Abstract
LHQK is a patented Traditional Chinese Medicine (TCM) which is clinically used for acute tracheobronchitis, cough, and other respiratory diseases. Recent studies have proved that LHQK exhibits excellent clinical efficacy in the treatment of acute lung injury (ALI). However, the corresponding mechanisms remain largely unexplored. In this study, we investigated the effects and the underlying mechanisms of LHQK on lipopolysaccharide (LPS)‐induced ALI in mice. The pathological examination, inflammatory cytokines assessments, and mucus secretion evaluation indicated that administration of LHQK ameliorated LPS‐induced lung injury, and suppressed the secretion of Muc5AC and pro‐inflammatory cytokines (IL‐6, TNF‐α, and IL‐1β) in plasma and BALF. Furthermore, the results of cell‐free DNA level showed that LHQK significantly inhibited LPS‐induced NETs formation. Western blot revealed that LHQK effectively inhibited LPS‐triggered pyroptosis in the lung. In addition, RNA‐Seq data analysis, relatively bioinformatic analysis, and network pharmacology analysis revealed that LHQK and relative components may play multiple protective functions in LPS‐induced ALI/acute respiratory distress syndrome (ARDS) by regulating multiple targets directly or indirectly related to NETs and pyroptosis. In conclusion, LHQK can effectively attenuate lung injury and reduce lung inflammation by inhibiting LPS‐induced NETs formation and pyroptosis, which may be regulated directly or indirectly by active compounds of LHQK.
Keywords: acute respiratory distress syndrome (ARDS), pulmonary inflamation, Traditional Chinese Medicine (TCM)
Abbreviations
- ALI
acute lung injury
- ARDS
acute respiratory distress syndrome
- BALF
bronchoalveolar lavage fluid
- BP
biological process
- CitH3
citrullinated histone H3
- C‐T
compound‐target
- DEGs
differentially expressed genes
- GSDMD
gasdermin D
- H&E
hematoxylin and eosin
- LHQK
Lianhua Qingke
- LPS
lipopolysaccharide
- MPO
myeloperoxidase
- NETs
neutrophil extracellular traps
- RNA‐Seq
RNA sequencing
- TCM
Traditional Chinese Medicine
INTRODUCTION
Despite decades of efforts to improve the diagnosis and therapy, acute lung injury (ALI) and acute respiratory distress syndrome (ARDS) are still life‐threatening refractory diseases with high mortality. 1 , 2 Lung‐protective ventilation has been proven to decrease mortality in ARDS, but the 28‐day mortality remains static at 30%–40%. 3 Therefore, effective treatments for ALI/ARDS are urgently needed.
NETosis and pyroptosis can aggravate lung inflammation and are key features of ARDS. 4 , 5 , 6 Neutrophil is one of the main members of the innate immune system in the body. Upon infection, activated neutrophils can release decondensed chromatin to the outside of the cell to form neutrophil extracellular traps (NETs), which may trigger a regulated cell death process termed NETosis. During the formation of NETs, fused nucleic acid substances and granular proteins are released extracellularly by a ruptured cytoplasmic membrane, unfolding to form web‐like structures decorated with granular proteins that can trap pathogens. 7 However, excessive NETs formation is associated with various pathologic processes, including inflammation, tissue damage, and micro‐thrombosis, and leads to multiple organ dysfunction and even death. 4 , 8 , 9
Pyroptosis is a form of lytic programmed cell death that is distinct from apoptosis and necrosis. 10 Accumulating evidence has indicated that pyroptosis is involved in the pathogenesis of ALI/ARDS in a variety of lung cells, including macrophages, dendritic cells, epithelial cells, and endothelial cells, 6 , 11 , 12 , 13 triggered by pro‐inflammatory signals. Mechanistically, pyroptosis is mediated by activated caspases including caspase‐1, murine caspase‐11, and human caspase‐4/5, among which, caspase‐1 is activated by the canonical inflammasomes assembled by the nucleotide‐binding oligomerization domain (NOD) leucine‐rich repeat (LRR)‐containing protein receptors (NLRs), such as NLRP1, NLRP3, NLRC4, and absent in melanoma 2 (AIM2), 6 while the caspase‐4/5/11 are activated directly by cytosolic LPS from Gram‐negative bacteria. 6 , 14 Subsequently, activated caspase‐1 and caspase‐11 can cleave pro‐IL‐1β and pro‐IL‐18 into mature IL‐1β and IL‐18, whereas activated caspases also actuates the formation of activated N‐terminal domain of gasdermin D (GSDMD‐N), 15 which can rapidly target the membrane fraction and form large permeability pores triggering the release of IL‐1β, IL‐18, and other inflammatory cytokines, promoting pyroptosis. 16 , 17 Pyroptosis occurs faster and leads to more pro‐inflammatory factors release than apoptosis. Additionally, pyroptosis can be augmented by NETs. 5 , 18
