Abstract
Topoisomerase II catalyzes the passage of one DNA helix through another via a transient double-stranded break. The essential nature of this enzyme in cell proliferation and its mechanism of action make it an ideal target for cytotoxic agents. Saccharomyces cerevisiae topoisomerase II has been frequently used as a model for testing potential inhibitors of eukaryotic topoisomerase II as antitumor agents. The standard in vivo method of estimating the sensitivity of S. cerevisiae to the antitopoisomerase drugs is via inhibition or kill curves which rely on viable-cell counts and is labor intensive. We present an alternative to this, a high-throughput in vivo screen. This method makes use of a drug-permeable S. cerevisiae strain lacking endogenous topoisomerase II, which is modified to express either human topoisomerase IIα or IIβ or S. cerevisiae topoisomerase II carried on plasmids. Each modified strain expresses a full-length topoisomerase II enzyme, as opposed to the more commonly used temperature-sensitive S. cerevisiae mutant expressing yeast or yeast/human hybrid enzymes. A comparison of this new method with a plating-and-counting method gave similar drug sensitivity results, with increased accuracy and reduced manual input for the new method. The information generated has highlighted the sensitivities of different topoisomerase II enzymes and isoenzymes to several different classes of topoisomerase II inhibitor.
Eukaryotic topoisomerase II enzymes are essential for efficient chromosome DNA segregation in both mitosis and meiosis (10, 19), and this makes them attractive targets for cytotoxic agents (3, 8). All topoisomerase II enzymes catalyze the passage of one DNA double helix through another via a transient double-stranded break in DNA. The topoisomerase II reaction requires the binding of the enzyme as a dimer and the creation of a 4-bp staggered break in the DNA via the formation of a covalent bond between each enzyme monomer and the 5′-DNA ends of a G (gate) segment of DNA. Another DNA segment, the T (transported) segment, is then captured by an ATP-operated clamp and passed through the broken gate strand, which is then religated (2).
Topoisomerase II is inhibited by a variety of antitumor drugs. For example, doxorubicin, m-AMSA (amsacrine), epipodophyllotoxins, and mitoxantrone all interfere with the breakage and religation of the G segment of DNA, forming structures which favor DNA strand breakage often referred to as “cleavable complexes.” In the absence of antitumor agents, such structures are usually short-lived. The presence of antitumor agents induces a large number of cleavable complexes, which if unresolved ultimately lead to cell death (8). ICRF-159, a bisdioxopiperazine derivative which “locks” the ATP-operated clamp of the enzyme (18), and merbarone (7), a thiobarbiturate derivative which acts via an as yet unknown mechanism, also inhibit DNA topoisomerases and are cytotoxic agents.
In contrast to what is found for many other eukaryotes, there are two isoforms of human topoisomerase II, topoisomerase IIα and topoisomerase IIβ. The α-isozyme form has a monomeric molecular mass of 170 kDa and is encoded by a gene on chromosome 17q21-22 (21), whereas the β isoform has a molecular mass of 180 kDa and is encoded by a gene on chromosome 3p24 (12). Although it is known that both human isoenzymes can be inhibited by antitumor agents such as etoposide, m-AMSA, and merbarone in vitro (6), the extent to which inhibition of either topoisomerase IIα or IIβ is cytotoxic in vivo is unclear. Topoisomerase IIα is known to be preferentially expressed during mitosis, whereas topoisomerase IIβ shows little variation in levels during the cell cycle (26). One would speculate from these data that topoisomerase IIα is the major target of cytotoxic agents. However, drug-resistant cell lines have shown altered levels of either or both topoisomerase isoforms, suggesting some drug selectivity for α or β isoforms (11, 24, 25), and there have been some in vitro studies suggesting that α and β isoforms respond differently to different topoisomerase inhibitors (7, 15). The exact nature of such selectivity has, however, been difficult to determine due to the problems associated with the isolation and separation of the two isoforms for both in vivo and in vitro studies.
Saccharomyces cerevisiae has a single form of topoisomerase II which has been frequently used as a eukaryotic model in functional studies and in the study of antitumor agents (17, 23). An S. cerevisiae mutant temperature sensitive for topoisomerase II in combination with yeast/human hybrid topoisomerases has been used as a model to study the relative sensitivities of human α and β topoisomerase II enzymes to a variety of topoisomerases II inhibitors both in vitro and in vivo (4). Sensitivities to the antitopoisomerase drugs were estimated following a short contact inhibition assay (15) based on viable-cell counts. Such methods are highly labor intensive and can have quite large margins of error.
