Abstract
In this work, luminescent carbon dots with gardenia seeds as carbon precursors (GCDs) were synthesized using a one-step mild pyrolysis process and were then used as probes for imaging of bacterial (Escherichia coli). The GCDs showed a strong emission at 430 nm when excited at 370 nm. The relative fluorescence quantum yield of GCDs was found to be 1.13% in an aqueous medium. Rapid internalization of the GCDs by bacteria was confirmed by three colors (blue, green, and yellow) images that were obtained using confocal fluorescence microscopy. In addition, GCDs were noted to exhibit potent scavenging activities against DPPH˙, ˙OH, and ˙O2− free radicals. GCDs were also assayed as antioxidants in an oil sample by volumetric determination of the peroxide value. Thus, GCDs exhibited good antioxidant properties both in aqueous and oil media. In addition, a free fatty acid quantification kit in the presence of GCDs showed enhanced fluorescence detection of palmitic acid with a remarkably good limit of detection of 0.08 μM, which is lower than that in the absence of GCDs (0.76 μM). The proposed fluorescence method was then successfully used to determine the concentration of palmitic acid spiked in milk powder samples, with spiked recoveries of 82.6–109.6% and relative standard deviations of 0.9–4.6%.
Gardenia seed carbon dots: bacterial imaging and antioxidant activity.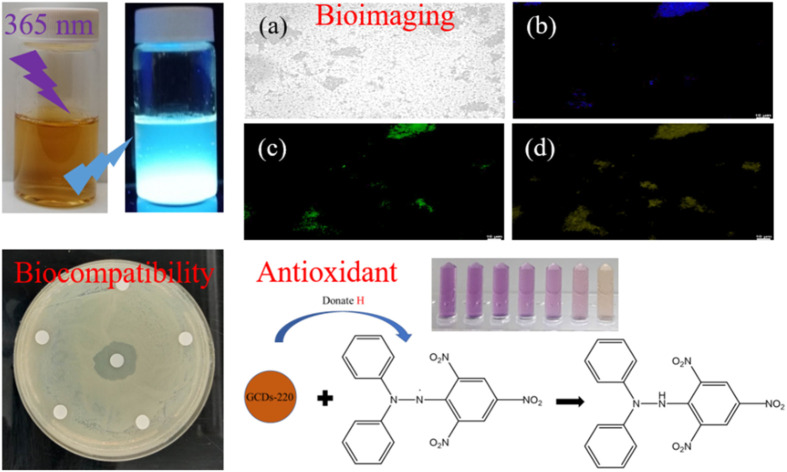
Introduction
The unique luminescent properties, chemical stability, low toxicity, and minimal environmental impact of carbon dots (CDs) have garnered significant attention.1–4 CDs can be synthesized using hydrothermal methods from various natural resources, including albumin, chitosan, gelatin, fruit peels, grains, and plant matter, as well as vegetable waste such as apple or grape pomace.5–7 The choice of precursor material played a crucial role in determining the yields and properties of the CDs, thereby making them promising candidates for biomedical applications.8–10 This approach allows for producing “green” nanoparticles, as highlighted in several comprehensive reviews focused on synthesizing CDs from biomass waste sources.11 These reviews also discuss surface modification techniques to enhance the physicochemical properties of CDs and explore their wide-ranging applications in bioimaging, biosensing, catalysis, forensics, and optoelectronics.12–14
Fluorescence imaging experiments were conducted using the prepared CDs, which exhibited accumulation in various cellular compartments such as cell membranes, cytoplasm, and nucleus.15–17 For instance, Kasibabu et al. utilized fluorescent CDs synthesized via a hydrothermal method using pomegranate fruits (Punica granatum) as the precursors.18 These CDs served as effective probes for imaging bacterial cells (Pseudomonas aeruginosa) and fungal cells (Fusarium avenaceum). Excitation at 383 nm resulted in a strong emission at 453 nm from the CDs. Confocal fluorescence microscopy revealed the rapid internalization of CDs by the cells, as evidenced by two-color (green and red) images. In another study, Goel's research group reported the synthesis of luminescent carbon quantum dots (CQDs) derived from watermelon juice.19 These CQDs were functionalized and used as nanomaterials for the detection of toxic lead (Pb2+) ions in contaminated water and cancer cells. By incorporating surface passivating ligands such as ethanolamine (EA) and ethylenediamine (ED) into watermelon juice, watermelon–ethanolamine (WMEA)-CQDs and watermelon–ethylenediamine (WMED)-CQDs were obtained. Compared to non-doped WM-CQDs, WMEA-CQDs exhibited an approximately ten-fold increase in fluorescence intensity, while WMED-CQDs showed an approximately six-fold increase. In an aqueous medium, the relative fluorescence quantum yields of WMEA-CQDs and WMED-CQDs were measured to be 8% and 7%, respectively. The functionally modified WMED-CQDs demonstrated the ability to detect Pb2+ metal ions in polluted water and a human cervical cancer cell line (HeLa). These findings highlight the potential of eco-friendly nanomaterials as diagnostic tools in environmental and biomedical research fields.
In addition to their bioimaging applications, CDs have gained attention for their antioxidant activities.20–22 For example, Murru et al. conducted a study where CDs were prepared from vegetable wastes using a hydrothermal method.23,24 The obtained CDs exhibited excellent antioxidant activity. Similarly, Yun et al. synthesized CQDs from red cabbage through hydrothermal means and tested their efficacy against DPPH˙, ˙OH, and KMnO4 radicals.25 The red cabbage CQDs demonstrated remarkable scavenging activities against all tested radicals. The antioxidant properties of CDs are influenced by various factors, including sp2 hybrid carbon domains, hydrogen donor behavior, electron transport, unpaired electrons resulting from vacancies and defects, types of doping elements, and surface functional groups.26 However, it is worth noting that the hydrothermal method has certain limitations, such as the requirement for a Teflon-coated autoclave and a longer reaction time.
