Abstract
Genome-wide association studies indicate that allele variants in MIR137, the host gene of microRNA137 (miR137), confer an increased risk of schizophrenia (SCZ). Aberrant expression of miR137 and its targets, many of which regulate synaptic functioning, are also associated with an increased risk of SCZ. Thus, miR137 represents an attractive target aimed at correcting the molecular basis for synaptic dysfunction in individuals with high genetic risk for SCZ. Advancements in nanotechnology utilize lipid nanoparticles (LNPs) to transport and deliver therapeutic RNA. However, there remains a gap in using LNPs to regulate gene and protein expression in the brain. To study the delivery of nucleic acids by LNPs to the brain, we found that LNPs released miR137 cargo and inhibited target transcripts of interest in neuroblastoma cells. Biodistribution of LNPs loaded with firefly luciferase mRNA remained localized to the mouse prefrontal cortex (PFC) injection site without circulating to off-target organs. LNPs encapsulating Cre mRNA preferentially co-expressed in neuronal over microglial or astrocytic cells. Using quantitative proteomics, we found miR137 modulated glutamatergic synaptic protein networks that are commonly dysregulated in SCZ. These studies support engineering the next generation of brain-specific LNPs to deliver RNA therapeutics and improve symptoms of central nervous system disorders.
Keywords: microRNA, lipid nanoparticle, prefrontal cortex, synaptic proteins, schizophrenia, glutamate networks, neuron, brain delivery
Graphical abstract
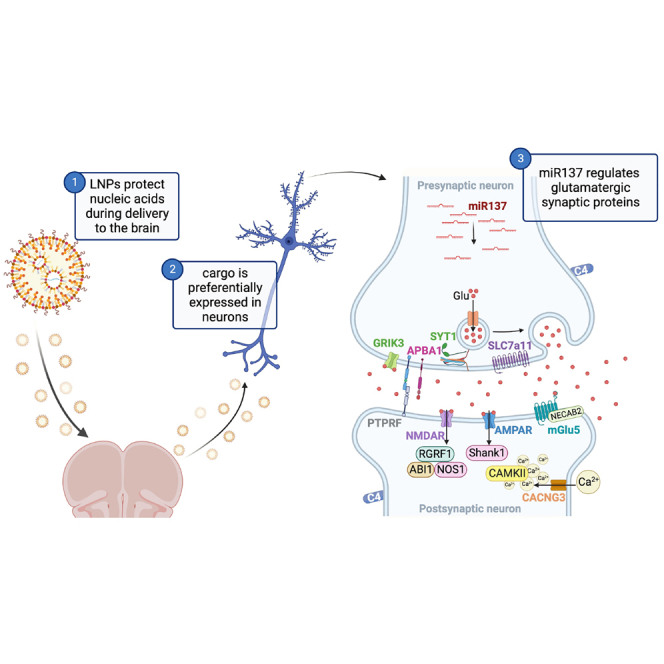
Janowsky and colleagues investigated microRNA137 and its association to proteins involved in schizophrenia risk. It is unknown if lipid nanoparticles can deliver therapeutic RNAs to the brain. The authors determined lipid nanoparticles can express nucleic acid cargo in prefrontal cortex neurons to modulate glutamatergic synaptic protein networks linked to schizophrenia.
Introduction
MicroRNAs (miRNAs) are ∼22-nucleotide, single-stranded, noncoding RNAs that modulate developmental processes and cellular function. miRNAs target specific sets of messenger RNAs (mRNAs) and are key translational silencers of gene expression.1 Up to 80% of human genes are regulated by miRNAs, and individual miRNA sequences can regulate complex networks of tens to hundreds of gene targets.2,3 MicroRNA137 (miR137) is enriched in the embryonic midbrain and forebrain across neurodevelopment.4 In adulthood, miR137 is located in the synapto-dendritic compartment of cortical, subcortical, and hippocampal areas,5,6 where it targets a number of synaptic and developmental mRNAs. Accordingly, over-expression of miR137 in adult mice alters synaptogenesis, synaptic ultrastructure, and synaptic function.7 Genetic deletion of miR137 results in postnatal lethality, while miR137 heterozygous mice have disrupted synaptic and dendritic growth, repetitive behavior, and impaired learning and social behavior.8 Thus, miR137 dysregulation may be a factor in brain disorders associated with synaptic dysfunction.9,10
Schizophrenia (SCZ) is a highly heritable,11 polygenic neuropsychiatric disorder characterized by impaired cognition,12 deficits in sensory processing,13 negative symptoms, and episodic psychosis.14 These symptoms result from synaptic pathology,15 and there is substantial overlap of synaptic genes that increase the risk of SCZ and those that are regulated by miR137.16,17,18,19,20 Furthermore, single-nucleotide polymorphisms (SNPs) in the MIR137 gene are also associated with an increased risk of SCZ.21,22 MIR137 allele variants are linked to SCZ clinical endophenotypes, including earlier age of onset, more severe psychotic symptoms, decreased cognitive function,6,23,24 and alterations in brain structure and activity.6,25 Importantly, SNPs in the noncoding region of MIR137 reduce the expression of miR137.26 Since miR137 expression is decreased in some individuals with SCZ, exogenous replacement of miR137 represents an attractive therapeutic target. However, free-floating miRNAs are subject to degradation, so a delivery vehicle to protect and target the miRNA is needed.
The lipid nanoparticles (LNPs) encapsulating the COVID-19 mRNA vaccine demonstrated that nanocarriers can be used to traffic nucleic acids for previously unmet therapeutic needs. LNP delivery vectors exhibit the following desirable properties: (1) protection of nucleic acids from nucleases, (2) customization for tissue and cell selectivity, (3) high payload delivery, (4) low toxicity, and (5) feasibility for large-scale production.27 LNP-delivered small nucleic acid therapeutics, including small interfering RNAs (siRNAs), are approved for clinical trials.28 Previous studies delivering miRNAs by LNPs to the brain were limited to therapeutic agents for gliomas.29 However, the therapeutic effect of LNP-delivered miRNA on synaptic protein networks remains a large gap in neuro-nanomedicine.
As a result, we developed an LNP-based platform to deliver miR137 to neurons, thereby modulating synaptic proteins. Our in vitro studies demonstrated that LNPs were effective nucleic acid delivery vehicles and confirmed the release of miR137 cargo and subsequent inhibition of target transcripts. Mouse in vivo studies showed that LNPs remained localized to the prefrontal cortex injection site and preferentially expressed tdTomato in Ai9 mice following Cre-recombination in neurons relative to other cell types. Delivery of miR137 to the mouse prefrontal cortex significantly modulated a diverse network of pre-and post-synaptic glutamatergic proteins. This work demonstrates that LNPs offer a unique delivery mechanism for nucleic acid transportation to the brain to modulate key synaptic proteins.
Results
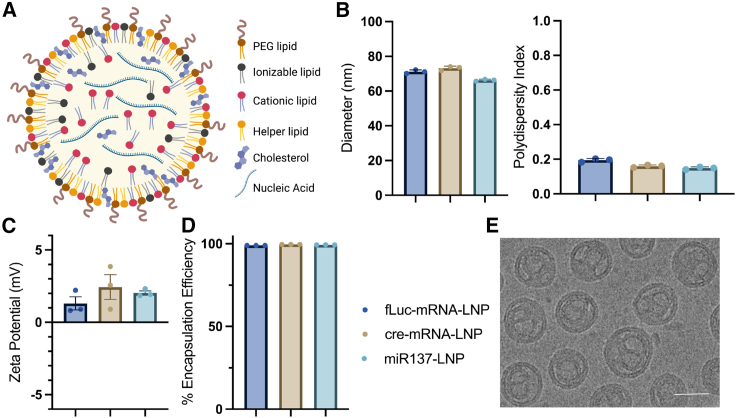
We first synthesized and characterized LNPs to modulate synapses in the brain. Initial testing of LNP formulations compared LNPs composed of MC3 (ionizable) only, SM102 (ionizable cationic) only, SM102 and 1,2-dioleoyl-3-trimethylammoniumpropane (DOTAP) (cationic), or MC3 and DOTAP lipids. We found LNP formulations combining MC3 and DOTAP lipids were the most efficient at cargo release (data not shown). Subsequent formulations used MC3 and DOTAP lipids to self-assemble into LNPs with nucleic acid cargo via intermolecular interactions using microfluidics along with structural lipids (DSPC), sterols (cholesterol), and PEG-lipids (DMG-PEG2k) (Figure 1A). LNPs with three nucleic acid cargos were tested with consistent batch results: firefly luciferase mRNA (fLuc-mRNA-LNP), Cre mRNA (cre-mRNA-LNP), and microRNA137 (miR137-LNP). Dynamic light scattering analysis of LNP formulations demonstrated homogeneous particle sizes (∼70 nm) with low polydispersity index (<0.2) (Figure 1B), slightly positive charge (∼2 mV) (Figure 1C), and high encapsulation efficiencies (>98%) (Figure 1D). One-way ANOVA determined no significant variation in LNP zeta potential (F (2, 6) = 1.024, p = 0.4142). Cryo-transmission electron microscopy (cryo-TEM) images of miR137-LNPs showed concentric multilamellar “onion-like” structures with internal aqueous space (Figure 1E). Overall, reliable LNP batch consistencies suggest this preparation method is suitable for various nucleic acid cargos.
Figure 1.
LNP characterization
(A) Schematic of an LNP composed of lipid components and an encapsulated nucleic acid. DLS analysis characterizing LNPs with (B) hydrodynamic size (nm) and particle heterogeneity (polydispersity index), (C) zeta potential (mV), and (D) nucleic acid EE (%). (E) Cryo-TEM image of miR137-LNP. Scale bar, 50 μm. n = 3 with triplicate determinations, mean ± SEM. Firefly luciferase mRNA (fLuc-mRNA-LNP), Cre mRNA (cre-mRNA-LNP), and microRNA137 (miR137-LNP).
