Abstract
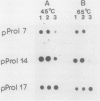
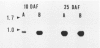
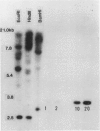
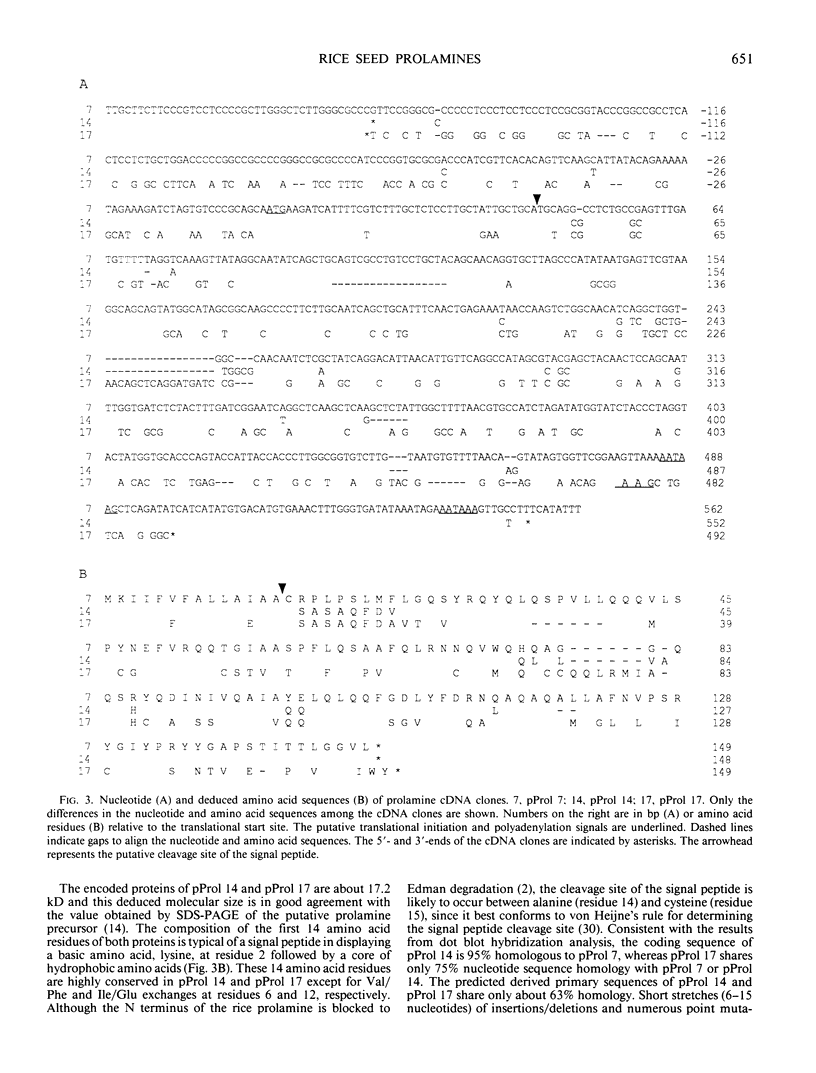
By screening two rice (Oryza sativa L.) seed cDNA libraries, recombinant cDNA clones encoding the rice prolamine seed storage protein were isolated. Based on cross-hybridization and restriction enzyme map analyses, these clones can be divided into two homology classes. All clones contain a single open reading frame encoding a putative rice prolamine precursor (molecular weight = 17,200) possessing a typical 14-amino acid signal peptide. The deduced primary structures of both types of prolamine polypeptides are devoid of repetitive sequences, a feature prevalent in other cereal prolamines. Clones of these two homology classes diverge mainly by insertions/deletions of short nucleotide stretches and point mutations. An isolated genomic clone about 15.5 kilobases in length displays a highly conserved 2.5-kilobase EcoRI fragment, repeated in tandem four times, each containing the prolamine coding sequence. Close homology is exhibited by the coding segments of the genomic and cDNA sequences, although the 5′ ends of the untranslated regions are widely divergent. The sequence heterogeneity displayed by these genomic and cDNA clones and large gene copy number (∼80-100 copies/haploid genome) indicate that the rice prolamines are encoded by a complex multigene family.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett M. D., Smith J. B. Nuclear dna amounts in angiosperms. Philos Trans R Soc Lond B Biol Sci. 1976 May 27;274(933):227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- Bietz J. A. Cereal prolamin evolution and homology revealed by sequence analysis. Biochem Genet. 1982 Dec;20(11-12):1039–1053. doi: 10.1007/BF00498931. [DOI] [PubMed] [Google Scholar]
- Cathala G., Savouret J. F., Mendez B., West B. L., Karin M., Martial J. A., Baxter J. D. A method for isolation of intact, translationally active ribonucleic acid. DNA. 1983;2(4):329–335. doi: 10.1089/dna.1983.2.329. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Heidecker G., Messing J. Sequence analysis of zein cDNAs obtained by an efficient mRNA cloning method. Nucleic Acids Res. 1983 Jul 25;11(14):4891–4906. doi: 10.1093/nar/11.14.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Higuchi W., Fukazawa C. A rice glutelin and a soybean glycinin have evolved from a common ancestral gene. Gene. 1987;55(2-3):245–253. doi: 10.1016/0378-1119(87)90284-8. [DOI] [PubMed] [Google Scholar]
- Kim W. T., Okita T. W. Nucleotide and primary sequence of a major rice prolamine. FEBS Lett. 1988 Apr 25;231(2):308–310. doi: 10.1016/0014-5793(88)80839-1. [DOI] [PubMed] [Google Scholar]
- Krishnan H. B., Okita T. W. Structural Relationship among the Rice Glutelin Polypeptides. Plant Physiol. 1986 Jul;81(3):748–753. doi: 10.1104/pp.81.3.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lizardi P. M. Methods for the preparation of messenger RNA. Methods Enzymol. 1983;96:24–38. doi: 10.1016/s0076-6879(83)96006-8. [DOI] [PubMed] [Google Scholar]
- Rafalski J. A. Structure of wheat gamma-gliadin genes. Gene. 1986;43(3):221–229. doi: 10.1016/0378-1119(86)90210-6. [DOI] [PubMed] [Google Scholar]
- Reeves C. D., Krishnan H. B., Okita T. W. Gene Expression in Developing Wheat Endosperm : Accumulation of Gliadin and ADPglucose Pyrophosphorylase Messenger RNAs and Polypeptides. Plant Physiol. 1986 Sep;82(1):34–40. doi: 10.1104/pp.82.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spena A., Viotti A., Pirrotta V. Two adjacent genomic zein sequences: structure, organization and tissue-specific restriction pattern. J Mol Biol. 1983 Oct 5;169(4):799–811. doi: 10.1016/s0022-2836(83)80137-5. [DOI] [PubMed] [Google Scholar]
- Yamagata H., Sugimoto T., Tanaka K., Kasai Z. Biosynthesis of storage proteins in developing rice seeds. Plant Physiol. 1982 Oct;70(4):1094–1100. doi: 10.1104/pp.70.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]