Abstract
The metallo-β-lactamase L1 from Stenotrophomonas maltophilia was cloned, overexpressed, and characterized by spectrometric and biochemical techniques. Results of metal analyses were consistent with the cloned enzyme having 2 mol of tightly bound Zn(II) per monomer. Gel filtration chromatography demonstrated that the cloned enzyme exists as a tightly held tetramer with a molecular mass of ca. 115 kDa, and matrix-assisted laser desorption ionization and time-of-flight mass spectrometry indicated a monomeric molecular mass of 28.8 kDa. Steady-state kinetic studies with a number of diverse penicillin and cephalosporin antibiotics demonstrated that L1 effectively hydrolyzes all tested compounds, with kcat/Km values ranging between 0.002 and 5.5 μM−1 s−1. These characteristics of the recombinant enzyme are contrasted to those previously reported for metallo-β-lactamases isolated directly from S. maltophilia.
β-Lactamases hydrolyze penicillins and cephalosporins, rendering a species that is no longer an inhibitor of bacterial transpeptidases. There are four classes of β-lactamases: functional groups 1, 2, and 4, which consist of enzymes that use an active-site serine for nucleophilic attack on the β-lactam carbonyl, and group 3, which consists of enzymes that use a Zn(II) center for the hydrolysis of antibiotics (3). Recently, Rasmussen and Bush have further divided the group 3 β-lactamases into three subclasses (a, b, and c) based on their preferential hydrolysis of carbapenems relative to that of penicillins and cephalosporins (20). Currently, the serine β-lactamases are the most prevalent; however, there are available clinically useful compounds that inhibit many of the serine β-lactamases and that have been successfully employed in therapeutic regimens.
Recently, biochemical and genetic results have suggested structural and mechanistic hetereogeneity among the metallo-β-lactamases, with the most distinct of the enzymes being from Stenotrophomonas maltophilia (L1) and from several Aeromonas strains. Rasmussen and Bush have noted that the metallo-β-lactamases from the Aeromonas strains are functionally distinct from other group 3 β-lactamases, warranting their inclusion in the b subclass (20, 21). The group 3b β-lactamases appear to hydrolyze carbapenems preferentially over other antibiotics. The S. maltophilia metallo-β-lactamase is functionally similar to the enzymes from Bacillus cereus and Bacteroides fragilis; however, L1 has some distinct structural differences. For example, the pI for L1 is different from those of the other two enzymes, the L1 enzyme has been reported to exist as a tetramer while the other two are thought to be monomers, L1 is the only metallo-β-lactamase yet sequenced that does not have a cysteine [a Zn(II) binding ligand] at position 168, and the monomeric molecular mass of L1 is reported to be 4 to 5 kDa larger than that of the other two enzymes (22, 27). Even more importantly, the crystal structures of the B. cereus and Bacteroides fragilis enzymes implicated several amino acids in having functional roles in metallo-β-lactamases (5). The position of Lys171, with respect to the docked substrate’s position, suggested that this residue forms an electrostatic interaction with the carboxylate on the five- or six-member rings of penicillins or cephalosporins (5). In addition, Asp152 was implicated in orienting the metal binding ligands in the Bacteroides fragilis enzyme (5). Neither of these amino acids is conserved in the L1 enzyme, suggesting some major structural differences in the group 3a enzymes. These differences may explain the subtle differences among the group 3a enzymes in the steady-state kinetic constants and the differing interactions with mercaptoacetate compounds (19). The study of Payne et al. (19) suggests that the heterogeneity of the metallo-β-lactamases may prevent one compound from inhibiting all metallo-β-lactamases.
In order to rationally design and prepare compounds that will inhibit the metallo-β-lactamases, it is necessary to characterize several enzymes in hopes of uncovering structural or mechanistic similarities between the enzymes. Many of these studies require large amounts of pure, active enzyme and the ability to prepare site-directed mutants of the enzyme. We report here the overexpression, purification, and characterization of the cloned L1 enzyme from S. maltophilia.
MATERIALS AND METHODS
General.
All antibiotics used in this study were purchased from Sigma, except the following, which were kind gifts from the manufacturers: biapenem (Lederle-Japan); clavulanic acid, ceftizoxime, and BRL42715 (Smith Kline Beecham); and cefadroxil, cefaprozil, and cefepime (Bristol-Myers Squibb). Nitrocefin was purchased from Becton Dickinson. A bicinchoninic acid kit was purchased from Pierce Chemical Company, and gel filtration standards were purchased from Pharmacia Biotech and used according to the manufacturer’s instructions. All chromatographic steps were carried out on a Pharmacia fast-performance liquid chromatography system at 4°C.
Metal analyses were performed on a Varian inductively coupled plasma emission spectrometer. Calibration curves with five standards and correlation coefficients of better than 0.9999 were used. The final dialysis buffer was used as a blank. Three trials of each sample were made, and the results were averaged. The following absorbance wavelengths for the indicated metal ions were used to ensure the lowest detection limit possible: 213.856 nm for Zn, 324.754 nm for Cu, 321.604 nm for Ni, 238.892 nm for Co, 259.940 nm for Fe, 257.610 nm for Mn, and 228.802 nm for Cd.