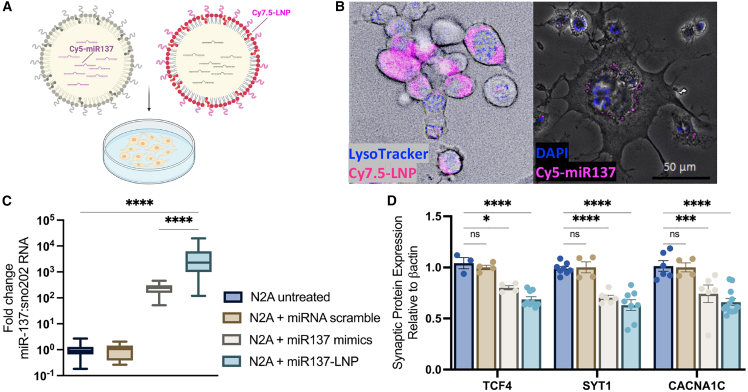
By using a fluorophore tagged to PEG lipids or the mature miR137 sequence, we visualized LNP uptake, intracellular trafficking, and endosomal escape of miR137 in live cells (Figure 2A). Mouse neuroblastoma Neuro2A (N2A) cells endocytosed Cy7.5 tagged-LNPs (Cy7.5-LNP, magenta), which did not sequester to the lysosomal compartment (LysoTracker, blue) at 6 h. Importantly, miR137 cargo (Cy5-miR137, magenta) was visualized in the cytosol and perhaps was released in the cytoplasm through endosomal escape (nuclear stain, DAPI, blue) (Figure 2B). Treating N2A cells with increasing concentrations of miR137-LNPs was non-toxic with high cell survival (Figure S1). At 24 h, a one-way ANOVA determined that adding 200 nM scramble miRNA in Lipofectamine to N2A cells did not change the expression of miR137 levels compared to untreated cells (p > 0.999). Adding miR137 loaded in LNPs significantly increased miR137 levels (F (3, 61) = 11.18, p < 0.0001) and improved the transfection efficiency compared to adding the miR137 mimics in Lipofectamine (F (3, 61) = 11.18, p < 0.0001) (Figure 2C). A two-way ANOVA demonstrated an effect of treatment that miR137 expression persisted for 96 h post treatment (F (3, 39) = 6.011, p = 0.0018) (Figure S2A) and was dose dependent (Figure S2B).
Figure 2.
LNP delivery and miR137 release in cell culture
(A) Schematic of fluorophore tagging of LNPs (Cy7.5-LNP) or miR-137 cargo (Cy5-miR137) in Neuro2A cells. (B) Live-cell imaging of LNP uptake (Cy7.5-LNP, magenta) at 6 h without lysosomal compartment sequestering (LysoTracker, blue) and cytoplasmic release of miR137 (Cy5-miR137, magenta) cargo (nuclear stain, DAPI, blue). Scale bars, 50 μm. (C) RT-qPCR fold-change miR137 expression compared to sno202RNA in N2A cells at 24 h after treatment with 200 nM miRNA scramble in Lipofectamine, 200 nM miR137 mimic in Lipofectamine, or miR137-LNPs. (D) Translational inhibition of target synaptic proteins transcription factor 4 (TCF4), synaptotagmin 1 (SYT1), and voltage-gated calcium channel α1c (CACNA1C) after treatment with 200 nM miR137 mimic in Lipofectamine, miRNA scramble in Lipofectamine, or miR137-LNPs compared to untreated N2A cells. n = 3–8 with triplicate determinations, mean ± SEM. ns, not significant, p > 0.05; ∗p < 0.05; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001.
Compared to N2A untreated cells, a two-way ANOVA with Dunnett’s multiple comparisons test determined treatment with miR137 mimics in Lipofectamine significantly inhibited translation of several miR137 synaptic protein targets: transcription factor 4 (TCF4) (F (3, 62) = 45.35, p = 0.0177), synaptotagmin 1 (SYT1) (F (3, 62) = 45.35, p < 0.0001), and the voltage-gated calcium channel a1C (CACNa1C) (F (3, 62) = 45.35, p = 0.0003) (Figure 2D) (Figure S3), in addition to other synaptic protein targets such as the glutamate receptor 1 (GluA1) (F (1, 48) = 87.33, p = 0.0141), synaptophysin (Syn) (F (1, 48) = 87.33, p = 0.0245), complexin 1 (CPLX1) (F (1, 48) = 87.33, p < 0.0001), and the fragile X mental retardation protein (FMRP) (F (1, 48) = 87.33, p = 0.0008) (Figures S4A and S4B). Treatment with miR137-LNPs showed an even larger inhibition of TCF4 (F (3, 62) = 45.35, p < 0.0001), SYT1 (F (3, 62) = 45.35, p < 0.0001), and CACNa1C (F (3, 62) = 45.35, p < 0.0001), while miRNA scramble had no effect on TCF4 (F (3, 62) = 45.35, p = 0.9171), SYT1 (F (3, 62) = 45.35, p = 0.9971), or CACNa1C (F (3, 62) = 45.35, p = 0.9953) (Figure 2D). Importantly, expression of a protein that is not a target of miR137, post-synaptic density 95 (PSD95), was unaffected (F (1, 48) = 87.33, p > 0.9999) by miR137 mimic or miR137-LNPs, demonstrating the specificity of miR137 to target specific transcripts. From this, we concluded that LNPs endocytosed into neuroblastoma cell cultures and released functional miR137 cargo capable of targeting downstream transcripts.
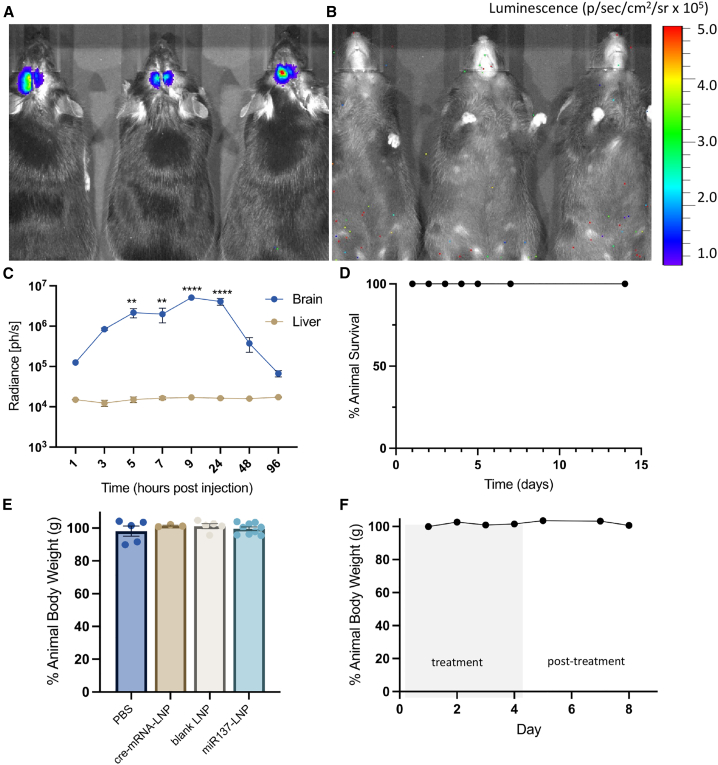
To study the in vivo biodistribution and pharmacokinetics of LNPs in the brain, we bilaterally injected firefly luciferase mRNA encapsulated in LNPs (fLuc-mRNA-LNP) into the mouse prefrontal cortex (PFC). A two-way ANOVA revealed robust luminescent radiance near the injection site with no detectable expression in the liver (F (1, 26) = 89.99, p < 0.0001) or other off-target organs (Figure 3A-3B). The same experimental animals were repeatedly measured, and radiance peaked between 9 and 24 h (F (7, 26) = 10.77, p < 0.0001) post injection, and returned to baseline by 96 h (Figure 3C). Thus, LNP nucleic acid cargo is bilaterally and locally expressed at the direct PFC injection site without detectable off-target expression. We did not detect any overt toxicity as all experimental animals survived until the experimental endpoint, 2 weeks following LNP injection (Figure 3D) and did not lose more than 5% of their original body weight (grams) (Figure 3E). In separate experiments, we found that delivery of miR137-LNPs every day for 5 days did not result in weight loss (grams) (Figure 3F) or cellular anatomical disruptions determined by neural cell density counts30 (unpaired t test, t(12) = 1.175, p = 0.2629) (Figure S5), demonstrating the potential use of repeated LNP treatments in the brain.
Figure 3.
LNP-mediated transfection and toxicity in vivo
Representative (A) dorsal and (B) ventral images of luciferase bioluminescence in mouse brain and other tissues 24 h post a single injection in the prefrontal cortex with firefly luciferase mRNA LNPs. (C) Time curve (hours post injection) of luciferase radiance (photons per second) in the brain and the liver. n = 3, mean ± SEM. ∗∗p < 0.01 and ∗∗∗∗p < 0.0001. (D) Percentage animal survival after a single bilateral intracerebral injection with LNPs. n = 6, mean ± SEM. (E) Percentage change in animal body weight after single bilateral intracerebral injection with PBS, cre-mRNA-LNP, blank LNP, or miR137-LNPs. n = 3–10, mean ± SEM. In separate experiments, animals received five consecutive bilateral intracerebral injections with miR137-LNPs (day 1–5) and (F) the percentage change in animal body weight was measured up to 8 days. n = 2, mean ± SEM.
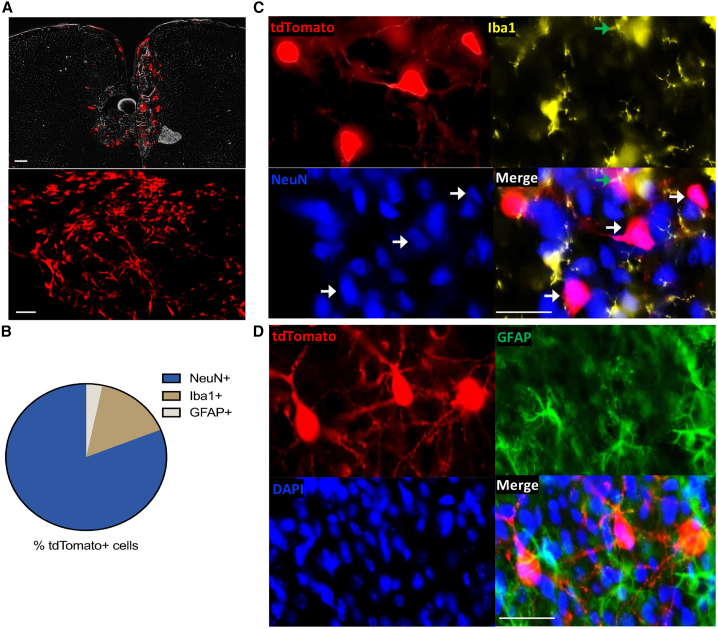
Using immunohistochemistry to multiplex neuron (NeuN+), microglia (Iba1+), and astrocyte (GFAP+) markers in sectioned PFC tissue, we determined LNP cargo expression in specific cell types. We delivered cre-mRNA-LNPs to Ai9 mice, a Cre reporter strain with a loxP-flanked STOP cassette preventing transcription of a CAG promoter within the ubiquitously expressed ROSA26 locus. Ai9 mice express robust tdTomato fluorescence following Cre-mediated recombination. In this case, there is no fluorophore tagging of the LNP itself, thus there is no way to visualize the uptake of LNPs. Instead, we rely on visualizing the expression of the Cre mRNA as it is translated to the tdTomato reporter protein. To focus our analysis on LNP-induced effects, rather than tissue damage and microglial recruitment to the needle tract, we quantified tdTomato expression in the adjacent 100–200 μm from the injection site (Figures S6A and S6B).31 Low-magnification fluorescent microscopy images showed robust bilateral tdTomato+ (red) cells at the PFC injection site (Figure 4A). Of the tdTomato+ cells, 81.11% were NeuN+, 15.64% of tdTomato+ cells were Iba1+, and 3.25% of tdTomato+ cells were GFAP+. One-way ANOVA indicated a significant effect of cell marker (NeuN/Iba1/GFAP) on tdTomato+ expression (F (2, 66) = 427.4, p < 0.0001) (Figures 4B–4D). Post hoc analysis using Tukey’s multiple comparisons test indicated that tdTomato expression differed significantly by cell marker (NeuN vs. Iba1, p < 0.0001), (NeuN vs. GFAP, p < 0.0001), and (GFAP vs. Iba1, p < 0.0001). These data suggest LNP nucleic acid cargo is preferentially expressed in neurons in the mouse PFC.
