Significance
Identifying the factors that shape microbe consortia is essential to mitigate the devastating consequences of infections by multiple pathogens, often triggered by viruses, like influenza A virus or Human Immunodeficiency Virus. Such a polymicrobial infection initiated by the OsHV-1 Herpes virus has spread worldwide and dramatically affects the oyster production. We show here that OsHV-1 shapes the assemblage of Vibrio species in OsHV-1-infected oysters. Vibrio harveyi are key colonizers that dampen oyster cellular defenses and facilitate nutrient-uptake by other Vibrio that are cheaters in the polymicrobial community. Thereby they create a more favorable niche for microbes and accelerate disease progression. These findings may have practical implications for mitigating fatal polymicrobial synergy between viruses and bacteria in human and animal diseases.
Keywords: microbiota, mollusk, immune suppression, T3SS, iron uptake
Abstract
Polymicrobial infections threaten the health of humans and animals but remain understudied in natural systems. We recently described the Pacific Oyster Mortality Syndrome (POMS), a polymicrobial disease affecting oyster production worldwide. In the French Atlantic coast, the disease involves coinfection with ostreid herpesvirus 1 (OsHV-1) and virulent Vibrio. However, it is unknown whether consistent Vibrio populations are associated with POMS in different regions, how Vibrio contribute to POMS, and how they interact with OsHV-1 during pathogenesis. By connecting field-based approaches in a Mediterranean ecosystem, laboratory infection assays and functional genomics, we uncovered a web of interdependencies that shape the structure and function of the POMS pathobiota. We show that Vibrio harveyi and Vibrio rotiferianus are predominant in OsHV-1-diseased oysters and that OsHV-1 drives the partition of the Vibrio community observed in the field. However only V. harveyi synergizes with OsHV-1 by promoting mutual growth and accelerating oyster death. V. harveyi shows high-virulence potential and dampens oyster cellular defenses through a type 3 secretion system, making oysters a more favorable niche for microbe colonization. In addition, V. harveyi produces a key siderophore called vibrioferrin. This important resource promotes the growth of V. rotiferianus, which cooccurs with V. harveyi in diseased oysters, and behaves as a cheater by benefiting from V. harveyi metabolite sharing. Our data show that cooperative behaviors contribute to synergy between bacterial and viral coinfecting partners. Additional cheating behaviors further shape the polymicrobial consortium. Controlling cooperative behaviors or countering their effects opens avenues for mitigating polymicrobial diseases.
A number of polymicrobial diseases impact human and animal species (1). They are defined as diseases that result from infections by multiple pathogens (2). Complex microbe communities that form a cohesive entity with the potential to cause disease in polymicrobial diseases can be referred to as a “pathobiota” (3). Within pathobiota, microbes synergize to cause disease: Their interactions enhance disease progression compared to infection with the single microbes (1). Fatal polymicrobial synergy has been reported between viruses and bacteria including influenza A virus and Streptococcus pneumonia (4, 5) or Human Immunodeficiency Virus (HIV) and Mycobacterium tuberculosis (6–8). Synergy has also been reported between viruses such as Herpes virus simplex and HIV (9, 10). Pathobiota show cell-level cooperative capacities that include the production of public goods (e.g., shared metabolites), division of labor, resource transport, and creation and maintenance of the extracellular environment, as in other examples of multicellular organization (11). Dissecting the interactions within pathobiota is needed to understand and possibly control disease establishment, progression, and symptoms. Theoretical models have been developed in response to these challenges to predict what microbial behaviors are favored during such complex interactions (12, 13). In addition, a series of animal models (both vertebrates and invertebrates) have been used to mimic polymicrobial diseases and validate theoretical assumptions (1). Still, natural pathobiota remain poorly explored.
We recently described a typical example of a polymicrobial disease, the Pacific Oyster Mortality Syndrome (POMS). This disease is caused by the Ostreid herpesvirus OsHV-1 and opportunistic bacteria (14) and has devastating consequences for the aquaculture of Crassostrea gigas oysters worldwide. The bacterial genera colonizing oysters during POMS are conserved across environments suggesting functional complementarity within the pathobiota (15). Members of the Vibrionaceae family are the best characterized bacteria in the POMS pathobiota (16–20). Several Vibrio species have been shown to have virulence functions in this disease (19, 21). Vibrio crassostreae (Splendidus superclade) uses cytotoxicity to evade oyster cellular immune responses, leading to systemic infection (19, 21). This virulent Vibrio species is highly prevalent in OsHV-1-infected oysters on the French Atlantic coast, often associated with other Vibrio species from the superclade Splendidus (17). A number of studies suggest that Vibrio species found in OsHV-1-infected oysters vary worldwide (22–24). These species include Vibrio harveyi (superclade Harveyi). A strain of V. harveyi was isolated in 2003 from C. gigas oyster spat during a mortality episode in the Thau lagoon, which is a major Mediterranean area used for oyster farming, and is pathogenic to oysters (25). However, we still know little about V. harveyi in POMS and whether V. crassostreae or other populations colonize OsHV-1-infected oysters in different ecosystems. In addition, how Vibrio colonizes oysters and how Vibrio interacts with the OsHV-1 virus during pathogenesis remains unclear.
Here, we characterized the complex interactions between OsHV-1 virus and Vibrio species as well as between the different Vibrio species that assemble in the POMS pathobiota in the Thau lagoon. Specifically, we investigated how cooperative behaviors could influence the polymicrobial consortium via shared interests (i.e., when a self-serving behavior benefits another individual within the consortium) (26). For that, we examined i) the capacity of POMS pathobiota members to enhance their collective expansion and accelerate disease progression, ii) their potential to alleviate the antagonistic environment of the niche, and iii) their ability to engage in the sharing of public goods. We also tested whether species within the polymicrobial consortium, who engage in cooperation, can be vulnerable to exploitation by other members, as predicted by evolutionary theory (27, 28). We find that two species of the Harveyi superclade – namely V. harveyi and Vibrio rotiferianus – cooccur in OsHV-1-infected oysters in the Thau lagoon. Using mesocosm experiments, we find that OsHV-1 infection favors stable colonization by V. harveyi and V. rotiferianus but no other Vibrio from the Harveyi superclade. However, only V. harveyi synergizes with OsHV-1 by promoting their mutual growth. A series of functional assays, including gene knockouts, indicate that strains of V. harveyi act cooperatively by dampening host cellular defenses through cytotoxic mechanisms and by producing vibrioferrin, a key siderophore required for Vibrio growth in iron-poor environments. We also find that V. rotiferianus behaves as a cheater by using vibrioferrin secreted by V. harveyi to promote its own growth and to spread in the pathobiota without paying the vibrioferrin production costs itself. Overall, our data show the involvement of both cooperative and cheating behaviors in shaping Vibrio assemblages in OsHV-1-infected oysters.
Results
Specific Vibrio Populations Assemble in Oysters Infected by OsHV-1 Virus.
We first asked whether Vibrio populations that naturally assemble in oysters infected by OsHV-1 were conserved across farming environments. With this objective in mind, we performed a field experiment in the Thau lagoon (South of France). Vibrio populations have been suggested to be distinct in this Mediterranean ecosystem from those previously found in the Atlantic (bay of Brest, northwest of France) (25). We performed our study during an episode of oyster mortality. Specific pathogen-free (SPF) juvenile oysters were immersed in September 2015 in the Thau lagoon, which hosts significant oyster farming activity (352 acres). Oysters tested positive for OsHV-1 after 1 mo (October 2015), indicating an ongoing episode of POMS (SI Appendix, Table S1). We characterized the population structure of the Vibrio isolated from a pool of oysters and from the surrounding seawater. A total of 472 isolates were sampled on Vibrio-selective medium from infected oyster tissues and from the water column (SI Appendix, Table S2). Partial hsp60 sequences were obtained for 437 isolates. Assignment to the Vibrio genus was confirmed for 304 sequences (67.8 %), which exhibited ≥95% identity with the hsp60 sequence from a Vibrio type-strain (SI Appendix, Table S2). Isolates with hsp60 sequence identity below this threshold were not included in the study. We observed contrasting population structures in oyster tissues and in the water column. Oyster tissues were dominated by three populations of the V. Harveyi superclade (V. harveyi, V. rotiferianus, and V. owensii), representing 54/55 isolates (Fig. 1A). The water column showed a higher diversity of Vibrio with only 53/249 isolates (21%) falling into the Harveyi superclade (Fig. 1A). These water column isolates were dominated by V. jasicida (39/53) (Fig. 1A). Taxonomic assignments of Harveyi-related isolates were confirmed by multilocus sequence typing (MLST) phylogeny based on 4 Vibrio genes (hsp60, rctB, topA, and mreB) (Fig. 1B). A total of 101 isolates could be assigned to V. harveyi (n = 63), V. rotiferianus (n = 17), V. jasicida (n = 15) and V. owensii (n = 6) (Fig. 1B). Among them, V. harveyi and V. rotiferianus showed a positive association with oyster tissues (Fisher exact test, P < 0.001) whereas V. jasicida was almost exclusively associated with the water column (Fisher exact test, P < 0.001) (Fig. 1B). The high relative abundance of V. harveyi in POMS-diseased oysters from the Thau lagoon was confirmed by three subsequent samplings in 2016 to 2017: V. harveyi was isolated during but not outside of POMS episodes, almost exclusively from oysters infected with OsHV-1 (SI Appendix, Table S1). This result contrasts with the preferential association of the species V. crassostreae with POMS-diseased oysters on the French Atlantic coast (17).
Fig. 1.