Mass spectra were acquired on a Bruker Reflex II time-of-flight (TOF) mass spectrometer operating in the linear mode. Ions were produced by matrix-assisted laser desorption ionization (MALDI) with the 355-nm line of a New Wave Research MiniLase-10 neodymium: yttrium aluminum garnet laser. The matrix solution for MALDI was saturated α-cyano-4-hydroxycinnamic acid in a solvent system of acetonitrile-water (70:30, vol/vol) with 0.01% trifluoroacetic acid. The enzyme sample solution (11 μM) was mixed with the matrix solution at a ratio of about 2:5. In order to increase the mass accuracy, bovine serum albumin was added to sample solutions as an m/z calibrator at levels of ca. 10−5 M. The [M+2H]2+ and [M+3H]3+ m/z of the calibror (m/z 33,216 and 22,144, respectively) bracketed the enzyme sample’s measured m/z of 28,844. Subunit stoichiometry was determined by gel filtration chromatography, where a Sephacryl S200 column (1.6 by 60 cm) was used in a running buffer of 50 mM Tris, pH 7.5, containing 100 mM NaCl and at flow rate of 1 ml/min. RNase A, albumin, ovalbumin, chymotrypsinogen, and aldolase were used as molecular weight standards, and blue dextran was used to determine the column void volume.
Construction of overexpression plasmid.
The L1 gene, contained within a 1.6-kb EcoRI insert (27), was subcloned into plasmid pUB5811. Novel restriction sites (NdeI and HindIII) were introduced directly before and after the gene encoding L1 by PCR. The primers used to introduce these restriction sites, reading 5′ to 3′, were the N-terminal primer, GGGCATATGCGTTCTACCCTGCTCGCCTTCGCCCTG, and the C-terminal primer, GGGAAGCTTAGCGGGCCCCGGCCGTTTCCTTGGCCAG. The PCR products were phosphorylated with kinase and ATP and ligated into pUC18 to create pUB5831. The resulting plasmid was used to transform DH5α Escherichia coli cells. Plasmid pUB5831 was digested with NdeI and HindIII and ligated into pET26b to create pUB5832, which is the expression plasmid. This plasmid was transformed into BL21(DE3)pLysS E. coli cells and plated onto Luria-Bertani (LB) agar plates containing 25 μg of kanamycin per ml and 25 μg of chloramphenicol per ml.
Purification of L1.
The expression plasmid, pUB5832, was used to transform BL21(DE3)pLysS E. coli cells. A 10-ml overnight culture of these cells in LB medium was used to inoculate 1 liter of LB medium containing 25 μg of kanamycin per ml. The cells were allowed to grow at 37°C with shaking until the cells reached an optical density at 600 nm of 0.6. Protein production was induced by making the culture 1 mM in isopropyl-β-d-thiogalactopyranoside (IPTG), and the cells were allowed to shake at 37°C for 2 h. The cells were collected by centrifugation and resuspended in 20 ml of 30 mM Tris, pH 7.5, containing 600 mM NaCl. The cells were ruptured by two passages through a French press at 16,000 lb/in2, and the cell debris was collected by centrifugation. The cleared supernatant was dialyzed versus 30 mM Tris, pH 8.5, overnight at 4°C, centrifuged to remove insoluble matter, and loaded onto an equilibrated Q-Sepharose column (1.5 by 12 cm with a 25-ml bed volume). The protein was eluted with a 0 to 500 mM NaCl gradient in 30 mM Tris, pH 8.5, at 2 ml/min. The L1 enzyme eluted at <100 mM NaCl. Fractions (6 ml) containing L1 were pooled and concentrated with an Amicon ultrafiltration cell with a YM-10 membrane. Protein purity was ascertained by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. L1 was quantitated by a bicinchoninic acid assay according to the manufacturer’s directions and also by amino acid analysis (Commonwealth Biotechnologies Inc., Richmond, Va.). Four different preparations of the enzyme yielded an extinction coefficient at 280 nm (ɛ280) of 1.9 ml/mg · cm. This value was used to quantitate enzyme with the absorbance at 280 nm. N-terminal amino acid sequence analysis was performed by the Biosynthesis and Sequencing Facility at Johns Hopkins University. Circular dichroism spectra of 30 μM L1 in 50 mM phosphate, pH 7.0, or in 50 mM cacodylate, pH 7.0, were collected by Commonwealth Biotechnologies, Inc.
Steady-state kinetic studies.
Steady-state kinetic assays were conducted at 25°C in 50 mM phosphate buffer, pH 7.0, or in 50 mM cacodylate buffer, pH 7.0, containing 100 μM ZnCl2 on a Hewlett-Packard model 5480A diode array UV-visible light spectrophotometer. The molar absorptivities (Δɛ) of the antibiotics were evaluated by measuring the maximal changes in absorbance before and after enzymatic hydrolysis at 25°C. The Δɛ (per molar per centimeter) used to quantitate product were as follows: Δɛ485 = 17,400 for nitrocefin, Δɛ260 = −5,140 for cephalosporin C, Δɛ270 = −18,700 for moxalactam, Δɛ265 = −6,980 for cephaloridine, Δɛ265 = −7,050 for cefmetazole, Δɛ265 = −8,790 for cephalothin, Δɛ280 = −5,180 for cefprozil, Δɛ265 = −6,940 for cefadroxil, Δɛ260 = −9,970 for cefepime, Δɛ262 = −5,330 for cephradine, Δɛ265 = −7,000 for cefoxitin, Δɛ280 = −6,410 for cefaclor, Δɛ260 = −4,420 for ceftizoxime, Δɛ260 = −9,320 for cefuroxime, Δɛ260 = −7,040 for cefotaxime, Δɛ262 = −8,990 for cefsulodin, Δɛ215 = −3,920 for azlocillin, Δɛ235 = −936 for piperacillin, Δɛ235 = −673 for ticarcillin, Δɛ293 = −8,630 for biapenem, Δɛ215 = −5,120 for clavulanic acid, Δɛ235 = −809 for ampicillin, Δɛ358 = 1,140 for BRL42715, and Δɛ235 = −936 for penicillin G. When possible, substrate concentrations were varied between 0.1 and more than 10 times the Km value. Steady-state kinetic constants, the Km and the catalytic constant (kcat), were determined by fitting data for initial velocity versus substrate concentration directly to the Michaelis equation with CurveFit. The reported errors reflect fitting uncertainties. All steady-state kinetic studies were performed in triplicate with recombinant L1 from three different enzyme preparations.