Figure 4.
LNP cell-type expression in the brain
Mice received bilateral intracerebral injections in the prefrontal cortex with cre-mRNA-LNPs that express tdTomato (red) following Cre-recombination. (A) High (top) and low (bottom) magnification of tdTomato florescence and injection placements in the prefrontal cortex. Scale bars, 200 μm. (B) Quantification of tdTomato+ cells preferentially co-expressed with neuronal (NeuN+, blue) over microglial (Iba1+, yellow) or astrocyte (GFAP+, green) markers (nuclear stain, DAPI, blue). (C) TdTomato+ cells overlap slightly with Iba1+ cells (green arrows), but mostly co-express with NeuN+ cells (white arrows). (D) TdTomato+ cells do not co-label with GFAP+ cells. Scale bars, 25 μm. n = 4, mean ± SEM.
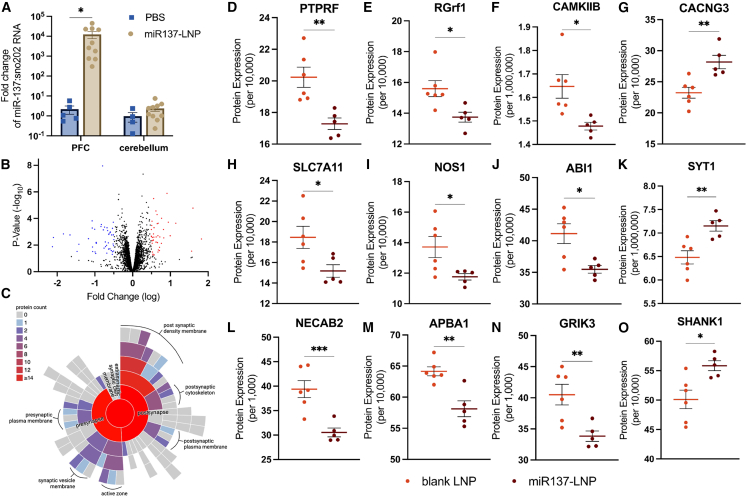
Following confirmation that LNPs can effectively deliver nucleic acids to PFC neurons in vivo, we sought to determine if miR137-LNPs can alter synaptic protein expression. Mice received bilateral injections of miR137-LNPs or PBS into the PFC. After 24 h, a two-way ANOVA with Tukey’s test for post hoc comparisons determined miR137 levels were significantly upregulated in the PFC compared to small nucleolar RNA 202 (sno202) with no detectable changes in cerebellar tissue (F (1, 25) = 2.865, p = 0.0391) (Figure 5A). Unbiased isobaric-labeling quantitative proteomics of PFC biopsy punches was used to compare 4,807 proteins after 72 h from three experimental groups (n = 5–6/group): PBS, blank LNPs, or miR137-LNPs. This experimental design allowed us to determine the proteomic effects of the LNPs alone (PBS vs. blank LNP) and of the miR137 alone (blank LNP vs. miR137-LNP). Comparing differentially expressed proteins (DEPs) from blank LNP and miR137-LNP treatments indicated that 224 proteins were altered with 105 upregulated (red) and 119 downregulated (blue) (Figure 5B). The STRING,32 Gene Ontology (GO), SynGo,33 and FUMA GENE2FUNC34 databases generated functional and structural classifications of known protein-protein network interactions based on the DEPs from miR137-LNP compared to blank LNP treatments. The databases confirmed that 52% of the total DEPs from miR137-LNPs were involved in neuronal development (GO: 0032502), including neurogenesis and neuron differentiation (Figure S7), although the experimental animals were adults, where we might expect the majority of neuronal development to have already occurred.35 Additionally, 23% DEPs from miR137-LNPs were in neuronal synapses (GOCC: 0045202) (Figure S8A). miR137-LNPs affected both pre-and post-synaptic proteins (Figure 5C), with the majority of the DEPs translated in the neuropil (dendrites and axons) compared to the soma (Figure S8B),36 suggesting local translation of synaptic proteins. Further investigation determined 67% of synaptic DEPs from miR137-LNP were glutamatergic (GOCC: 0098978). Our findings support previous work demonstrating that miR137 modulates genes and proteins involved in neuronal development17 and glutamatergic synaptic function.37
Figure 5.
LNP delivery and proteomic analysis of miR137 in the brain
Mice received bilateral intracerebral injections in the prefrontal cortex with miR137-LNPs or PBS. (A) RT-qPCR fold change miR137 expression compared to sno202RNA at 24 h. n = 4–10, mean ± SEM, ∗p < 0.05. TMT proteomics comparing prefrontal cortex biopsies of animals who received miR137-LNP or blank LNPs at 72 h. (B) Volcano plot of DEPs between blank LNP and miR137-LNP (log2fold change and adjusted p values <0.05). (C) Pre- and post-synaptic location of DEPs by count following miR137-LNP treatment. Treatment with miR137-LNPs disrupts glutamatergic synaptic proteins compared to blank LNPs including (D) receptor-type tyrosine-protein phosphatase F (PTPRF), (E) Ras-specific guanine nucleotide-releasing factor 1 (RGrf1), (F) calcium/calmodulin dependent protein kinase II beta (CAMKIIB), (G) voltage-dependent calcium channel gamma-3 (CACNG3), (H) solute carrier family 7 member 11 (SLC7A11), (I) nitric oxide synthase 1 (NOS1), (J) Abl interactor 1 (ABI1), (K) synaptotagmin 1 (SYT1), (L) N-terminal EF-hand calcium-binding protein 2 (NECAB2), (M) amyloid beta A4 precursor protein (APBA1), (N) glutamate receptor ionotropic kainate 3 (GRIK3), and (O) SH3 (SHANK1). n = 5–6/group, mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
A two-sample t test found specific glutamatergic synaptic proteins were significantly inhibited in miR137-LNP treatments compared to blank LNPs (Figures 5D–5O), including receptor-type tyrosine-protein phosphatase F (PTPRF) (t(9) = 3.778, p = .0044) (Figure 5D), Ras-specific guanine nucleotide-releasing factor 1 (RGrf1) (t(9) = 2.877, p = .0183) (Figure 5E), calcium/calmodulin dependent protein kinase II beta (CAMKIIB) (t(9) = 2.937, p = .0166) (Figure 5F), solute carrier family 7 member 11 (SLC7A11) (t(9) = 2.5, p = .0338) (Figure 5H), nitric oxide synthase 1 (NOS1) (t(9) = 2.487, p = .0346) (Figure 5I), Abl interactor 1 (ABI1) (t(9) = 3.123, p = .0123) (Figure 5J), N-terminal EF-hand calcium-binding protein 2 (NECAB2) (t(9) = 4.241, p = .0022) (Figure 5L), amyloid beta A4 precursor protein (APBA1) (t(9) = 4.303, p = .0020) (Figure 5M), and the glutamate receptor ionotropic kainate 3 (GRIK3) (t(9) = 3.356, p = .0084) (Figure 5N). Three glutamatergic synaptic proteins were upregulated in miR137-LNP treatments compared to blank LNPs, including the voltage-dependent calcium channel gamma-3 (CACNG3) (t(9) = 3.611, p = .0056) (Figure 5G), synaptotagmin 1 (SYT1) (t(9) = 3.606, p = .0057) (Figure 5K), and SH3 (SHANK1) (t(9) = 3.027, p = .0143) (Figure 5O). Many of the altered glutamatergic proteins effected by miR137-LNPs are in the post-synaptic density, including CACNG3 (GO: 0099061), PTPRF (GO: 0099061), NOS1 (GO: 0099092), CAMKIIB (GO: 0014069), ABI1 (GO: 0014069), and SHANK1 (GO: 0014069). These data suggest miR137 delivered to neurons using LNPs modulates the expression of proteins involved in synaptic glutamate neurotransmission.
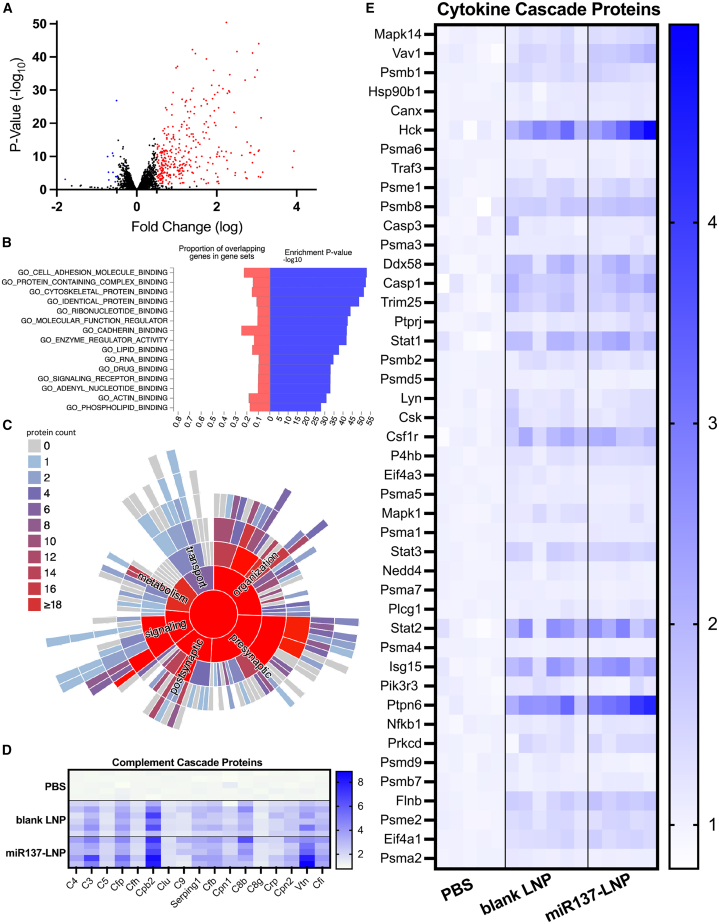
Last, we investigated the effect of blank LNPs on the proteome. Compared to PBS, we found blank LNPs induced differential expression of 1,536 proteins (923 upregulated [red] and 613 downregulated [blue]) (Figure 6A). Based on the FUMA GENE2FUNC34 database, the top GO molecular functions from DEPs following blank LNP treatment included binding of cell adhesion molecules, protein complexes, RNA, and lipids (Figure 6B). Additionally, the SynGo database confirmed the cellular function of DEPs from blank LNPs include synaptic organization, cellular signaling, metabolism, and transport (Figure 6C). This finding was validated though the biological pathway analysis, which determined an upregulation of proteins involved in lipid metabolism (GO: 0006629, GO: 0044255, GO: 0006644), response to lipids (GO: 0033993), and lipid transport (GO: 0006869) (Table S2). The Reactome pathways indicated activation of 44% of the complement (MMU-166658) (Figure 6D) and 16.54% of the cytokine (MMU-1280215) cascades (Figure 6E) (Table S1). It appears the cytokine and complement activation are driven by the LNP itself, as the blank LNP and miR137-LNP treatments show similar trends in protein activation compared to PBS. However, the DEPs involved in these immune pathways only made up 1.45% and 4.14% of the total DEPs in blank LNPs, respectively. To determine if LNPs activate microglia, we quantified five proteins known to be upregulated in activated microglia.38,39 A two-way ANOVA with Sidak’s multiple comparisons found enrichment of activated microglia proteins: cluster of differentiation 44 (CD44) (F (4, 45) = 3.224, p = 0.0004), moesin (MSN) (F (4, 45) = 3.224, p = 0.0036), profilin 1 (PFN1) (F (4, 45) = 3.224, p = 0.6957), myosin heavy chain 9 (MYH9) (F (4, 45) = 3.224, p = 0.0011), and cluster of differentiation 11 (CD11) (F (4, 45) = 3.224, p < 0.0001) after blank LNP treatment compared to PBS (Figure S6C). Of note, this analysis does not differentiate between microglia recruited to sites of damaged tissue due to the injection and cells specifically affected by LNPs. Finally, in agreement with previous proteomic analysis using the cationic DOTAP lipid in the LNP formulation,40 our proteomics data showed elevation of vitronectin, fibrinogen gamma chain, fibrinogen beta chain, clusterin, and alpha-s1-casein. Our mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRoteomics IDEntification (PRIDE) partner repository41 with the dataset identifier PXD041648.