V. harveyi and V. rotiferianus are the most prevalent Vibrio species in OsHV-1-infected oysters. The population structure of Vibrionaceae was determined in seawater and oyster flesh during an episode of POMS (Thau lagoon, October 2015). (A) shows the distribution (%) of Vibrio isolated from oysters and seawater. A total of 304 isolates whose hsp60 sequence displayed ≥95% identity with a Vibrio type strains were included. A high prevalence of the Harveyi super-clade (V. harveyi –Vh, V. owensii –Vo, and V. rotiferianus –Vr) is observed in oysters. (B) shows the high prevalence of V. harveyi and V. rotiferianus in oysters after taxonomic affiliation shown in A was validated by an MLST analysis. Four marker genes (hsp60, rctB, topA and mreB) were used. A phylogenetic tree was constructed with the concatenated sequences of the four markers. The following reference strains were used in the analysis: V. azureus NBRC 104587, V. campbellii CAIM 519, V. harveyi NBRC 15634, V. hyugaensis 090810a, V. jasicida CAIM 1864, V. owensii CAIM 1854, V. rotiferianus CAIM 577, V. sagamiensis NBRC 104589. Vibrio sagamiensis type strain was used as an outgroup to root the tree. Bootstrap values (>50%) are indicated by circles on branches. Color boxes indicate whether a strain was isolated from seawater (blue boxes) or from oysters (purple boxes). Stars indicate strains whose genome was sequenced in this study, and filled stars those used for comparative genomics. Red indicates strains used in mesocosm experiments (Fig. 2).
Preferential Association of V. harveyi and V. rotiferianus with OsHV-1-Infected Oysters.
As a number of interdependent environmental variables (e.g., temperature, salinity, season, viral infection) may influence Vibrio assemblages in oysters in the field, we next tested how Vibrio populations partition and the role of OsHV-1 in this partitioning. To achieve this aim, we used mesocosms, which allow controlled infection experiments of juvenile oysters through natural routes with microorganisms relevant to POMS. We developed a synthetic Vibrio community composed of a mixture of 20 isolates from the Harveyi superclade, in the presence (VO) or absence (V) of OsHV-1 (Fig. 2A). Vibrio representative of the four populations isolated from the Thau lagoon were used: V. harveyi and V. rotiferianus (positively associated with oysters), V. jasicida (negatively associated with oysters) and V. owensii (neutral). In parallel, oysters were exposed to OsHV-1 virus only (O) or were kept in tanks devoid of introduced pathogens (control). Oysters were collected in the first 48 h, before mortalities occurred. We first examined which Vibrio populations colonized oysters in the presence/absence of the OsHV-1 virus by comparing the V and VO conditions. To discriminate between the four populations introduced in the mesocosm, we developed an amplicon sequencing method based on the rctB polymorphic gene (SI Appendix, Fig. S1). In the absence of OsHV-1, the Harveyi-related population assemblage remained stable in oysters over time, as shown by rctB-barcoding (SI Appendix, Fig. S2). In contrast, coinfection with OsHV-1 had a significant effect on the structure of the assemblage, as observed after 48 h (Permutational multivariate analysis of variance, P = 0.001) (Fig. 2B and SI Appendix, Figs. S2). The species V. harveyi and V. rotiferianus were significantly enriched in oyster flesh in the presence of OsHV-1 (P < 0.05, Welch’s t test, P-value corrected with Benjamini–Hochberg FDR) (Fig. 2C). In contrast, V. owensi was equally abundant in the presence/absence of OsHV-1, and V. jasicida was more abundant in oyster flesh in the absence of OsHV-1 (P < 0.001) (Fig. 2C). The positive effect of OsHV-1 on oyster colonization by V. harveyi/V. rotiferianus but not V. jasicida/V. owensii at 48 h was confirmed by qPCR monitoring of pathogen loads (multiple t test, P < 0.01) (SI Appendix, Fig. S3). Altogether, our experimental results show i) that OsHV-1 is necessary to reproduce the distribution of Vibrio community observed in the field during a POMS episode, and ii) that only V. harveyi and V. rotiferianus efficiently colonize OsHV1-infected oysters.
Fig. 2.
OsHV-1 synergizes with V. harveyi and/or V. rotiferianus to kill oysters. (A) Mesocosm experiment to test OsHV-1 synergy with Vibrio species (Design 1). Specific pathogen-free oysters were placed in contact with seawater containing OsHV-1 (108 genomic units mL−1) or Vibrio (107 CFU mL−1) or both during 6 d at 20 °C. (O): seawater containing only OsHV-1. (V): 4 species of the Vibrio Harveyi superclade (V. harveyi, V. rotiferianus, V. owensii, V. jasicida) (VO): both OsHV-1 and Vibrio. Controls refer to oysters not exposed to pathogens. (B) Monitoring of the OsHV-1-induced dysbiosis by rctB-barcoding. Principal component analysis (PCoA) ordination plots of Bray–Curtis dissimilarities for the Harveyi-related community associated with oysters. PCoA results are depicted for each time point (i.e., 4, 24, and 48 h). Each dot represents the Vibrio microbiome of one oyster. Colors refer to the experimental condition (blue for V, red for VO). Ellipses represent the 95% CI for each group. PERMANOVA between the two experimental conditions and the three time-points indicates significant differences P = 0.001 (SI Appendix, Fig. S2). Statistical differences between V and VO conditions are observed at 48 h. (C) Colonization of diseased oysters by V. harveyi and V. rotiferianus at 48 h. The left barplot represents the mean proportion of the species V. harveyi, V. rotiferianus, V. owensii, V. jasicida in oysters from the V and VO conditions at T = 48 h. The right dot plot represents the difference in mean proportion of each species by STAMPS analysis. Statistical differences were obtained from Welch’s t test (P-value corrected with Benjamini–Hochberg FDR). (D) Synergy between OsHV-1 and V. harveyi/V. rotiferianus. Left panel shows a significant increase in the mortality rate for oysters exposed to both Harveyi and OsHV-1 (in red) compared to OsHV-1 only (in black) (Kaplan–Meier survival curves, log-rank test, P = 0.0018). Right panel shows that oyster colonization by Vibrio is favored by OsHV-1 (Kruskal–Wallis test, P value < 0.001).
OsHV-1 Virus Synergizes with V. harveyi and/or V. rotiferianus in Oyster Mortality.
We next tested the effect of the Vibrio/OsHV-1 interaction on POMS progression. We used the same synthetic community of Vibrio (Fig. 2A) to monitor oyster mortality and pathogen loads under different experimental conditions. Mortalities were only observed in tanks containing the OsHV-1 virus, indicating that the synthetic Vibrio community alone (V) was not lethal to oysters through natural infection routes (Fig. 2 D, Left). Mortalities started at day 2 in tanks containing OsHV-1 (both O and VO conditions). Still, mortalities progressed significantly more rapidly in oyster tanks containing both OsHV-1 and the synthetic Vibrio community (VO) with 90% mortalities in 3 d as opposed to 6 d for oysters exposed to OsHV-1 only (O) (Kaplan–Meier survival curves, log-rank test, P = 0.0018) (Fig. 2 D, Left). Therefore, introducing the synthetic Vibrio community accelerated the OsHV-1-induced disease. As expected from our previous study (14), the total Vibrio load increased constantly over 48 h in OsHV-1-infected oysters (O). Remarkably, the increase in Vibrio load was significantly higher in oysters exposed to both the Vibrio community and OsHV-1 (VO) than in oysters exposed to Vibrio only (O) at 48 h, i.e. at the onset of mortality (Kruskal–Wallis test, P < 0.001) (Fig. 2 D, Right), with a significant contribution of V. harveyi and V. rotiferianus (SI Appendix, Fig. S3). In oysters exposed to Vibrio only (V), i.e. oysters that did not die, Vibrio colonization tended to be transient with a peak between 4 h to 24 h (Fig. 2 D, Right). Altogether, this indicates that in the absence of OsHV-1, oysters tolerate transiently high loads of the synthetic Vibrio community but Vibrio colonization is not stable and ultimately decreases without causing mortality. In contrast, OsHV-1 infection favors the proliferation and persistent colonization of Vibrio such as V. harveyi and V. rotiferianus, which exacerbate pathogenesis, an effect not seen with V. jasicida and V. owensii. These data show that OsHV-1 and specific populations of Vibrio act in synergy to accelerate oyster death.
V. harveyi and OsHV-1 Reciprocally Promote Inside-Host Growth.
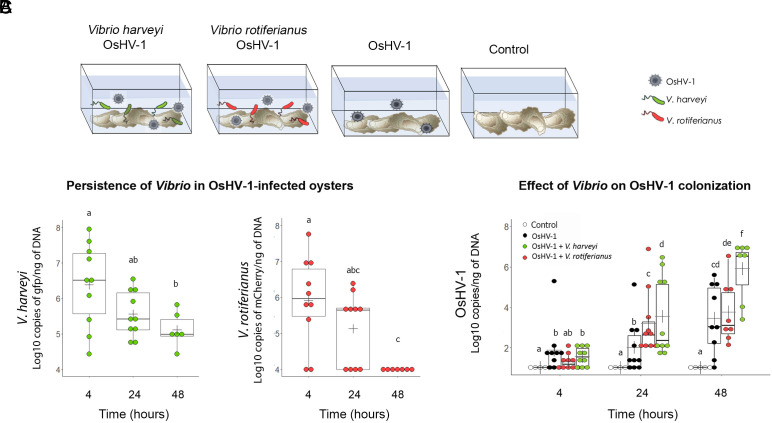
To get insight into the synergistic process, we next tested whether OsHV-1 and strains of V. harveyi or V. rotiferianus affect one another’s growth in coinfections. To facilitate pathogen monitoring, SPF juvenile oysters were exposed to OsHV-1 and fluorescent Vibrio strains representing each population (Fig. 3A). Infection with OsHV-1 only was used as a control (Fig. 3A). As in the previous mesocosm experiment (Fig. 2), live oysters were sampled at 0, 4, 24, and 48 h, before the onset of mortalities (SI Appendix, Fig. S4) to monitor pathogen loads in every individual (Fig. 3). First, we compared V. harveyi and V. rotiferianus colonization in oysters infected with OsHV-1. Only V. harveyi had the ability to colonize OsHV-1-infected oysters efficiently. Indeed, V. harveyi remained present at high doses (105 to 5 × 106 copies/ng of DNA) in live oyster tissues throughout the time course. In contrast, V. rotiferianus loads decreased rapidly over the same period and were undetectable (<104 copies/ng of DNA) in most individuals after 48 h (Fig. 3B). Second, we analyzed the effect of Vibrio strains on OsHV-1 growth. Remarkably, the viral load was 100-fold higher at 48 h in oysters coinfected with V. harveyi and OsHV-1 than in oysters infected with OsHV-1 only (t test, P < 0.05). Such an increase in viral load was not observed in coinfections with V. rotiferianus (Fig. 3C), consistent with the rapid elimination of V. rotiferianus from host tissues (Fig. 3D). Altogether, these results indicate that V. harveyi and OsHV-1 cooperate by increasing mutual growth during pathogenesis, an effect not observed with V. rotiferianus.