We have previously shown that a topoisomerase II deletion strain of S. cerevisiae can be fully complemented by either of the two human topoisomerase II isozymes expressed from full-length cDNAs (13). Using this system we are now able to distinguish from one another the effects of different chemical classes of topoisomerase inhibitor on yeast topoisomerase II and on either of the full-length human α or β isoenzymes. In addition, this study uses a rapid throughput microwell assay, which is shown to be comparable to cell counting, to monitor the effects of drugs on yeast growth. Using this novel assay to generate data, we present a study which uses full-length human cDNAs expressed in a drug-permeable S. cerevisiae topoisomerase II deletion strain to determine the relative sensitivities of two human topoisomerases to a variety of anti-tumor drugs.
MATERIALS AND METHODS
Materials.
Yeast extract-Bacto peptone-glucose (YPD) medium components and carbohydrates were purchased from Gibco. Yeast-nitrogen base and amino acid supplements were purchased from Bio 101. Mineral oil (nuclease free, molecular biology grade) was purchased from Sigma, Poole, United Kingdom. Teniposide was obtained from Bristol-Meyers, Wallingford, Conn. Mitoxantrone and doxorubicin were kind gifts from L. Patterson, De Montfort University, Leicester, United Kingdom. 2-methyl-9-Hydroxyellipticine (2M9HE) was a kind gift from E. Lescot, Institut Gustave-Roussy, Villejuif, France. Merbarone and m-AMSA were kind gifts from the National Cancer Institute, Bethesda, Md. ICRF-159 was a kind gift from I. Hickson, Imperial Cancer Research Fund, Oxford, United Kingdom. Etoposide was purchased from Sigma. All drugs were dissolved in dimethyl sulfoxide and stored at −20°C prior to use in the assay.
Yeast strains.
The following strains were used: JN394 MATa ura3 leu2 trp1 top2-4 rad52::LEU2 ISE2, BJ201 (pHT173) (Schizosaccharomyces pombe TOP2/URA3-ARS/CEN) MATa ura3 trp1 leu2 pep4::HIS3 can1 top2::TRP1 GAL, and JJ700 (JN394 × BJ201, backcrossed to BJ201) (pHT173) ura3 trp1 leu2 ISE2 Δtop2::TRP1.
Media.
Yeast cells were grown in YPD medium (1% yeast extract, 2% Bacto peptone, 2% glucose) or synthetic media comprising yeast-nitrogen base, glucose, and amino acid mixtures lacking uracil (−URA media) or leucine (−LEU media). Solid medium contained 2% Bio/Agar. 5-Fluoro-orotic acid (Sigma; 1 mg/ml) was used to counterselect against the URA3 plasmid carrying the S. pombe topoisomerase II gene. Yeast transformations were performed by a modified lithium acetate method; for yeast protocols see reference 20.
Plasmids.
Low-copy-number CEN-ARS/LEU2 expression was from the following constitutive triose phosphate isomerase plasmids: pYTO300 (wild-type S. cerevisiae topoisomerase II gene); pHT300 (human topoisomerase IIα gene [cDNA]); pHT400 (human topoisomerase IIβ gene [cDNA]). The multicopy 2μm-based URA3 GAL1 expression plasmid was YEpWob6 (fusion; contains the promoter region and the first five amino acids of S. cerevisiae topoisomerase II followed by human topoisomerase IIα from amino acid 29 onwards [23]).
Microwell assay.
Yeast strains were grown on −LEU or −URA medium agar plates containing 2% glucose at 30 or 35°C (for deletion or temperature-sensitive strains, respectively). Small samples were picked from growing colonies of each strain and diluted into appropriate media containing 4% glucose to produce seeding cultures containing approximately 105 to 106 cells/ml. Microtiter plates (96 well, flat bottomed; Costar) were primed with 180-μl volumes of similar media and test compounds diluted across columns or down rows either by directly diluting media between wells or by adding fixed volumes of drug diluted in sterile TE buffer (10 mM Tris-HCl [pH 7.5], 0.5 mM EDTA). Plates were inoculated with 10 μl of seeding culture, wells were overlaid with 2 drops (≈50 μl) of mineral oil, and the optical density at 630 nm (OD630) of each well was read (Bio-Tek EL340 microplate reader). For continuous studies, plates were left at 30°C in the plate reader, with a maximum of 80 OD630 readings taken at set time intervals. Alternatively, initial OD630 readings were taken and plates were then transferred to an incubator at 35 or 30°C until the OD630 of control wells had reached approximately 0.3. Aqueous well contents were then carefully mixed with a multichannel pipette, and the OD630 was recalculated for each well. The 50% inhibitory dose (ID50) for test compounds was defined as the dose of drug in media which results in a 50% decrease in the number of cells as measured by determining the OD630 at a given time point. All experiments to determine ID50s were performed in triplicate. Values of the percentage of control OD630 increase were plotted against the log of the concentration of the drug to generate dose-response curves, and best-fit sigmoid curves were generated with Mac Curvefit software (Kevin Raner Software, Mt. Waverley, Australia); ID50s were calculated as the antilogs of the x-axis values of the inflection points. Error values in the inflection were automatically calculated by the software.
