RXRA, SLC4A2, SULT2A1, LBP, and CD40) to determine their relative mRNA expression. Three housekeeping genes (GAPDH, β-actin, and PGK1) with high relative stability, calculated using the GeNorm software, were used as internal standard controls (Vandesompele et al., 2002). The specific sequences are listed in Appendix Table 4. The relative mRNA expression was determined using the SYBR Premix Ex Taq kit (TaKaRa, China) and Cycler IQTM5 system (Bio-Rad, USA). The procedures were performed as previously described (Zheng et al., 2020). All data were normalized to the geometric mean of the housekeeping genes, and calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
2.9. Statistical analysis
Regarding the metabolomic analysis, the differential metabolites in MRDS vs. LRDS, HRDS vs. LRDS, and HRDS vs. MRDS were identified by combining variable importance in the projection (VIP) obtained from OPLS-DA and the 2-tailed Student's t test. Significantly different metabolites were defined as VIP > 1.5 and P < 0.05, and metabolites with a tendency to differ were defined as VIP > 1.5 and 0.05 ≤ P < 0.10. Statistical analysis of pathway enrichment was performed using Fisher's test, and significantly different pathways were defined as those with P < 0.05 (Zhou et al., 2019b). Regarding the WBC count of the mammary artery blood samples, the differences among the 3 groups were analyzed using the one-way ANOVA model of SPSS 20.0 (Chicago, IL, USA). Duncan's multiple comparison test was used to determine the significance of the differences between treatments. Regarding the transcriptomic analysis, the DEG with a false discovery rate (FDR) value that is smaller than 0.05 and a fold change (FC) greater than 2 were screened using DESeq2 of the R statistical software package. Significantly enriched GO terms and KEGG pathways were defined by a P value that is smaller than 0.05. The difference in gene expression in the liver quantified by Quantitative Real-time PCR between LRDS and HRDS treatments was analyzed using the Student's t test. Data related to WBC count and Quantitative Real-time PCR are shown as the mean with SEM. Significance was considered at P < 0.05, and trends were defined at 0.05 ≤ P < 0.10.
3. Results
3.1. Metabolomic analysis
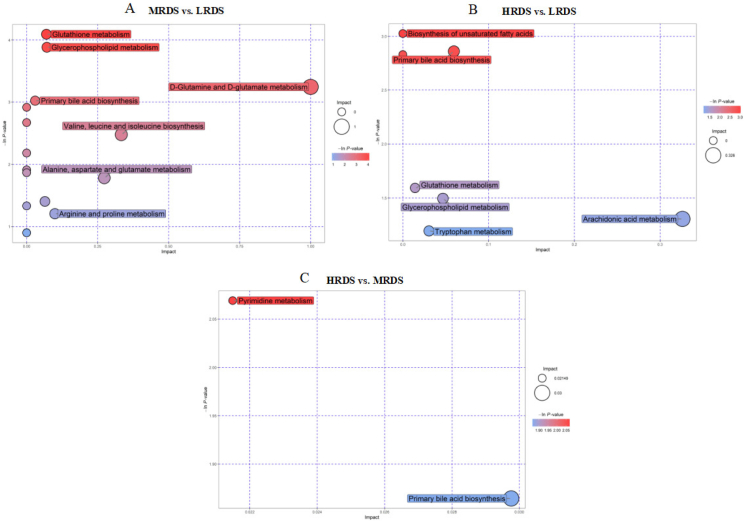
Two thousand three hundred and forty-five metabolites were identified in the LRDS, MRDS, and HRDS groups. In total, 15 differential metabolites were identified between the MRDS and LRDS groups. There were 12 metabolites with differential trends were identified between the MRDS and LRDS groups. Detailed information on the VIP, FC, and P-values of the above-mentioned metabolites is presented in Appendix Table 5. KEGG analysis of the above-mentioned metabolites between MRDS and LRDS groups were enriched in glutathione metabolism, glycerophospholipid metabolism, D-glutamine metabolism, D-glutamate metabolism, and primary bile acid biosynthesis (Fig. 1A, P < 0.05). In total, 17 differential metabolites were identified between the HRDS and LRDS groups. There were 16 metabolites with differential trends were identified between the HRDS and LRDS groups. KEGG analysis of the above-mentioned metabolites between HRDS and LRDS treatments were enriched in the biosynthesis of unsaturated fatty acids and primary bile acid biosynthesis (Fig. 1B, P < 0.05). In total, 15 differential metabolites were identified between the HRDS and LRDS groups. There were 6 metabolites with differential trends were identified between the HRDS and MRDS groups. Detailed information on the VIP, FC, and P values of the above-mentioned metabolites is presented in Appendix Table 5. Pyrimidine metabolism and primary bile acid biosynthesis tended to be enriched between HRDS and MRDS groups (Fig. 1C, 0.05 ≤ P < 0.10). Moreover, the relative abundances of differential metabolites related to the synthesis of primary bile acids decreased with increasing RDS levels (Table 1). Compared with the LRDS group, the relative abundance of taurocholic acid in the MRDS group was reduced (P = 0.04). Furthermore, the relative abundance of taurocholic acid was lower in the HRDS group than in the LRDS group (P = 0.03). The relative abundance of glycolic acid tended to decrease in the HRDS group compared with those in the LRDS group (P = 0.08). Compared with the MRDS group, the relative abundance of glycocholic acid in the HRDS group tended to decrease (P = 0.09).