Phosphate inhibition studies were conducted with 50 mM cacodylate buffer, pH 7.0, at 25°C with nitrocefin as the substrate and sodium phosphate, pH 7.0, as the inhibitor. Nitrocefin concentrations were varied between 3 and 75 μM, and phosphate concentrations were varied between 0 and 400 mM phosphate. The mode of inhibition was determined by generating Lineweaver-Burk plots of the data, and the Ki of phosphate was determined by replotting the data for slope and intercept versus phosphate concentration (23, 26).
RESULTS
Overexpression and purification of L1.
In order to isolate large quantities of metallo-β-lactamase (L1) and to allow the use of site-directed mutagenesis to study the structure and function of the enzyme, efforts were made to overexpress and purify L1 from E. coli. The L1 gene was previously cloned as a 1.6-kb EcoRI fragment and sequenced (27). In order to place the L1 gene close to the ribosomal binding site and promoter of a commercially available overexpression plasmid, PCR with nondegenerate primers was used to introduce novel, overexpression plasmid-compatible restriction sites. The PCR product was blunt end ligated into pUC18, to allow facile sequencing of the gene, which confirmed a successful PCR experiment. No mutations that altered the translation product were identified. The L1 gene was then restriction digested with HindIII and NdeI and ligated directly into pET26b to create pUB5832. The insertion of L1 into pET26b allowed production of L1 to be under the control of a strong T7 promoter, the expression of which was enhanced by 1 mM IPTG. Attempts to express L1 directly in E. coli in the absence of such a promoter were unsuccessful. Small test cultures (25 ml) of pUB5832 in BL21(DE3)pLysS E. coli cells demonstrated that L1 was overexpressed >100-fold over basal amounts. Localization studies clearly demonstrated that modified L1 is periplasmic.
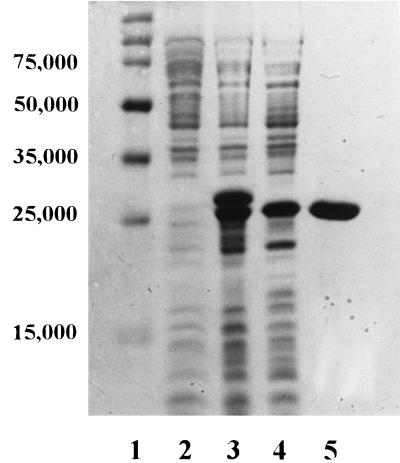
By Q-Sepharose chromatography, the L1 enzyme was isolated to >95% purity as described in Materials and Methods and as indicated by the SDS-polyacrylamide gel in Fig. 1. Unlike early reports on the recombinant Bacteroides fragilis metallo-β-lactamase (CcrA) (6, 30), L1 is produced as a soluble protein. The overall yield of active L1 after chromatography is 15 to 20 mg/liter of culture. N-terminal amino acid sequence analysis of purified, recombinant L1 revealed the sequence A-E-V-P-L.
FIG. 1.
SDS-polyacrylamide gel of L1 purification. Lane 1, Novagen perfect protein molecular weight markers; lane 2, boiled cell fraction of BL21(DE3)pLysS containing pUB5832 before induction; lane 3, boiled cell fraction of BL21(DE3)pLysS containing pUB5832 after a 2-h induction with 1 mM IPTG; lane 4, crude protein after French press; lane 5, purified L1. Molecular weights are noted at the left.
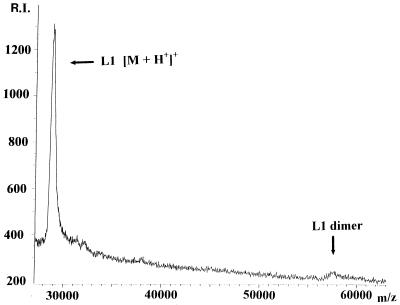
MALDI-TOF mass spectrometry was used to determine the molecular mass of recombinant, overexpressed L1 (Fig. 2). The peak at 28,844 m/z was assigned as the [M+H]+ peak for L1. This value represents an error of 0.014% in the error predicted from the DNA sequence. Including the masses of two Zn(II) atoms in the predicted mass increases the error to 0.43%, suggesting that the Zn(II) ions may not remain bound during the MALDI process. The spectrum also suggests the presence of some dimer in the sample, with a small peak at 57,735 m/z (Fig. 2), and there were no peaks at higher m/z values.
FIG. 2.
MALDI-TOF mass spectrogram of recombinant L1. The labeled peaks exhibit values of 28,844 m/z for the L1 monomer ([M+H+]+) and 57,735 m/z for the L1 dimer ([2M+H+]+). See Materials and Methods for sample conditions. R.I., relative intensity.
Gel filtration chromatography of purified L1, with a Sephacryl S200 column, resulted in a peak exhibiting a mobility consistent with a molecular mass of 110 kDa, suggesting that L1 exists as a tetramer under the conditions used to run the gel filtration studies. This result is supported by sedimentation studies which revealed a solution molecular mass for L1 of 115 kDa (24a).