Viable-cell counts of yeast cells.
Cells were grown in microwell plates in a fashion identical to that described above. At the beginning of each assay a sample of the seeding culture was diluted, plated onto agar plates, and incubated for 36 h and viable colonies were counted. At the end of the assay period, OD630 values were recorded and then samples of the media were similarly plated for viable-cell counts. Each assay was performed in triplicate, and three different initial-plating dilutions were used for each replicate. Count data were expressed as percentages of cell growth relative to control values, and ID50s were calculated as described above.
RESULTS
Growth of yeast cells in microwell plates as monitored by measuring the OD630.
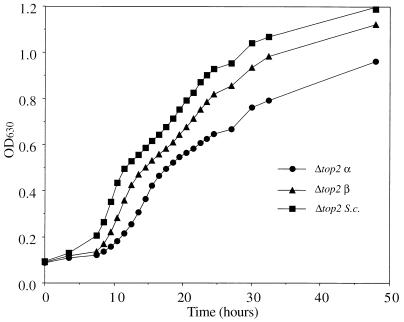
Figure 1 shows an example of the growth curves of JJ700 yeast strains carrying plasmid pHT300, pHT400, or pYTO300; these plasmids encode full-length human topoisomerase IIα and IIβ and S. cerevisiae topoisomerase II, respectively. The strains will be referred to as Δtop2 α, Δtop2 β, and Δtop2 S.c. hereafter. There is an initial lag phase, followed by rapid growth to an OD630 value of approximately 0.4; after this point an inflection is seen, and then growth continues at a slower rate. Further experiments at equal seeding levels showed that Δtop2 α grows at a slightly slower rate than Δtop2 β and Δtop2 S.c., which showed very similar rates of growth (data not shown). The temperature-sensitive JN394 (YEpWoB6) strain, referred to hereafter as top2(Ts) α, expressing a fusion topoisomerase IIα, grew extremely slowly at the nonpermissive temperature, typically taking two to three times longer than the deletion strains to reach a suitable OD630 for drug studies (data not shown).
FIG. 1.
Growth of yeast cells in microwell plates as determined by monitoring OD630. Yeast cells were seeded into microplate wells and transferred to an incubator at 30°C as described in Materials and Methods. The OD630 was measured at 3- to 4-h intervals until significant increases were seen. The plate was then transferred to the reader at 30°C, and readings were taken hourly for 17 h. To conserve lamp time, readings were then taken at 2- to 5-h intervals.
Effects of topoisomerase inhibitors on yeast strain growth.
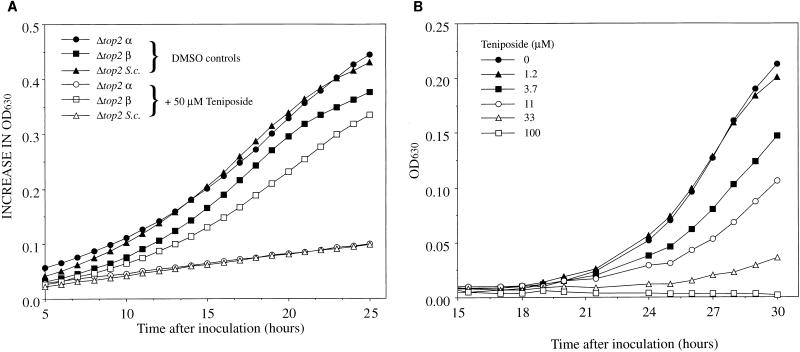
In order to investigate the effects of antitumor agents, strains were incubated with potential inhibitors as described in Materials and Methods. Figure 2a shows the effects of 50 μM teniposide (a concentration determined from earlier experiments to significantly affect the growth of Δtop2 S.c.) on the growth rates of the three strains which express different topoisomerases. Teniposide has a differential effect on strains containing different topoisomerase subtypes: Δtop2 β is more resistant to the drug than is Δtop2 α or Δtop2 S.c.; it exhibits a growth rate similar to that of the untreated control. Figure 2b shows the effects of a series of teniposide concentrations on the growth of Δtop2 α. Increasing the concentration of teniposide decreases the growth of this strain in a dose-dependent fashion. Experiments with tightly controlled and varied initial seeding values were performed and showed that best results for determining the effects of topoisomerase inhibitors were obtained by seeding at low levels and reading when the OD630 was less than 0.4, i.e., near the end of the initial exponential-growth phase. With a seeding number of approximately 105/ml, plates were read after 20 to 24 h for the deletion strains and after approximately 50 h for the temperature-sensitive strain.