Fig. 1.
KEGG pathway analysis of different metabolites in the mammary artery plasma. (A) The KEGG pathway analysis of differential metabolites in the mammary artery plasma between MRDS and LRDS groups. (B) The KEGG pathway analysis of differential metabolites in the mammary artery plasma between HRDS and LRDS groups. (C) The KEGG pathway analysis of differential metabolites in the mammary artery plasma between HRDS and MRDS groups. LRDS = low rumen degradable starch treatment (RDS = 20.52% DM); MRDS = medium rumen degradable starch treatment (RDS = 22.15% DM); HRDS = high rumen degradable starch treatment (RDS = 24.88% DM).
Table 1.
Differential metabolites related to primary bile acid biosynthesis in the mammary artery plasma.1
| Metabolite | MRDS vs. LRDS |
HRDS vs. LRDS |
HRDS vs. MRDS |
||||||
|---|---|---|---|---|---|---|---|---|---|
| VIP | FC2 | P-value | VIP | FC2 | P-value | VIP | FC2 | P-value | |
| Taurocholic acid | 1.88 | 0.49 | 0.04 | 2.26 | 0.40 | 0.03 | |||
| Glycocholic acid | 2.00 | 0.11 | 0.08 | 1.83 | 0.32 | 0.09 | |||
VIP = variable importance in projection.
LRDS = low rumen degradable starch treatment (RDS = 20.52% DM); MRDS = medium rumen degradable starch treatment (RDS = 22.15% DM); HRDS = high rumen degradable starch treatment (RDS = 24.88% DM).
FC = fold change, calculated as the mean value of the peak area obtained from the treatment group/mean value of the peak area obtained from the control group.
3.2. White blood cells count and differential
As presented in Table 2, the WBC count was increased in the HRDS group compared with that in the LRDS and MRDS groups (P < 0.05). The count of LYMPH and NEUT tended to be higher in goats fed with the HRDS diet (P = 0.09 and 0.08, respectively). The different RDS treatments had no effect on the MONO, EO, BASO, and ALY counts.
Table 2.
White blood cells count in the mammary artery blood (109/L).
| Item | Treatment1 |
SEM | P-value | ||
|---|---|---|---|---|---|
| LRDS | MRDS | HRDS | |||
| WBC | 8.90b | 9.63b | 14.17a | 0.809 | <0.01 |
| LYMPH | 8.15 | 8.03 | 11.19 | 2.127 | 0.089 |
| MONO | 0.30 | 0.49 | 0.59 | 0.070 | 0.247 |
| NEUT | 0.39 | 1.07 | 1.05 | 0.136 | 0.076 |
| EO | 0.30 | 0.29 | 0.21 | 0.049 | 0.778 |
| BASO | 0.04 | 0.04 | 0.06 | 0.008 | 0.192 |
| ALY | 0.21 | 0.27 | 0.42 | 0.040 | 0.125 |
SEM = standard error of mean; WBC = white blood cell; LYMPH = lymphocyte; MONO = monocyte; NEUT = neutrophil; EO = eosinophil; BASO = basophils; ALY = atypical lymphocyte.
a, bMeans within same row with different superscripts differ (P < 0.05).
LRDS = low rumen degradable starch treatment (RDS = 20.52% DM); MRDS = medium rumen degradable starch treatment (RDS = 22.15% DM); HRDS = high rumen degradable starch treatment (RDS = 24.88% DM).