Fig. 3.
OsHV-1 and V. harveyi cooperate to colonize oysters. (A) Simplified mesocosm experiment using fluorescent strains of V. harveyi and V. rotiferianus (Design 2). In order to identify putative cooperative behaviors between OsHV-1 and Vibrio, oysters were immersed at 20 °C in seawater containing OsHV-1 (104 copy µL−1) and fluorescent Vibrio (either gfp-labeled V. harveyi Th15_O_G11 or mCherry-labeled V. rotiferianus Th15_O_G05) (each Vibrio at 107 CFU mL−1). Exposure to OsHV-1 only or immersion in seawater without introduced pathogen was used as a control. Oysters were collected at 4 h, 24 h, and 48 h, i.e. before mortalities occurred, to monitor pathogen load in oyster tissues. (B) Higher persistence of V. harveyi in OsHV-1-infected oysters. Vibrio loads were determined by qPCR by quantifying gfp and mCherry copies in total DNA extracted from oyster flesh in OsHV-1/V. harveyi and OsHV-1/V. rotiferianus coinfections, respectively. Each dot represents an individual oyster (t test, P < 0.05). Only V. harveyi is detected at >105 copies/ng DNA over the time course of the experiment. Detection limit: 104 copies/ng DNA. (C) V. harveyi promotes OsHV-1 replication. OsHV-1 load was measured by qPCR in the flesh of oysters exposed to OsHV-1 and gfp-labeled V. harveyi Th15_O_G11 (green) or OsHV-1 and mCherry-labeled V. rotiferianus Th15_O_G05 (red), or OsHV-1 only (black). Each dot represents an individual. A significant increase in OsHV-1 load is observed in the presence of V. harveyi (t test, P < 0.05).
V. harveyi Actively Dampens Oyster Immune Defenses.
We next investigated Vibrio traits that may facilitate host colonization and favor polymicrobial synergy with OsHV-1. We first compared the virulence potential of V. harveyi and V. rotiferianus (successful colonizers, Figs. 1B and 2C) and V. jasicida and V. owensii (poor colonizers). Virulence potential was tested in vivo by direct injection of Vibrio isolates into juvenile oyster adductor muscle. V. harveyi isolates showed strong virulence potential as revealed by an average of 50% oyster mortality one day after injection, as compared to no mortality in sham-injected controls (Fig. 4A). However, mortalities remained below 15% on average after injection of V. rotiferianus, V. jasicida, and V. owensii (Fig. 4A). Not only did V. harveyi have a significantly higher virulence potential than other species (Kruskal–Wallis test, P < 0.001), but it also showed greater cytotoxicity toward oyster immune cells (hemocytes) in vitro (Fig. 4B). Here, we compared the cytotoxic activity of two strains per Vibrio species among the four Harveyi-related species from our study. Upon exposure to V. harveyi strains, 67 to 76% of hemocytes underwent lysis (P < 0.001, One-way ANOVA, and Tukey post-hoc test) (Fig. 4B). The three other Vibrio species were not cytotoxic, with the percentage of lysed cells similar to controls (7 to 23%). We further confirmed the ability of V. harveyi, but not V. rotiferianus, to damage hemocytes using fluorescent Vibrio strains. After a 2-h exposure to V. harveyi, hemocytes were massively damaged and many extracellular V. harveyi were observed (Fig. 4C). In contrast, no cellular damage was observed when hemocytes were exposed to V. rotiferianus, with most bacteria being phagocytized. Thus, V. harveyi strains are equipped with specific virulence/cytotoxicity mechanisms that can dampen oyster cellular defenses. To uncover genomic features that might explain V. harveyi virulence/cytotoxicity, we sequenced and analyzed 17 Harveyi-related genomes from our collection (4 V. harveyi, 5 V. owensii, 4 V. jasicida and 4 V. rotiferianus isolates; SI Appendix, Table S6). We found that the V. harveyi genome contained the most candidate virulence factors of the four Vibrio species in the present study. Candidate virulence genes in the V. harveyi genome included a type 3 secretion system (T3SS) and its associated effectors as well as three to four different type 6 secretion systems (T6SS) (SI Appendix, Table S7). The exsA gene of our V. harveyi strains showed 98.5 to 99.65% nucleotide sequence identity with that of V. harveyi ORM4 controlling type 3 secretion system (T3SS) expression and virulence in this strain highly pathogenic for abalone (29). Without any growth impact (SI Appendix, Fig. S5), knock out of the exsA sequence was sufficient to completely abolish both the virulence potential (Fig. 4D) and the cytotoxicity (Fig. 4E) of V. harveyi, indicating that T3SS plays a key role in V. harveyi virulence and cytotoxicity to oyster immune cells.
Fig. 4.
V. harveyi strains show high virulence potential and cytotoxicity toward oyster immune cells. (A) V. harveyi is significantly more virulent than other Harveyi-related species in oyster experimental infection. Oyster mortality rate (%) was measured at 24 h following an injection of Harveyi-related isolates (108 CFU animal−1) or sterile seawater (sham-injected controls). Mortality rates were compared for each Vibrio species using the Kruskal–Wallis test with post hoc Dunn Test. Significant differences between mean values are represented by different letters (P-value < 0.0001). V. harveyi (n = 63), V. rotiferianus (n = 16), V. jasicida (n = 15), V. owensii (n = 6) (Kruskal–Wallis test, P < 0.001). V. harveyi isolates induced significantly higher mortalities than other strains. (B) V. harveyi is significantly more cytotoxic to oyster hemocytes than other species. Vibrio cytotoxicity was determined on monolayers of hemocytes adherent to the bottom of microplates and monitored using Sytox green labeling. Cells were incubated with bacteria at a MOI of 50:1. The maximum percentage of hemocyte lysis (%) caused by Vibrio is displayed. Error bars represent the standard deviation of the mean, different letters represent significant differences between means in a multiple comparison test (P < 0.05, One-way ANOVA with post hoc Tukey HSD Test). Controls were challenged with sterile seawater only. Strains of V. harveyi were significantly more cytotoxic than other strains. (C) V. harveyi but not V. rotiferianus causes damage to oyster hemocytes. The Vibrio effect on hemocytes was observed by epifluorescence microscopy. Monolayers of hemocytes adherent to a glass coverslip were incubated with fluorescently-labeled V. harveyi Th15_O_G11 or V. rotiferianus Th15_O_G05 at a MOI of 50:1 for 2 h. Actin was stained with Fluorescent-phalloidin (Red), Chromatin was stained with DAPI. Vibrio strains expressing fluorescent proteins are shown in Green. Cell damage was only observed with V. harveyi. (D and E) V. harveyi virulence and cytotoxicity implies exsA. (D) Oyster mortality rate (%) was measured at 24 h following an injection of Harveyi-related isolates. Mortality rates were compared for each Vibrio species using the Kruskal–Wallis test with post hoc Dunn Test. (E) Vibrio cytotoxicity was determined on monolayers of hemocytes and monitored using Sytox green labeling as detailed above. Different letters represent significant differences between means in a multiple comparison test (P < 0.05, One-way ANOVA with post hoc Tukey HSD Test).