FIG. 2.
Inhibition of yeast cell growth by teniposide. (A) Microplate wells were seeded as described in Materials and Methods with yeast strains only or with yeast strains plus 50 μM teniposide. After an initial lag phase the cells were transferred to a microplate reader at 30°C, and OD630 readings were taken at 30-min intervals. (B) Microplate wells were seeded as described in Materials and Methods with Δtop2 α. Teniposide was present in the media at the concentrations shown. The lag phase in these experiments is increased due to seeding at low levels. DMSO, dimethyl sulfoxide.
Calculation of ID50s by microwell assay.
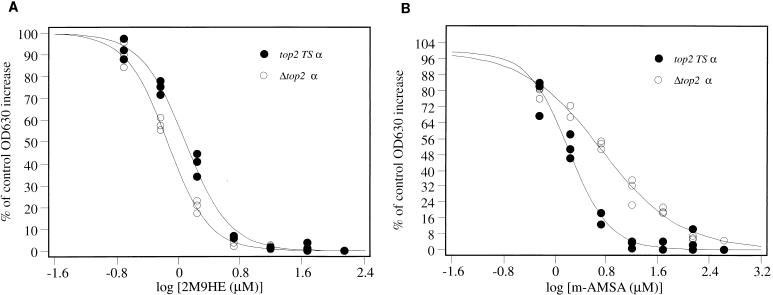
Experiments similar to that shown in Fig. 2b were repeated; three replicates at six drug concentrations were performed. Once cells had grown such that the OD630 values were approximately 0.3, well contents were mixed and ID50s were calculated as described in Materials and Methods. Figure 3 shows the results of experiments for Δtop2 α and top2(Ts) α when challenged with 2M9HE and m-AMSA. ID50s were calculated from best-fit curves to plots containing three data points for each drug concentration. As shown in Fig. 3, this increased the accuracy of estimation of the ID50. top2(Ts) α and Δtop2 α give similar, but not identical, ID50s for each drug, and the values are shown in Table 1. Figure 4 shows the effect of teniposide on all three Δtop2 strains; results are shown as average values ± one standard deviation. The resistance of Δtop2 β to teniposide is manifest by the shift of the dose-response curve to the right. ID50s for each compound against each strain are shown in Table 1.
FIG. 3.
Dose-response curves of Δtop2 α and top2(Ts) α challenged with 2M9HE (A) and m-AMSA (B). Microplate wells were seeded as described in Materials and Methods, with one plate for each yeast strain. All experiments were carried out in triplicate. ID50s were obtained by calculation of the antilog of the x-axis value at the inflection point of each sigmoid-curve fit.
TABLE 1.
ID50s calculated for topoisomerase II inhibitors against yeast strains harboring different eukaryotic topoisomerase II enzymes
| Drug | ID50 (μM) (1 SD) for yeast strain:
|
|||
|---|---|---|---|---|
| Δtop2 α | Δtop2 β | Δtop2 S.c. | top2(Ts) α | |
| m-AMSA | 5.4 (0.49) | 60 (5.2) | 38 (1.1) | 1.6 (0.1) |
| Etoposide | 22 (1.2) | 85 (7.2) | 25 (3.8) | 26 (3.0) |
| Teniposide | 16 (0.97) | 66 (3.0) | 15 (0.5) | 8.6 (0.9) |
| Mitoxantrone | 20 (2.4) | 40 (2.3) | 36 (4.5) | 27 (2.0) |
| Doxorubicin | 13 (0.59) | 16 (0.6) | 22 (0.7) | 1.7 (0.1) |
| ICRF-159 | 20 (2.8) | 250 (23) | 350 (19) | 18 (1.5) |
| Merbarone | 170 (15) | 120 (7.7) | 100 (4.4) | 180 (40) |
| 2M9HE | 0.72 (0.06) | 0.64 (0.05) | 0.43 (0.2) | 1.5 (0.1) |
FIG. 4.