3.3. Transcriptomic analysis of the liver from LRDS and HRDS groups
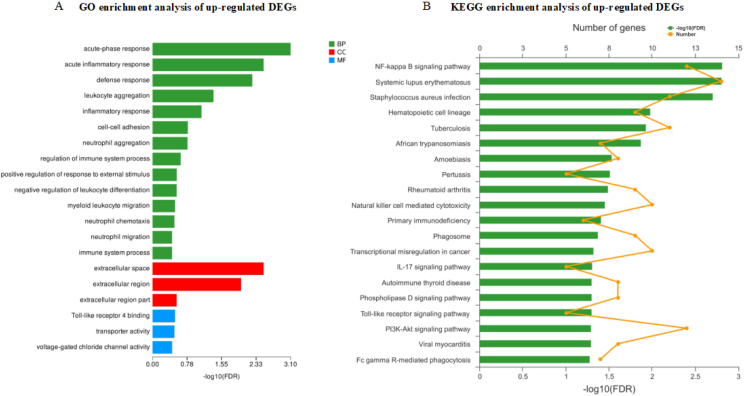
The total number of paired-end reads per sample, the percentage of total reads uniquely mapped to the C. hircus genome, and the total number of expressed genes after normalization are shown in Appendix Table 5. A total of 569 DEG (250 genes upregulated and 319 genes downregulated) were identified in the hepatic transcriptomes between the HRDS and LRDS groups. The GO enrichment analysis of upregulated genes in the liver between HRDS and LRDS groups showed that most of the enriched GO terms were related to the immune process (Fig. 2A), including the acute-phase response (P < 0.01; 7 genes: ITIH4, AGP, LOC102168428, LBP, LOC102168699, PTGER3, LOC102176354), acute inflammatory response (P < 0.01; 7 genes: ITIH4, AGP, LOC102168428, LBP, LOC102168699, PTGER3, LOC102176354), defense response (P < 0.01; 21 genes: S100A8, ITIH4, AGP, LOC102168428, CD40, ACOD1, CFD, CD74, LOC108636006, C1QB, SLFN11, LOC102168412, LBP, S-LZ, LOC102168699, CLEC6A, LY96, PTGER3, LOC102176354, S100A9, CXCL8), and Toll-like receptor 4 binding (P < 0.01; 2 genes: S100A8, S100A9).
Fig. 2.
Enrichment analysis of GO and KEGG of up-regulated DEG in the liver between HRDS and LRDS groups. (A) GO enrichment analysis of up-regulated DEG in the liver between HRDS and LRDS groups. (B) KEGG enrichment analysis of up-regulated DEG in the liver between HRDS and LRDS groups. GO = Gene Ontology; KEGG = Kyoto Encyclopedia of Genes and Genomes; DEG = differentially expressed genes; LRDS = low rumen degradable starch treatment (RDS = 20.52% DM); HRDS = high rumen degradable starch treatment (RDS = 24.88% DM); BP = biological process; CC = cellular component; MF = molecular function.
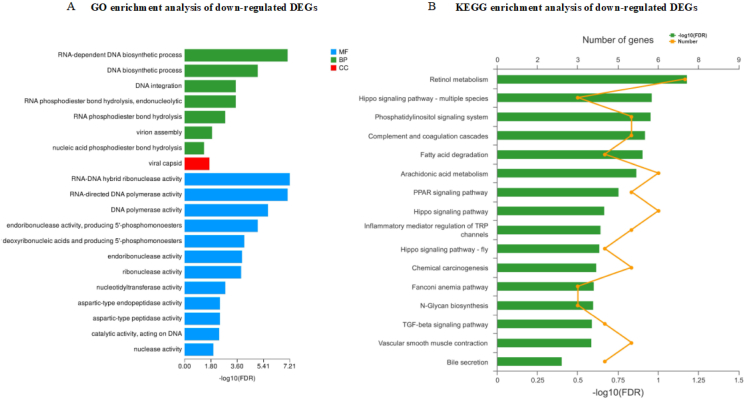
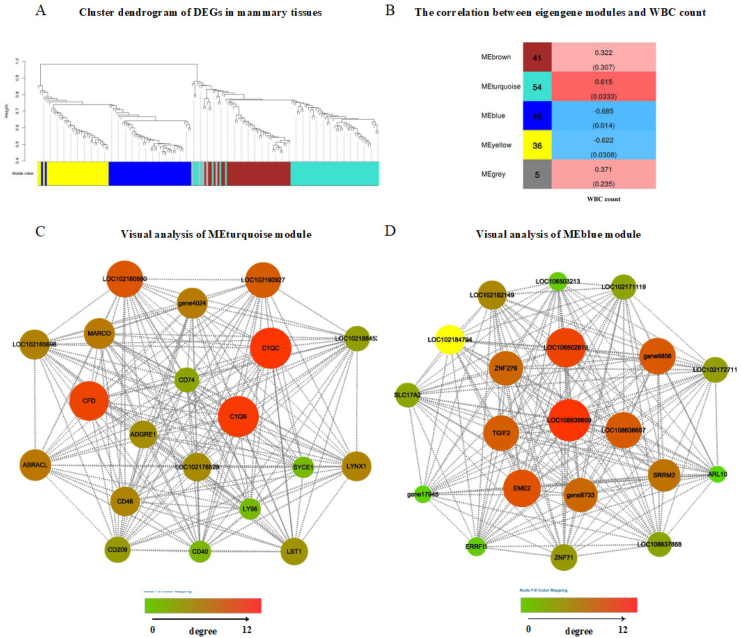
Moreover, the expression of LPS-binding protein (LBP) was also increased in the HRDS group. KEGG pathway analysis of the 250 up-regulated DEG in the liver between HRDS and LRDS groups showed that the most-enriched pathway was the NF-kappa B signaling pathway (P < 0.01; 11 genes: LOC106503980, LOC108638497, CD40, LOC102180880, LOC102170970, LOC102175177, LBP, LOC102168903, LOC102185461, LY96, CXCL8) (Fig. 2B). The GO enrichment analysis of 319 down-regulated genes in the liver between HRDS and LRDS groups indicated that the enriched GO terms included RNA–DNA hybrid ribonuclease activity (P < 0.01; 8 genes: LOC108637619, LOC108636266, LOC102177135, LOC108636107, LOC108633770, LOC108635625, LOC108635654, LOC108636278) and the RNA-dependent DNA biosynthetic process (P < 0.01; 12 genes: LOC108637619, LOC108636266, LOC102177135, LOC108636107, LOC108633770, LOC108636657, LOC108635625, LOC108636664, LOC108635654, LOC108636278, LOC108636661, LOC108636659) (Fig. 