The objective of this study was to develop CDs using gardenia seeds as carbon sources (abbreviated to be GCDs) through a one-step mild pyrolysis process. Gardenia seeds possess various potential medical functions, including antioxidant, anti-inflammatory, liver protection, sedative and calming effects, digestive aid, antibacterial and antiviral activities, and blood pressure regulation.27,28 Despite their medicinal value, research about the use of gardenia seeds as carbon sources for CDs preparation remains limited.29,30 This study conducted comprehensive investigations to analyze the morphology, spectroscopic properties, and electronic properties of the GCDs. The antibacterial activity was assessed using optical methods and colony culture assays, while the antioxidant activity was evaluated through DPPH˙ assays in aqueous media. Furthermore, the study demonstrated the practical applications of GCDs as green antioxidant additives in mineral base oil samples and as fluorescent enhancers in milk powder samples. The results highlighted the bio-friendly properties of GCDs, their efficacy as cell-imaging probes, their favorable antioxidant properties in oil-based media, and their synergistic effects in the purchased free fatty acid quantification kit. The study also elucidated the potential mechanisms underlying the radical scavenging activity of GCDs in both aqueous and oil environments. Leveraging the unique optical characteristics, excellent photostability, low toxicity, and environmentally friendly nature of GCDs, this research provides a solid foundation for the development of innovative luminescent GCDs with enhanced antioxidant properties. These GCDs hold great promise for a wide range of applications in bioimaging and the food industry.
Results and discussion
Characterization of GCDs
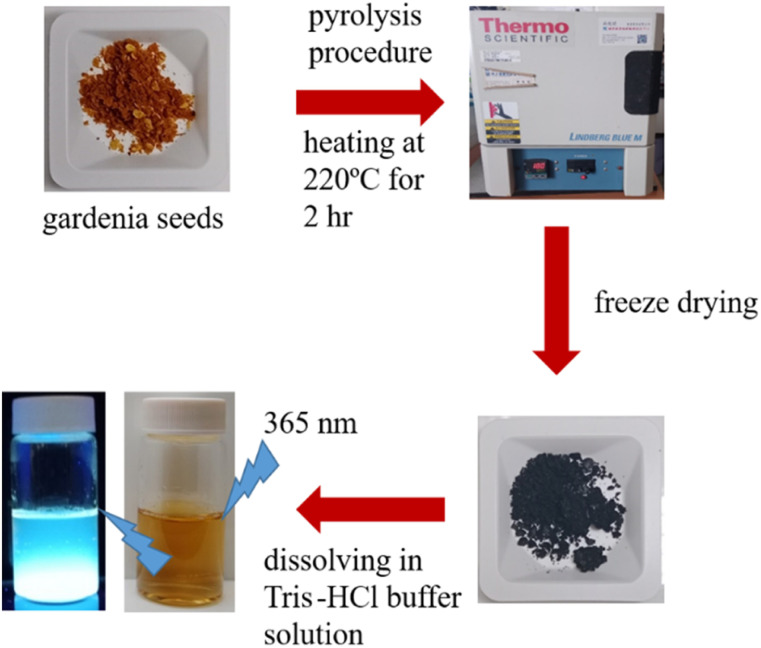
Scheme 1 provides a schematic illustration of the synthesis of luminescent GCDs using gardenia seeds as a precursor. The resulting GCDs, synthesized at different pyrolysis temperatures (180, 200, 220, and 240 °C), exhibit transparency in daylight and emit blue luminescence when excited at 365 nm (Fig. 1A). The UV-visible absorption spectra of the GCDs prepared at these temperatures show similar characteristics, with a peak at 265 nm attributed to π → π* transitions of sp2 hybridized groups (C C bonds), and another peak at 335 nm associated with n → π* transitions of C O groups or C–OH in sp3 hybridized bonds (Fig. 1B).31
Scheme 1. Schematic procedure for preparation of luminescent carbon dots using gardenia seeds as a carbon source through a one-step mild pyrolysis process.
Fig. 1. (A) Photographic images of GCDs synthesized at various temperatures (180, 200, 220, and 240 °C) under daylight conditions and their corresponding fluorescence properties when excited with UV light at a wavelength of 365 nm. (B) UV-Vis spectra and (C) PL spectra of GCDs prepared at different temperatures.
The PL spectra of the GCDs were investigated using an excitation wavelength of 365 nm (Fig. 1C). The PL intensity gradually increases with increasing pyrolysis temperature from 180 to 220 °C, indicating optimal conditions for achieving high quantum yield (QY) of the GCDs. However, the PL intensity decreases significantly at 240 °C due to excessive carbonization of the gardenia seeds. Consequently, it is concluded that the optimum pyrolysis temperature for producing GCDs with high QY is 220 °C. The PL characteristics of GCDs-220 were further examined at various excitation wavelengths ranging from 300 to 420 nm (Fig. S1†). The PL spectrum of GCDs-220 exhibits a bathochromic shift, with a gradual redshift towards longer wavelengths accompanied by a decrease in PL intensity.32,33 The QY of GCDs-220 was determined to be 1.13% (quinine sulfate as the standard in 0.1 M H2SO4 (QY of 54%)).34 The decay curve of the PL emission of GCDs-220 (Fig. S2†) reveals measured lifetimes (τ) of τ1 = 0.001 ns (76.94%) and τ2 = 7.535 ns (23.06%) with an average lifetime of 1.8375 ns. PL decay curves for GCDs-220 exhibited multi-exponential behaviour, indicating the coexistence of different fluorescence mechanisms. Due to the percentage of τ1 higher than that of τ2, we considered that GCDs-220 exhibited a fast radiative recombination of excited electron–hole pairs. This is often associated with quantum confinement effects or efficient surface passivation that minimizes non-radiative recombination pathways. Thus, the fluorescence mechanism of GCDs-220 may involve mainly radiative recombination. These findings suggest that the PL lifetime is influenced by nanostructured carbon particles and comparable values were obtained for C-dots derived from various food sources.35
To assess their photostability, the prepared GCDs-220 was subjected to continuous UV light exposure for up to 2 hours, demonstrating no significant change in PL intensity (Fig. S3A†). Furthermore, after one month of storage in a domestic refrigerator, GCDs-220 remained stable. The pH-dependent stability of GCDs-220 was evaluated by testing their PL in different pH solutions ranging from 2.0 to 12.0, revealing stable PL within these pH ranges (Fig. S3B†).