Dose-response curves of all Δtop2 yeast strains challenged with teniposide. Microplate wells were seeded as described in Materials and Methods, with one plate for each yeast strain. All experiments were carried out in triplicate. Values of the percentage of control growth are plotted as the averages of each set of three experiments, and error bars indicate one standard deviation.
Comparison of microwell and viable-count methods for determination of ID50.
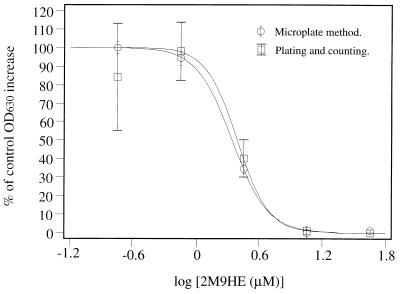
Figure 5 shows a direct comparison of data collected by plating out cells from microwells for viable counts to that collected by measuring OD630 values from Δtop2 α challenged with 2M9HE. While the two methods give almost identical ID50s, the variance in the viable-count data is larger than that for the microwell method, and the former method is also more time consuming and labor intensive.
FIG. 5.
Comparison of viable-count and microwell plate methods. Microplate wells were seeded as described in Materials and Methods, with Δtop2 α and 2M9HE in the media at five fourfold dilutions. All experiments, including control experiments, were performed in triplicate. The percentage increase in cell number relative to controls was estimated by viable-count and spectrophotometric methods. Error bars indicate one standard deviation.
DISCUSSION
The primary aim of this study was to differentiate between responses of eukaryotic topoisomerase II enzymes to topoisomerase II inhibitors. We describe a method for rapid screening of topoisomerase II inhibitors in vivo, based upon the responses of drug-permeable yeast topoisomerase II deletion strains complemented by plasmid-encoded eukaryotic topoisomerase II. The response of the yeast was measured via a microwell assay based upon the increase of OD630 as yeast growth progressed. Evaporation of media from the wells was successfully avoided by overlaying them with mineral oil, but such an overlay may create a potentially anaerobic environment for growth and may therefore alter the response of the organism to the drug. It is, however, known that very small amounts of dissolved oxygen in media will trigger aerobic growth in S. cerevisiae, and therefore we assume that the yeast strains in this assay grew in an aerobic fashion at first, by using dissolved oxygen, and switched to anaerobic growth when such resources were depleted, possibly giving rise to the inflection in the growth curve in Fig. 1.
The most useful data for drug assays are acquired in the early exponential phase of growth, and by altering seeding concentrations of yeast cells to approximately 105/ml, plates could be left overnight before checking for sufficient growth and estimation of cell number by mixing and measuring OD630 values (Fig. 2b). The use of a microwell format allows for reproducibility and accuracy through the use of replicates, and examples of data used to calculate ID50s are shown in Fig. 3 and 4. These data compare favorably to data gathered from kill curves for estimation of yeast cell number. Figure 5 shows the results of a direct comparison of the two methods. Due to the time-consuming nature of the viable-count method fewer drug concentrations were used, but three replicates were retained for each point. The ID50s calculated for Δtop2 α against 2M9HE by the two methods are almost identical, with the microplate method showing much less variation in the data values.
This is the first example of the use of full-length human topoisomerase II enzymes in a yeast topoisomerase II deletion strain to study the differential drug sensitivities of eukaryotic topoisomerase II enzymes. It is important to note that although the hybrid strains used in earlier studies will complement a temperature-sensitive strain (15, 23), it has subsequently been shown that the topoisomerase IIα hybrid enzyme will not complement a deletion strain (13). It may therefore be possible that such top2(Ts) strains express a low level of yeast topoisomerase II, which may alter the response of the organism to drugs. In addition this study enables the separation of full-length human topoisomerase IIα and IIβ in vivo for drug studies, and the methodology has the potential to be utilized for any eukaryotic topoisomerase which will complement a topoisomerase II deletion yeast strain.
The data in Fig. 1 and 2 show the response of the assay to yeast cell growth and the inhibition of growth by topoisomerase II inhibitors. The growth of Δtop2 α was always slower than that of Δtop2 β and Δtop2 S.c. for a given inoculum size. This phenomenon has been observed elsewhere (9) and suggests that the β isoenzyme is more efficient at substituting for the yeast topoisomerase than is the α isoenzyme and hence may be more similar to the yeast enzyme in its cellular role. Traditionally topoisomerase IIβ has been thought of as the “housekeeping” enzyme, while topoisomerase IIα is a more specialized enzyme essential in chromosome condensation and disjunction (16). Yeast possesses only one topoisomerase II gene, which presumably performs all of the roles of the two human isoenzymes (22). It is therefore possible that topoisomerase IIα is not as efficient at housekeeping functions, resulting in a slower-growing strain.