Metal content of L1.
In order to determine the metal-enzyme stoichiometry of recombinant L1, metal analyses were performed. After purification of the enzyme, L1 was dialyzed versus 3× 1 liter of freshly prepared, Chelexed 50 mM HEPES buffer, pH 7.5, for 3 days at 4°C to remove any loosely bound metal ions. Metal analyses were performed on four different preparations of the enzyme, and the data indicate that recombinant L1 binds 1.9 ± 0.2 Zn(II) ions per monomer and does not contain any appreciable amounts of Co(II), Cu(II), Ni(II), Mn(II), or Fe. When purified L1 is dialyzed versus 50 mM acetate, pH 4.6, for 2 days at 4°C, the enzyme retains >90% of its bound Zn(II). These results suggest that L1 tightly binds two Zn(II) ions per monomer, in agreement with the results of a previous report (2).
Steady-state kinetic studies.
In phosphate buffer, recombinant L1 effectively hydrolyzes and exhibits saturation kinetics for all penicillins, carbapenems, and cephalosporins tested (Table 1). In general, there was a slightly higher kcat for penicillins, warranting L1’s inclusion in functional subclass a of group 3 β-lactamases (20). Kinetic studies of these compounds resulted in kcat/Km values ranging from 0.002 to 5.5 μM−1 s−1. The large relative errors in Km values for a few of the compounds reflect our inability to fully saturate the enzyme with these compounds in 50 mM phosphate buffer, pH 7.0, due primarily to substrate inhibition at high concentrations of the substrate.
TABLE 1.
Steady-state kinetic constants of recombinant L1 and metallo-β-lactamase isolated from S. maltophilia ULA-511
| Compound | Recombinant L1
|
ULA-511
|
||||||
|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | Buffera | Reference | |
| Piperacillin | 200 ± 20 | 420 ± 20 | 2.1 | 20 ± 1 | 140 | 7.1 | B | 9 |
| Ampicillin | 300 ± 15 | 580 ± 20 | 1.9 | 40 ± 3 | 175 | 4.4 | A | 10 |
| Cefuroxime | 130 ± 40 | 53 ± 9 | 0.41 | 30 ± 3 | 80 | 2.7 | A | 10 |
| BRL42715 | 600 ± 70 | 170 ± 10 | 0.28 | 270 ± 10 | 0.55 | 0.0020 | D | 16 |
| Penicillin G | 75 ± 10 | 410 ± 20 | 5.5 | 50 ± 6 | 1,100 | 22 | A | 10 |
| Cefprozil | 100 ± 20 | 15 ± 1 | 0.15 | |||||
| Cefotaxime | 160 ± 20 | 140 ± 9 | 0.88 | 26 ± 3 | 66 | 2.5 | A | 10 |
| Ticarcillin | 490 ± 100 | 440 ± 70 | 0.90 | 140 ± 2 | 375 | 2.7 | B | 9 |
| Biapenem | 75 ± 11 | 64 ± 4 | 0.85 | 56 ± 1 | 65 | 1.2 | C | 12 |
| Nitrocefin | 12 ± 1 | 31 ± 1 | 2.6 | 7 ± 1 | 20 | 2.9 | A | 10 |
| Cephalothin | 43 ± 7 | 65 ± 3 | 1.5 | 90.9 | ||||
| Cefaclor | 31 ± 3 | 16 ± 1 | 0.52 | |||||
| Cephaloridine | 170 ± 30 | 67 ± 5 | 0.39 | 300 ± 50 | 28 | 0.093 | A | 10 |
| Cephradine | 260 ± 40 | 16 ± 1 | 0.062 | 50 ± 5 | 12.5 | 0.25 | B | 9 |
| Ceftizoxime | 7.9 ± 1.6 | 2.7 ± 0.1 | 0.34 | |||||
| Cefoxitin | 3.3 ± 0.4 | 2.2 ± 0.1 | 0.67 | 2 ± 0.05 | 1.10 | 0.55 | A | 10 |
| Cefmetazole | 4.1 ± 0.2 | 4.7 ± 0.1 | 1.1 | |||||
| Cephalosporin C | 180 ± 18 | 280 ± 13 | 1.6 | 25 ± 1 | 62 | 2.5 | E | 11 |
| Clavulanic acid | 22 ± 2 | 11 ± 1 | 0.50 | |||||
| Moxalactam | 5 ± 1 | 0.15 ± 0.01 | 0.030 | 1 ± 0.05 | 0.29 | 0.29 | A | 10 |
| Cefadroxil | 250 ± 50 | 23 ± 2 | 0.092 | |||||
| Cefepime | 130 ± 30 | 0.33 ± 0.04 | 0.025 | >1,000 | >15.0 | B | 9 | |
| Azlocillin | 59 ± 6 | 37 ± 2 | 0.63 | 6 ± 0.1 | 75 | 13 | B | 9 |
| Cefsulodin | 32 ± 5 | 0.086 ± 0.005 | 0.0027 | 5 ± 0.3 | 7.5 | 1.5 | B | 9 |
Buffer A was 50 mM sodium cacodylate, pH 7.0, containing 100 μM Zn(II) at 35°C. Buffer B was 30 mM sodium cacodylate, pH 6.5, containing 100 μM Zn(II) at 30°C. Buffer C was 30 mM sodium cacodylate, pH 6.5, containing 100 μM Zn(II) at 30°C. Buffer D was 50 mM sodium cacodylate, pH 7.0, containing 100 μM Zn(II) at 35°C. Buffer E was 30 mM sodium cacodylate, pH 7.0, containing 50 μM Zn(II) at 30°C.