V. harveyi Produces Vibrioferrin, Which Can Be used by V. rotiferianus.
We next used comparative genomics to examine how V. harveyi and V. rotiferianus both colonize OsHV-1-infected oysters but make distinct contributions to pathogenesis (SI Appendix, Table S6). Using genome comparisons, 15 genes were identified that are exclusively shared by V. harveyi and V. rotiferianus (good colonizers; SI Appendix, Table S8). Of these, iron acquisition systems differed between species (SI Appendix, Table S7). We paid particular attention to genes involved in the vibrioferrin pathway, a siderophore whose uptake system was only found in strains of V. harveyi and V. rotiferianus (Fig. 5A). Vibrioferrin is a tricarboxylic acid siderophore derived from citric acid; it was shown to be shared as a public good within populations of V. splendidus (30). Remarkably, we found that V. harveyi harbors genes responsible for the biosynthesis and uptake of vibrioferrin whereas V. rotiferianus only carries the vibrioferrin uptake system (Fig. 5A). This suggests that by secreting vibrioferrin, V. harveyi could facilitate iron uptake by V. rotiferianus. To determine whether V. rotiferianus is able to use vibrioferrin produced by V. harveyi, we compared the growth of V. rotiferianus in the presence/absence of increasing amounts of V. harveyi culture supernatant. V. harveyi was able to grow in iron-depleted medium (2,2′-bipyridine used as iron chelator, SI Appendix, Fig. S6), in agreement with its ability to produce siderophores. In contrast, growth of V. rotiferianus was impaired upon iron depletion (P < 0.05, Mann–Whitney U test) (SI Appendix, Fig. S6). Growth of V. rotiferianus was rescued in a dose-dependent manner by the addition of cell-free culture supernatant from V. harveyi (Kruskal-Wallis, P < 0.05) (Fig. 5B). To determine whether this rescue is linked to vibrioferrin, we synthesized the siderophore (SI Appendix, Fig. S7). Synthetic vibrioferrin was sufficient to rescue V. rotiferianus growth in iron-depleted medium (Kruskal-Wallis, P < 0.05) (Fig. 5C). Finally, we constructed a V. rotiferianus mutant strain which lacks the two genes encoding the PvuA1-A2 receptor that are required for vibrioferrin uptake (SI Appendix, Fig. S5). Vibrioferrin failed to rescue the growth of the V. rotiferianus ΔpvuA1-A2 mutant (Fig. 5C). This demonstrates that V. rotiferianus is able to acquire vibrioferrin, an important resource produced by V. harveyi (and potentially other Vibrio within the microbiota), to grow in iron-poor environments. Vibrioferrin uptake was actually critical for V. rotiferianus fitness in competition assays with V. harveyi. Indeed, whereas wild-type V. rotiferianus was as fit as V. harveyi in iron-poor conditions, its fitness was reduced by half upon pvuA1-A2 deletion (t test, P < 0.05) (Fig. 5D). This significant loss of fitness was not observed when iron was not limiting, i.e., when siderophores are not needed for growth. Instead, the wild-type V. rotiferianus, which does not invest into siderophore production, tended to be more fit than V. harveyi (vibrioferrin producer), suggesting a cost of vibrioferrin production. Overall, our data show that V. rotiferianus behaves as a cheater by using a siderophore produced by V. harveyi. Thus unlike V. harveyi and the OsHV-1 virus, which behave cooperatively, V. rotiferianus successfully colonizes oysters taking advantage of public goods contributed by the microbiota without providing benefit to the POMS-associated microbiota.
Fig. 5.
V. rotiferianus depends on V. harveyi vibrioferrin production for growth and fitness in iron-poor conditions. (A) Possible cheating on vibrioferrin uptake in V. rotiferianus. Comparative genomics between strains of V. harveyi (Th15_O_G11, Th15_F5_F11, Th15_O_D03, Th15_O_A01) and V. rotiferianus (Th15_O_G05, Th15_O_G10, Th15_O_B06, Th15_O_E12) point to a possible cheating behavior for vibrioferrin uptake in V. rotiferianus, which has receptor for vibrioferrin but does not produce it. (B) V. rotiferianus growth in iron-depleted medium is rescued by V. harveyi culture supernatant (sup). Iron-depletion was obtained by adding 150 µM 2,2′-bipyridine (BIP) to the minimal culture medium. Dose-dependent growth rescue of V. rotiferianus strain Th15_O_G05 was achieved by adding V. harveyi Th15_O_G11 culture supernatant (either 1× or 2× concentrated by lyophilization). (C) V. rotiferianus growth in iron-depleted medium is rescued by vibrioferrin and requires the iron-vibrioferrin receptor PvuA. Dose-dependent growth rescue of V. rotiferianus WT strain Th15_O_G05 (dark grey) was achieved by 8.75 to 70 µM vibrioferrin. Growth of V. rotiferianus Th15_O_G05 ΔpvuA1-A2 (light grey) in iron-depleted medium was not rescued by addition of vibrioferrin up to 70 µM (see SI Appendix, Fig. S7 for vibrioferrin synthesis). Letters indicate significant differences between conditions (P < 0.05, Kruskal-Wallis). (D) V. rotiferianus relative fitness in iron-depleted medium is dependent on vibrioferrin uptake. Relative fitness of V. rotiferianus in coculture with V. harveyi (ratio of V. rotiferianus and V. harveyi growth rates in coculture) is significantly reduced in iron-depleted medium upon deletion of the iron-vibrioferrin receptor (P < 0.05, t test). Vf stands for vibrioferrin.
Discussion
Here, we studied the interactions that shape Vibrio assemblages in a polymicrobial disease, the POMS, which is initiated by an OsHV-1 viral infection. By focusing on a number of players ecologically relevant for this natural pathosystem, our data highlight a complex web of interdependencies within the POMS pathobiota. We also provide mechanistic insight in the cooperative traits accelerating disease progression as well as in incidental cheating traits, which contribute to shaping the Vibrio community associated with OsHV-1-infected oysters.
We showed here that the population structure of Vibrio, a genus consistently found in the POMS pathobiota (31), varies across French Atlantic and Mediterranean oyster-farming environments. Indeed, we found V. harveyi and V. rotiferianus (Harveyi superclade) positively associate with OsHV-1-infected oysters during field mortalities in the Mediterranean. This finding contrasts with a previous analysis of Atlantic oysters where V. crassostreae (Splendidus superclade) dominated (17). Experimentally, two out of four different species from the same clade were recruited from seawater, suggesting that the environment serves as a reservoir for Vibrio recruitment during POMS. Differences in Vibrio species associated with OsHV-1-infected oysters are therefore likely related to different environmental distributions of Vibrio species. Indeed, while V. crassostreae is present at high latitudes, e.g., in the North sea, Germany (32), V. harveyi preferentially grows in warm waters (33), such as those found for half the year in the Thau lagoon (16 to 30 °C) (34). Consistently, V. harveyi has been associated with disease outbreaks under rising sea surface temperatures (35). Remarkably, the data from both locations (Atlantic/Mediterranean) converge in that the OsHV-1 virus is associated with specific populations of Vibrio in diseased oysters only. Our data indicate that OsHV-1 is key to enabling colonization by such specific Vibrio species.
By focusing on Vibrio that naturally coinfect oysters with OsHV-1, we observed a first level of interdependence and polymicrobial synergy occurring during POMS. Building on earlier demonstrations that the bacterial and viral components of POMS are both needed to induce oyster mortalities (14), we showed that the combined effects of OsHV-1 and Vibrio triggered faster host death than that observed when the microorganisms were used in isolation to infect oysters. While both V. harveyi and V. rotiferianus successfully colonized OsHV-1-infected oysters, polymicrobial synergy was specifically observed between OsHV-1 and V. harveyi. The benefits of coinfection were not only observed for OsHV-1 but also for the whole Vibrio community: both showed significantly greater expansion prior to oyster death when OsHV-1 and V. harveyi were coinfected. Whether other bacterial genera conserved among POMS consortia (e.g., Arcobacter, Marinomonas) (15, 31, 36) also contribute to this polymicrobial synergy or play complementary roles in the polymicrobial consortium remains to be established.
A key mechanism underlying polymicrobial synergy between OsHV-1 and Vibrio is the dampening of oyster cellular defenses, which makes the local environment less hostile for the entire microbiota. This manipulation of cellular immunity is key in a number of polymicrobial infections. For instance, induction of IL-10-producing macrophages by M. tuberculosis favors HIV-1 replication and spread (8). Similarly, the apoptosis of macrophages, neutrophils, dendritic cells, and NK cells contributes to the fatal outcome of influenza A virus/S. pneumoniae coinfections (5). Importantly, cytotoxicity to host immune cells is a phenotypic trait conserved in Vibrio species that cooccur with OsHV-1 in diseased oysters. Indeed, cytotoxic effects toward oyster hemocytes is a functional trait conserved in Mediterranean strains of V. harveyi, as well as in strains of V. crassostreae, V. tasmaniensis and V. splendidus (19, 21, 37) isolated from POMS-diseased oysters in the Atlantic (17, 20, 25, 38, 39). Here, we have shown that for virulence/cytotoxicity, V. harveyi relies on a T3SS whereas it is T6SS in V. crassostreae and V. tasmaniensis (19, 21). We previously showed that cytotoxicity toward immune cells is essential for Vibrio to colonize oyster tissues (21). This indicates that Vibrio species that harbor similar functions are likely replaceable within the POMS bacterial consortia, independently of the molecular mechanisms involved. Our mesocosm experiment validated the preferential association of V. harveyi with OsHV-1-infected oysters under controlled conditions, showing that cytotoxic Vibrio species can be recruited from the ecosystems where they circulate. In the mutualistic association between the mollusk Euprymna scolopes and its symbiont Vibrio fischeri, it was also shown that Vibrio phenotypic traits determine their capacity to be recruited from the environment (40). Overall, our data show that similar to OsHV-1 (14), cytotoxic Vibrio species modify their extracellular environment by targeting oyster immunity. This results in eased proliferation of coinfecting partners, as shown here for OsHV-1 and specific Vibrio species. This fundamental cooperation taking place within the POMS pathobiota has benefits for the entire polymicrobial consortium.