Table 1 lists the ID50s calculated for eight compounds against both the top2(Ts) and Δtop2 strains. A comparison of the Δtop2 α and top2(Ts) α values shows that the hybrid enzyme has slightly different in vivo sensitivities to the compounds tested. It is possible that the different growth rates previously mentioned have an effect on the drug sensitivities of the strains. However, we think this unlikely as the very slow-growing top2(Ts) α strain shows sensitivities to most drugs similar to those of the much faster growing Δtop2 α strain. It is difficult to directly compare these data with those previously reported for the yeast/human hybrid enzymes (15) as these experiments were performed by using a short-contact assay, with cells only in contact with the drug for 4 h, and in general give higher ID50s. It is reported that the yeast/human hybrid enzyme is identical to the human enzyme isolated from HeLa cells in its in vitro sensitivities to antitumor agents (23), and our studies have shown that the full-length topoisomerase II from Δtop2 α also has drug sensitivities in vitro similar to those of human enzyme isolated from HeLa cells (data not shown). Hence, one would assume that the in vitro characteristics of topoisomerase II from Δtop2 α and top2(Ts) α are similar. The explanation of different sensitivities in vivo may therefore lie in the fate of the cleavable complex once formed. It is interesting to note that the two compounds showing statistically similar ID50s are not cleavable-complex formers and that all but one of the cleavable-complex formers show different sensitivities between strains. The hybrid enzyme contains an altered N terminus, and the response of the cell to the cleavable complex may be different if it is the N terminus of the complex which is recognized by other nuclear proteins. In addition, the incorporation of a rad52 mutation in the top2(Ts) α strain is reported to increase the sensitivity of yeast to DNA-damaging agents (17).
Whatever the differences between strains containing topoisomerase IIα, these are found to have no significant effect when either the top2(Ts) α or Δtop2 α ID50s are compared to those for Δtop2 β. Topoisomerase IIα is as likely, or more likely, to induce cell death upon interaction with all but one of the compounds tested. If topoisomerase IIα is primarily responsible for the cytotoxic nature of these compounds in vivo, then this raises the question of whether the sensitivity to these compounds is manifest via an increased drug affinity for topoisomerase IIα or via an increased cellular toxicity of topoisomerase IIα-enzyme-DNA complexes over that of topoisomerase IIβ-enzyme-DNA complexes. Comparisons of these in vivo data with in vitro sensitivities of the purified enzyme are not straightforward. Human topoisomerase IIβ has been reported to be less sensitive than human topoisomerase IIα to etoposide in vitro (6), but the same report also showed topoisomerase IIβ to be less sensitive in vitro to merbarone. Our in vivo data show similar patterns of sensitivity to etoposide, but opposite patterns with merbarone, an observation also apparent in previous studies (15). Our own in vitro studies with isolated enzymes have shown that the direct comparison of data is not simple, as the concentrations of drug needed to elicit in vitro responses are very different from those used in vivo (data not shown).
Studies with resistant cell lines have shown that down regulation of topoisomerase IIα, with normal levels of topoisomerase IIβ (as measured by determining mRNA levels), results in resistance to cleavable-complex forming drugs and especially increased resistance to etoposide (24), suggesting that topoisomerase IIβ is indeed more resistant to etoposide. A similar cell line with down regulated topoisomerase IIβ was also resistant to all inhibitors of topoisomerase II tested (24), suggesting that topoisomerase IIβ is a target for antitumor agents along with topoisomerase IIα. Studies with human breast cancer cells reported increased sensitivity to m-AMSA in cells with overexpressed topoisomerase IIα and sensitivity to mitoxantrone when either enzyme isoform was overexpressed, data which support our findings (11). However, cells showing an increase in topoisomerase IIβ enzyme levels were more sensitive to etoposide, a finding which seems to contradict our data. There are many contradictions in data concerning resistance and isoenzyme expression, and is therefore probable that many factors affect the mechanisms of resistance to topoisomerase II inhibitors in whole-cell systems.