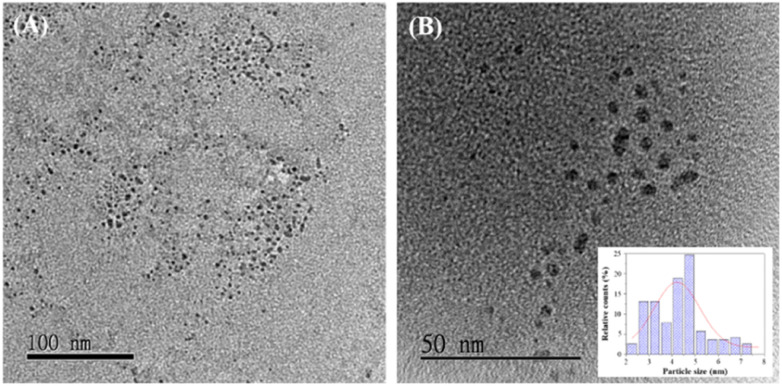
Transmission electron microscopy (TEM) images of GCDs-220 (Fig. 2) revealed spherical GCDs with an average diameter of approximately 4.6 ± 1.8 nm. The surface functional groups of GCDs-220 were identified using FT-IR analysis (Fig. S4A†). Broad absorption bands in the range of 3000–3750 cm−1 were attributed to O–H and N–H stretching vibrations. Specific stretching vibrations of C O and C–O were observed at 1634 and 1112 cm−1, respectively, while the band at 1415 cm−1 indicated C C stretching vibrations, suggesting the carbonization of organic compounds into graphite-like nanostructures. These results provide strong evidence of the functionalization of GCDs-220 with hydroxyl, carbonyl, and carboxylic acid groups. These surface functional groups offer valuable insights into the PL mechanisms of GCDs-220, making them suitable and environmentally friendly probes for biochemical research.
Fig. 2. TEM images of GCDs-220 dispersed in water, with scale bars of (A) 100 nm and (B) 50 nm, and an inset showing the particle size distribution.
The X-ray diffraction (XRD) pattern of GCDs-220 (Fig. S4B†) displayed a broad reflection peak at 2θ = 25°, which could be assigned to the (002) crystal plane diffraction peak, showing the presence of amorphous carbon.36,37 The prepared GCDs-220 also lacked polymer characterization based on the MALDI-MS results (Fig. S4C†). The signal values of m/z at 619.376 and 913.496 obtained from the MS spectra will be studied in detail in the future.
To further investigate the surface functionalities, X-ray photoelectron spectroscopy (XPS) was utilized. The full XPS spectrum of GCDs-220 (Fig. S5A†) exhibited peaks at 29, 63, 284, 496, 531, 975, and 1070 eV, corresponding to Na2p, Na2s, C1s, NaKLL, O1s, OKLL, and Na1s, respectively. High-resolution spectra of C1s (Fig. S5B†) revealed three distinct chemical environments: C C at 284.5 eV, C–C/C–H at 285 eV, C–OH/C–O–C at 286.5 eV, and O–C O at 289 eV. Similarly, high-resolution spectra of O1s (Fig. S5C†) exhibited three different chemical environments: C O at 531 eV, O–(C O)–C at 531.6 eV, C–OH at 533.6 eV, and O–(C O) at 534.8 eV.
Bacterial imaging
GCDs-220 exhibited strong and durable PL, coupled with excellent photostability and biocompatibility, making them highly desirable for a broad range of applications in biomedical imaging, sensing, and optoelectronics.
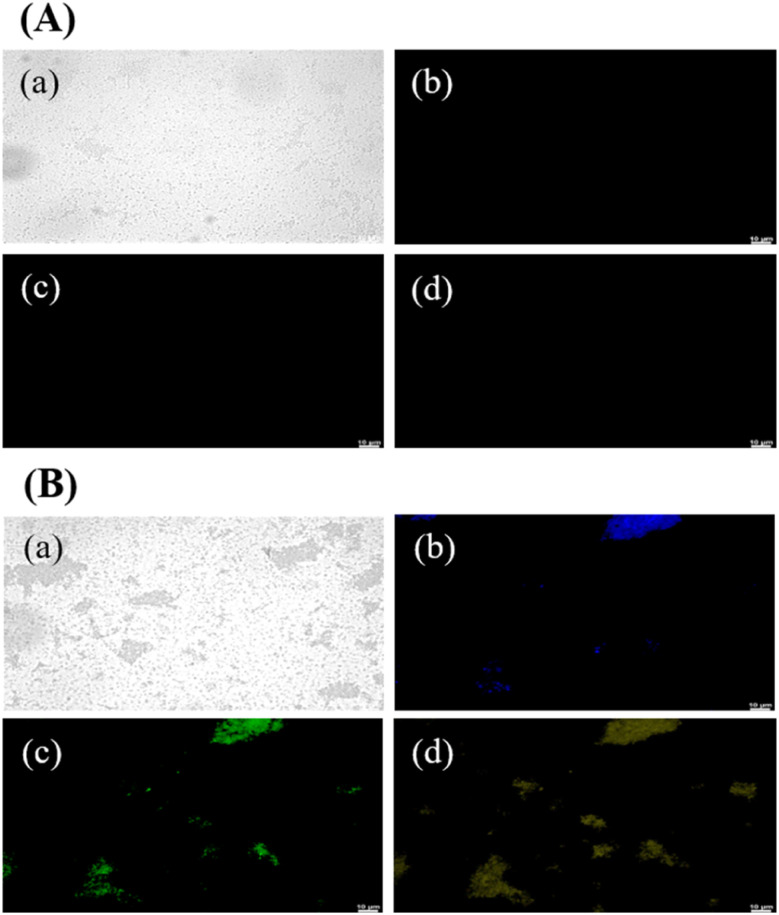
Fig. 3 presents PL images of GCDs-220-labeled bacterial (E. coli) cells. Laser excitations at 365, 475, and 532 nm were employed to activate the internalized GCDs, resulting in PL emissions in three distinct colors: blue, green, and yellow. The illuminated regions coincided with the cell locations, indicating the uptake and retention of GCDs-220 within the cells. GCDs possess favorable attributes, including a small average size, surface functional groups, and biocompatibility, which facilitate their efficient internalization by cells.38 It's important to note that the specific mechanisms of carbon dot transport across the bacterial cell wall can vary depending on factors such as the size, surface properties, and composition of the carbon dots, as well as the type of bacteria and the conditions of the surrounding environment.38 In our case, small-sized and highly water-soluble GCDs-220 may be able to passively diffuse through the pores and channels present in the bacterial cell wall. In addition, GCDs-220 can be actively taken up by bacterial cells through endocytosis or other energy-dependent processes. Bacterial cells have mechanisms for internalizing various molecules, including nanoparticles. The GCDs-220 may bind to cell surface receptors and transported into the cell. As depicted in Fig. 3A, bacteria without GCDs-220 did not exhibit any PL emission, confirming the easy passage of GCDs-220 through the cell membranes and their subsequent uptake by the cells. These observations strongly suggest the accumulation of the synthesized GCDs-220 in both the cell membranes and cytoplasm.39,40
Fig. 3. Confocal laser microscopic images of E. coli cells (A) without and (B) with GCDs-220 (25.6 mg mL−1) after incubation at 37 °C for 6 h: (a) bright field and fluorescence mode at excitation wavelengths of (b) 365, (c) 475, and (d) 532 nm.