3A). The KEGG analysis of down-regulated DEG in the liver between HRDS and LRDS groups showed that 16 pathways were enriched (Fig. 3B). Among them, the pathway related to bile secretion was downregulated (P = 0.04; 4 genes: MDR1, RXRA, AE2, SULT2A1). In addition, the KEGG analysis of down-regulated DEG in the liver between HRDS and LRDS groups showed that the pathways related to lipid metabolism were also enriched, including the PPAR signaling pathway (P < 0.01; 5 genes: CPT1B, LOC108633240, RXRA, LOC108635109, LOC102178903), arachidonic acid metabolism (P < 0.01; 6 genes: LOC108633240, gene17945, PLA2G2F, LOC108635109, LOC102185056, LOC102178903), and fatty acid degradation (P < 0.01; 4 genes: CPT1B, LOC108633240, LOC108635109, LOC102178903). Detailed information on the genes related to enriched GO terms and KEGG pathways can be seen in Appendix Table 6 and Appendix Table 7. The interaction analysis between DEG in the liver tissue was performed using the R package's “WGCNA,” in which 5 modules were clustered: brown (41 genes), turquoise (54 genes), blue (46 genes), yellow (36 genes), and gray (5 genes) (Fig. 4A). Based on the correlation analysis between modules and phenotype, the Meturquoise and MEblue modules were positively and negatively correlated with the number of WBC, respectively. MEyellow was also negatively correlated, but the correlation coefficient was lower than that of MEblue (Fig. 4B). Therefore, the gene co-expression networks of the turquoise (Fig. 4C) and blue modules (Fig. 4D) were selected for visualization. Based on the GO database, most of the top 20 nodes of the turquoise module gene co-expression network were related to the immune response, including C1QC, C1QB, LOC102180880, CFD, CD40, CD48, CD74, LST1, and LY96. In addition, C1QC had the highest degree of connectivity in the gene co-expression network of the turquoise module. The highly connected nodes in the gene co-expression network of the blue module were related to biosynthetic processes of RNA-dependent DNA (LOC108636657 and LOC108636659), RNA processing and modification (EME2), transcription regulation (TGIF2), and nucleic acid binding (ZNF276).
Fig. 3.
Enrichment analysis of GO and KEGG of down-regulated DEG in the liver between HRDS and LRDS groups. (A) GO enrichment analysis of down-regulated DEGs in the liver between HRDS and LRDS groups. (B) KEGG enrichment analysis of down-regulated DEGs in the liver between HRDS and LRDS groups. GO = Gene Ontology; KEGG = Kyoto Encyclopedia of Genes and Genomes; DEG = differentially expressed genes; LRDS = low rumen degradable starch treatment (RDS = 20.52% DM); HRDS = high rumen degradable starch treatment (RDS = 24.88% DM); BP = biological process; CC = cellular component; MF = molecular function.