Figure 6.
Proteomic analysis of blank LNPs in the brain
Quantitative proteomics comparing prefrontal cortex biopsies of animals that received PBS or blank LNPs. (A) Volcano plot of DEPs between PBS and blank LNPs (log2fold change and adjusted p values <0.05). (B) Gene Ontology (GO) molecular functions of enriched proteins by count from blank LNPs. (C) Functional enrichment of DEPs from blank LNPs. Heatmap of activated protein expression in PBS, blank LNP, and miR137-LNP groups involved in (D) complement and (E) cytokine cascade pathways normalized to PBS. n = 5–6/group.
Discussion
The risk for developing schizophrenia (SCZ) is linked to SNPs in MIRNA137, which decreases miR137 expression, causing dysregulation of target synaptic proteins.25,26 Many miR137 target transcripts are implicated in genome-wide association studies (GWASs) and linked to an increased risk of SCZ.42 Polygenic disorders such as SCZ may benefit from the therapeutic application of miRNAs as broad-spectrum genetic regulators. Delivering replacement miRNA, or miRNA cocktails designed to tune synaptic function, is a path toward treatment of SCZ. Development of a transportation vehicle is required to protect and deliver therapeutic nucleic acid cargo to the appropriate location and cell type. LNPs are attractive delivery vectors due to their customizable size, charge, cargo loading, high bioavailability, and targeting capability.43,44
Here we evaluated LNP delivery of nucleic acids to the prefrontal cortex (PFC) and the effects of miR137 on protein expression in the mouse brain. After confirming that LNP nanocarriers could successfully deliver miR137 to neuroblastoma cells in vitro, we showed that miR137 modulated downstream synaptic proteins. In follow-up studies in vivo, we administered several types of nucleic acid cargo to the brain, including small molecules (miR137) and large gene constructs (firefly luciferase mRNA and Cre mRNA). We injected LNPs containing firefly luciferase mRNA into the PFC and found that the LNPs remained in the brain. Additional studies found Cre mRNA delivered by LNPs predominantly expressed their cargo in neurons and to a lesser extent in microglia. Last, quantitative proteomic analysis of PFC tissues indicated that LNPs containing miR137 modulated glutamatergic synaptic proteins.
One key aspect of SCZ pathology is glutamatergic synaptic dysfunction in cortical neurons.45,46 Disrupted excitatory neurotransmission due to aberrant expression of miR137 could affect synaptic function and result in the behavioral abnormalities found in SCZ.47 However, no tool currently exists to restore the synaptic glutamatergic circuits to normal levels. Our proteomic analysis determined miR137 delivered by LNPs altered pre-and post-synaptic glutamatergic proteins involved in neurotransmitter and receptor gating, binding, release, trafficking, and glutamate-dependent synaptic plasticity. Interestingly, a subset of the glutamatergic synaptic proteins, whose expression was modulated by miR137, are identified in GWASs that increase the risk for SCZ, including PTPRF, SHANK1, CAMKIIB, C4, GRIK3, and CACNAα1C.42,48,49,50 Unexpectedly, some known targets of miR137 did not respond in vivo as they did in vitro (including TCF4, GluA1, Syn, CPLX1, and FMRP), suggesting that the pattern of synaptic protein modulation in vivo may be different than in vitro. One possibility is that the timing of protein homeostasis in vivo could be driven by exogenously inflating the miRNA machinery.51 Future studies varying the dose or timing of miRNA treatments will be required to establish long-term neural network or behavioral effects.
A complementary hypothesis in SCZ pathology suggests aberrant neuronal glutamate release leads to microglial activation through the complement cascade, implicating the immune system.46 In some individuals, modulation of complement genes may be therapeutic, as mutations in complement C4 genes have been linked to excessive synaptic pruning in SCZ.52,53 We found that ∼15% of the mRNA cargo delivered by LNPs was expressed in microglia, which play a role in synaptic pruning. It is possible the mRNA cargo expressed in microglia may be due to LNP phagocytosis at the injection site. To determine if LNPs are recruiting and activating microglia, future experiments could isolate cells expressing LNP cargo and measure (1) whether the acceptor cells express activated microglial proteins, and (2) whether activated microglia are pro- or anti-inflammatory. Additionally, further studies could analyze microglial morphological changes (e.g., ramified, bushy, ameboid microglia) due to LNP exposure. We found stimulation of cytokine and complement proteins was driven by the LNP itself, although these proteins accounted for less than 5% of all DEPs. Our findings warrant further studies on the long-term biological consequences of the LNP immunogenicity54 in the brain. Current genetic modulators, such as adeno-associated viruses (AAVs), significantly activate the innate immune response,55 but additional studies are required to directly compare LNP immune activation to AAVs. Nonetheless, future research directed at reducing immunogenic responses—for example, by decorating the particle surface with anti-inflammatory agents or endogenous cell membranes from neuronal tissue—would be valuable.56
Gene therapies in lipid-based nanoplatforms are US Food and Drug Administration (FDA) approved to treat hereditary transthyretin-mediated amyloidosis with siRNA57 and SARS-CoV-2 with mRNA vaccines.58 However, only a few studies have evaluated the safety and efficiency of LNP-based RNA therapies for central nervous system disorders. Previous studies have systemically injected selective organ targeting (SORT) LNPs and showed targeted lung, spleen, and liver tropism.59 Improving on first-generation SORT LNPs, we formulated LNPs with a mixture of DOTAP and Dlin-MC3 ionizable lipids and selectively showed high brain expression for ∼24 h with no detectable clearance to systemic organs.31,60,61,62 Our LNPs have distinct onion-like morphologies, possibly due to the cationic lipids, which could be highly solvated and promote the formation of aqueous internal domains. Another possible reason is that the arrangement of concentric phospholipid bilayers enclosing an aqueous core can form an onion-like multilayer structure.63 Maintaining exogenous long-term RNA expression in the brain could require multiple weekly treatments with potential risk for inflammation or cytotoxicity.64 However, our studies with single or multiple doses of miR137-LNP administrations found no toxicity in terms of body weight loss or change in tissue integrity. To design the next generation of noninvasive gene therapies, future studies will investigate the systemic delivery of targeted LNPs to cross the blood-brain barrier (BBB). However, traversing the BBB will require careful evaluation of the risk that off-target tissues could be affected. To target specific cell types, LNPs could be coated with ligands, antibodies, or peptides,65 which would significantly improve drug delivery systems and therapies for brain disorders.
Overall, the evidence presented here encourages further research of miRNAs in pathological conditions. Using delivery of miR137 by LNPs, we have modulated neuronal glutamatergic synaptic protein networks connected to SCZ risk. These studies provide promising support for using LNPs as customizable tools for gene and protein therapies in the brain.
Materials and methods
Materials
Mature mmu-miR137-3p mimic (assay ID MC10513) and miRNA scramble control (4464059) were purchased from Thermo Fisher Scientific (Waltham, MA). Cy5-mmu-miR137-3p was purchased from Creative Biogene (Shirley, NY) and Cy7.5-PEG-LNP Avanti Polar Lipids (Alabaster, AL). Ionizable lipid Dlin-MC3-DMA (MC3) was purchased from BioFine International, (BC, Canada). Cationic lipid DOTAP (chloride salt) and 2-distearoyl-sn-glycero-3-phosphocholine (18:0 PC, DSPC) were acquired from Avanti Polar Lipids (Alabaster, AL). Cholesterol and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2k) were purchased from Sigma Aldrich (St. Louis, MO).
Nanoparticle formulation and characterization
To encapsulate cargo into LNPs, CleanCap Cre mRNA (5-methoxyuridine [5-moU]) (L-7211) and CleanCap firefly luciferase (Fluc) mRNA (Fluc-L-7602) were purchased from Trilink Biotechnologies (San Diego, CA). LNPs were formulated via microfluidic mixing of one part ethanol phase (containing the lipids) and three parts aqueous phase (containing the nucleic acid cargo). The ethanol phase contains the MC3:DOTAP:DSPC:cholesterol:DMG-PEG2k at a molar ratio of 25:50:5:19:1.0, respectively.66,67,68 The aqueous phase consists of the nucleic acid cargo in 50 mM citrate buffer at pH 4. Following microfluidic mixing in the NanoAssmblr Benchtop (product code NIT0055, Precision NanoSystem, BC, Canada) at a 1:3 (ethanol to aqueous) flow ratio, the LNPs were subjected to dialysis for 4 h at room temperature with PBS (pH 7.2) using a 10-kDa Slide-A-Lyzer (Thermo Fisher Scientific, Waltham, MA) dialysis bag before being transferred to fresh PBS solution overnight at 4°C. LNPs were then concentrated using pre-washed Amicon Ultra-15 100k MWCO (EMD Millipore) centrifugal filter tubes (Burlington, MA). The nanoparticles were stored at 4°C and used for in vitro and in vivo studies. The LNPs were characterized for hydrodynamic radius and polydispersity index (PDI) using dynamic light scattering (DLS) (Zetasizer Nano ZSP, Malvern Instruments, Malvern, UK). The cholesterol content of the LNPs was determined using Amplex Red Cholesterol Assay Kit (catalog # A12216, Thermo Fisher Scientific, Waltham MA)69,70 according to manufacturer’s protocol. This assay is based on an enzyme-coupled reaction that detects the free cholesterol. For each sample, background fluorescence was corrected by subtracting the values derived from the no-cholesterol control. The plate was analyzed using Infinite 200 PRO plate reader (TECAN, Männedorf, Switzerland) using an excitation of 560 nm and emission of 590 nm. The calculated total amount of cholesterol in LNPs was 0.45 μg/μL. Therefore theoretically, within the lipid blend, calulated based on the molar percentage used in the formulation,the other lipid concentrations in the final LNP solution will be: 0.99 μg/μL of MC3, 2.15 μg/μL of DOTAP, 0.24 μg/μL of DSPC, and 0.15 μg/μL of DMG-PEG-2k.