Traditional Chinese Medicine (TCM) has long been used for treating pulmonary diseases, such as cough, cold, acute tracheitis and bronchitis‐related ALI/ARDS for thousands of years. 19 , 20 , 21 , 22 Lianhua Qingke (LHQK) is a patented Traditional Chinese Medicine composed of Ephedra sinica Stapf, Forsythia suspensa (Thunb.) Vahl, Scutellaria baicalensis Georgi, Morus Alba L., Prunus sibirica L., Peucedanum praeruptorum Dunn, Pinellia ternate (Thunb.) Breit., Citrus reticulata Blanco, Fritillaria thunbergii Miq., Arctium lappa L., Lonicera hypoglauca Miq., Rheum palmatum L., Platycodon grandiflorus (Jacq.) A. DC., Glycyrrhiza uralensis Fisch., and Gypsum fibrosum. In 2010, LHQK was approved by the State Food and Drug Administration (SFDA No. 2010L00120) for treating acute tracheitis and bronchitis‐related ALI, especially those accompanied by cough and sputum. Experimental and clinical evidence have proved that LHQK exhibits excellent clinical efficacy in the treatment of ALI, including bacteria and smoking‐induced ALI, and pneumonia caused by HCoVs (HCoV‐229E and SARS‐CoV‐2). 23 , 24 , 25 , 26 Wang et al., 24 analyzed and identified 14 major active compounds of LHQK from 22 common characteristic fingerprints, including neochlorogenic acid, chlorogenic acid, cryptochlorogenic acid, isoforsythiaside, phillygenin, hesperidin, baicalin, arctiin, aloe‐emodin, glycyrrhizic acid ammonium salt, rhein, emodin, 1,8‐dihydroxy‐3‐methylanthraquinone, and physcion. However, the underlying mechanisms of LHQK in ALI remain largely unexplored. In this study, lipopolysaccharide (LPS)‐induced ALI mice were used to explore the therapeutic benefits and related mechanisms of LHQK.
METHODS
Reagent preparation
The LHQK tablets materials (Lot No. A1401001) was provided by Shijiazhuang Yiling Pharmaceutical Co., Ltd. LHQK tablets were dissolved in phosphate‐buffered saline (PBS) before use.
LHQK was extended from two TCM prescriptions: Maxing Shigan decoction and Qingjin Huatan decoction. 27 , 28 The recipe of LHQK is composed of E. sinica Stapf, F. suspensa (Thunb.) Vahl, S. baicalensis Georgi, M. Alba L., P. sibirica L., P. praeruptorum Dunn, P. ternate (Thunb.) Breit., C. reticulata Blanco, F. thunbergii Miq., A. lappa L., L. hypoglauca Miq., R. palmatum L., P. grandiflorus (Jacq.) A. DC., G. uralensis Fisch., and G. fibrosum. The standard characteristic fingerprints of LHQK consisted of 22 common characteristic peaks, 14 of which were identified as: neochlorogenic acid, chlorogenic acid, cryptochlorogenic acid, isoforsythiaside, phillygenin, hesperidin, baicalin, arctiin, aloe‐emodin, glycyrrhizic acid ammonium salt, rhein, emodin, 1,8‐dihydroxy‐3‐methylanthraquinone, and physcion. 24
Animals
Eight‐week‐old male C57BL/6 mice (20–25 g) were routinely bred in the animal facility of Zhongshan Hospital at Fudan University. The mice were divided into Control, LPS (L9143, Sigma), LPS + LHQK LD (low‐dose), and LPS + LHQK HD (high‐dose) groups. LPS (2.5 mg/kg) was instilled intratracheally in LPS, LPS + LHQK LD, and LPS + LHQK HD groups, while mice in the control group received an equal amount of PBS. 2 h following LPS instillation, low‐dose (2 g/kg) and high‐dose (8 g/kg) LHQK was administered orally once a day for 3 days in LPS + LHQK LD and LPS + LHQK HD groups, and an equal amount of PBS was gavaged in Control and LPS groups. The mice were killed on Day 3.
Hematoxylin and eosin (H&E) staining
The right upper lobes of the lung were fixed in 4% paraformaldehyde overnight and embedded in paraffin. Lung sections were stained with H&E following standard histological staining protocols. As in our previous study, 29 lung injury scores were calculated.