Fig. 4.
Weighted gene co-expression network analysis of DEG in the liver between HRDS and LRDS groups. (A) Hierarchical cluster tree conducted by weighted gene co-expression network analysis showing co-expression modules. Each leaf represents a gene, and each module corresponds to branches marked by different colors. (B) The correlation between eigengene modules and WBC count. The MEturquoise and MEblue modules were significantly positively and negatively correlated with the WBC, and the values in parentheses represent significance (P < 0.05). (C) Gene co-expression network of the turquoise module. The size and color of the nodes represent the degree of connectivity of the corresponding genes. (D) Gene co-expression network of the blue module. The size and color of the nodes represent the degree of connectivity of the corresponding genes. DEG = differentially expressed genes; WBC = white blood cell.
3.4. Quantitative analysis of mRNA
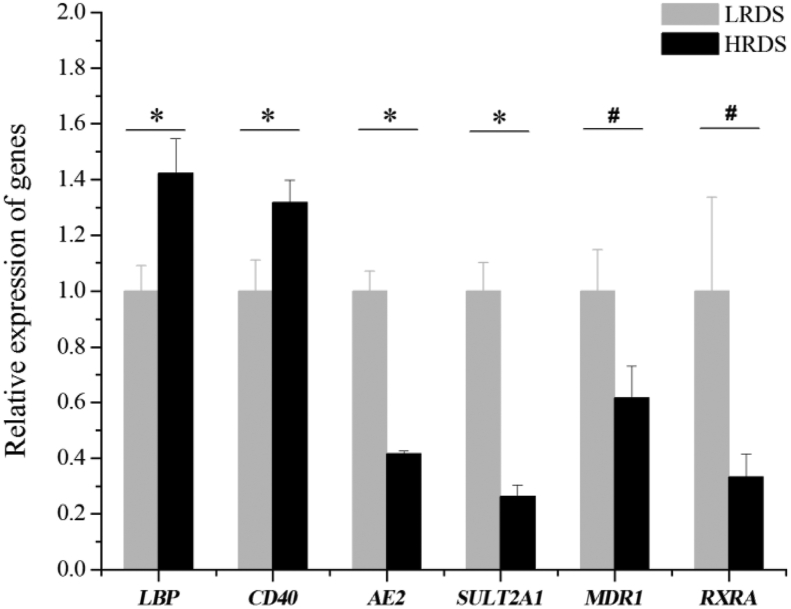
The results of the mRNA quantification are shown in Fig. 5. To validate the RNA-Seq results, qPCR analysis was performed on 6 DEG: 2 upregulated genes related to the inflammatory response (LBP, CD40) and 4 downregulated genes related to bile secretion (MDR1, RXRA, AE2, SULT2A1). Differential expression detected by RNA-Seq for LBP, CD40, AE2, and SULT2A1 was confirmed by qPCR (P < 0.05). The expression of MDR1 and RXRA in the HRDS group, detected by qPCR, showed a downward trend (0.05 ≤ P < 0.10).
Fig. 5.
Relative expression of DEG in the liver between HRDS and LRDS groups. The results of qPCR validation of RNA-sequencing data. ∗ Represents a significant difference in the relative expression of the genes (P < 0.05). # Represents the trend of differences in the relative expression of genes (0.05 ≤ P < 0.01). DEG = differentially expressed genes; LRDS = low rumen degradable starch treatment (RDS = 20.52% DM); HRDS = high rumen degradable starch treatment (RDS = 24.88% DM).
4. Discussion
For ruminants, starch is often used to improve rumen fermentation, optimizing digestion of carbohydrates and increasing protein flow to the small intestine. Starch in ruminant diets can be divided into RDS and rumen bypass starch. Our previous studies showed that HRDS reduced rumen acetate-to-propionate ratio, the relative abundances of primary bile acids in mammary vein plasma, and milk fat production in dairy goats (Shen et al., 2020b; Zheng et al., 2020). To investigate the metabolic mechanisms of the decrease of primary bile acids, metabolomic analysis of the peripheral blood and transcriptomic analysis of the liver were performed in this study.
4.1. The HRDS diet induced blood bile acids profile changes
According to the metabonomic analysis of peripheral blood, HRDS disturbed primary bile acid biosynthesis, which was determined by decreased levels of plasma taurocholic acid, glycolic acid and glycocholic acid. These results were consistent with our previous observations, in which HRDS reduce chenodeoxycholate, cholic acid, and taurocholate in mammary vein plasma (Zheng et al., 2020). Primary bile acids are synthesized in the liver and secreted into the duodenum (Axelson et al., 2000). Once bile acids reach the basolateral membrance of enterocytes, they are transported out to the blood. The composition and abundance of bile acids in plasma could be influenced by the primary bile acid synthesis and secretion in the liver.