Nucleic acid encapsulation efficiency (EE) was determined by RiboGreen assay using a modified Quant-iT RiboGreen RNA reagent (Life Technologies, Carlsbad, CA), which can be used to quantify both mRNA and miRNA. For the quantification process, nucleic acid and LNP stock solutions were appropriately prepared in two distinct buffers: 1× TE buffer and 2% Triton X-100 in TE buffer with RNase-free water. The assay was performed according to the manufacturer’s protocol in a black, clear-bottom, 96-well plate and analyzed using Infinite 200 PRO plate reader (TECAN. Männedorf, Switzerland) at an excitation of 480 nm and emission of 520 nm. The background fluorescence was subtracted from each sample using PBS control during analysis. The nucleic acid concentration was determined by fitting our LNP sample data to the nucleic acid standard curve. The percentage EE for each sample was calculated using the following equation:
Cryo-TEM
Cryo-TEM images were captured using Falcon III and K3 Summit cameras (Gatan, Pleasanton, CA) with DED at 300 kV. A copper lacey carbon film-coated cryo-EM grid (Quantifoil, R1.2/1.3 300 Cu mesh) was plunge-frozen using the Vitrobot Mark IV system (FEI, Waltham, MA). To freeze the samples, 2 μL of LNPs were dispensed onto the glow-discharged grids in the Vitrobot chamber, which was maintained at 23°C and 100% relative humidity. After 30 s, the samples were blotted with filter paper for 3 s before being immersed in liquid ethane cooled by liquid nitrogen. The frozen grids were carefully examined for flaws and then clipped and assembled into cassettes. The images were captured at a nominal magnification of 45,000 at an electron dose of 15–20 e−/Å2 then processed and analyzed using Fiji (ImageJ 1.53t, National Institute of Health, USA).
Cell culture
Neuro2A (N2A) cells (ATCC, CCL-131) are neuroblasts with neuronal and ameboid stem cell morphology isolated from mouse brain tissue. N2A cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FetalClone serum (Thermo Fisher Scientific, Waltham, MA) and maintained in a humidified incubator with 10% CO2. Two days before the treatment, cells were plated in 12-well plates with 1 × 106 cells/well. Cells were treated with a final concentration of 200 nM miR137-3p mimic or 200 nM miRNA scramble control with equal volume of Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA) in 0.5 μL of Opti-Mem medium (Thermo Fisher Scientific, Waltham, MA). When cells were treated with LNPs, miR137-3p mimic was loaded into the LNPs and applied to cells to a final concentration of 200 nM. In all conditions, after 24 h, the medium was replaced with complete DMEM medium, and the cells were harvested 24, 48, 72, or 96 h after treatment.
Toxicity
In vitro cell toxicity after LNP treatment was determined by MTT cell proliferation kit and processed according to manufacturer’s protocol (AB211091, Abcam, Cambridge, UK). In vivo LNP toxicity was determined by weighing animals pre and post LNP injection as well as by total animal survival rate.
RNA isolation, cDNA, RT-qPCR
Live Neuro2A cells or flash frozen brain tissue was cut at 300 μm on a cryostat and 1 mm bilateral prefrontal cortex or cerebellar biopsy punches (Ted Pella, Redding, CA, USA) were homogenized mechanically using a pellet pestle motor (DWK Life Sciences, Millville, NJ). Total RNA was extracted following manufacturers protocol using the MagMax mirVana miRNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA). Total RNA concentration and quality was measured on a NanoDrop spectrophotometer, and 10 ng of RNA was transcribed to cDNA using specific mmu-miR137-3p and Sno202 RNA (endogenous control) primers with MultiScribe Reverse Transcriptase (Applied Biosystems, Waltham, MA). Samples were run in duplicated with universal TaqMan real-time qPCR (StepOnePlus, Applied Biosystems, Waltham, MA) and fold change levels of miR137 were analyzed using the delta-delta Ct calculation procedure (Applied Biosystems, Waltham, MA). Statistical comparisons were made using a two-way ANOVA followed by Sidak’s multiple comparisons post hoc test in GraphPad Prism 9.5.0. A p value <0.05 was considered significant.
Immunoblotting
Neuro2A cell samples were lysed and homogenized in 1× RIPA buffer containing 1× protease inhibitor (Roche, Branchburg, NJ) and then centrifuged at 14,500 × g for 10 min at 4°C. Supernatants were collected for use in the BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). Ten milligrams of protein for each sample was subjected to SDS-PAGE through a 12% Bis-Tris precast gel (Bio-Rad, Hercules, CA) against Precision Plus Protein Dual Color Standards and run at 180 V for 1 h with XT MOPS (Bio-Rad) running buffer. The gel was transferred onto a polyvinylidene fluoride (PVDF) membrane with transfer buffer containing 10% methanol at 30 mV overnight at 4°C then washed three times for 5 min with Tris-buffered saline with 0.05% Tween (TBST). Membranes were blocked with 5% nonfat dry milk in TBST at room temperature for 30 min. The blots were incubated overnight at 4°C with primary antibodies diluted in TBST: rabbit anti-GluA1 (1:250, AB1504, Millipore Sigma, Burlington, MA), rabbit anti-β-actin (1:1,000, AB6276, Abcam, Cambridge, UK), rabbit anti-PSD95 (1:1,000, AB238135, Abcam, Cambridge, UK), rabbit anti-TCF4 (1:1,000, 185736, Abcam, Cambridge, UK), mouse anti-SYT1 (1:1,000, 105011, Synaptic Systems, Gottingen, Germany), mouse anti-CACNA1C (1:1,000, AC84814, Abcam, Cambridge, UK), rabbit anti-CPLX1 (1:1,000, 10246-2-AP, ProteinTech, Rosemont, IL), and rabbit anti-FMRP (1:1,000, AB17722, Abcam, Cambridge, UK). The next day, after washing 3 times for 10 min with TBST, the blots were incubated with donkey anti-mouse or anti-rabbit horseradish peroxidase (HRP) secondary antibody (1:10,000, Jackson ImmunoResearch, West Grove, PA) in 5% milk in TBST solution for 1 h at room temperature. After antibody incubation, the blots were washed again three times for 5 min with TBST and visualized using Clarity Western ECL Substrate (Bio-Rad, Hercules, CA) and the ChemiDoc MP Imaging detection system (Bio-Rad). Bands were analyzed by densitometry using ImageJ (National Institutes of Health, USA). The proteins of interest were normalized to β-actin as an internal control. Statistical analysis comparing relative protein expression from each treatment group was performed using a two-way ANOVA in GraphPad Prism 9.5.0. A p value <0.05 was considered significant.
Animals
Male and female C57BL/6J or Ai9 mice (strain #007909) (Jackson Labs, Bar Harbor, ME) were maintained on a 12-h light/12-h dark cycle with lights on at 7 am. Food and water were available ad libitum. All experimental procedures followed the protocols approved by the Institutional Animal Care and Use Committee at Oregon Health & Science University and the Veterans Affairs Portland Health Care System Institutional Animal Care and Use Committee. Mice were deeply anesthetized with 3% vaporized isoflurane in oxygen (1 L/min) until sedated and sacrificed using rapid cervical dislocation, and brains were rapidly extracted, flash frozen with liquid nitrogen, and stored at −80°C for further processing.
Injections
Mice were deeply anesthetized with 3% vaporized isoflurane in oxygen (1 L/min) until sedated and placed in a stereotaxic apparatus. For the duration of the surgery, isoflurane was maintained at 1%, and mice were kept warm on a heating pad. Carprofen (0.11 mL/25 g body weight) and dexium (0.01 mL/1 g body weight) were injected subcutaneously before the surgery. Lidoject (0.2 mL/incision) was administered at the surgery site before making an incision. Mice were injected with LNPs containing firefly luciferase mRNA (0.0023 mg/kg, L-7602, Trilink Biotechnologies), mmu-miR137-3p (0.0018 mg/kg, Life Technologies, Carlsbad, CA), or Cre mRNA (0.0014 mg/kg, L-7211,Trilink Biotechnologies, San Diego, CA) at three locations within the medial PFC bilaterally (±0.4 medial/lateral; +1.94 anterior/posterior; 1.0, 1.6, and 2.6 depth below brain surface; 0.5 μL of nanoparticle per injection site), at a rate of 100 nL per minute.
In vivo bioluminescent imaging and quantification
Mice were injected intraperitoneally with 150 mg of Pierce D-Luciferin (Thermo Fisher Scientific, Waltham, MA) per kilogram of body weight according to the manufacturer’s protocol. Bioluminescent imaging was conducted on the same animals at 1, 3, 5, 7, 9, 24, 48, and 96 h post LNP injection on the IVIS Spectrum In Vivo Imaging System (PerkinElmer, Waltham, MA). The mouse skull was shaved prior to surgery, but no other parts of the animal required shaving. Image analysis for region of interest (ROI) measurement was performed on Living Image Software (PerkinElmer, Waltham, MA) and was reported as average radiance (the sum of the radiance from each pixel inside the ROI/number of pixels or super pixels; photons/s/cm2/sr). Statistical analysis comparing average radiance of fLuc in the brain and the liver at each time point was performed using a two-way ANOVA in GraphPad Prism 9.5.0. A p value <0.05 was considered significant.
Immunohistochemistry
Brains were extracted and immediately post-fixed overnight in 4% paraformaldehyde in 1× PBS then cryoprotected in 30% sucrose in 1× PBS until the tissue sank. Then 40-μm sections were cut on a cryostat and free-floating sections were stored in 1× PBS at 4°C until used for immunohistochemistry experiments. Sections were incubated with Tris-EDTA for 20 min at room temperature then permeabilized three times for 5 min with 1× PBS and 0.3% Triton X-. Tissue was then blocked in 1× PBS, 0.3% Triton-X, and 5% normal goat serum for 1 h at room temperature. Primary antibodies (mouse anti-NeuN, 1:1,000, AB104224, Abcam, Cambridge, UK) and rabbit anti-Iba1 (1:1,000, AB178846, Abcam, Cambridge, UK) were added to 1× PBS and 0.3% Triton-X and incubated overnight at 4°C. The next day, tissue was washed in 1× PBS and 0.3% Triton-X three times for 5 min and secondary antibodies (1:1,000, Alexa 488 goat anti-rabbit, AB150077, Abcam, Cambridge, UK) and Alexa 647 goat anti-mouse (1:1,000, AB150115, Abcam, Cambridge, UK) were added for 2 h at room temperature. Sections were washed with 1× PBS, mounted, coverslipped, and dried overnight before imaging. Negative control samples were run with no primary antibodies.