Our present study shows that cooperation through dampening host defenses contributes to the shaping of the POMS pathobiota assembly. This supports recent studies highlighting that cooperative interactions within animal microbiota participate in the shaping of community composition and functioning (for review see ref. 41). This complements previous knowledge on the key role of bacterial cooperation in beneficial interactions with their animal hosts (42). Importantly, pathogenic microbes cooperating to manipulate their host may be outcompeted by other members of the microbiota that do not invest in cooperation. Manipulation is therefore only expected to be favored if its benefits predominantly fall back on the manipulator (43). One important benefit for OsHV-1 and V. harveyi observed here is a higher load for both manipulators. Another likely benefit is increased pathogen transmission due to accelerated host death (Fig. 6). From a general point of view, accelerating host death is not predicted to be favorable to pathogens as this may drive the host, and consequently themselves, to extinction, unless transmissibility is also increased (44). This is particularly true for pathogens with narrow host spectra like V. harveyi and V. crassostreae, but also for OsHV-1, which are almost exclusively found in oysters (this study; ref. 17). POMS is typically transmitted from oyster to oyster by a massive release of pathogens into the seawater, which in turn infects neighboring oysters through filter-feeding (36, 45). We can therefore hypothesize that polymicrobial synergy leading to accelerated release of OsHV-1 and Vibrio into seawater is advantageous in terms of group selection.
Fig. 6.
A systemic view of POMS polymicrobial synergy. Oysters are infected by OsHV-1 µvar, which impairs host immune defenses making the environment less hostile for stable bacterial colonization. Secondary bacterial colonization is enabled. The subset of cooperative bacteria that exhibit cytotoxicity toward hemocytes further dampen oyster immune defenses. This dampening is beneficial to the whole microbial community (bacteria and the OsHV-1 virus) and leads to dysbiosis. Cooperative bacteria also secrete siderophores that can be used by other members of the community who do not invest in costly mechanisms of colonization (cheaters). Cooperation between the virus and cytotoxic bacteria accelerates host death leading to the shedding of bacteria and viruses that can infect new hosts.
Beyond cooperative traits conserved among successful Vibrio colonizers, we find evidence that cheating is an efficient strategy that Vibrio use to colonize oysters affected by POMS. Indeed, unlike V. harveyi, V. rotiferianus does not invest resources in virulence nor cytotoxicity. Moreover, our data argue that V. rotiferianus lacks the costly pathways to synthesize vibrioferrin. Instead, this species imports this siderophore, produced by V. harveyi, to promote its own growth. In contrast, the unsuccessful colonizers V. owensii and V. jasicida lack the vibrioferrin uptake machinery, which may contribute to their competitive exclusion (46).
Remarkably, vibrioferrin biosynthetic pathways are highly conserved between V. harveyi and V. crassostreae (47), the two main species associated with POMS in the Mediterranean and the Atlantic. Both Vibrio species can, therefore, supply similar public goods to the POMS pathobiota. Metabolite cross-feeding, which enables a bacterium to consume metabolites produced by another community member, can mediate synergy in multispecies infections (for review see ref. 1). Cheating behavior regarding iron-acquisition was earlier described in Vibrio by Cordero et al. (30). The authors showed that within ecologically cohesive clusters of closely related Vibrio, only some genotypes were able to produce siderophores. Meanwhile, nonproducers had selectively lost siderophore biosynthetic pathways. We observe traces of selective loss in V. rotiferianus strains: while V. rotiferianus and V. harveyi both harbor the genomic region that contains vibrioferrin receptors, the former species specifically lacks the vibrioferrin biosynthetic genes.
The cheating behavior arguably coevolves with ecological adaptation of Vibrio toward association with larger particles in the water column, consistent with efficient siderophore sharing where local densities of bacteria are high (30). Because oysters host dense populations of bacteria in their body fluids [>107 culturable bacteria mL−1 during episodes of POMS (48)], they constitute microhabitats where cheating can occur. Oysters could, therefore, provide a favorable niche for Vibrio strains that can import iron-loaded siderophores. Importantly, the present study shows that cheating for iron acquisition occurs interspecifically and coincides with the cooccurrence of V. harveyi and V. rotiferianus in POMS-diseased oyster. Indeed, our 2-y Mediterranean field survey showed that V. harveyi is repeatedly associated with diseased oysters. V. harveyi cooccurs with V. rotiferianus in oyster flesh. These host-associated states provide conditions where V. rotiferianus could have evolved cheating behavior. Thus, social interactions through siderophore-sharing appear to further structure the assembly of bacterial species in oysters affected by POMS.
Conclusion
By disentangling the complex interactions at play in the POMS, we have shown that cooperation is key in the functioning of this natural pathosystem. Cooperation and cheating seem to drive the assembly of the pathobiota. The former manifests as polymicrobial synergy between the OsHV-1 virus, the etiological agent of POMS and secondary Vibrio colonizers. Dampening of oyster cellular defenses and siderophore sharing, both of which make the host environment more favorable for microbial proliferation, are two cooperative traits conserved in the main POMS-associated Vibrio species, namely V. harveyi and V. crassostreae, across oyster farming environments. This knowledge opens avenues for the control of polymicrobial diseases by interfering with polymicrobial assembly. Implementation of ecological principles, such as interfering with cooperative behavior within the microbiome (e.g., siderophore sharing) or altering the local environment of the POMS pathobiota (i.e., stimulating host defenses) are promising solutions for future exploration. We have shown recently that eliciting oyster antiviral defenses through so-called immune priming is sufficient to prevent colonization by OsHV-1 and subsequent disease development (49, 50). Similarly, exposure to microbial communities at early developmental stages (referred to as biological embedding) was also protective against POMS (51). Such solutions, which make the host a less favorable niche for the OsHV-1 virus (50) and/or bacteria (51), are promising avenues for preventing or reducing the assembly of microbial consortia that produce POMS.
Materials and Methods
Oyster Production.
For field immersion and virulence assays, batches of Crassostrea gigas oysters were produced in 2015, 2017 and 2019, in Ifremer hatchery (Argenton, Bretagne, France) from wild seed broodstocks sampled in farming and nonfarming areas as described previously (14). While multiparental oyster batches were used for field experiments, a biparental family was produced at the Ifremer facilities in La Tremblade (Charente-Maritime, France) for experimental infections in mesocosms. This family was selected for its susceptibility to OsHV-1 infection. Spawn occurred in June 2017, and larval and spat cultures were performed as described (52, 53). Oysters used in the different experiments correspond to juveniles (6 to 10 mo old; 3 to 4 cm) to ensure susceptibility to POMS (53).
Oyster and Seawater Sampling.
Juveniles (6 mo old) were immersed in the Thau lagoon (Occitanie, France) and sampled at four timepoints between October 2015 and March 2017. Sampling coincided with mortality events and in the absence of any observed mortality (see SI Appendix, Tables S1 and S2 for details). Of note, 40 L of seawater was collected and size-fractionated by sequential filtration (from >60 µm to 0.2 µm) as described by Bruto et al. (17).
Bacterial Sampling.
Oysters and large seawater particle fractions (>60 µm) were ground up with Ultra-Turrax (IKA) and 100 µL was plated on Vibrio selective media (thiosulfate-citrate-bile salts sucrose agar, TCBS). Filters of 5-, 1-, and 0.22-µm porosity were directly placed on TCBS agar and incubated at 20 °C for 2 d. About 100 colonies per sample were randomly picked then restreaked first on TCBS and then on Zobell agar (4 g L−1 bactopeptone, 1 g L−1 yeast extract and 15 g L−1 agar in sterile seawater, pH 7.4). Stock cultures were stored at −80 °C in Zobell containing 15% glycerol (v/v).
Nucleic Acid Extractions.
Total DNA was extracted from oyster flesh, seawater samples, and bacterial cultures using standard protocols (SI Appendix, Material and Methods).
Population Structure Analysis.
Isolates from October 2015 were genotyped by partial hsp60 sequencing (54, 55) (SI Appendix, Table S3) generating a total of 437 hsp60 sequences (56). hsp60 sequence ambiguities were corrected using four peaks and Seaview software (http://nucleobytes.com/index.php/4peaks; ref. 57) and received a taxonomic affiliation if the best BLAST-hit displayed an identity greater than 95% with a type-strain. Fisher-exact tests were performed with a 2 × 2 contingency table using the computing environment R (58) for statistical validation of the ecological preferences of populations and the distribution of bacterial populations in oyster tissues and seawater. Significance was assessed using P-value ≤ 0.05.
MLST Genotyping.
To validate hsp60 sequence-based taxonomic assignments to the Harveyi superclade, three additional protein-coding genes were sequenced (rctB, topA and mreB). First, Harveyi isolates were screened by PCR using rctB-F/rctB-R primers designed to specifically hybridize to Harveyi-related rctB gene sequences (SI Appendix, Table S3). The PCR program was: 2 min at 95 °C; 30 cycles of 30 s at 95 °C, 1 min at 53 °C and 1.45 min at 72 °C; 5 min at 72 °C. As a result of this analysis, 143 rctB+ isolates were considered to belong to the Harveyi superclade. Then topA and mreB sequences were amplified using VtopA400F/VtopA1200R and VmreB12F/VmreB999R primers, respectively (59, 60) (SI Appendix, Table S3). All three genes were amplified using the Gotaq G2 flexi polymerase (Promega) following the manufacturer’s instructions and sequenced using the reverse PCR primer at GATC Biotech. Next, hsp60, rctB, topA, and mreB sequences were aligned with 8 Harveyi superclade type strains using Muscle (61). Alignments were concatenated with Seaview (57). Phylogenetic trees for each marker were reconstructed with RAxML using a GTR model of evolution and Gamma law of rate heterogeneity. Bootstrap values were calculated for 100 replicates. All other options were left at default values.
Bacterial Growth Conditions.
Bacteria were cultured using standard protocols (SI Appendix, Material and Methods).
Virulence Potential of Vibrio Strains.