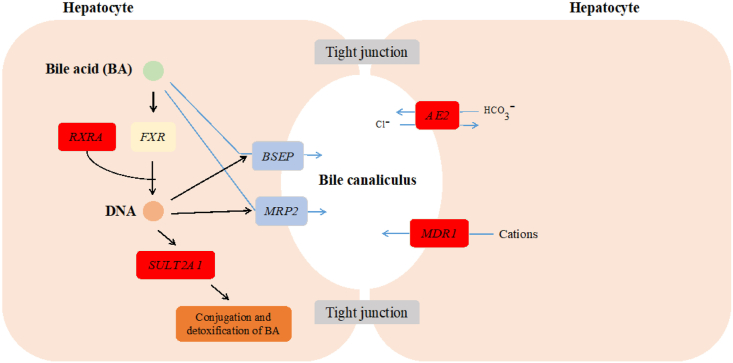
In the present study, transcriptome analysis of liver tissues showed that no significant changes in the mRNA expression of genes related to primary bile acid synthesis in the HRDS group. However, HRDS group reduced MDR1, RXRα, AE2, SULT2A1 mRNA expression, which were related with bile acid secretion (Fig. 6). RXRα encodes an important factor that regulates the activity of bile salt excretory pumps (Cheng et al., 2007). In humans, mutations in the bile salt excretory pump gene can induce particularly low levels of bile acid secretion (Lam et al., 2005). The downregulation of RXRα in the HRDS group may induce a decrease in bile salt excretory pump promoter activity, which in turn interferes with primary bile acids secretion and reduces the relative abundance of primary bile acids in plasma. SULT2A1 is highly expressed in the liver and is mainly responsible for the sulfonation of bile acids. Additionally, the expression of SULT2A1 was related to cholestasis. In liver samples from human patients with cholestatic liver disease, the expression of SULT2A1 was significantly reduced (Jin et al., 2015). In the present study, the decrease in the relative abundance of taurocholic acid in the plasma of HRDS group is likely related to the downregulation of SULT2A1. In mice, SULT2A1 expression was suppressed during LPS-induced acute liver inflammation (Kim et al., 2004). From this, decreased secretion of primary bile acids may be caused by inflammation of the liver.
Fig. 6.
The role of down-regulated genes related to bile acid secretion in the process of bile acid secretion. The red box contains significantly down-regulated genes related to bile acid secretion. The blue arrows denote that the substances are transported form the hepatocyte to bile canaliculus. The black arrows represent a regulatory effect.
4.2. The HRDS diet induced hepatic inflammatory response may interfere with bile acid metabolism in liver
In the present study, the increased WBC in the peripheral blood proved that inflammatory response was induced in HRDS group. Our previous studies showed that HRDS increased free LPS concentrations in the rumen liquid and plasma, indicating that more LPS translocate into the bloodstream in HRDS group. LPS can activate immune responses and promote the secretion of proinflammatory cytokines. In the present study, the transcriptomic analysis of liver showed that HRDS group upregulated DEG enriched in inflammation-related GO terms, such as acute-phase response and defense response. Furthermore, hepatic LBP expression, which is a key biomarker of the inflammatory response induced by LPS, was significantly upregulated in HRDS group. LBP binds with LPS and CD14 to form the LPS–LBP-CD14 complex, activates Toll-like receptor 4 and then mediates a series of intracellular signal transductions, activating NF-κB signaling (Triantafilou et al., 2001; Panaro et al., 2010; Cronin et al., 2012). The genes S100A8 and S100A9, which mainly mediate intracellular inflammatory signal transduction pathways by binding and activating Toll-like receptor 4, were significantly upregulated in HRDS group (Ehrchen et al., 2009). The KEGG analysis showed that the up-regulated DEG in the HRDS groups enriched in NF-κB signaling pathway. According to the results of WGCNA analysis, C1QB, C1QC, and CFD are hub genes with a high degree of connectivity, and interacted with multiple genes involved in the complement system, which regulate immunity and mediate tissue injury responses. C1QB and C1QC belong to the C1q family and can engage a variety of self- and non-self-ligands, including LPS, to induce inflammation and a cascade of complement system activation processes (Ghai et al., 2007). Those results suggest that HRDS induced hepatic inflammatory response in dairy goats, which may be mediated by activating the Toll-like receptor 4 binding and NF-kappa B signaling pathway after free LPS translated into the liver.