Neuronal cell measurements
Mice (n = 2/treatment) were treated every day for 5 days with PBS or miR137-LNPs. On day 8, brains were extracted and immediately post-fixed in 4% paraformaldehyde then cryoprotected in 30% sucrose. Then 40-μm sections were cut on a cryostat and free-floating sections were processed based on manufacturer’s protocol using a hematoxylin and eosin staining kit (Vector Laboratories, Newark, CA).71 Prefrontal cortex tissues sections were imaged on a Keyence BZ-X800 fluorescent microscope. Three sections per animal were analyzed for neuronal cell density based on the prominent nucleus and pale cytoplasm staining. An unpaired t test was performed using mean neuronal cell densities in PBS or miR137-LNP animals by area field of view (μm2).
Image analysis
For in vitro live-cell imaging, cells were washed with PBS and imaged on a Keyence BZ-X800 fluorescent microscope (Itasca, IL) at 37°C with 10% humidity and 5% CO2 and imaged at 2, 4, 6, 24, and 48 h post treatment. For immunohistochemistry imaging, slides were imaged on a Zeiss ApoTome.2 Axio Imager (Oberkochen, Germany) with standardized exposure levels for each fluorescent channel. The ApoTome imaging system generates three fluorescent images in one optical section, which helps maintain higher homogeneous resolution across the field of view compared to a laser scanning confocal microscope.72 Four animals with six images per animal were analyzed for each condition. All images were quantified at 20× magnification. Due to the potential microglial or astrocyte recruitment to the needle tract injection site, the brain tissue directly adjacent (100–200 μm)31 to the injection site was quantified so overlapping cell type expression would not be skewed. The expression of NeuN (a neuron-specific nuclear marker), Iba1 (a microglia-specific protein marker), GFAP (an astrocyte-specific cell body and extension marker), and tdTomato (a cytoplasmic reporter protein specific to cells where CremRNA is translated) was determined as overlapping if the same cell labeled one or more indicated cellular markers. Co-expression was determined by separating each channel and counting fluorescence that overlapped for each cell. Using GraphPad Prism 9.5.0, an ordinary one-way ANOVA with a post hoc Tukey’s multiple comparisons test was used to compare conditions. A p value <0.05 was considered significant.
Quantitative proteomics sample preparation
Mass spectrometric analysis was performed by the OHSU Proteomics Shared Resource. Whole-brain mouse tissue samples were flash frozen 72 h after treatment (PBS n = 6, blank LNP n = 6, miR137-LNP n = 6; three males/three females per group). Blank LNPs include all the same lipid components as the miR137-LNPs but do not contain any nucleic acid. The brain tissue was cut at 300 μm on a cryostat and 1-mm bilateral prefrontal cortex (Ted Pella, Redding, CA) samples were homogenized in 200 μL of 5% SDS, 50 mM triethyl ammonium bicarbonate buffer using Bioruptor Pico and heated to 95°C for 5 min. Samples were cooled and centrifuged for 5 min at 16,000 × g and supernatant transferred to an Eppendorf LoBind tube. Protein concentrations were determined using the Pierce BCA assay. Fifty micrograms per sample was digested using trypsin (1:16, trypsin:protein) overnight using S-Trap micro cartridge (Protifi, Farmingdale, NY) after reduction with dithiothreitol and alkylation with iodoacetamide. Peptides were eluted from the S-Trap using 40 μL each of 50 mM TEAB, 0.2% aqueous formic acid, and 50% acetonitrile (ACN) containing 0.2% formic acid, then pooled and dried in a speedvac. Each sample was suspended in 100 μL of high-pressure liquid chromatography (HPLC) water and a peptide assay was done using Pierce Quantitative Colorimetric Peptide Assay Kit (Thermo Fisher Scientific, Waltham, MA).
TMT labeling and mass spectrometric analysis
In preparation for tandem mass tag (TMT) labeling, 18 dried unfractionated peptide samples (15 μg/sample) were dissolved in 20 μL of 100 mM triethylammonium bicarbonate buffer, and TMT 18-plex reagents (Thermo Fisher Scientific, Waltham, MA) were dissolved at a concentration of 9.6 μg/μL in anhydrous ACN. Each of the samples was then labeled by adding 12 μL (115 μg) of an individual TMT reagent, followed by shaking at room temperature for 1 h. Two microliters of each labeled sample in each group were then pooled, 2 μL of 5% hydroxylamine added, and the samples were incubated for 15 min at room temperature, dried by vacuum centrifugation, dissolved in 33.8 μL of 5% formic acid, and 2 μg of peptides were analyzed by a single 40-min liquid chromatography-tandem mass spectrometry (LC-MS/MS) method using an Orbitrap Fusion as described below. This run was performed to normalize the total reporter ion intensity of each multiplexed sample and to check labeling efficiency. After the normalization and efficiency run, the remaining unmixed samples were then combined in adjusted volumes to yield equal summed reporter ion intensities during the subsequent two-dimensional liquid chromatography/mass spectrometry analysis (2DLC/MS). Following volume-based normalization, the combined samples (45.6 μg) were dried by vacuum centrifugation, and TMT-labeled samples were reconstituted in 40 μL of 10 mM ammonium formate, pH 9, and separated by two-dimensional nano reverse-phase liquid chromatography/mass spectrometry (2D-LC/MS) using a Dionex NCS-3500RS UltiMate RSLCnano ultra-performance liquid chromatography (UPLC) and Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific, Waltham, MA). TMT-labeled peptides were eluted from the first-dimension high-pH column using sequential injections of 20-μL volumes of 17%, 20%, 21%, 22%, 23%, 24%, 25%, 26%, 27%, 28%, 29%, 30%, 31%, 32%, 33%, 34%, 35%, 40%, 50%, and 90% ACN in 10 mM ammonium formate (pH 9) at a 3 μL/min flow rate. Eluted peptides were diluted at a tee with a mobile phase containing 0.1% formic acid at a 24 μL/min flow rate. Peptides were delivered to an Acclaim PepMap 100 μm × 2 cm NanoViper C18, 5-μm trap on a switching valve. After 10 min of loading, the trap column was switched on-line to a PepMap RSLC C18, 2 μm, 75 μm × 25 cm EasySpray column (Thermo Scientific, Waltham, MA). Peptides were then separated at low pH in the second dimension using a 5%–25% ACN gradient over 100 min in mobile phase containing 0.1% formic acid at a 300 nL/min flow rate. Tandem mass spectrometry data were collected using an Orbitrap Fusion Tribrid instrument (Thermo Scientific, Waltham, MA). Peptides were separated using the instrument’s EasySpray NanoSource, survey scans performed in the Orbitrap mass analyzer, and data-dependent MS2 scans performed in the linear ion trap using collision-induced dissociation following isolation with the instrument’s quadrupole. Reporter ion detection was performed in the Orbitrap mass analyzer using MS3 scans following synchronous precursor isolation in the linear ion trap, and higher-energy collisional dissociation in the instrument’s ion-routing multipole.
Proteomic data analysis
The protein sequences searched were canonical mouse reference FASTA sequences (21,968 proteins) downloaded on December 26, 2022, from www.UniProt.org. Common contaminants (175 sequences) were added, and sequence-reversed entries were concatenated for a final protein FASTA file of 44,286 sequences.
The 20-fraction TMT 18-plex sample produced 20 instrument files that were processed with the PAW pipeline73 (https://github.com/pwilmart/PAW_pipeline). Binary files were converted to text files using MSConvert.74 Python scripts extracted TMTpro reporter ion peak heights and fragment ion spectra in MS2 format.75 There were 334,353 linked MS2/MS3 scans acquired. The Comet search engine (version 2016.03)76 was used: 1.25-Da monoisotopic peptide mass tolerance, 1.0005-Da monoisotopic fragment ion tolerance, fully tryptic cleavage with up to two missed cleavages, variable oxidation of methionine residues, static alkylation of cysteines, and static modifications for TMTpro labels (at peptide N-termini and at lysine residues).
Top-scoring peptide spectrum matches (PSMs) were filtered to a 1% false discovery rate (FDR) using an interactive graphical user interface application to set thresholds in delta-mass histograms and conditional peptide-prophet-like linear discriminant function77 score histograms where incorrect delta-mass and score histogram distributions were estimated using the target/decoy method.78 The 115,725 filtered PSMs were assembled into protein lists using basic and extended parsimony principles and required two distinct peptides per protein per plex. The final list of identified proteins, protein groups (indistinguishable peptide sets), and protein families (highly homologous peptide sets) were used to define unique and shared peptides for quantitative use. Total (summed) reporter ion intensities were computed from the PSMs associated with all unique peptides (final grouped protein context) for each protein.
The protein intensity values for each biological sample in each biological condition were compared for differential protein expression using the Bioconductor package edgeR79 within Jupyter notebooks. Result tables contained typical proteomics summaries, reporter ion intensities, and statistical testing results. Further investigation comparing proteomic distributions across biological groups determined two outlier samples (PBS, miR137-LNP). A Grubb’s test was used to determine putative sample outliers based on the relative protein abundance levels of housekeeping genes (β-tubulin, GAPDH, DJ-1, actin).80 Subsequent data were interpreted with the removal of these two samples from further analysis. A two-sample t test was used to compare abundance differences of individual a priori proteins of interest between biological conditions.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRoteomics IDEntification (PRIDE) repository41 with the dataset identifier PXD041648.
Biological network analysis
Following tandem TMT mass spectrometry analysis of samples treated with miR137-LNP, blank LNPs, or PBS (n = 5–6/group), public databases were used to determine differentially expressed protein cellular location and biological process enrichment. The STRING 11.532 database creates networks of known and predicted protein-protein interactions from over 24.5 million proteins. Following input of DEPs, we sought to determine network scale protein enrichment in functional classifications defined by GO, including biological process (https://www.informatics.jax.org/vocab/gene_ontology/GO:0008150), subcellular components (https://www.informatics.jax.org/vocab/gene_ontology/GO:0005575), and molecular function (https://www.informatics.jax.org/vocab/gene_ontology/GO:0003674). Similarly, SynGO33 was used to investigate subcellular compartmentalization of DEPs and where genes of interest are translated, either in the neuropil or the soma.36 Finally, when comparing DEPs after miR137 treatment to GWAS hits for SCZ risk, the FUMA 1.5.3 GENE2FUNC was cross-referenced.34
Data and code availability
All data associated with this study are available in the main text or the supplemental material. All quantitative proteomics data have been submitted to the ProteomeXchange Consortium via the PRIDE partner repository41 with the dataset identifier PXD041648.