To test virulence potential, Vibrio were grown under shaking at 20 °C for 18 h in Zobell liquid medium before adjustment to OD600 = 0.7. A volume of 40 μL was injected intramuscularly into 20 specific pathogen-free (SPF) 6-mo-old juvenile C. gigas oysters (16) previously anesthetized during 2 h in hexahydrate MgCl2 bath (50 g L−1, 100 oysters L−1). An injection with sterile filtered seawater was used as a negative control (sham injection). After injection, animals were transferred to aquaria (20 oysters per 1 L aquarium) containing 400 mL of aerated seawater at 20 °C and kept under static conditions. Mortalities were recorded 24 h post injection.
In Vitro Cytotoxicity Assays.
Cytotoxicity of vibrios was measured on hemocyte monolayers according to procedures described in SI Appendix, Material and Methods. Statistical analysis was performed using one-way ANOVA and a post hoc Tukey test for pairwise comparison of maximum cytolysis values for each condition. Significance was assessed using P-value ≤ 0.05.
Fluorescence Microscopy.
Hemocytes were plated onto glass coverslips in a 24-well plate to obtain monolayers of 5 × 105 cells per well. Adherent hemocytes were exposed to GFP or mCherry-expressing Vibrio that had been washed with sterile seawater and opsonized in plasma for 1 h, were then added to the wells at a MOI of 50:1. Vibrio (SI Appendix, Table S4) were added at a multiplicity of infection of 50:1, as in ref. 62. Binding of bacteria to hemocytes was synchronized by centrifugation for 5 min at 400 g. After a 2-h incubation, the cell monolayers (coverslips placed at the bottom of the wells) were fixed with 4% paraformaldehyde for 15 min. Coverslips were then washed in PBS and stained with 0.25 μg mL−1 DAPI (Sigma) and 0.5 μg mL−1 Phalloidin-TRITC or FITC (Sigma). Fluorescence imaging was performed using a Zeiss Axioimager fluorescence microscope and a Zeiss 63× Plan-Apo 1.4 oil objective equipped with a Zeiss MRC black and white camera for image acquisition.
Experimental Infection in Mesocosm.
For experimental infections, juvenile oysters (10 mo) were infected with OsHV-1 virus, Vibrio, or both, by natural infection routes, and samples were collected as described in details in SI Appendix, Material and Methods.
Vibrio Genome Sequencing and Assembly.
Individual genomic libraries were prepared from 1 ng of bacterial DNA at the Bio-Environment platform (University of Perpignan) using the Nextera XT DNA Library Prep Kit (Illumina) according to the manufacturer’s instructions. The quality of the libraries was checked using High Sensitivity DNA chip (Agilent) on a Bioanalyzer. Pooled libraries were sequenced in 2 × 150 paired-end mode on a NextSeq 550 instrument (Illumina). Reads were assembled de novo using Spades software. Computational prediction of coding sequences together with functional assignments and comparative genomics were performed using the MaGe MicroScope (63). The genome sequence assemblies have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under project accession no. PRJEB49488 (SI Appendix, Table S6).
OsHV-1 Detection and Quantification.
The ground oyster flesh (1.5 mL) was centrifuged for 10 min at 4 °C and 2,200 g and genomic DNA was extracted from 50 µL of supernatant using phenol:chloroform:isoamyl alcohol (25:24:1) and isopropanol precipitation. Detection and quantification of OsHV-1 µVar DNA was performed using quantitative PCR targeting a predicted DNA polymerase catalytic subunit (DP) using OsHVDPFor/OsHVDPRev primers (SI Appendix, Table S3) as in Webb et al. (64) using the protocol previously described by ref. 49. All amplification reactions were performed in duplicate using a Roche LightCycler 480 Real-Time thermocycler (qPHD-Montpellier GenomiX platform, Montpellier University). Oyster flesh samples exhibiting a viral load greater than 100 genome units per ng of total DNA (GU/ng) were considered to be infected by OsHV-1.
Vibrio Quantification.
16S rDNA sequences were used to quantify total Vibrio present in oysters by extracting 25 ng of DNA from tissue or crude extracts from seawater (see above). Amplification reactions were carried out in duplicate, in a total volume of 20 μL on Mx3005 Thermocyclers (Agilent) using Brilliant III Ultra-Fast SyberGreen Master Mix (Agilent), and 567F and 680R primers at 0.3 µM (65) (SI Appendix, Table S3). Absolute quantification of Vibrio genomes in oyster samples was estimated using standards from 102 to 109 genome copies of Vibrio (SI Appendix, Material and Methods).
V. harveyi-V. rotiferianus and V. owensii-V. jasicida were quantified based on 25 ng of DNA extracted from tissue or crude extracts from seawater (see above) through detection of a specific chemotaxis protein and ompA, respectively (SI Appendix, Table S3). As described above, amplification reactions were performed in duplicate using a Roche LightCycler 480 Real-Time thermocycler, SYBR Green I Master mix (Roche) and primers at 0.3 and 0.2 µM, respectively (final concentration). For absolute quantification, standard curves of known concentration of V. harveyi, V. rotiferianus, V. jasicida, V. owensii genomes were used (SI Appendix, Material and Methods).
rctB Metabarcoding.
Locus-specific PCR primers, including Illumina overhang adaptors, were designed to amplify a 573 bp region of the rctB gene in all our Harveyi strains (rctB-Fw-I and rctB-Rv-I primers, SI Appendix, Table S3). PCR analysis of total DNA extracted from oysters (N = 60) used the high-fidelity Q5 polymerase (New England Biolabs) in a total volume of 50 μL under the following conditions: 98 °C for 25 s followed by 35 cycles of 98 °C for 10 s, 51 °C for 25 s and 72 °C for 30 s. Final extension was performed at 72 °C for 2 min. Presence of the 573 bp amplicon was validated by 1.5% gel electrophoresis. Libraries were constructed with the Two-Step amplicon sequencing approach using Illumina dual indexes (ref. 15044223) and sequenced on a MiSeq instrument to produce paired end reads 2x300 bp, by the GenSeq platform, University of Montpellier (ISEM), France. Sequencing data were processed using the SAMBA pipeline v3.0.1 (66). All bioinformatics processes used the next-generation microbiome bioinformatics platform QIIME 2 (67) (version 2020.2) and grouped sequences in ASV (Amplicon Sequence Variants) using DADA2 v1.14 (68). The resulting ASVs were annotated against an in-house database containing the Harveyi rctB sequences and filtered for low abundance ASVs to limit the prevalence of putative artifacts due to sequencing errors. To do this, we only retained ASVs showing at least four reads in at least four samples. Statistical analyses were performed with R (58) using the R packages Phyloseq v1.38.0 (69) and Vegan v2.6-2 (70). Principal coordinate analyses (PCoA) based on Bray–Curtis distances at each kinetic point were used to assess variation in the composition of Harveyi communities. Putative differences between groups were assessed by statistical analyses [Permutational Multivariate Analysis of Variance (PERMANOVA)] using the function adonis2 implemented in vegan (70). Finally, we used DESeq2 v1.36.0 and STAMP software (71) to identify ASVs with significant variation in abundance.
Mutagenesis.
Deletion of pvuA1 (THOG05_v1_100041) and pvuA2 (THOG05_v1_100042) in V. rotiferianus Th15_O_G05 was achieved through double homologous recombination between the pLP12 suicide plasmid (SI Appendix, Table S5) and the bacterial chromosome (29). Briefly, two fragments of around 800 bp flanking the target region were amplified, assembled by GeneArt, and cloned into the pLP12 plasmid (72). The suicide plasmid (named pAM010) was transferred by conjugation between an Escherichia coli β3914 donor (73) and V. rotiferianus Th15_O_G05 recipient using a triparental mating procedure (SI Appendix, Tables S4). The first and second recombination events leading to pAM010 integration and elimination were selected following a recently published method (29). Mutants were screened by PCR using primers del-pvuA1-A2-OG05-F and del-pvuA1-A2-OG05-R (SI Appendix, Table S3). A V. rotiferianus Th15_O_G05 mutant strain deleted for the pvuA1-2 genes was stored in glycerol at −80 °C (strain V. rotiferianus Th15_O_G05 ∆pvuA1-2).
Deletion of exsA (THOG11_v1_10133) in V. harveyi Th15_O_G11 was realised using the pSW7848T suicide plasmid (SI Appendix, Table S5). Briefly, Vibrio strains were grown at 25 °C in LB NaCl medium. Escherichia coli strains were grown at 37 °C in LB medium for cloning and conjugation experiments. Chloramphenicol (Cm) at 10 μg mL−1, thymidine (0.3 mM), and diaminopimelate (0.3 mM) were added as supplements when necessary. Induction of the Pbad promoter was achieved by the addition of 0.2% L-arabinose to the growth media, and conversely, was repressed by the addition of 1% D-glucose. Gene deletion was performed by allelic exchange using the pSW7848T suicide plasmid (73, 74). To this end, two ≈500 bp fragments flanking the target gene were amplified (see primer details in SI Appendix, Table S3), cloned into pSW7848T as previously described (75) and transferred by conjugation from E. coli as a donor to Vibrio as a recipient. Subsequently, the first and second recombinations leading to pSW7848T integration and elimination were selected on Cm/glucose and arabinose-containing media, respectively. Deletion mutants were screened by PCR using external primers flanking the target gene (SI Appendix, Table S3). Two V. harveyi Th15_O_G11 mutant strains deleted for the exsA gene were stored in glycerol at −80 °C.
Vibrio Growth in Iron-Depleted Media and Rescue.