To evaluate their toxicity, we investigated the effects of GCDs-220 and BA on the viability and inhibition zone of E. coli and Staphylococcus aureus (Fig. S6†). The results demonstrated that the synthesized GCDs-220 did not impede the growth of E. coli and Staphylococcus aureus, indicating their compatibility with bacterial cells. As a result, GCDs offer promising alternatives to conventional organic dyes and semiconductor quantum dots, presenting opportunities as probes for imaging various types of bacteria.
Free radical scavenging abilities of GCDs-220
We evaluated the antioxidant activity of GCDs-220 using the radical scavenging colorimetric test, a widely recognized method for assessing the antioxidant capacity of different substances.41
Fig. S7† illustrates that GCDs-220 displayed a dose-dependent DPPH˙ radical scavenging activity, indicating its effective ability to neutralize free radicals. The percentage of the DPPH˙ scavenging effect increased as the concentration of GCDs-220 rose, indicating that higher concentrations of GCDs-220 resulted in more substantial scavenging effects on DPPH˙ radicals. At a concentration of 6.4 mg mL−1, GCDs-220 exhibited a DPPH˙ radical scavenging activity of 67.7%. Furthermore, the EC50 value of GCDs-220 was calculated to be 4.51 mg mL−1, representing the concentration required to reduce 50% of the initial DPPH concentration. Thus, the results from the DPPH˙ radical scavenging colorimetric test confirm the notable dose-dependent antioxidant activity of GCDs-220, where higher concentrations demonstrate enhanced scavenging effects on DPPH˙ radicals. The calculated EC50 value of 4.51 mg mL−1 further supports the significant antioxidant activity of GCDs-220, suggesting its potential application in diverse fields, including medicine and the food industry. In addition, the Fenton reaction produced ˙OH radicals, and their absorbance was observed at 510 nm (Fig. S8A†). And a possible scavenging mechanism of GCDs-220 on ˙OH radicals is also given. The functional groups on the surface of GCDs-220 could contribute hydrogen atoms to ˙OH radical reactions.
The ability of GCDs-220 to scavenge ˙O2− radicals was demonstrated by comparing the absorbance change in the presence and absence of GCDs-220 (Fig. S8B†). The absorbance at 600 nm reduced while the scavenging ability increased in the presence of GCDs-220. These findings suggest that GCDs-220 have the ability to scavenge ˙O2− radicals. The production of the more stable molecules in this situation may be caused by an electron transfer from GCDs-220 to ˙O2− radicals, which is a plausible scavenging process.
Application of GCDs as antioxidant additives in oil samples
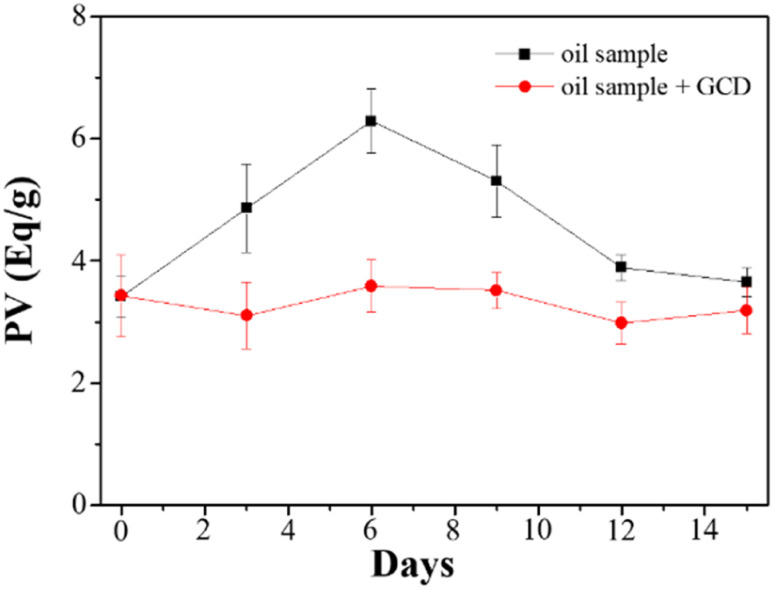
Mineral oils are widely utilized as lubricants across various industries, and their properties are influenced by the specific source of crude oil and the refining process. These variations stem from factors such as chemical composition, sulfur content, and viscosity. However, during the course of their usage, oils undergo oxidation, leading to several undesirable effects including increased friction, wear, energy dissipation as heat, and reduced efficiency of the mechanical system. The oxidation process is initiated by factors such as oxygen, moisture, heat, and metals, resulting in the formation of hydroperoxide radicals. To assess the formation of hydroperoxides, PV is commonly employed, and there are multiple available methods for its determination. In this study, we evaluated GCDs-220 as a potential additive for lubricant oils, specifically as an antioxidant and radical scavenger. The measurement of PV was performed using the iodometric titration method.