IL‐6, TNF‐α, and IL‐1β enzyme‐linked immunoassay (ELISA)
IL‐6, TNF‐α, and IL‐1β in bronchoalveolar lavage fluid (BALF) were measured using ELISA kits (R&D Systems) according to the manufacturer's instructions. The catalog numbers of kits were as follows: DY406, DY410, and DY401.
Cell free (cf)‐DNA in BALF
We used Quant‐iTTM PicoGreen® dsDNA reagent (P11496, Invitrogen), an ultrasensitive fluorescent nucleic acid stain, to quantitate double‐stranded DNA (dsDNA) in BALF, following the manufacturer's protocol.
CitH3‐DNA ELISA
Referring to previous literature, we detected citrullinated histone H3 (CitH3)‐DNA complexes as a measure of NETs in BALF. 30 We used CitH3 primary antibody (ab5103, Abcam) coated onto 96‐well plates overnight at 4°C as the capture antibody, and a peroxidase‐labeled anti‐DNA antibody (Cell Death Detection ELISA Kit, Roche) as the detection antibody. Absorbance was measured at 405‐nm wavelength, and quantification of soluble NET formation were presented as a percentage increase in absorbance above control.
Immunofluorescence staining for NETs
The right upper lobes of the lungs were fixed in 4% paraformaldehyde overnight and then embedded in paraffin. Briefly, we used myeloperoxidase (MPO, NBP1‐51148; Novus) and citrullinated histone H3 (ab5103; Abcam) antibodies as primary antibodies with Alexa Fluor 647‐conjugated anti‐rabbit and Alexa Fluor 488‐conjugated anti‐mouse secondary antibodies. DAPI (Thermo Fisher Scientific) was used as a nuclear counterstain. Images were captured using confocal microscopy (Leica SP8).
Western blot analysis
Lung tissue protein was extracted using a RIPA Lysis Kit (Beyotime Biotechnology) and quantified by BCA protein assay kit (Beyotime Biotechnology) according to the manufacturer's protocol. Equal amounts of protein from each sample were loaded onto polyacrylamide gels and then transferred to polyvinylidene fluoride membranes. The membranes were blocked at room temperature for 1 h and then incubated at 4°C overnight with primary antibodies against caspase‐1 (22915‐1‐AP, Proteintech Group), GSDMD (ab209845, Abcam), IL‐1β (AF5103, Affinity Biosciences), and β‐actin (4970, Cell Signaling Technology). After washing three times, the membranes were incubated with an appropriate HRP‐conjugated secondary antibody at room temperature for 1 h. The bands were analyzed by an Imaging System (Bio‐Rad) with enhanced electro‐chemiluminescence reagents (Beyotime Biotechnology).
RNA isolation and library preparation
Lung samples were collected from each group of mice and preserved at −80°C until used. Total RNA was isolated using the TRIzol reagent (Invitrogen). RNA purity, quantification and integrity were evaluated with the NanoDrop 2000 spectrophotometer (Thermo Scientific) and the Agilent 2100 Bioanalyzer (Agilent Technologies). The construction of libraries using TruSeq Stranded mRNA LT Sample Prep Kit (Illumina), as well as transcriptome sequencing and analysis, was conducted by OE Biotech Co., Ltd.
RNA sequencing
The libraries were sequenced using an Illumina HiSeq X Ten platform to generate 150 bp paired‐end reads. The raw data (raw reads) were first processed in fastq format. To obtain high‐quality clean reads, then quality control was performed, and low‐quality reads were removed using Trimmomatic 31 software. The clean reads were aligned to the mouse genome using HISAT2. 32 Cufflinks 33 was used to calculate Fragments Per Kilobase per Million mapped reads (FPKMs) 34 of each gene and FPKMs between each group were compared with a false discovery rate (FDR) adjusted p value < 0.05 and foldchange > 2 or foldchange < 0.5, which were identified as differentially expressed genes (DEGs). DEGs were used for gene ontology (GO) and KEGG pathway analysis.