In order to ascertain the effect of different assay conditions on the steady-state kinetic constants of recombinant L1, studies examining the effects of added Zn(II) and elevated temperature were conducted. In studies using nitrocefin as the substrate, the inclusion of 100 μM Zn(II) in the phosphate buffer resulted in an increase in the kcat by a factor of 1.4, with no effect on the Km. When the temperature of the kinetic assays was raised from 25 to 35°C, the value of kcat increased by a factor of 2, with no effect on Km. The effects of added Zn(II) and elevated temperatures on L1 activity would have resulted in even higher kcat values than those listed in Table 1 and demonstrate that recombinant L1 is as active as the ULA-511 metallo-β-lactamase.
To test the effects of differing buffers, steady-state kinetic studies of recombinant L1 with cacodylate buffer were conducted (Table 2). For the five compounds tested, L1 exhibited Km values two to five times lower in cacodylate buffer than in phosphate buffer. In fact, the Km values for L1 in cacodylate buffer compare favorably to those reported for the metallo-β-lactamase isolated from strain ULA-511. The kcat values for L1 in cacodylate buffer were slightly raised (factor of ∼1.2), except when ampicillin was used as the substrate, which resulted in a decrease. The increased kcat values can be explained by the added Zn(II) in the buffer (see above).
TABLE 2.
Steady-state kinetic constants of recombinant L1 and other metallo-β-lactamases isolated from S. maltophilia in cacodylate buffer
| Compound | Recombinant L1a
|
Metallo-β-lactamase isolated from S. maltophilia
|
|||
|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | Km (μM) | kcat (s−1) | Reference | |
| Piperacillin | 38 ± 3 | 440 ± 10 | 20 ± 1 | 140 | 10 |
| Ampicillin | 55 ± 5 | 520 ± 10 | 40 ± 3 | 175 | 10 |
| BRL42715 | 240 ± 20 | 210 ± 10 | 270 ± 10 | 0.55 | 16 |
| Penicillin G | 22 ± 3 | 480 ± 10 | 50 ± 6 | 1,100 | 10 |
| Nitrocefin | 4 ± 1 | 41 ± 1 | 7 ± 1 | 20 | 10 |
The buffer used for studies with recombinant L1 was 50 mM sodium cacodylate, pH 7.0, containing 100 μM Zn(II) at 25°C.
To further probe why elevated Km values are observed in phosphate buffer, inhibition studies using phosphate as the inhibitor and nitrocefin as the substrate were conducted. Analysis of the data demonstrates that phosphate is a weak, competitive inhibitor of L1, with a Ki of 30 ± 1 mM. Circular dichroism spectra of L1 demonstrate that no large, buffer-induced structural change occurs when the enzyme is in phosphate and cacodylate buffers.
DISCUSSION
The initial purification of L1 directly from strain IID 1275 involved protein induction with penicillin G and protein purification with various anion-exchange and size exclusion columns (2, 22). Dufresne and coworkers reported successful cloning and expression of L1 in E. coli; however, the expression levels were apparently too low for purification (7). All subsequent purification protocols were performed on metallo-β-lactamase that had been isolated directly from the S. maltophilia clinical strain ULA-511. Enzyme production was induced with imipenem, and the purification protocol involved isolation of periplasmic proteins and two anion-exchange columns (10). The work presented here describes the preparation of an overexpression system in E. coli that has L1 production under the control of a T7 promoter. The overexpressed enzyme can be purified with one anion-exchange chromatography step, and the overall yield is 15 to 20 mg of >95% pure, soluble enzyme per liter of growth culture.
The biochemical properties of recombinant L1 are very similar to those of the enzymes purified from clinical strain IID 1275 (2, 22) and from several other clinical strains (17). The molecular mass of metallo-β-lactamase from S. maltophilia has been reported to be 26 (17) and 31.6 (2) kDa. The predicted molecular mass of unmodified, monomeric L1, determined from analysis of the DNA sequence, is 30.8 kDa (27). N-terminal amino acid sequence analysis of purified, recombinant L1 revealed the sequence A-E-V-P-L, indicating that a 21-amino-acid leader sequence is removed from the primary translation product during processing in vivo (27). The removal of such a leader sequence is consistent with L1 being a secreted enzyme, which is true of most β-lactamases. Therefore, the predicted molecular mass of modified L1 is 28,840 Da. The MALDI-TOF mass spectrum of recombinant L1 shows a major peak with an m/z of 28,844, representing an error of 0.014% from the predicted molecular mass of monomeric L1. This mass also suggests that L1 does not retain its metal ions during the MALDI process.
Using gel filtration chromatography, Saino et al. (22) and Paton et al. (17) reported that the subunit stoichiometry of L1 is tetrameric and that it has an overall molecular mass of 118 and 96 kDa, respectively. A similar stoichiometry was reported by Bicknell and coworkers (2). MALDI-TOF mass spectrometry was used to probe the subunit stoichiometry of recombinant L1. A small, reproducible peak was observed at 57,735 m/z and is assigned as the L1 dimer. This m/z value is not twice that of the monomer [M+H+]+ peak; however, this discrepancy can be explained by the poor signal-to-noise ratio for the dimer peak and by the possibility of the dimer retaining some of its bound Zn(II) during ionization. Spectra were also taken at higher m/z values; however, there were no observed peaks. The conditions used for the MALDI experiment may have lowered the levels of other oligomers held together by noncovalent interactions. The internal energy deposited in the sample by MALDI may cleave weak bonds that hold oligomeric proteins together. In addition, recent evidence indicates that the cinnamic acid matrix used in our studies may destroy noncovalent interactions (14); however, covalent linkages such as dissulfide bonds should be stable in the matrix as well as during the MALDI experiment. A more gentle matrix solution was not used because the use of such matrices often leads to less intense signals.