Analysis of the obtained data (Fig. 4) revealed that in the absence of GCDs-220, the PV exhibited an initial increase over the first six days due to oil oxidation. However, this was followed by a sudden decrease, likely attributed to the termination of free radical chain reactions. Conversely, oil samples containing GCDs-220 demonstrated improved oxidative stability, suggesting the potential of GCDs-220 as an effective antioxidant additive for lubricant oils. Furthermore, given the growing concerns surrounding environmental issues, the utilization of environmentally acceptable materials has become imperative.
Fig. 4. Study of the aging of oil samples with and without GCDs-220 additives.
Determination of fatty acid in milk powder samples
Palmitic acid served as the target molecule to evaluate the sensitivity of a free fatty acid quantification kit in the presence and absence of GCDs. The addition of GCDs led to an enhanced emission intensity at 535 nm, as demonstrated in Fig. S9.† By introducing carboxyl groups to the GCDs, their affinity for palmitic acid increased through hydrophilic interaction. Additionally, GCDs displayed notable scavenging activities against DPPH˙ free radicals, while their composites with palmitate demonstrated improved oxidative stability.42 The chemical reagents in the kit catalyzed the formation of palmitate-GCDs composites, increasing fluorescence intensity. The inset in Fig. S9† illustrated that the fluorescence intensity varied with the concentration of palmitic acid. To determine the detection limit for palmitic acid, fluorescence titration studies were conducted, with the fluorescence emission spectra measured in triplicate for each concentration. The calculated detection limits at signal-to-noise of 3.0 for palmitic acid were 0.76 μM in the absence of GCDs-220 and 0.08 μM in their presence.
To evaluate the practicality of the approach, different concentrations of palmitic acid (2.0 to 10.0 μM) spiked in milk powder samples were examined with GCDs-220. Each sample underwent three parallel tests, with relative standard deviations (RSDs) of less than 4.6%. The recovery of palmitic acid spiked in milk samples ranged from 82.6% to 109.6%, demonstrating the potential applicability of the free fatty acid quantitation kit in real-world scenarios when GCDs are utilized.
Experimental
Chemicals
All reagents were purchased from Sigma-Aldrich (Milwaukee, WI, USA). Ultrapure water (18.2 MΩ cm−1) from a Milli-Q ultrapure system was used in this study. Gardenia seeds were a gift from Dr. Chiu-Lan Hsieh (National Changhua University of Education, Changhua).
Characterization
The UV-visible spectra of GCDs were collected using an Evolution 200 UV-Vis spectrophotometer (ThermoFisher, NY, USA). X-ray diffraction (XRD) patterns of GCDs were obtained using a LabX XRD-6000 X-ray diffractometer (SHIMADZU, Kyoto, Japan) with Cu Kα radiation (λ = 0.15418 nm). The Fourier transfer-infrared (FT-IR) spectra of GCDs were measured at room temperature using an FT-IR spectrometer (Agilent Cary 600; Agilent, California, USA). The morphology and microstructure of GCDs were studied using high-resolution transmission electron microscopy (HRTEM) on a JEOL-1200EX II TEM (JEOL, Tokyo, Japan) with a 200 kV accelerating voltage. The photoluminescence (PL) spectra were acquired using a Synergy H1 Hybrid Multi-Mode Microplate Reader (Biotek Instruments, Inc., Winooski, VT, USA). X-ray photoelectron spectroscopy (XPS) was used to validate the surface status of GCDs utilizing an electron spectroscopy instrument (VG ESCA210; VG Scientific, West Sussex, UK). The time-resolved fluorescence spectra of GCDs were measured on a time-correlated photon-counting spectrometer (HORIBA MIRA Limited, Nuneaton, Warwickshire, UK). A Carl Zeiss 510 LSM laser scanning confocal microscope was used to obtain cell images. The matrix-assisted laser desorption/ionization-time of flight mass spectrometer (MALDI-TOF-MS, Bruker Daltonics, Bremen, Germany) experiment was used to determine the MS spectra of GCDs.
Synthesis of GCDs
GCDs were synthesized from 500 mg of gardenia seeds heated in an oven at 180–240 °C for 3 h to yield dark black residues. These residues were then cooled to room temperature (∼25 °C) and dissolved in 5 mL of DMSO. Each solution was then sonicated (DC200H, Honeywell, Charlotte, NC, USA) at room temperature for 1 h and subsequently centrifuged at a relative centrifugation force (RCF) of 35 000g for 1 h to remove the large particles. A freeze-drying method was utilized to determine the concentration of purified GCDs in the solution (in terms of mg mL−1).
Bacterial culture, survival rate, and imaging
A single colony of bacterial (E. coli strain BRBC 12438 and Staphylococcus aureus (wild)) was carefully selected from solidified agar plates and introduced into a 1.0 mL culture of Luria Bertani (LB) medium. The bacterial cultures were then subjected to incubation at a temperature of 37 °C while continuously shaking at 200 rpm until the optical density at a wavelength of 600 nm reached a value of 1.0. This measurement was performed using an optical path length of 1.0 cm. Following this, each bacterial suspension underwent centrifugation at a speed of 3000g for a duration of 10 minutes at a temperature of 25 °C. The resulting bacterial pellets were subsequently washed three times using a phosphate-buffered saline (PBS) solution.
To facilitate the formation of bacterial–GCDs conjugates, the bacterial cells were treated with a 70% (v/v) ethanol solution. This step, aimed at promoting the internalization of GCDs-220 by the cells, was conducted at a temperature of 4 °C for a period of 5 minutes. Subsequently, the cells were stained by dispersing them in a 100 mM PBS solution containing GCDs-220 at a concentration of 25.6 mg mL−1. The staining process was carried out for a duration of 10 minutes at room temperature. After staining, the cell–GCD conjugates underwent a thorough washing procedure, involving three washes using double-distilled water. Finally, a volume of 20 mL of the resulting conjugate solution was transferred onto a glass slide to facilitate fluorescence measurements. Fluorescence images of the cells were captured using laser excitations at wavelengths of 365, 475, and 532 nm.