In summary, hepatic inflammatory response in dairy goats which was induced by HRDS diet may interfere with bile acid metabolism in liver. The disordered hepatic metabolism may be an important reason for the reduction in the relative abundances of taurocholic acid and glycocholic acid in the arteriovenous plasma of the mammary gland. However, bile acids are synthesized from cholesterol in the liver and further metabolized by the gut microbiota into secondary bile acids. Diets with different RDS levels may change the microflora related to bile acid metabolism in the intestinal tract of dairy goats, thereby changing the profile of bile acid in the blood. More research will be carried out on the relationship between intestinal microorganisms and bile acids in the future.
5. Conclusion
In summary, the HRDS diet induced a hepatic inflammatory response may be mediated by activating the Toll-like receptor 4 binding and NF-kappa B signaling pathway after free LPS entered the liver, and triggered inflammation and complement responses. The changes of bile acids profile in blood and the down-regulation of 4 key genes (MDR1, RXRα, AE2, SULT2A1) involved in bile secretion in liver are probably related to liver inflammation.
Author contributions
Lixin Zheng, Jing Shen, Xiaoying Han and Junhu Yao: conceived and designed the experiments. Lixin Zheng, Jing Shen, Xiaoying Han, Chunjia Jin and Xiaodong Chen: mainly performed the experiments. Lixin Zheng: analyzed the data and wrote the manuscript. Xiaodong Chen and Junhu Yao: participated in the revision of the manuscript.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
This research was financially supported by the National Key Research and Development Program of China (award number: 2017YFD0500500) and National Natural Science Foundation of China (award number: 32072761). Appreciation is extended to the staff of the Innovative Research Team of Animal Nutrition & Healthy of Northwest A&F University for providing valuable assistance and care of the goats.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2023.04.008.
Appendix supplementary data
The following is the Supplementary data to this article:
References
- Abaker J.A., Xu T.L., Jin D., Chang G.J., Shen X.Z. Lipopolysaccharide derived from the digestive tract provokes oxidative stress in the liver of dairy cows fed a high-grain diet. J Dairy Sci. 2017;100(1):666–678. doi: 10.3168/jds.2016-10871. [DOI] [PubMed] [Google Scholar]
- Axelson M., Ellis E., Mrk B., Garmark K., Einarsson C. Bile acid synthesis in cultured human hepatocytes: support for an alternative biosynthetic pathway to cholic acid. Hepatology. 2000;31(6):1305–1312. doi: 10.1053/jhep.2000.7877. [DOI] [PubMed] [Google Scholar]
- Cheng X., Buckley D., Klaassen C.D. Regulation of hepatic bile acid transporters ntcp and bsep expression. Biochem Pharmacol. 2007;74(11):1665–1676. doi: 10.1016/j.bcp.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang L.J.Y. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang L.J.Y. Bile acid metabolism and signaling. Compr Physiol. 2013;3(3):1191–1212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin J.G., Turner M.L., Goetze L., Bryant C.E., Sheldon I.M. Toll-like receptor 4 and MYD88-dependent signaling mechanisms of the innate immune system are essential for the response to lipopolysaccharide by epithelial and stromal cells of the bovine endometrium. Biol Reprod. 2012;86:51. doi: 10.1095/biolreprod.111.092718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrchen J.M., Sunderkötter C., Foell D., Vogl T., Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009;86(3):557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- Ghai R., Waters P., Roumenina L.T., Gadjeva M., Kojouharova M.S., Reid K.B.M., et al. C1q and its growing family. Immunobiology. 2007;212(4–5):253–266. doi: 10.1016/j.imbio.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Guo J., Chang G., Zhang K., Xu L., Jin D., Bilal M.S., et al. Rumen-derived lipopolysaccharide provoked inflammatory injury in the liver of dairy cows fed a high-concentrate diet. Oncotarget. 2017;8(29) doi: 10.18632/oncotarget.18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail Z.B., Jawasreh K., Al-Majali A. Effects of xylazine–ketamine–diazepam anesthesia on blood cell counts and plasma biochemical values in sheep and goats. Comp Clin Pathol. 2010;19:571–574. [Google Scholar]