Gel filtration chromatography was, therefore, used to detect the presence of any oligomers held together by noncovalent interactions. A molecular mass of 109 kDa was observed for L1, suggesting, within the error of the technique, a tetrameric structure for recombinant L1. The predicted molecular mass of the tetramer including the mass of eight Zn(II) ions is 115,900 Da, which is similar to the molecular mass determined by sedimentation studies (24a).
Previously, it was reported that L1 tightly binds two Zn(II) ions per monomer (2, 22). However, recent reports suggest that all metallo-β-lactamases are not dinuclear Zn(II) enzymes. Specifically, the crystal structure of the B. cereus enzyme indicated only one Zn(II) ion (4), and recent evidence suggests that the Aeromonas enzyme may tightly bind only one Zn(II) ion (24a, 25). The study of Carfi et al., however, may have described experiments on an enzyme that did not contain its full complement of metal (8). In addition, the crystal structure of the Bacteroides fragilis enzyme indicated a closely spaced, dinuclear Zn(II) binding site (5). Since the S. maltophilia enzyme is the only metallo-β-lactamase that does not have all the putative metal binding ligands conserved (Cys168 is replaced by Ser), the metal content of the purified L1 was assessed. Recombinant L1 tightly binds two Zn(II) ions per monomer, suggesting an active-site structure similar to that of the enzyme isolated directly from strain IID 1275.
Initial steady-state kinetic studies demonstrated that L1 from strain IID 1275 hydrolyzes penicillins ≥2 orders of magnitude faster than cephalosporins, and several compounds, including cefoxitin, cefmetazole, and moxalactam, were reported to be inhibitors, with micromolar Ki values (22). Other kinetic studies of metallo-β-lactamase from S. maltophilia demonstrate that the ULA-511 metallo-β-lactamase effectively hydrolyzes all compounds tested, including cefoxitin, cefmetazole, and moxalactam (9, 10, 12, 16). In the last-mentioned studies, no consistent assay conditions were reported. For example, 50 mM Tris at pH 8.0, 50 mM phosphate at pH 7.0 (17), 50 mM cacodylate at pH 7.0 (10, 16), and 30 mM cacodylate at pH 6.5 (9, 12) and pH 7.0 (11) have been used as buffers (assay conditions for previous studies are listed in Table 1). In addition, the previous studies used buffers containing 0 to 100 μM Zn(II) and performed the experiments at temperatures ranging from 30 to 35°C (9–12, 16, 17). Previously, we and others studied the metallo-β-lactamase (CcrA) from Bacteroides fragilis, and kinetic studies were conducted in 50 mM phosphate buffer, pH 7.0 (6, 30). Having no consensus, we selected 50 mM phosphate, pH 7.0, as our buffer to facilitate comparison to our previous work on CcrA (Table 1).
In comparison to the results of the previous kinetic studies of the ULA-511 enzyme, recombinant L1 exhibited higher kcat values for all compounds tested except cefuroxime, penicillin G, moxalactam, cefepime, azlocillin, and cefsulodin (9–12, 16). However, in 50 mM phosphate buffer, pH 7.0, recombinant L1 exhibited higher Km values for all tested compounds except cephaloridine and cefepime (9–12, 16). Even though recombinant L1 generally exhibited higher kcat values at low temperatures and was shown to tightly bind two Zn(II) ions per monomer, the high Km values may suggest improper folding of the overexpressed enzyme. Experiments were conducted to address the effects of different assay conditions on the steady-state kinetic constants for recombinant L1.
Assays at higher temperatures and the inclusion of Zn(II) in kinetic buffers resulted in higher kcat values for recombinant L1. Most enzymes are more active at elevated temperatures, as long as the enzyme is stable at the tested temperature (13). The increase in kcat by a factor of 2 with a 10°C increase in temperature suggests that recombinant L1 is stable at 35°C. The inclusion of Zn(II) into the assay buffers for L1 resulted in an average increase in kcat of a factor of 1.4. Previously, Zn(II) has apparently been included in buffers to stabilize the enzyme and to ensure that the enzyme contains the proper stoichiometry of Zn(II), etc. Recombinant L1 is stable and is as active as the ULA-511 metallo-β-lactamase in the absence of added Zn(II).
In order to better compare the steady-state kinetic constants determined for recombinant L1 to those determined for the ULA-511 enzyme, additional kinetic studies were performed with 50 mM cacodylate, pH 7.0, containing 100 μM Zn(II). In this cacodylate buffer, L1 exhibits kcat values similar to those determined when 50 mM phosphate, pH 7.0, containing 100 μM Zn(II) was used; however, the Km values for L1 in the cacodylate buffer are significantly lower (Table 2) and are similar to those reported for the ULA-511 metallo-β-lactamase.
To probe this buffer-induced lowering of Km, inhibition studies were conducted with phosphate as the inhibitor, nitrocefin as the substrate, and 50 mM cacodylate, pH 7.0, as the buffer. Analysis of the data revealed that phosphate is a weak, competitive inhibitor of L1, with a Ki value of 30 mM. A competitive inhibitor is expected to increase the apparent Km and to leave the maximum rate of hydrolysis (Vmax) unchanged. This increased apparent Km may explain why previous studies using cacodylate buffer did not report problems with substrate inhibition; the enzyme could be saturated at lower concentrations of the substrate. With this in mind, we will conduct all future kinetic studies on metallo-β-lactamases in cacodylate buffer.