To assess the antibacterial activity of the synthesized GCDs-220 against E. coli, a suspension containing 5.0 × 103 colony-forming units (CFU) per milliliter was evenly spread onto solidified LB agar plates. Subsequently, six paper ingots were placed on the surface of the agar plate. Different concentrations of GCDs, ranging from 0 to 32 mg mL−1, were deposited onto the paper ingots. A positive control group was included, utilizing a concentration of 32 mg mL−1 of benzoic acid (BA). Following an incubation period of 24 hours, the size of the inhibition zone formed around the paper ingots was measured and compared among the various treatment groups.
Determination of free radical scavenging activity
In individual test tubes, 1 mL of each GCDs-220 aqueous solution (ranging from 0.4 to 12.8 mg mL−1) was combined with 1 mL of the 0.2 mM DPPH (2,2-diphenyl-1-picrylhydrazyl) solution. Each test tube was thoroughly mixed to ensure a proper reaction. The tubes were then allowed to stand for 20 minutes to facilitate the reaction between GCDs-220 and DPPH. After the completion of the reaction period, the absorbance of the resulting solution was measured using an Evolution 200 UV-Vis spectrophotometer. The absorbance was recorded at a wavelength of 525 nm. All tests were performed in triplicate and ultrapure water was used as a blank control. The scavenging efficiency of the GCDs-220 for DPPH was calculated using eqn (1):
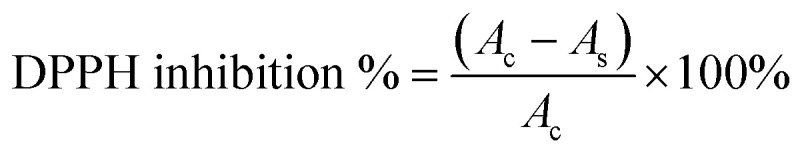 |
1 |
where Ac is the absorbance of the control and As is the absorbance of the sample.
The scavenging activity of GCDs-220 against hydroxyl free radicals (˙OH) was determined based on the Fenton reaction. Briefly, 200 μL of FeSO4 solution (7.5 mM) and 600 μL of ethanolic salicylic acid solution (1.8 mM) were mixed with 100 μL of GCDs-220 (6.4 mg mL−1); this was followed by the addition of 200 μL of H2O2 (200 mM). Then, ultrapure water was added to make up the volume to 2 mL. The absorbance at 510 nm was measured after allowing the reaction to take place for 15 min at 37 °C.
The scavenging activity of CDs against superoxide anion radicals (˙O2−) was determined using the methods of Bae and Suh with some modification.43 500 μL of xanthine (0.4 mM) was mixed with 500 μL of 0.24 mM nitro blue tetrazolium chloride; this was followed by mixing with 1 mL of xanthine oxidase (0.049 unit per mL). Then, 500 μL of GCDs-220 (12.8 mg mL−1) was added. After incubation at 37 °C for 30 min, the absorbance was measured at 550 nm.
Determination of peroxide value in oil samples
The peroxide value (PV) is an indicator of the extent of peroxidation in a sample, reflecting the total amount of peroxides that react with potassium iodide (KI).44 The liberated iodine resulting from the peroxides is then quantitatively determined through an iodometric titration using a standard solution of sodium thiosulfate. The equivalence point of the titration provides the basis for PV determination.45
Two oil samples (250.0 mL) were prepared for analysis: one containing 0.25% w/v of GCDs-220 and the other without GCDs-220 (used as a blank). Both samples were subjected to a heating period of 15 days under an oxygen atmosphere with moderate stirring. The temperature was gradually increased to 75 °C to facilitate the slow formation of reactive chemical species. For the PV measurement, an accurate weight of 10.0 g was taken for each sample. To this, 1.0 mL of a saturated solution of KI was added, followed by 20.0 mL of a mixture solution of n-hexane and glacial acetic acid in a volumetric ratio of 2 : 3 (v/v). Subsequently, 1.0 mL of a starch solution (0.67% w/v) was added to each sample. The mixtures were gently stirred until the titration analysis using a standard solution of sodium thiosulfate (0.01 M) was completed. The same procedure was carried out for the others without GCDs-220. The PV value was calculated using eqn (2):
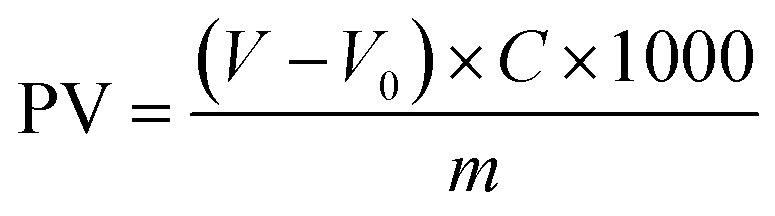 |
2 |
where V and V0 are the volumes of the sodium thiosulfate standard solution additions to the analyzed oil sample and the blank, respectively. C is the molar concentration of the standard solution of sodium thiosulfate and m is the weight (g) of the oil sample.
Determination of fatty acid in milk powder samples
Following the formulation of the purchased free fatty acid quantification kit, 5.0 μL of a 12.8 mg mL−1 GCDs-220 aqueous solution is added first. Then, palmitic acid standard, with concentrations of 0.1 mM, is added in different volumes: 0 μL, 1.0 μL, 2.0 μL, 3.0 μL, 4.0 μL, and 5.0 μL. The mixture is then supplemented with fatty acid assay buffer to a final volume of 50 μL. Next, enzyme, mix enhancer, and fatty acid probe in DMSO, each in a volume of 2 μL, are added to the mixture, and the mixture is placed in an oven for a 30 minute incubation at 37 °C. For the control group, the GCDs-220 aqueous solution is replaced with fatty acid assay buffer. Others are the same as mentioned above. After the incubation, the samples are ready for measurement using a Synergy H1 Hybrid Multi-Mode Microplate Reader. The samples are excited with light at 535 nm, and the signal value of the emitted fluorescence at 590 nm is collected. For the spiked recovery experiment aimed at detecting free fatty acids in milk powder samples, all procedures remained the same as mentioned above, with the only modification being the dissolution of 5.0 g of milk powder in 50.0 mL of water as the matrix.