Growth experiments in iron-depleted medium were performed with V. rotiferianus Th15_O_G05 and its ∆pvuA1-2 isogenic derivative. Isolates were grown overnight at room temperature in Artificial Sterile Seawater (ASW: NaCl 40 mM, KCl 20 mM, MgSO4 5 mM, CaCl2 2 mM) with 0.3 % (wt/vol) casamino acids and vitamins (0.1 μg/L vitamin B12, 2 μg/L biotin, 5 μg/L calcium pantothenate, 2 μg/L folic acid, 5 μg/L nicotinamide, 10 μg/L pyridoxin hydrochloride, 5 μg/L riboflavin, 5 mg/L thiamin hydrochloride). Cultures were pelleted (2 min at 15,000 g) and washed in ASW. Cells were then inoculated (1:100) into minimal media with (iron-poor) or without (iron-replete) the iron-specific chelator 2,2′-bipyridin (150 µM). Bacteria were grown in 96-well plates with orbital shaking in a Tecan microplate reader (Infinite M200) at 25 °C for 24 h. The OD600 was recorded at 30 min intervals. Rescue was performed either by adding freeze-drying concentrated V. harveyi Th15_O_G11 cell-free supernatant or by adding synthetic vibrioferrin (8.7 to 70 µM) to cultures. Vibrioferrin was synthesized according to Takeuchi et al. (76). NMR and HRMS spectroscopic data confirmed the structure (SI Appendix, Fig. S7) and were consistent with the literature (76).
Competition Assay.
Two-way competition assays were performed to investigate the performance of V. rotiferianus (able to import vibrioferrin without producing it) and its ΔpvuA1-A2 isogenic mutant (enable to import vibrioferrin) in presence of V. harveyi (vibrioferrin producer). Basically, strains of V. rotiferianus (wild-type or ΔpvuA1-A2) expressing mCherry were mixed with GFP-expressing V. harveyi at equal initial frequencies in minimal medium with (iron-poor) or without (iron-replete) the iron-specific chelator 2,2′-bipyridin (BIP 150 µM). Cultures were seeded at 1/100 with equal amounts of V. harveyi and V. rotiferianus. Growth was estimated at 20 °C, on triplicates, in CLARIOstar plate reader (BMG Labtech) by monitoring ƛex 470-15 nm/ƛem 515-20 nm for gfp-tagged strain and ƛex 570-15 nm/ƛem 620-20 nm for mCherry-tagged strain. The relative fitness (w) of each V. rotiferianus strain was calculated from the ratio of each strain’s 20-h Malthusian growth parameter (exponential growth rate) on independent experiments by comparing the relative fluorescence of each strain (77). Fitness differences between cheaters were determined by carrying out one-sample t tests.
Ethics Approval.
The animal (oyster Crassostrea gigas) testing followed all regulations concerning animal experimentation. The authors declare that the use of genetic resources fulfill the French regulatory control of access and EU regulations on the Nagoya Protocol on Access and Benefit-Sharing (ABSCH-IRCC-FR-259502-1).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Jean-François Allienne at the Bio-environment platform (University Perpignan Via Domitia, https://bio-environnement.univ-perp.fr/) for supporting NGS library preparation and sequencing as well as Valentin Outrebon and Anna Amouroux for crucial technical help. We thank Montpellier RIO Imaging (https://www.mri.cnrs.fr) for access to microscopy facilities and the qPHD platform/Montpellier genomix for access to qPCR. We thank Montpellier Exaaqt plateform (EXpérimentation Animale AQuaTique) for experimental pathology facilities. We thank the Ifremer hatchery team EMMA (PMMLT, PMMB) of La Tremblade and Bouin for the production of Pacific oysters. We thank the Regional Committee of Mediterranean Shellfish Aquaculture (CRCM) and the Ifremer for access to the shellfish tables and for boats. We thank the GENSEQ platform (http://www.labex-cemeb.org/fr/genomique-environnementale-2) from the labEx CeMEB for nucleotide sequencing. This work was funded by the Agence Nationale de la Recherche (DECICOMP, ANR-19-CE20-0004 and REVENGE, ANR-16-CE32-0008-01), IFREMER, the European Union’s Horizon 2020 Research and Innovation Program Grant Vivaldi 678589. F.D. was awarded the “Rising Star” SMIDIDI project from the French Pays de la Loire Region. A.L. was awarded a “Research Quality Grant” from the University of Perpignan (PURIVIB project). This study falls within the framework of the “Laboratoires d’Excellence (LABEX)” Tulip (ANR-10-LABX-41) and CEMEB (ANR-10-LABX-0004).
Author contributions
D.O., A.L., G.M., F.L.R., G.M.C., M.-A.T., and D.D.-G. designed research; D.O., A.L., P.H., A.M., Y.L., S.d.L.F.D., F.D., B.M., J.-M.E., B.P., L.D., D.T., L.-L.P., M.L., O.R., J.P., G.M.C., M.-A.T., and D.D.-G. performed research; N.I. contributed new reagents/analytic tools; D.O., A.L., M.B., P.H., A.M., Y.L., Y.D., S.d.L.F.D., F.D., C.M., B.M., E.T., C.C., J.-M.E., Y.G., J.V.-D., J.d.L., B.P., L.D., G.M., F.L.R., G.M.C., M.-A.T., and D.D.-G. analyzed data; and D.O., A.L., J.-M.E., G.M., F.L.R., G.M.C., M.-A.T., and D.D.-G. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Marie-Agnès Travers, Email: Marie.Agnes.Travers@ifremer.fr.
Delphine Destoumieux-Garzón, Email: Delphine.destoumieux-garzon@cnrs.fr.
Data, Materials, and Software Availability
Targeted gene sequences (hsp60, rctB, topA and mreB) and all sequence files with associated metadata generated in mesocosm experiments are available in Ifremer Oceanic database [https://doi.org/10.12770/173c0414-a3ca-4a79-b6b2-cd424ee90593 (56) and https://doi.org/10.12770/63b02659-cd9d-4834-8e6d-8adfa736755d (78)]. Genome assemblies have been deposited in the European Nucleotide Archive (ENA) under project accession no. PRJEB49488. Original R statistic scripts for metagenomics analyses and the phyloseq table are available https://doi.org/10.5281/zenodo.7599486.
Supporting Information
References
- 1.Murray J. L., Connell J. L., Stacy A., Turner K. H., Whiteley M., Mechanisms of synergy in polymicrobial infections. J. Microbiol. 52, 188–199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brogden K. A., Guthmiller J. M., “Polymicrobial diseases: Current and future research” in Polymicrobial Diseases (American Society for Microbiology Press, 2002), pp. 401–410. [Google Scholar]
- 3.Bartoli C., et al. , In situ relationships between microbiota and potential pathobiota in Arabidopsis thaliana. ISME J. 12, 2024–2038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCullers J. A., Rehg J. E., Lethal synergism between influenza virus and Streptococcus pneumoniae: Characterization of a mouse model and the role of platelet-activating factor receptor. J. Infect. Dis. 186, 341–350 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Mikušová M., Tomčíková K., Briestenská K., Kostolanský F., Varečková E., The contribution of viral proteins to the synergy of Influenza and bacterial co-infection. Viruses 14, 1064 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diedrich C. R., Flynn J. A. L., HIV-1/Mycobacterium tuberculosis coinfection immunology: How does HIV-1 exacerbate tuberculosis? Infect. Immun. 79, 1407–1417 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rottenberg M. E., Pawlowski A., Jansson M., Sko M., Tuberculosis and HIV co-infection. PLoS Pathog. 8, e1002464(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Souriant S., et al. , Tuberculosis exacerbates HIV-1 infection through IL-10/STAT3-dependent tunneling nanotube formation in macrophages. Cell Rep. 26, 3586–3599.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corey L., Synergistic copathogens—HIV-1 and HSV-2. N. Engl. J. Med. 356, 854–856 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Van De Perre P., et al. , Herpes simplex virus and HIV-1: Deciphering viral synergy. Lancet Infect. Dis. 8, 490–497 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Athena Aktipis C., et al. , Cancer across the tree of life: Cooperation and cheating in multicellularity. Philos. Trans. R. Soc. B Biol. Sci. 370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coyte K. Z., Schluter J., Foster K. R., The ecology of the microbiome: Networks, competition, and stability. Science 350, 663–666 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Sofonea M. T., Alizon S., Michalakis Y., Exposing the diversity of multiple infection patterns. J. Theor. Biol. 419, 278–289 (2017). [DOI] [PubMed] [Google Scholar]
- 14.de Lorgeril J., et al. , Immune-suppression by OsHV-1 viral infection causes fatal bacteraemia in Pacific oysters. Nat. Commun. 9, 4215–4215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clerissi C., et al. , A core of functional complementary bacteria infects oysters in Pacific Oyster Mortality Syndrome. Anim. Microbiome 5, 26 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petton B., et al. , Crassostrea gigas mortality in France: The usual suspect, a herpes virus, may not be the killer in this polymicrobial opportunistic disease. Front. Microbiol. 6, 1–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruto M., et al. , Vibrio crassostreae, a benign oyster colonizer turned into a pathogen after plasmid acquisition. ISME J. 11, 1043–1052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Roux F., Diagnosis of vibriosis in the era of genomics: Lessons from invertebrates. Rev. Sci. Tech. 35, 259–269 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Piel D., et al. , Selection of Vibrio crassostreae relies on a plasmid expressing a type 6 secretion system cytotoxic for host immune cells. Environ. Microbiol. 22, 4198–4211 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Lemire A., et al. , Populations, not clones, are the unit of vibrio pathogenesis in naturally infected oysters. ISME J. 9, 1523–1531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubio T., et al. , Species-specific mechanisms of cytotoxicity toward immune cells determine the successful outcome of Vibrio infections. Proc. Natl. Acad. Sci. U.S.A. 116, 14238–14247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lasa A., et al. , Dynamics of the Pacific oyster pathobiota during mortality episodes in Europe assessed by 16S rRNA gene profiling and a new target enrichment next-generation sequencing strategy. Environ. Microbiol. 21, 4548–4562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King W. L., Jenkins C., Seymour J. R., Labbate M., Oyster disease in a changing environment: Decrypting the link between pathogen, microbiome and environment. Marine Environ. Res. 143, 124–140 (2019). [DOI] [PubMed] [Google Scholar]
- 24.King W. L., et al. , Characterisation of the Pacific oyster microbiome during a summer mortality event. Microb. Ecol. 77, 502–512 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Saulnier D., et al. , A large-scale epidemiological study to identify bacteria pathogenic to Pacific oyster Crassostrea gigas and correlation between virulence and metalloprotease-like activity. Microbiol. Ecol. 59, 787–798 (2010). [DOI] [PubMed] [Google Scholar]