We cannot discount other possibilities that might explain the differences between our reported kinetic constants and those from other sources. For example, L1 was cloned from an S. maltophilia IID 1275 strain while most of the other kinetic studies were conducted with an S. maltophilia ULA-511 enzyme. There is no certainty that the metallo-β-lactamases from these strains are identical. Even more significantly, variations in data handling and fitting may account for some of the observed differences. For example, substrate inhibition at high concentrations of the substrate can appear to be saturation of the enzyme; therefore, lower values for the maximum rate of hydrolysis (Vmax) and Km would be reported.
The metallo-β-lactamases have assumed increasing clinical significance due to their ability to hydrolyze carbapenems such as imipenem and meropenem, which, apart from a few exceptions, are poorly hydrolyzed by serine β-lactamases. These enzymes hydrolyze all known penicillin- and cephalosporin-based compounds and are also resistant to all commercially available serine-β-lactamase inhibitors. Dissemination of these enzymes throughout bacterial populations is a key issue in therapeutic strategies. Plasmid-mediated metallo-β-lactamases have now been identified in key pathogens such as Klebsiella pneumoniae, Serratia marcescens, Pseudomonas aeruginosa, and Bacteroides fragilis (1, 15, 28, 29). Furthermore, the metallo-β-lactamase from S. marcescens, IMP1, has been mobilized on a Tn9106-like integron and has been identified in Pseudomonas putida, K. pneumonia, and Alcaligenes spp. (24). Metallo-β-lactamases have also been identified and characterized from emerging pathogens such as Aeromonas spp., S. maltophilia, and Burkholderia cepacia and have been the subject of recent reviews (18, 20).
The increasing prevalence of metallo-β-lactamases in major pathogenic organisms will gravely affect the antibiotic therapies offered to combat bacterial infections. Given the large population of immunocompromised patients (from age, AIDS, and cancer treatment, etc.), even bacterial infections due to minor pathogens are becoming problematic. Previous studies of metallo-β-lactamases have demonstrated structural and kinetic heterogeneity among the enzymes, and an inhibition study using mercaptoacetic acid thiol ester derivatives suggested that one inhibitor will probably not be a clinically useful inhibitor for all of the metallo-β-lactamases (19). It is for this reason that structural and mechanistic characterizations of several group 3 β-lactamases are required. A recombinant source of L1 now allows production of large quantities of enzyme, without the need to grow large amounts of S. maltophilia, and of site-directed mutants. Structural and mechanistic studies of L1 are now in progress.
ACKNOWLEDGMENTS
This work was supported by the Pharmaceutical Researchers and Manufacturers of America Foundation, the Ohio Board of Regents Research Challenge Program, and Miami University (support to M.W.C.).
We thank SmithKline Beecham, Lederle-Japan, and Bristol-Myers Squibb for the antibiotics used in the steady-state kinetic studies. We also thank Jaran Jai-nhuknan for his assistance in obtaining the MALDI-TOF mass spectrometry data and Scott Holmstrom for his assistance in obtaining the inductively coupled plasma emission spectrometry data.
REFERENCES
- 1.Bandoh K, Watanabe K, Muto Y, Tanaka Y, Kato N, Ueno K. Conjugal transfer of imipenem resistance in Bacteriodes fragilis. J Antibiot. 1992;45:542–547. doi: 10.7164/antibiotics.45.542. [DOI] [PubMed] [Google Scholar]
- 2.Bicknell R, Emanuel E L, Gagnon J, Waley S G. The production and molecular properties of the zinc β-lactamase of Pseudomonas maltophilia IID 1275. Biochem J. 1985;229:791–797. doi: 10.1042/bj2290791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carfi A, Pares S, Duee E, Galleni M, Duez C, Frere J M, Dideberg O. The 3-D structure of a zinc metallo-β-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995;14:4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Concha N O, Rasmussen B A, Bush K, Herzberg O. Crystal structure of the wide-spectrum binuclear zinc β-lactamase from Bacteroides fragilis. Structure. 1996;4:823–836. doi: 10.1016/s0969-2126(96)00089-5. [DOI] [PubMed] [Google Scholar]
- 6.Crowder M W, Wang Z, Franklin S L, Zovinka E P, Benkovic S J. Characterization of the metal binding sites of the β-lactamase from Bacteroides fragilis. Biochemistry. 1996;35:12126–12132. doi: 10.1021/bi960976h. [DOI] [PubMed] [Google Scholar]
- 7.Dufresne J, Vezina G, Levesque R C. Molecular cloning and expression of the imipenem-hydrolyzing β-lactamase gene from Pseudomonas maltophilia in Escherichia coli. Rev Infect Dis. 1988;10:806–817. doi: 10.1093/clinids/10.4.806. [DOI] [PubMed] [Google Scholar]
- 8.Fabiane S M, Sohi M, Wan T, Payne D J, Bateson J H, Mitchell T, Sutton B J. Crystal structure of a metallo-β-lactamase II from B. cereus at 2.5 Å. International Union of Crystallography, XVII Congress and General Assembly, August 8–17, Seattle, Wash. 1996. [Google Scholar]