Conclusions
In this study, we successfully developed a one-step mild pyrolysis process using gardenia seeds as a precursor to synthesize carbon dots, specifically referred to as GCDs. These GCDs were effectively employed as luminescent probes for imaging E. coli bacteria. Notably, the GCDs did not impede the growth of E. coli and Staphylococcus aureus, demonstrating their excellent biocompatibility and non-toxic nature. Additionally, the GCDs displayed remarkable scavenging activities against DPPH˙, ˙OH, and ˙O2− free radicals, indicating their potential as antioxidants. Capitalizing on their antioxidant properties, GCDs-220 was utilized as additives for protecting lubricating oil. The findings from our study strongly suggest that these GCDs exhibit considerable potential for safeguarding oil samples against oxidative damage. Furthermore, our research highlights the unique opportunity presented by CDs derived from gardenia seeds, allowing us to investigate the intrinsic properties of the starting materials, particularly in terms of their antioxidant activity. Moreover, the presence of GCDs significantly improved the fluorescence detection of palmitic acid using a free fatty acid quantitation kit. This improvement can be attributed to the formation of stable composites between palmitic acid and GCDs through hydrophilic interaction. Consequently, GCDs demonstrate their efficacy as valuable luminescent probes for imaging bacteria, antioxidants in oil samples, and detecting palmitic acid in milk powder samples.
Author contributions
Conceptualization: H.-W. Tsai, C.-L. Hsieh, Y.-W. Lin; data curation: H.-W. Tsai; formal analysis: H.-W. Tsai, S.-F. Fu, M.-Y. Wu, Y.-W. Lin; funding acquisition: Y.-W. Lin; investigation: H.-W. Tsai, S.-F. Fu, Y.-W. Lin; methodology: H.-W. Tsai, C.-L. Hsieh, Y.-W. Lin; project administration: Y.-W. Lin; resources: C.-L. Hsieh, S.-F. Fu, Y.-W. Lin; supervision: T. Wu, Y.-W. Lin; validation: M.-Y. Wu, Y.-W. Lin; visualization: H.-W. Tsai, Y.-W. Lin; writing – original draft: H.-W. Tsai, M.-Y. Wu, and Y.-W. Lin; writing – review & editing: T. Wu, Y.-W. Lin.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This study was supported by the National Science and Technology Council (NSTC) under contracts (111-2113-M-018-011 and 112-2113-M-018-005).
Electronic supplementary information (ESI) available: Fig. S1. Excitation-dependent emissions spectra of GCDs-220. Fig. S2. The decay curves of GCDs-220. Fig. S3. (A) The plot of the PL intensity at 450 nm of GCDs-220 under continuous irradiation of the 365 nm light. (B) pH dependence of PL response of GCDs-220. Fig. S4. (A) FT-IR spectrum, (B) XRD pattern and (C) MS spectra of GCDs-220. Fig. S5. (A) Full range XPS analysis of GCDs-220. High-resolution XPS spectrum of the (B) C1s and (C) O1s region of GCDs-220. Fig. S6. (A) Bacterial viability (%) of E. coli and photograph images of the inhibition zone of GCDs-220 against (B) E. coli and (C) Staphylococcus aureus. Fig. S7. UV-Vis spectra of GCDs-220 in the presence of DPPH. Fig. S8. Antioxidant activities of GCD-220 against ˙OH, and ˙O2− free radicals. Fig. S9. Fluorescence intensity of palmitic acid by a free fatty acid quantitation kit with/without GCDs-220. See DOI: https://doi.org/10.1039/d3ra06293g
References
- Xu D. Lin Q. Chang H. T. Small Methods. 2020;4:1900387. [Google Scholar]
- Chahal S. Macairan J.-R. Yousefi N. Tufenkji N. Naccache R. RSC Adv. 2021;11:25354–25363. doi: 10.1039/d1ra04718c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansuriya B. D. Altintas Z. Nanomaterials. 2021;11:2525. doi: 10.3390/nano11030578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Li R. Yang B. ACS Cent. Sci. 2020;6:2179–2195. doi: 10.1021/acscentsci.0c01306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barve K. Singh U. Yadav P. Bhatia D. Mater. Chem. Front. 2023;7:1781–1802. [Google Scholar]
- Das R. Bandyopadhyay R. Pramanik P. Mater. Today Chem. 2018;8:96–109. [Google Scholar]
- Dias C. Vasimalai N. Sárria M. P. Pinheiro I. Vilas-Boas V. Peixoto J. Espiña B. Nanomaterials. 2019;9:199. doi: 10.3390/nano9020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Xiong M. Zhang J. He C. Ma X. Zhang H. Kuang Y. Yang M. Huang Q. Microchem. J. 2021;160:105604. [Google Scholar]
- Bag P. Maurya R. K. Dadwal A. Sarkar M. Chawla P. A. Narang R. K. Kumar B. ChemistrySelect. 2021;6:2774–2789. [Google Scholar]
- Ghosal K. Ghosh A. Mater. Sci. Eng., C. 2019;96:887–903. doi: 10.1016/j.msec.2018.11.060. [DOI] [PubMed] [Google Scholar]
- de Oliveira B. P. da Silva Abreu F. O. M. Mater. Lett. 2021;282:128764. [Google Scholar]