- Jin C., Feng X., Zhang L., Chen S., Cheng Y., He X., et al. Hepatic expression of detoxification enzymes is decreased in human obstructive cholestasis due to gallstone biliary obstruction. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0120055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.S., Shigenaga J., Moser A., Grunfeld C., Feingold K.R. Suppression of dhea sulfotransferase (sult2a1) during the acute-phase response. Am J Physiol Endocrinol Metab. 2004;287(4):e731–e738. doi: 10.1152/ajpendo.00130.2004. [DOI] [PubMed] [Google Scholar]
- Kim Y.H., Nagata R., Ohkubo A, Ohtani N., Kushibiki Ichijo T., Sato S. Changes in ruminal and reticular pH and bacterial communities in Holstein cattle fed a high-grain diet. BMC Vet Res. 2018;14(1):310. doi: 10.1186/s12917-018-1637-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P., Wang R., Ling V. Bile acid transport in sister of P-glycoprotein (ABCB11) knockout mice. Biochemistry. 2005;44(37):12598–12605. doi: 10.1021/bi050943e. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Minuti A., Ahmed S., Trevisi E., Piccioli-Cappelli F., Bertoni G., Jahan N., et al. Experimental acute rumen acidosis in sheep: consequences on clinical, rumen, and gastrointestinal permeability conditions and blood chemistry. J Anim Sci. 2014;92(9):3966–3977. doi: 10.2527/jas.2014-7594. [DOI] [PubMed] [Google Scholar]
- Ohtaki T., Ogata K., Kajikawa H., Sumiyoshi T., Asano S., Tsumagari S., et al. Effect of high-concentrate corn grain diet-induced elevated ruminal lipopolysaccharide levels on dairy cow liver function. J Vet Med Sci. 2020;82(7):971–977. doi: 10.1292/jvms.20-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaro M.A., Gagliardi N., Saponaro C., Calvello R., Mitolo V., Cianciulli A. Toll-like receptor 4 mediates lps-induced release of nitric oxide and tumor necrosis factor-α by embryonal cardiomyocytes: biological significance and clinical implications in human pathology. Curr Pharm Des. 2010;16(7):766–774. doi: 10.2174/138161210790883624. [DOI] [PubMed] [Google Scholar]
- Shen J., Han X.Y., Zheng L.X., Liu S.M., Jin C.J., Liu T., et al. 3. High rumen-degradable starch diet promotes hepatic lipolysis and disrupts enterohepatic circulation of bile acids in dairy goats. J Nutr. 2020;150(10) doi: 10.1093/jn/nxaa238. [DOI] [PubMed] [Google Scholar]
- Shen J., Zheng L.X., Chen X.D., et al. Metagenomic analyses of microbial and carbohydrate-active enzymes in the rumen of dairy goats fed different rumen degradable starch. Front Microbiol. 2020;11(1003) doi: 10.3389/fmicb.2020.01003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou K., Triantafilou M., Dedrick R.L. A CD14-independent LPS receptor cluster. Nat Immunol. 2001;2:338–345. doi: 10.1038/86342. [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest P.J., Robertson J.B., Lewis B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. 1991;74:3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- Yu S.H., Zhu K.Y., Chen J., Liu X.Z., Xu P.F., Zhang W., et al. JMJD3 facilitates C/EBPbeta-centered transcriptional program to exert oncorepressor activity in AML. Nat Commun. 2018;9:3369. doi: 10.1038/s41467-018-05548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebeli Q., Mansmann D., Steingass H., Ametaj B.N. Balancing diets for physically effective fiber and ruminally degradable starch: a key to lower the risk of sub-acute rumen acidosis and improve productivity of dairy cattle. Livest Sci. 2010;127:1–10. [Google Scholar]
- Zebeli Q., Dijkstra J., Tafaj M., Steingass H., Ametaj B.N., Drochner W. Modeling the adequacy of dietary fiber in dairy cows based on the responses of ruminal pH and milk fat production to composition of the diet. J Dairy Sci. 2008;91:2046–2066. doi: 10.3168/jds.2007-0572. [DOI] [PubMed] [Google Scholar]
- Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zheng L.X., Wu S.R., Shen J., Han X.Y., Jin C.J., Chen X.D., et al. High rumen degradable starch decreased goat milk fat via trans-10, cis-12 conjugated linoleic acid-mediated downregulation of lipogenesis genes, particularly, INSIG1. J Anim Sci Biotech. 2020;11(30) doi: 10.1186/s40104-020-00436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Liu L., Lan X., Cohen D., Zhang Y., Ravindran A.V., et al. Polyunsaturated fatty acids metabolism, purine metabolism and inosine as potential independent diagnostic biomarkers for major depressive disorder in children and adolescents. Mol Psychiatry. 2019;24:1478–1488. doi: 10.1038/s41380-018-0047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Yang H., Yan Q., Ren A., Kong Z., Tang S., et al. Evidence for liver energy metabolism programming in offspring subjected to intrauterine undernutrition during midgestation. Nutr Metab. 2019;16:20. doi: 10.1186/s12986-019-0346-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.