- 9.Felici A, Amicosante G. Kinetic analysis of extension of substrate specificity with Xanthomonas maltophilia, Aeromonas hydrophila, and Bacillus cereus metallo-β-lactamases. Antimicrob Agents Chemother. 1995;39:192–199. doi: 10.1128/aac.39.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felici A, Amicosante G, Oratore A, Strom R, Ledent P, Joris B, Fanuel L, Frere J M. An overview of the kinetic parameters of class B β-lactamases. Biochem J. 1993;291:151–155. doi: 10.1042/bj2910151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felici A, Perilli M, Fransceschini N, Rossolini G M, Galleni M, Frere J-M, Oratore A, Amicosante G. Sensitivity of Aeromonas hydrophila carbapenemase to Δ3-cephems: comparative study with other metallo-β-lactamases. Antimicrob Agents Chemother. 1997;41:866–868. doi: 10.1128/aac.41.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felici A, Perilli M, Segatore B, Franceschini N, Setacci D, Oratore A, Stefani S, Galleni M, Amicosante G. Interactions of biapenem with active-site serine and metallo-β-lactamases. Antimicrob Agents Chemother. 1995;39:1300–1305. doi: 10.1128/aac.39.6.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fersht A. Enzyme structure and mechanism. 2nd ed. New York, N.Y: W. H. Freeman and Co.; 1985. [Google Scholar]
- 14.Glocker M, Bauer S H J, Kast J, Volz J, Przybylski M. Characterization of specific noncovalent protein complexes by UV matrix-assisted laser desorption ionization mass spectrometry. J Mass Spectrometry. 1996;31:1221–1227. doi: 10.1002/(SICI)1096-9888(199611)31:11<1221::AID-JMS410>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 15.Ito H, Arakawa Y, Ohsuka S, Wacharotayankun R, Kato N, Ohta M. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob Agents Chemother. 1995;39:824–829. doi: 10.1128/aac.39.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matagne A, Ledent P, Monnaie D, Felici A, Jamin M, Raquet X, Galleni M, Klein D, Francois I, Frere J M. Kinetic study of interaction between BRL42715, β-lactamases, and d-alanyl-d-alanyl peptidases. Antimicrob Agents Chemother. 1995;39:227–231. doi: 10.1128/aac.39.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paton R, Miles R S, Amyes S G B. Biochemical properties of inducible β-lactamases produced from Xanthomonas maltophilia. Antimicrob Agents Chemother. 1994;38:2143–2149. doi: 10.1128/aac.38.9.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Payne D J. Metallo-β-lactamases—a new therapeutic challenge. J Med Microbiol. 1993;39:93–99. doi: 10.1099/00222615-39-2-93. [DOI] [PubMed] [Google Scholar]
- 19.Payne D J, Bateson J H, Gasson B C, Proctor D, Khushi T, Farmer T H, Tolson D A, Bell D, Skett P W, Marshall A C, Reid R, Ghosez L, Combret Y, Marchand-Brynaert J. Inhibition of metallo-β-lactamases by a series of mercaptoacetic acid thiol ester derivatives. Antimicrob Agents Chemother. 1997;41:135–140. doi: 10.1128/aac.41.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasmussen B A, Bush K. Carbapenem-hydrolyzing β-lactamases. Antimicrob Agents Chemother. 1997;41:223–232. doi: 10.1128/aac.41.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossolini G M, Walsh T, Amicosante G. The Aeromonas metallo-β-lactamases: genetics, enzymology, and contribution to drug resistance. Microb Drug Resist. 1996;2:245–252. doi: 10.1089/mdr.1996.2.245. [DOI] [PubMed] [Google Scholar]
- 22.Saino Y, Kobayashi F, Inoue M, Mitsuhashi S. Purification and properties of inducible penicillin β-lactamase isolated from Pseudomonas maltophilia. Antimicrob Agents Chemother. 1982;22:564–570. doi: 10.1128/aac.22.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segel I H. Enzyme kinetics. New York, N.Y: John Wiley and Sons, Inc.; 1993. [Google Scholar]
- 24.Senda K, Arakawa Y, Nakashima K, Ito H, Ichiyama S, Shimokata K, Kato N, Ohta M. Multifocal outbreaks of metallo-β-lactamase producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob Agents Chemother. 1996;40:349–353. doi: 10.1128/aac.40.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Spencer, J. Unpublished data.
- 25.Valladares M H, Felici A, Weber G, Adolph H W, Zeppezauer M, Rossolini G M, Amicosante G, Frere J M, Galleni M. Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-β-lactamase activity and stability. Biochemistry. 1997;36:11534–11541. doi: 10.1021/bi971056h. [DOI] [PubMed] [Google Scholar]
- 26.Vincent J B, Crowder M W, Averill B A. Spectroscopic and kinetics studies of a high-salt stabilized form of the purple acid phosphatase from bovine spleen. Biochemistry. 1991;30:3025–3034. doi: 10.1021/bi00226a007. [DOI] [PubMed] [Google Scholar]
- 27.Walsh T R, Hall L, Assinder S J, Nichols W W, Cartwright S J, MacGowan A P, Bennett P M. Sequence analysis of the L1 metallo-β-lactamase from Xanthomonas maltophilia. Biochim Biophys Acta. 1994;1218:199–201. doi: 10.1016/0167-4781(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:147–151. doi: 10.1128/aac.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi H, Nukaga M, Sawai T. Sequence of the Klebsiella pneumoniae RDK4 metallo-β-lactamase. EMBO database accession no. D29636. 1994. [Google Scholar]
- 30.Yang Y, Rasmussen B A, Bush K. Biochemical characterization of the metallo-β-lactamase CcrA from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1992;36:1155–1157. doi: 10.1128/aac.36.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]