Antimicrobial resistance (AMR) is a global health challenge and has long been subject of numerous investigations toward the development of new mitigation strategies to combat multidrug (MDR) resistant bacteria. The unbridled use of antimicrobials in the human, animal, and environment interface has posed a critical threat to public health in global scale, while alternative antimicrobial therapeutics are running out, and treatment options are becoming limited.1,2 The search for an alternative to antibiotics has led scientists to give renewed attention to CRISPR-Cas9 therapy. Indeed, CRISPR-Cas9 has revolutionized medicine and public health by developing a broad arsenal to fight AMR3 and its principal vehicle of spreading plasmids. Understanding the role of plasmids in antimicrobial resistance is crucial for developing mitigation strategies to combat the spread of resistance.4 Efforts are being made to develop alternative treatment options, including a CRISPR-Cas9-based approach.
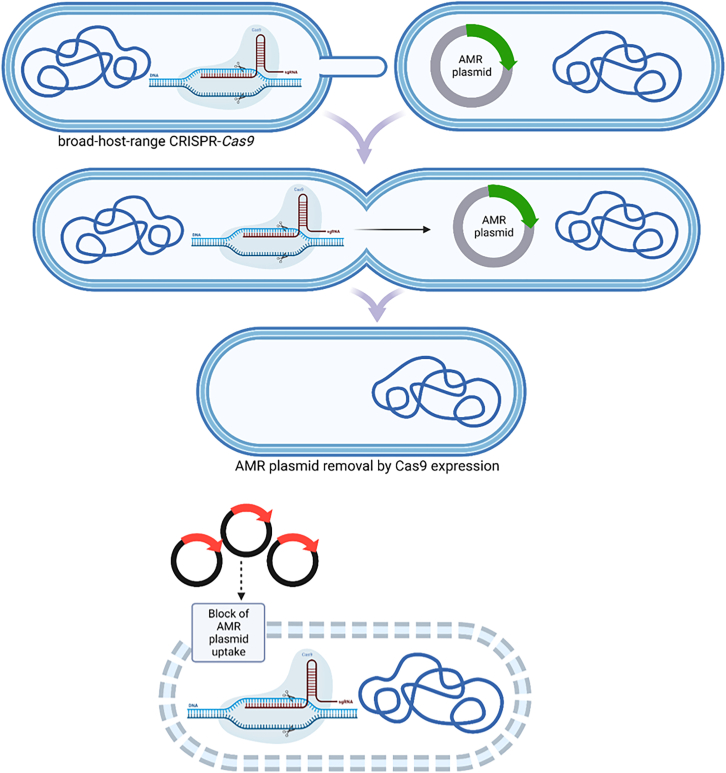
With this commentary, I want to highlight a recent paper by Walker-Sünderhauf and colleagues published in Microbiology, which reported the removal of AMR plasmids using a mobile, broad-host-range CRISPR-Cas9 delivery tool.5 The engineered IncP1-plasmid pKJK5 encoding cas9 programmed to target an AMR gene demonstrated the ability to block the uptake of AMR plasmids removing resident plasmids in multiple species (Figure 1).5
Figure 1.
Removal and uptake block of AMR plasmids using a mobile broad-host-range CRISPR-Cas9 delivery tool
The authors successfully engineered the fully conjugative plasmid pKJK5:csg, highlighting its ability to shield various host species from the acquisition of the targeted AMR plasmid pHERD30T. As a proof of concept, the conjugation of pKJK5:csg resulted in the elimination of pHERD30T from the recipient strain.
Such is the success of the CRISPR system that there is already removal of resident plasmids in multiple bacterial species. While these findings are promising, achieving similar results in natural environments becomes more challenging due to the presence of a complex microbiota community, where the exchange of mobile genetic elements contributes to the flux of genome size and content, and the responses could be different.
While continuous surveillance needs to be established as a priority to prevent the spread of AMR, this study is a perfect example of the development of a mitigation strategy to combat the intercontinental AMR dissemination facilitated by promiscuous plasmids.
Acknowledgments
Declaration of interests
The author declares no competing interests.
References
- 1.Levy S.B., Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 2004;10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 2.Monte D.F., Lincopan N., Fedorka-Cray P.J., Landgraf M. Current Insights on High Priority Antibiotic-Resistant Salmonella enterica in Food and Foodstuffs: A review. Curr. Opin. Food Sci. 2019;26:35–46. doi: 10.1016/j.cofs.2019.03.004. [DOI] [Google Scholar]
- 3.Gholizadeh P., Köse Ş., Dao S., Ganbarov K., Tanomand A., Dal T., Aghazadeh M., Ghotaslou R., Ahangarzadeh Rezaee M., Yousefi B., Samadi Kafil H. How CRISPR-Cas System Could Be Used to Combat Antimicrobial Resistance. Infect. Drug Resist. 2020;13:1111–1121. doi: 10.2147/IDR.S247271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larsson D.G.J., Flach C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022;20:257–269. doi: 10.1038/s41579-021-00649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker-Sünderhauf D., Klümper U., Pursey E., Westra E.R., Gaze W.H., van Houte S. Removal of AMR plasmids using a mobile, broad host-range CRISPR-Cas9 delivery tool. Microbiology (Reading, England) 2023;169 doi: 10.1099/mic.0.001334. [DOI] [PMC free article] [PubMed] [Google Scholar]