- Wei S.-C. Lin Y.-W. Chang H.-T. J. Food Drug Anal. 2020;28:558. doi: 10.38212/2224-6614.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.-W. Unnikrishnan B. Anand A. Lin Y.-W. Huang C.-C. J. Food Drug Anal. 2020;28:539. doi: 10.38212/2224-6614.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari L. Hallaj S. Hallaj T. Amjadi M. Colloids Surf., B. 2021;203:111743. doi: 10.1016/j.colsurfb.2021.111743. [DOI] [PubMed] [Google Scholar]
- Huang H.-H. Anand A. Lin C.-J. Lin H.-J. Lin Y.-W. Harroun S. G. Huang C.-C. Carbon. 2021;174:710–722. [Google Scholar]
- Anand A. Manavalan G. Mandal R. P. Chang H.-T. Chiou Y.-R. Huang C.-C. Curr. Pharm. Des. 2019;25:4848–4860. doi: 10.2174/1381612825666191216150948. [DOI] [PubMed] [Google Scholar]
- Yang C.-R. Lin Y.-S. Wu R.-S. Lin C.-J. Chu H.-W. Huang C.-C. Anand A. Unnikrishnan B. Chang H.-T. J. Colloid Interface Sci. 2023;634:575–585. doi: 10.1016/j.jcis.2022.12.076. [DOI] [PubMed] [Google Scholar]
- Kasibabu B. S. B. D'souza S. L. Jha S. Singhal R. K. Basu H. Kailasa S. K. Anal. Methods. 2015;7:2373–2378. [Google Scholar]
- Rawat K. S. Singh V. Sharma C. P. Vyas A. Pandey P. Singh J. Gupta N. M. Sachdev M. Goel A. J. Imaging. 2023;9:19. doi: 10.3390/jimaging9010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev A. Gopinath P. Analyst. 2015;140:4260–4269. doi: 10.1039/c5an00454c. [DOI] [PubMed] [Google Scholar]
- Wei X. Li L. Liu J. Yu L. Li H. Cheng F. Yi X. He J. Li B. ACS Appl. Mater. Interfaces. 2019;11:9832–9840. doi: 10.1021/acsami.9b00074. [DOI] [PubMed] [Google Scholar]
- Pal T. Mohiyuddin S. Packirisamy G. ACS Omega. 2018;3:831–843. doi: 10.1021/acsomega.7b01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murru C. Badía-Laíño R. Díaz-García M. E. Antioxidants. 2020;9:1147. doi: 10.3390/antiox9111147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Varillas S. Murru C. Díaz-García M. E. Badía-Laíño R. Antioxidants. 2022;11:2193. doi: 10.3390/antiox11112193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N. Das G. S. Yun K. Appl. Microbiol. Biotechnol. 2020;104:7187–7200. doi: 10.1007/s00253-020-10726-5. [DOI] [PubMed] [Google Scholar]
- Zhao L. Zhang M. Wang H. Devahastin S. Meat Sci. 2022;185:108715. doi: 10.1016/j.meatsci.2021.108715. [DOI] [PubMed] [Google Scholar]
- Dangoggo S. Muhammad A. Tsafe A. Aliero A. Itodo A. Arch. Appl. Sci. Res. 2011;3:485–492. [Google Scholar]
- Koo A. Li K. Am. J. Chin. Med. 1977;5:31–37. doi: 10.1142/s0192415x7700004x. [DOI] [PubMed] [Google Scholar]
- Saravanakumar K. Chelliah R. Shanmugam S. Varukattu N. B. Oh D.-H. Kathiresan K. Wang M.-H. J. Photochem. Photobiol., B. 2018;185:126–135. doi: 10.1016/j.jphotobiol.2018.05.032. [DOI] [PubMed] [Google Scholar]
- Kang D.-S. Jin D.-H. Oh D.-Y. Kim H.-S. J. Environ. Sci. Int. 2017;26:893–902. [Google Scholar]
- Arroyave J. Ambrusi R. Robein Y. Pronsato M. Brizuela G. Di Nezio M. Centurión M. Appl. Surf. Sci. 2021;564:150195. [Google Scholar]
- González-González R. B. González L. T. Madou M. Leyva-Porras C. Martinez-Chapa S. O. Mendoza A. Nanomaterials. 2022;12:298. doi: 10.3390/nano12030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba X.-X. Zhang L. Yin Y.-L. Jiang F.-L. Jiang P. Liu Y. J. Colloid Interface Sci. 2020;565:77–85. doi: 10.1016/j.jcis.2020.01.007. [DOI] [PubMed] [Google Scholar]
- Arkan E. Barati A. Rahmanpanah M. Hosseinzadeh L. Moradi S. Hajialyani M. Adv. Pharm. Bull. 2018;8:149. doi: 10.15171/apb.2018.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. N. Baker G. A. Angew. Chem., Int. Ed. 2010;49:6726–6744. doi: 10.1002/anie.200906623. [DOI] [PubMed] [Google Scholar]
- Jiang C. Wu H. Song X. Ma X. Wang J. Tan M. Talanta. 2014;127:68–74. doi: 10.1016/j.talanta.2014.01.046. [DOI] [PubMed] [Google Scholar]
- Wang Z. Liao H. Wu H. Wang B. Zhao H. Tan M. Anal. Methods. 2015;7:8911–8917. [Google Scholar]
- Li P. Sun L. Xue S. Qu D. An L. Wang X. Sun Z. SmartMat. 2022;3:226–248. [Google Scholar]
- Liu J.-H. Cao L. LeCroy G. E. Wang P. Meziani M. J. Dong Y. Liu Y. Luo P. G. Sun Y.-P. ACS Appl. Mater. Interfaces. 2015;7:19439–19445. doi: 10.1021/acsami.5b05665. [DOI] [PubMed] [Google Scholar]
- Luo P. G. Sahu S. Yang S.-T. Sonkar S. K. Wang J. Wang H. LeCroy G. E. Cao L. Sun Y.-P. J. Mater. Chem. B. 2013;1:2116–2127. doi: 10.1039/c3tb00018d. [DOI] [PubMed] [Google Scholar]
- Dong C. Wang S. Ma M. Wei P. Chen Y. Wu A. Zha Z. Bi H. Appl. Mater. Today. 2021;25:101178. [Google Scholar]
- Jin C. Wang L. Liu X. Lu Y. Yu N. Nie X. Ye Q. Meng X. Food Sci. Biotechnol. 2023:1–13. doi: 10.1007/s10068-023-01319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S.-H. Suh H.-J. LWT--Food Sci. Technol. 2007;40:955–962. [Google Scholar]
- Shang J. Wu X. Hu K. Huyan Z. Li Q. Yu X. Anal. Methods. 2018;10:3675–3679. [Google Scholar]
- Zhang N. Li Y. Wen S. Sun Y. Chen J. Gao Y. Sagymbek A. Yu X. Food Chem. 2021;358:129834. doi: 10.1016/j.foodchem.2021.129834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.