Network pharmacology and bioinformatics analysis
14 components identified from LHQK in the study by Wang et al. 24 were used for subsequent analysis. To improve reliability, the comprehensive target spectrum of 14 compounds was obtained by combing the known targets collected from DrugBank, 35 TTD, 36 ChEMBL, 37 PubChem, 38 and CTD. 39 The putative targets were predicted from STITCH, 40 SEA, 41 TargetNet, 42 SwissTargetPrediction, 43 ChEMBL_prediction, 44 and BATMAN‐TCM. 45 Genes closely related to the NETs and pyroptosis process were collected from the KEGG pathway, 46 GeneCards 47 and CTD database. 39 The gene symbols were normalized by UniProt. 48 TRRUST version 2, 49 a manually created transcriptional regulation network database, was used to predict the transcription factors (TFs) that cause differential gene changes (p < 0.05 as the threshold). The compound‐target (C‐T) network was constructed and visualized by Cytoscape v3.7.1. 50 The degree computed by Network Analyzer plugin 51 was used to evaluate the importance of the nodes. ClueGO plugin in Cytoscape was used to decipher functionally grouped biological process (BP) or KEGG pathways 52 of DEGs in RNA‐seq data.
Statistical analysis
Statistical tests were used based on the assumption that sample data are derived from a population following a Gaussian distribution based on a fixed set of parameters. T‐tests were used to determine statistical significance of differences between two groups. One‐way analysis of variance (ANOVA) was performed for multiple comparisons. A hypergeometric test was used for the overlap between two groups of genes. For all quantitative analysis, a minimum of three biological replicates were analyzed. The following values were considered to be statistically significant: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Calculations were done using the GraphPad Prism 8.0 software (GraphPad). The results were presented as the mean ± standard deviation (SD).
RESULTS
LHQK attenuated LPS‐induced lung injury
We first investigated the pharmaceutical effects of LHQK in a mouse model of LPS‐induced ALI. Morphology analysis by H&E staining and relative score assessment, including lung injury score, mean alveolar septal thickness (MAST), mean linear intercept (MLI), and destructive index (DI) were performed to evaluate the extent of lung injury. Morphology analysis showed that the LHQK‐treated group reduced peribranchial and perivascular inflammatory cell infiltration and kept more intact alveolar structure than the LPS group (Figure 1a–l). Furthermore, lung injury score demonstrated that LHQK could significantly decrease the lung‐injury score increased by LPS (Figure 1m). Additionally, the assessment of lung pathology also revealed that LHQK effectively attenuated LPS‐induced alveolar septum thickening, alveolar‐space broadening, and alveolar wall destruction (Figure 1n–p).
Figure 1.
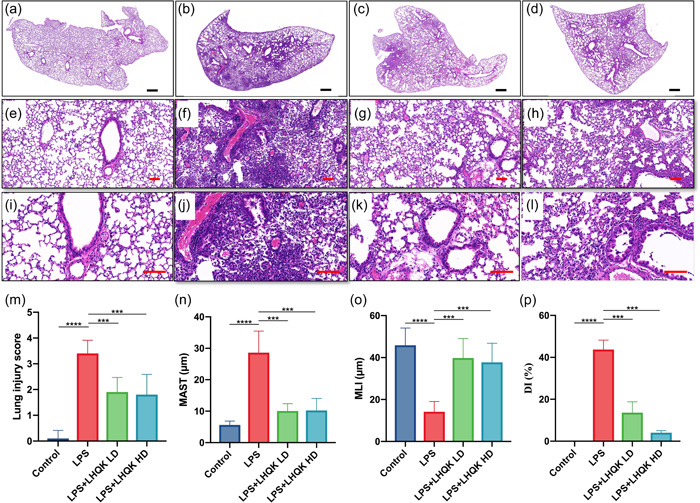
LHQK attenuated LPS‐induced lung injury. (a–l) H&E staining of lung sections with multiple magnifications (×2: a–d, ×10: e–h, ×20: i–l) of Control (a, e, and i), LPS (b, f, and j), LPS + LHQK LD (c, g, and k) and LPS + LHQK HD (d, h, and l). Scale bars in (a–d) are 500 μm and in (e–l) are 100 μm. (m) Lung injury scores. (n) MAST. (o) MLI. (p) DI. N = 6–8 in each group. The data are presented as the mean ± SD, and statistical differences were assessed by one‐way ANOVA. ***p < 0.001; ****p < 0.0001. ANOVA, analysis of variance; H&E, hematoxylin and eosin; LHQK, Lianhua Qingke; LPS, lipopolysaccharide; SD, standard deviation.