We believe that the toxic effects of topoisomerase II inhibitors are mediated via interaction with both topoisomerase IIα and IIβ. Although it has been shown in vitro that a degree of selectivity for different topoisomerase II isoforms exists within drug classes, the situation is not fully verified by in vivo studies (4, 6, 15). The determination of the toxicities of these compounds is complicated by the fact that the cellular response to topoisomerase IIα-enzyme-DNA complexes may be different than that for the topoisomerase IIβ-enzyme-DNA complexes. It should also be noted that topoisomerase IIα is more abundant in the mitotic nucleus than is topoisomerase IIβ (26) and hence may have a major role in determining toxicity simply via a concentration effect.
The response of the Δtop2 S.c. is in most cases similar to that for Δtop2 β. However, the response of the Δtop2 S.c. strain to the epipodophyllotoxins etoposide and teniposide is similar to that for Δtop2 α. The resistance of topoisomerase IIβ to etoposide and teniposide presents an interesting conundrum. If this resistance is manifest at the amino acid level, then a mutation must have occurred after the gene duplication event which created the human isoenzymes (5). If it is manifest via differential cellular response to the cleavable complexes, then it must be assumed that the cellular response is different for different drug-enzyme-DNA complexes. If it is the fate of the cleavable complex which regulates sensitivity of the yeast strains to topoisomerase II inhibitors, then the yeast cell must recognize and interact with the human topoisomerase II enzymes. Indeed, mouse topoisomerases IIα and IIβ are recognized and selectively localized by yeast cells, topoisomerase IIα is accumulated in the nucleus at mitosis, and topoisomerase IIβ is not localized in the nucleus at that time (1). Therefore, it is possible that topoisomerase IIα-containing clones are especially more sensitive to antitumor drugs, as the enzyme is localized in the nucleus at a crucial stage of cellular replication, a characteristic not shared by topoisomerase IIβ. Another alternative, with an evolutionary basis, is that topoisomerase IIβ and yeast topoisomerase are more like the original topoisomerase precursor, with some post-gene-duplication mutations (14). Topoisomerase IIα has evolved, after the gene duplication event, into a more specialized mammalian topoisomerase, which either happens also to be more sensitive to most antitumor agents or which creates a more toxic (i.e., less easily repaired) drug-enzyme-DNA complex.
ACKNOWLEDGMENTS
This work was supported by grants from the Leverhulme Trust and the Wellcome Trust (grant no. 042987/Z/94/Z). A. Maxwell is a Lister-Jenner Research fellow.
We thank Ian Hickson for his comments on this manuscript.
REFERENCES
- 1.Adachi N, Miyaike M, Kato S, Kanamaru R, Koyama H, Kikuchi A. Cellular distribution of mammalian DNA topoisomerase II is determined by its catalytically dispensable C-terminal domain. Nucleic Acids Res. 1997;25:3135–3142. doi: 10.1093/nar/25.15.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger J M, Gamblin S J, Harrison S C, Wang J C. Structure and mechanism of DNA topoisomerase-II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 3.Chen A Y, Liu L F. DNA topoisomerases: essential enzymes and lethal targets. Annu Rev Pharmacol Toxicol. 1994;34:191–218. doi: 10.1146/annurev.pa.34.040194.001203. [DOI] [PubMed] [Google Scholar]
- 4.Cornarotti M, Tinelli S, Willmore E, Zunino F, Fisher L M, Austin C A, Capranico G. Drug sensitivity and sequence specificity of human recombinant DNA topoisomerases II alpha (p170) and II beta (p180) Mol Pharmacol. 1996;50:1463–1471. [PubMed] [Google Scholar]
- 5.Coutts J, Plumb J A, Brown R, Keith W N. Expression of topoisomerase-II-alpha and topoisomerase-II-beta in an adenocarcinoma cell-line carrying amplified topoisomerase-II-alpha and retinoic acid receptor-alpha genes. Br J Cancer. 1993;68:793–800. doi: 10.1038/bjc.1993.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake F H, Hofmann G A, Bartus H F, Mattern M R, Crooke S T, Mirabelli C K. Biochemical and pharmocological properties of p170 and p180 forms of topoisomerase II. Biochemistry. 1989;28:8154–8160. doi: 10.1021/bi00446a029. [DOI] [PubMed] [Google Scholar]
- 7.Drake F H, Hofmann G A, Mong S-M, Bartus J, Hertzberg R P, Johns D B. In vitro and intracellular inhibition of topoisomerase II by the antitumour agent merbarone. Cancer Res. 1989;49:2578–2583. [PubMed] [Google Scholar]
- 8.Froelich-Ammon S J, Osheroff N. Topoisomerase poisons—harnessing the dark side of enzyme mechanism. J Biol Chem. 1995;270:21429–21432. doi: 10.1074/jbc.270.37.21429. [DOI] [PubMed] [Google Scholar]
- 9.Hickson, I. D. 1996. Personal communication.