Acknowledgments
This work was supported by funding from NIDA ADA 12013 (A.J.), FDA CDER-20-I-0546 (A.J.), DEA D-15-OD-002 (A.J.), NIAAA T32 AA007468 (A.S.), Department of Veterans Affairs 1IK6BX005754 (A.J.), NHLBI 1RO1HL146736-05 (G.S.), OHSU Physician Scientist Grant (A.I.A), and with partial support from NIH core and OHSU Knight Center Institute NCI Cancer Center Support grants P30EY010572 (A.P.R.), P30CA069533 (A.P.R.), S10OD012246 (A.P.R.). Illustrations were created with BioRender.com.
Author contributions
Conceptualization, M.C.P., M.G., A.I.A., D.L.M., G.S. and A.J.; investigation and methodology, M.C.P., M.G., K.K., P.A.W., A.P.R., A.S., X.S., D.L.M., A.I.A., G.S., and A.J.; visualization, M.C.P. and M.G.; resources, A.I.A., D.L.M., G.S., and A.J.; writing – original draft, M.C.P.; writing – review & editing, M.C.P., M.G., K.K., P.A.W., A.S., A.P.R., X.S., A.I.A., D.L.M., G.S., and A.J.; supervision, A.I.A., D.L.M., G.S., and A.J.; funding acquisition, A.J., A.S., A.I.A., A.P.R., and G.S.
Declaration of interests
D.L.M. is a consultant for Pfizer Inc. and for Endevica Bio, Inc. D.L.M. has received grants, holds equity, and is Chief Medical Officer for Endevica Bio, Inc. G.S. is a cofounder of EnterX Bio and has equity in the company.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2023.08.016.
Supplemental information
References
- 1.Catalanotto C., Cogoni C., Zardo G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 3.Friedman R.C., Farh K.K.-H., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollins S.L., Goldie B.J., Carroll A.P., Mason E.A., Walker F.R., Eyles D.W., Cairns M.J. Ontogeny of small RNA in the regulation of mammalian brain development. BMC Genomics. 2014;15:777. doi: 10.1186/1471-2164-15-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willemsen M.H., Vallès A., Kirkels L.A.M.H., Mastebroek M., Olde Loohuis N., Kos A., Wissink-Lindhout W.M., de Brouwer A.P.M., Nillesen W.M., Pfundt R., et al. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J. Med. Genet. 2011;48:810–818. doi: 10.1136/jmedgenet-2011-100294. [DOI] [PubMed] [Google Scholar]
- 6.Kuswanto C.N., Sum M.Y., Qiu A., Sitoh Y.-Y., Liu J., Sim K. The impact of genome wide supported microRNA-137 (MIR137) risk variants on frontal and striatal white matter integrity, neurocognitive functioning, and negative symptoms in schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2015;168B:317–326. doi: 10.1002/ajmg.b.32314. [DOI] [PubMed] [Google Scholar]
- 7.He E., Lozano M.A.G., Stringer S., Watanabe K., Sakamoto K., den Oudsten F., Koopmans F., Giamberardino S.N., Hammerschlag A., Cornelisse L.N., et al. MIR137 schizophrenia-associated locus controls synaptic function by regulating synaptogenesis, synapse maturation and synaptic transmission. Hum. Mol. Genet. 2018;27:1879–1891. doi: 10.1093/hmg/ddy089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng Y., Wang Z.-M., Tan W., Wang X., Li Y., Bai B., Li Y., Zhang S.-F., Yan H.-L., Chen Z.-L., et al. Partial loss of psychiatric risk gene miR-137 in mice causes repetitive behavior and impairs sociability and learning via increased Pde10a. Nat. Neurosci. 2018;21:1689–1703. doi: 10.1038/s41593-018-0261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao T., Zhen X.-C. Dysregulation of miRNA and its potential therapeutic application in schizophrenia. CNS Neurosci. Ther. 2018;24:586–597. doi: 10.1111/cns.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arakawa Y., Yokoyama K., Tasaki S., Kato J., Nakashima K., Takeyama M., Nakatani A., Suzuki M. Transgenic mice overexpressing miR-137 in the brain show schizophrenia-associated behavioral deficits and transcriptome profiles. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0220389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilker R., Helenius D., Fagerlund B., Skytthe A., Christensen K., Werge T.M., Nordentoft M., Glenthøj B. Heritability of Schizophrenia and Schizophrenia Spectrum Based on the Nationwide Danish Twin Register. Biol. Psychiatry. 2018;83:492–498. doi: 10.1016/j.biopsych.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 12.Abbas A.I., Sundiang M.J.M., Henoch B., Morton M.P., Bolkan S.S., Park A.J., Harris A.Z., Kellendonk C., Gordon J.A. Somatostatin Interneurons Facilitate Hippocampal-Prefrontal Synchrony and Prefrontal Spatial Encoding. Neuron. 2018;100:926–939.e3. doi: 10.1016/j.neuron.2018.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Javitt D.C., Siegel S.J., Spencer K.M., Mathalon D.H., Hong L.E., Martinez A., Ehlers C.L., Abbas A.I., Teichert T., Lakatos P., Womelsdorf T. A roadmap for development of neuro-oscillations as translational biomarkers for treatment development in neuropsychopharmacology. Neuropsychopharmacol. 2020;45:1411–1422. doi: 10.1038/s41386-020-0697-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.American Psychiatric Association, and American Psychiatric Association eds; 2013. Diagnostic and statistical manual of mental disorders. [Google Scholar]
- 15.OBI-NAGATA K., TEMMA Y., HAYASHI-TAKAGI A. Synaptic functions and their disruption in schizophrenia: From clinical evidence to synaptic optogenetics in an animal model. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2019;95:179–197. doi: 10.2183/pjab.95.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin J., Lin J., Luo X., Chen Y., Li Z., Ma G., Li K. miR-137: a new player in schizophrenia. Int. J. Mol. Sci. 2014;15:3262–3271. doi: 10.3390/ijms15023262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olde Loohuis N.F.M., Nadif Kasri N., Glennon J.C., van Bokhoven H., Hébert S.S., Kaplan B.B., Martens G.J.M., Aschrafi A. The Schizophrenia Risk Gene MIR137 Acts as a Hippocampal Gene Network Node Orchestrating the Expression of Genes Relevant to Nervous System Development and Function. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2017;73:109–118. doi: 10.1016/j.pnpbp.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakamoto K., Crowley J.J. A comprehensive review of the genetic and biological evidence supports a role for MicroRNA-137 in the etiology of schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2018;177:242–256. doi: 10.1002/ajmg.b.32554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegert S., Seo J., Kwon E.J., Rudenko A., Cho S., Wang W., Flood Z., Martorell A.J., Ericsson M., Mungenast A.E., Tsai L.H. The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nat. Neurosci. 2015;18:1008–1016. doi: 10.1038/nn.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maury E.A., Sherman M.A., Genovese G., Gilgenast T.G., Kamath T., Burris S.J., Rajarajan P., Flaherty E., Akbarian S., Chess A., et al. Schizophrenia-associated somatic copy-number variants from 12,834 cases reveal recurrent NRXN1 and ABCB11 disruptions. Cell Genom. 2023;3:100356. doi: 10.1016/j.xgen.2023.100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schizophrenia Psychiatric Genome-Wide Association Study GWAS Consortium. Sanders A.R., Kendler K.S., Levinson D.F., Sklar P., Holmans P.A., Lin D.-Y., Duan J., Ophoff R.A., Andreassen O.A., et al. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ripke S., O’Dushlaine C., Chambert K., Moran J.L., Kähler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M., et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cummings E., Donohoe G., Hargreaves A., Moore S., Fahey C., Dinan T.G., McDonald C., O’Callaghan E., O’Neill F.A., Waddington J.L., et al. Mood congruent psychotic symptoms and specific cognitive deficits in carriers of the novel schizophrenia risk variant at MIR-137. Neurosci. Lett. 2013;532:33–38. doi: 10.1016/j.neulet.2012.08.065. [DOI] [PubMed] [Google Scholar]
- 24.Lett T.A., Chakravarty M.M., Felsky D., Brandl E.J., Tiwari A.K., Gonçalves V.F., Gonçalves V.F., Daskalakis Z.J., Meltzer H.Y., Lieberman J.A., et al. The genome-wide supported microRNA-137 variant predicts phenotypic heterogeneity within schizophrenia. Mol. Psychiatry. 2013;18:443–450. doi: 10.1038/mp.2013.17. [DOI] [PubMed] [Google Scholar]
- 25.van Erp T.G.M., Guella I., Vawter M.P., Turner J., Brown G.G., McCarthy G., Greve D.N., Glover G.H., Calhoun V.D., Lim K.O., et al. Schizophrenia miR-137 locus risk genotype is associated with dorsolateral prefrontal cortex hyperactivation. Biol. Psychiatry. 2014;75:398–405. doi: 10.1016/j.biopsych.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guella I., Sequeira A., Rollins B., Morgan L., Torri F., van Erp T.G.M., Myers R.M., Barchas J.D., Schatzberg A.F., Watson S.J., et al. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J. Psychiatr. Res. 2013;47:1215–1221. doi: 10.1016/j.jpsychires.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J., Eygeris Y., Gupta M., Sahay G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021;170:83–112. doi: 10.1016/j.addr.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizk M., Tüzmen Ş. Update on the clinical utility of an RNA interference-based treatment: focus on Patisiran. Pharmgenomics. Pers. Med. 2017;10:267–278. doi: 10.2147/PGPM.S87945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiménez-Morales J.M., Hernández-Cuenca Y.E., Reyes-Abrahantes A., Ruiz-García H., Barajas-Olmos F., García-Ortiz H., Orozco L., Quiñones-Hinojosa A., Reyes-González J., Abrahantes-Pérez M.D.C. MicroRNA delivery systems in glioma therapy and perspectives: A systematic review. J. Control Release. 2022;349:712–730. doi: 10.1016/j.jconrel.2022.07.027. [DOI] [PubMed] [Google Scholar]
- 30.Liu S., Grigoryan M.M., Vasilevko V., Sumbria R.K., Paganini-Hill A., Cribbs D.H., Fisher M.J. Comparative Analysis of H&E and Prussian Blue Staining in a Mouse Model of Cerebral Microbleeds. J. Histochem. Cytochem. 2014;62:767–773. doi: 10.1369/0022155414546692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rungta R.L., Choi H.B., Lin P.J., Ko R.W., Ashby D., Nair J., Manoharan M., Cullis P.R., Macvicar B.A. Lipid Nanoparticle Delivery of siRNA to Silence Neuronal Gene Expression in the Brain. Mol. Ther. Nucleic Acids. 2013;2:e136. doi: 10.1038/mtna.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A.L., Fang T., Doncheva N.T., Pyysalo S., et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koopmans F., van Nierop P., Andres-Alonso M., Byrnes A., Cijsouw T., Coba M.P., Cornelisse L.N., Farrell R.J., Goldschmidt H.L., Howrigan D.P., et al. SynGO: An Evidence-Based, Expert-Curated Knowledge Base for the Synapse. Neuron. 2019;103:217–234.e4. doi: 10.1016/j.neuron.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semple B.D., Blomgren K., Gimlin K., Ferriero D.M., Noble-Haeusslein L.J. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 2013;106–107:1–16. doi: 10.1016/j.pneurobio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glock C., Biever A., Tushev G., Nassim-Assir B., Kao A., Bartnik I., Tom Dieck S., Schuman E.M. The translatome of neuronal cell bodies, dendrites, and axons. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2113929118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olde Loohuis N.F.M., Ba W., Stoerchel P.H., Kos A., Jager A., Schratt G., Martens G.J.M., van Bokhoven H., Nadif Kasri N., Aschrafi A. MicroRNA-137 Controls AMPA-Receptor-Mediated Transmission and mGluR-Dependent LTD. Cell Rep. 2015;11:1876–1884. doi: 10.1016/j.celrep.2015.05.040. [DOI] [PubMed] [Google Scholar]