- 26.West S. A., Griffin A. S., Gardner A., Diggle S. P., Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4, 597–607 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Diggle S. P., Griffin A. S., Campbell G. S., West S. A., Cooperation and conflict in quorum-sensing bacterial populations. Nature 450, 411–414 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Smith P., Schuster M., Public goods and cheating in microbes. Curr. Biol. 29, R442–R447 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Morot A., et al. , Virulence of Vibrio harveyi ORM4 towards the European abalone Haliotis tuberculata involves both quorum sensing and a type III secretion system. Environ. Microbiol. 23, 5273–5288 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Cordero O. X., Ventouras L. A., DeLong E. F., Polz M. F., Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc. Natl. Acad. Sci. U.S.A. 109, 20059–20064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clerissi C., et al. , Microbiota composition and evenness predict survival rate of oysters confronted to Pacific Oyster Mortality Syndrome. Front Microbiol. 11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wendling C. C., Mathias Wegner K., Adaptation to enemy shifts: Rapid resistance evolution to local Vibrio spp. in invasive Pacific oysters. Proc. Biol. Sci. 282, 20142244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou K., et al. , Effect of combined function of temperature and water activity on the growth of Vibrio harveyi. Braz. J. Microbiol. 43, 1365–1375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Joven C., et al. , Oyster farming, temperature, and plankton influence the dynamics of pathogenic vibrios in the Thau Lagoon. Front. Microbiol. 9, 2530–2530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montánchez I., et al. , Analysis of Vibrio harveyi adaptation in sea water microcosms at elevated temperature provides insights into the putative mechanisms of its persistence and spread in the time of global warming. Sci. Rep. 9, 289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richard M., et al. , In situ characterisation of pathogen dynamics during a Pacific oyster mortality syndrome episode. Mar. Environ. Res. 165, 105251 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Oyanedel D., et al. , Vibrio splendidus O-antigen structure: A trade-off between virulence to oysters and resistance to grazers. Environ. Microbiol. 22, 4264–4278 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Gay M., Renault T., Pons A. M., Le Roux F., Two Vibrio splendidus related strains collaborate to kill Crassostrea gigas: Taxonomy and host alterations. Dis. Aquat. Organ. 62, 65–74 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Bruto M., Labreuche Y., James A., Piel D., Ancestral gene acquisition as the key to virulence potential in environmental Vibrio populations. ISME J. 12, 2954–2966 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bongrand C., Ruby E. G., The impact of Vibrio fischeri strain variation on host colonization. Curr. Opin. Microbiol. 50, 15–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Figueiredo A. R. T., Kramer J., Cooperation and conflict within the microbiota and their effects on animal hosts. Front. Ecol. Evol. 8, 132–132 (2020). [Google Scholar]
- 42.McFall Ngai M., Henderson B., Ruby E. G., The Influence of Cooperative Bacteria on Animal Host Biology (Cambridge University Press, 2005). [Google Scholar]
- 43.Johnson K. V. A., Foster K. R., Why does the microbiome affect behaviour? Nat. Rev. Microbiol. 16, 647–655 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Lenski R. E., May R. M., The evolution of virulence in parasites and pathogens: Reconciliation between two competing hypotheses. J. Theor. Biol. 169, 253–265 (1994). [DOI] [PubMed] [Google Scholar]
- 45.Petton B., et al. , Fine-scale temporal dynamics of herpes virus and vibrios in seawater during a polymicrobial infection in the Pacific oyster Crassostrea gigas. Dis. Aquat. Organ. 135, 97–106 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Eickhoff M. J., Bassler B. L., Vibrio fischeri siderophore production drives competitive exclusion during dual-species growth. Mol. Microbiol. 114, 244–261 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thode S. K., Rojek E., Kozlowski M., Ahmad R., Haugen P., Distribution of siderophore gene systems on a Vibrionaceae phylogeny: Database searches, phylogenetic analyses and evolutionary perspectives. PLoS One 13, e0191860 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Lorgeril J., et al. , Inefficient immune response is associated with microbial permissiveness in juvenile oysters affected by mass mortalities on field. Fish Shellfish Immunol. 77, 156–163 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Lafont M., et al. , Long-lasting antiviral innate immune priming in the Lophotrochozoan Pacific oyster, Crassostrea gigas. Sci. Rep. 7, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lafont M., et al. , A sustained immune response supports long-term antiviral immune priming in the Pacific oyster, Crassostrea gigas. mBio 11, 1–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fallet M., et al. , Early life microbial exposures shape the Crassostrea gigas immune system for lifelong and intergenerational disease protection. Microbiome 10, 1–21 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dégremont L., et al. , Relative importance of family, site, and field placement timing on survival, growth, and yield of hatchery-produced Pacific oyster spat (Crassostrea gigas). Aquaculture 249, 213–229 (2005). [Google Scholar]
- 53.Azéma P., et al. , Genetic parameters of resistance to Vibrio aestuarianus, and OsHV-1 infections in the Pacific oyster, Crassostrea gigas, at three different life stages. Genet. Sel. Evol. 49, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goh S. H., et al. , HSP60 gene sequences as universal targets for microbial species identification: Studies with coagulase-negative staphylococci. J. Clin. Microbiol. 34, 818–823 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunt D. E., et al. , Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320, 1081–1085 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Lagorce A., Data from “Vibrio bacteria sampled from juvenile oysters and seawater collected in Thau Lagoon (Languedoc-Roussillon, France).” (2022). 10.12770/173c0414-a3ca-4a79-b6b2-cd424ee90593. Accessed 1 January 2023. [DOI]
- 57.Gouy M., Guindon S., Gascuel O., SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 (2010). [DOI] [PubMed] [Google Scholar]
- 58.R. Core Team, A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2020). [Google Scholar]
- 59.Sawabe T., et al. , Updating the Vibrio clades defined by multilocus sequence phylogeny: Proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front. Microbiol. 4, 414 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sawabe T., Kita-Tsukamoto K., Thompson F. L., Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 189, 7932–7936 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vanhove A. S., et al. , Outer membrane vesicles are vehicles for the delivery of Vibrio tasmaniensis virulence factors to oyster immune cells. Environ. Microbiol. 17, 1152–1165 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Vallenet D., et al. , MicroScope–An integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 41, D636–D647 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Webb S. C., Fidler A., Renault T., Primers for PCR-based detection of ostreid herpes virus-1 (OsHV-1): Application in a survey of New Zealand molluscs. Aquaculture 272, 126–139 (2007). [Google Scholar]
- 65.Thompson J. R., et al. , Diversity and dynamics of a north atlantic coastal Vibrio community. Appl. Environ. Microbiol. 70, 4103–4110 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Di Tommaso P., et al. , Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Bolyen E., et al. , Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Callahan B. J., et al. , DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McMurdie P. J., Holmes S., Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oksanen J., et al. , Vegan: Community ecology package, 2011 (R Package Version 1.17–18, 2011).
- 71.Parks D. H., Tyson G. W., Hugenholtz P., Beiko R. G., STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo P., He X., Liu Q., Hu C., Developing universal genetic tools for rapid and efficient deletion mutation in vibrio species based on suicide t-vectors carrying a novel counterselectable marker, vmi480. PLoS One 10, 1–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Le Roux F., Binesse J., Saulnier D., Mazel D., Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 73, 777–784 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Val M. E., Skovgaard O., Ducos-Galand M., Bland M. J., Mazel D., Genome engineering in Vibrio cholerae: A feasible approach to address biological issues. PLoS Genet. 8, e1002472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goudenège D., et al. , A single regulatory gene is sufficient to alter Vibrio aestuarianus pathogenicity in oysters. Environ. Microbiol. 17, 4189–4199 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Takeuchi Y., et al. , Synthesis and siderophore activity of vibrioferrin and one of its diastereomeric isomers. Chem. Pharm. Bull. 47, 1284–1287 (1999). [Google Scholar]
- 77.Stilwell P., Lowe C., Buckling A., The effect of cheats on siderophore diversity in Pseudomonas aeruginosa. J. Evol. Biol. 31, 1330–1339 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oyanedel D., Destoumieux-Garzon D., Travers M. A., Lagorce A., Data from “rctB metabarcoding sequencing of Vibrio Harveyi during oyster experimental infections.” (2022). 10.12770/63b02659-cd9d-4834-8e6d-8adfa736755d. Accessed 1 January 2023. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Targeted gene sequences (hsp60, rctB, topA and mreB) and all sequence files with associated metadata generated in mesocosm experiments are available in Ifremer Oceanic database [https://doi.org/10.12770/173c0414-a3ca-4a79-b6b2-cd424ee90593 (56) and https://doi.org/10.12770/63b02659-cd9d-4834-8e6d-8adfa736755d (78)]. Genome assemblies have been deposited in the European Nucleotide Archive (ENA) under project accession no. PRJEB49488. Original R statistic scripts for metagenomics analyses and the phyloseq table are available https://doi.org/10.5281/zenodo.7599486.