- 10.Holm C, Goto T, Wang J C, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 11.Houlbrook S, Addison C M, Davies S L, Carmichael J, Stratford I J, Harris A L, Hickson I D. Relationship between expression of topoisomerase-II isoforms and intrinsic sensitivity to topoisomerase-II inhibitors in breast-cancer cell-lines. Br J Cancer. 1996;74:1454–1461. doi: 10.1038/bjc.1995.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkins J R, Ayton P, Jones T, Davies S L, Simmons D L, Harris A L, Sheer D, Hickson I D. Isolation of cDNA clones encoding the b isozyme of human DNA topoisomerase II and localisation of the gene to chromosome 3p24. Nucleic Acids Res. 1992;20:5587–5592. doi: 10.1093/nar/20.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen S, Redwood C S, Jenkins J R, Andersen A H, Hickson I D. Human DNA topoisomerases II-alpha and II-beta can functionally substitute for yeast top2 in chromosome segregation and recombination. Mol Gen Genet. 1996;252:79–86. [PubMed] [Google Scholar]
- 14.Madhusudan K, Nagaraja V. Alignment and phylogenetic analysis of type-II DNA topoisomerases. J Biosciences. 1996;21:613–629. [Google Scholar]
- 15.Meczes E L, Marsh K L, Fisher L M, Rogers M P, Austin C A. Complementation of temperature-sensitive topoisomerase II mutations in Saccharomyces cerevisiae by a human TOP2 beta construct allows the study of topoisomerase II beta inhibitors in yeast. Cancer Chemother Pharmacol. 1997;39:367–375. doi: 10.1007/s002800050585. [DOI] [PubMed] [Google Scholar]
- 16.Meyer K N, Kjeldsen E, Straub T, Knudsen B R, Hickson I D, Kikuchi A, Kreipe H, Boege F. Cell cycle-coupled relocation of types I and II topoisomerases and modulation of catalytic enzyme activities. J Cell Biol. 1997;136:775–788. doi: 10.1083/jcb.136.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nitiss J L, Wang J C. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc Natl Acad Sci USA. 1988;85:7501–7505. doi: 10.1073/pnas.85.20.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roca J, Ishida R, Berger J M, Andoh T, Wang J C. Antitumour bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc Natl Acad Sci USA. 1994;91:1781–1785. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose D, Thomas W, Holm C. Segregation of recombined chromosomes in meiosis I requires DNA topoisomerase II. Cell. 1990;60:1009–1017. doi: 10.1016/0092-8674(90)90349-j. [DOI] [PubMed] [Google Scholar]
- 20.Schiestl R H, Geitz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 21.Tsai-Pflugelder M, Liu L F, Liu A A, Tewey K M, Whang-Peng J, Knutsen T, Huebner K, Croce C M, Wang J C. Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21-22. Proc Natl Acad Sci USA. 1988;85:7177–7181. doi: 10.1073/pnas.85.19.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J C. DNA topoisomerases: why so many? J Biol Chem. 1991;266:6659–6662. [PubMed] [Google Scholar]
- 23.Wasserman R A, Austin C A, Fisher L M, Wang J C. Use of yeast in the study of anticancer drugs targeting DNA topoisomerases: expression of a functional recombinant human DNA topoisomerase IIα in yeast. Cancer Res. 1993;53:3591–3596. [PubMed] [Google Scholar]
- 24.Withoff S, DeVries E, Keith W N, Nienhuis E F, VanderGraaf W, Uges D, Mulder N H. Differential expression of DNA topoisomerase II alpha and -beta in P- gp and MRP-negative VM26, mAMSA and mitoxantrone-resistant sublines of the human SCLC cell line GLC(4) Br J Cancer. 1996;74:1869–1876. doi: 10.1038/bjc.1996.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Withoff S, Keith W N, Knol A J, Coutts J C, Hoare S F, Mulder N H, Devries E. Selection of a subpopulation with fewer DNA topoisomerase II-alpha gene copies in a doxorubicin-resistant cell-line panel. Br J Cancer. 1996;74:502–507. doi: 10.1038/bjc.1996.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woessner R D, Mattern M R, Mirabelli C K, Johnson R K, Drake F H. Proliferation and cell cycle dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]