- 38.Rayaprolu S., Gao T., Xiao H., Ramesha S., Weinstock L.D., Shah J., Duong D.M., Dammer E.B., Webster J.A., Lah J.J., et al. Flow-cytometric microglial sorting coupled with quantitative proteomics identifies moesin as a highly-abundant microglial protein with relevance to Alzheimer’s disease. Mol. Neurodegener. 2020;15:28. doi: 10.1186/s13024-020-00377-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jurga A.M., Paleczna M., Kuter K.Z. Overview of General and Discriminating Markers of Differential Microglia Phenotypes. Front. Cell. Neurosci. 2020;14:198. doi: 10.3389/fncel.2020.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dilliard S.A., Cheng Q., Siegwart D.J. On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2109256118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D.J., Prakash A., Frericks-Zipper A., Eisenacher M., et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coyle J.T., Ruzicka W.B., Balu D.T. Fifty Years of Research on Schizophrenia: The Ascendance of the Glutamatergic Synapse. AJP. 2020;177:1119–1128. doi: 10.1176/appi.ajp.2020.20101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel S., Ryals R.C., Weller K.K., Pennesi M.E., Sahay G. Lipid nanoparticles for delivery of messenger RNA to the back of the eye. J. Control Release. 2019;303:91–100. doi: 10.1016/j.jconrel.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herrera-Barrera M., Ryals R.C., Gautam M., Jozic A., Landry M., Korzun T., Gupta M., Acosta C., Stoddard J., Reynaga R., et al. Peptide-guided lipid nanoparticles deliver mRNA to the neural retina of rodents and nonhuman primates. Sci. Adv. 2023;9 doi: 10.1126/sciadv.add4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uno Y., Coyle J.T. Glutamate hypothesis in schizophrenia. Psychiatry Clin. Neurosci. 2019;73:204–215. doi: 10.1111/pcn.12823. [DOI] [PubMed] [Google Scholar]
- 46.Parellada E., Gassó P. Glutamate and microglia activation as a driver of dendritic apoptosis: a core pathophysiological mechanism to understand schizophrenia. Transl Psychiatry. 2021;11:1–13. doi: 10.1038/s41398-021-01385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forsyth J.K., Lewis D.A. Mapping the Consequences of Impaired Synaptic Plasticity in Schizophrenia through Development: An Integrative Model for Diverse Clinical Features. Trends Cogn. Sci. (Regul. Ed. 2017;21:760–778. doi: 10.1016/j.tics.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trubetskoy V., Pardiñas A.F., Qi T., Panagiotaropoulou G., Awasthi S., Bigdeli T.B., Bryois J., Chen C.-Y., Dennison C.A., Hall L.S., et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–508. doi: 10.1038/s41586-022-04434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh T., Neale B.M., Daly M.J., Consortium, on behalf of the S.E.M.-A. (SCHEMA) 2020. Exome Sequencing Identifies Rare Coding Variants in 10 Genes Which Confer Substantial Risk for Schizophrenia. [DOI] [Google Scholar]
- 50.Fromer M., Pocklington A.J., Kavanagh D.H., Williams H.J., Dwyer S., Gormley P., Georgieva L., Rees E., Palta P., Ruderfer D.M., et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cursons J., Pillman K.A., Scheer K.G., Gregory P.A., Foroutan M., Hediyeh-Zadeh S., Toubia J., Crampin E.J., Goodall G.J., Bracken C.P., Davis M.J. Combinatorial Targeting by MicroRNAs Co-ordinates Post-transcriptional Control of EMT. Cell Syst. 2018;7:77–91.e7. doi: 10.1016/j.cels.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 52.Woo J.J., Pouget J.G., Zai C.C., Kennedy J.L. The complement system in schizophrenia: where are we now and what’s next? Mol. Psychiatry. 2020;25:114–130. doi: 10.1038/s41380-019-0479-0. [DOI] [PubMed] [Google Scholar]
- 53.Yilmaz M., Yalcin E., Presumey J., Aw E., Ma M., Whelan C.W., Stevens B., McCarroll S.A., Carroll M.C. Overexpression of schizophrenia susceptibility factor human complement C4A promotes excessive synaptic loss and behavioral changes in mice. Nat. Neurosci. 2021;24:214–224. doi: 10.1038/s41593-020-00763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohamed M., Abu Lila A.S., Shimizu T., Alaaeldin E., Hussein A., Sarhan H.A., Szebeni J., Ishida T. PEGylated liposomes: immunological responses. Sci. Technol. Adv. Mater. 2019;20:710–724. doi: 10.1080/14686996.2019.1627174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muhuri M., Maeda Y., Ma H., Ram S., Fitzgerald K.A., Tai P.W., Gao G. Overcoming innate immune barriers that impede AAV gene therapy vectors. J. Clin. Invest. 2021;131 doi: 10.1172/JCI143780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han Y., Gao C., Wang H., Sun J., Liang M., Feng Y., Liu Q., Fu S., Cui L., Gao C., et al. Macrophage membrane-coated nanocarriers Co-Modified by RVG29 and TPP improve brain neuronal mitochondria-targeting and therapeutic efficacy in Alzheimer’s disease mice. Bioact. Mater. 2021;6:529–542. doi: 10.1016/j.bioactmat.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang J. Patisiran for the treatment of hereditary transthyretin-mediated amyloidosis. Expert Rev. Clin. Pharmacol. 2019;12:95–99. doi: 10.1080/17512433.2019.1567326. [DOI] [PubMed] [Google Scholar]
- 58.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021;601 doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Campani V., Zappavigna S., Scotti L., Abate M., Porru M., Leonetti C., Caraglia M., De Rosa G. Hybrid lipid self-assembling nanoparticles for brain delivery of microRNA. Int. J. Pharm. 2020;588 doi: 10.1016/j.ijpharm.2020.119693. [DOI] [PubMed] [Google Scholar]
- 61.Brown R.A.M., Richardson K.L., Kalinowski F.C., Epis M.R., Horsham J.L., Kabir T.D., De Pinho M.H., Beveridge D.J., Stuart L.M., Wintle L.C., Leedman P.J. Evaluation of MicroRNA Delivery In Vivo. Methods Mol. Biol. 2018;1699:155–178. doi: 10.1007/978-1-4939-7435-1_12. [DOI] [PubMed] [Google Scholar]
- 62.Hald Albertsen C., Kulkarni J.A., Witzigmann D., Lind M., Petersson K., Simonsen J.B. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 2022;188 doi: 10.1016/j.addr.2022.114416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lombardo D., Kiselev M.A. Methods of Liposomes Preparation: Formation and Control Factors of Versatile Nanocarriers for Biomedical and Nanomedicine Application. Pharmaceutics. 2022;14:543. doi: 10.3390/pharmaceutics14030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verbeke R., Hogan M.J., Loré K., Pardi N. Innate immune mechanisms of mRNA vaccines. Immunity. 2022;55:1993–2005. doi: 10.1016/j.immuni.2022.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han L., Jiang C. Evolution of blood-brain barrier in brain diseases and related systemic nanoscale brain-targeting drug delivery strategies. Acta Pharm. Sin. B. 2021;11:2306–2325. doi: 10.1016/j.apsb.2020.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kinsey C., Lu T., Deiss A., Vuolo K., Klein L., Rustandi R.R., Loughney J.W. Determination of lipid content and stability in lipid nanoparticles using ultra high-performance liquid chromatography in combination with a Corona Charged Aerosol Detector. ELECTROPHORESIS. 2022;43:1091–1100. doi: 10.1002/elps.202100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu X., Yu C., Wu X., Cui Y., Liu X., Jin Y., Li Y., Wang L. Validation of an HPLC-CAD Method for Determination of Lipid Content in LNP-Encapsulated COVID-19 mRNA Vaccines. Vaccines. 2023;11:937. doi: 10.3390/vaccines11050937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mousli Y., Brachet M., Chain J.L., Ferey L. A rapid and quantitative reversed-phase HPLC-DAD/ELSD method for lipids involved in nanoparticle formulations. J. Pharm. Biomed. Anal. 2022;220 doi: 10.1016/j.jpba.2022.115011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lima T., Bernfur K., Vilanova M., Cedervall T. Understanding the Lipid and Protein Corona Formation on Different Sized Polymeric Nanoparticles. Sci. Rep. 2020;10:1129. doi: 10.1038/s41598-020-57943-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schultz M.L., Fawaz M.V., Azaria R.D., Hollon T.C., Liu E.A., Kunkel T.J., Halseth T.A., Krus K.L., Ming R., Morin E.E., et al. Synthetic high-density lipoprotein nanoparticles for the treatment of Niemann–Pick diseases. BMC Med. 2019;17:200. doi: 10.1186/s12916-019-1423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garman R.H. Histology of the central nervous system. Toxicol. Pathol. 2011;39:22–35. doi: 10.1177/0192623310389621. [DOI] [PubMed] [Google Scholar]
- 72.Weigel A., Schild D., Zeug A. Resolution in the ApoTome and the confocal laser scanning microscope: comparison. JBO. 2009;14 doi: 10.1117/1.3083439. [DOI] [PubMed] [Google Scholar]
- 73.Wilmarth P.A., Riviere M.A., David L.L. Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J. Ocul. Biol. Dis. Infor. 2009;2:223–234. doi: 10.1007/s12177-009-9042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chambers M.C., Maclean B., Burke R., Amodei D., Ruderman D.L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McDonald W.H., Tabb D.L., Sadygov R.G., MacCoss M.J., Venable J., Graumann J., Johnson J.R., Cociorva D., Yates J.R. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 76.Eng J.K., Jahan T.A., Hoopmann M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 77.Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 78.Elias J.E., Gygi S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 79.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiśniewski J.R., Mann M. A Proteomics Approach to the Protein Normalization Problem: Selection of Unvarying Proteins for MS-Based Proteomics and Western Blotting. J. Proteome Res. 2016;15:2321–2326. doi: 10.1021/acs.jproteome.6b00403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are available in the main text or the supplemental material. All quantitative proteomics data have been submitted to the ProteomeXchange Consortium via the PRIDE partner repository41 with the dataset identifier PXD041648.