LHQK ameliorated LPS‐induced inflammation and mucus secretion in the lung
Since uncontrolled pulmonary inflammation is a key feature of ALI/ARDS and previous studies improved the immunomodulatory capacities of LHQK, 25 , 53 we wonder whether LHQK has an impact on LPS‐induced lung inflammation. To address this question, we evaluated the concentration of total protein, the protein level of Mucin protein (Muc5AC), and the concentration of pro‐inflammatory cytokines (IL‐6, TNF‐α, and IL‐1β) in plasma and BALF of mice. The LPS group showed significantly higher levels of total protein concentration and pro‐inflammatory cytokines, including IL‐6, TNF‐α, and IL‐1β, in both plasma and BALF, while LHQK administration productively reduced the levels of total protein concentration and pro‐inflammatory cytokines compared with that in LPS group (Figure2a–g). Additionally, LHQK treatment significantly inhibited Muc5AC secretion triggered by LPS in a dose‐dependent manner (Figure 2h), confirming the improving of LHQK on respiratory symptoms.
Figure 2.
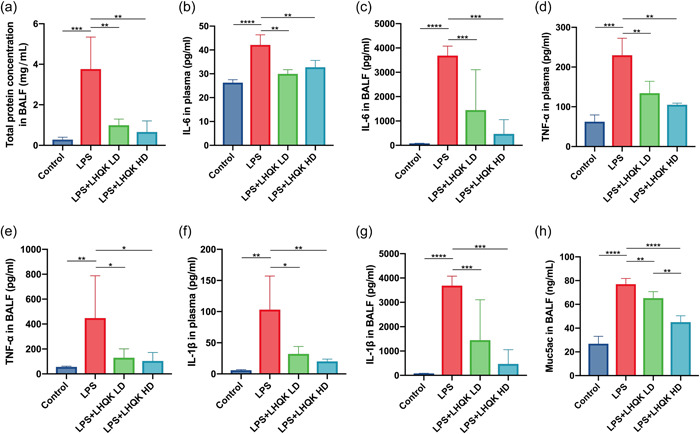
LHQK ameliorated LPS‐induced inflammation and mucus secretion in the lung. (a) Total protein levels in bronchoalveolar lavage fluid (BALF) were assessed. (b–h) Concentrations of IL‐6 (b and c), TNF‐α (d and e), IL‐1β (f and g) and Muc5AC (h) in plasma (b, d, and f) and BALF samples (c, e, g, and h) were measured by ELISA, respectively. N = 6–8 in each group. The data are presented as the mean ± SD, and statistical differences were assessed by one‐way ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ANOVA, analysis of variance; ELISA, enzyme‐linked immunoassay; H&E, hematoxylin and eosin; IL, interleukin; LHQK, Lianhua Qingke; LPS, lipopolysaccharide; SD, standard deviation; TNF‐α, tumor necrosis factor alpha.
LHQK inhibited the release of LPS‐induced NETs
Neutrophils are recruited into the alveolar spaces upon microbial infection. 7 , 54 It has been reported that LPS can induce NETs formation, resulting in NETosis. 54 , 55 To explore the effect of LHQK on NETs formation, we assessed the level of cf‐DNA and introduced a capture ELISA based on CitH3 (NETs‐specific biomolecules) associated with DNA. Our results indicated that cf‐DNA and CitH3‐DNA levels were significantly increased after the LPS challenge and effectively ameliorated by LHQK administration (Figure 3a,b). The results of immunofluorescence staining with anti‐MPO and anti‐CitH3 antibodies on lung sections were consistent (Figure 3c). We concluded that LHQK prohibited NETs formation in LPS‐induced ALI.
Figure 3.
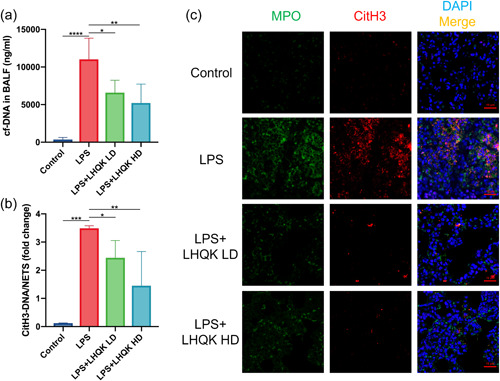
LHQK inhibited the release of LPS‐induced NETs. (a–b) cf‐DNA levels (a) and CitH3‐DNA levels (b) in BALF. (c) Immunofluorescence staining with anti‐MPO and anti‐CitH3 antibodies on lung sections. Scale bars: 10 μm. N = 6–8 in each group. The data are presented as the mean ± SD, and statistical differences were assessed by one‐way ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ANOVA, analysis of variance; BALF, bronchoalveolar lavage fluid; DAPI, 4′,6‐diamidino‐2‐phenylindole; H&E, hematoxylin and eosin; LHQK, Lianhua Qingke; LPS, lipopolysaccharide; MPO, myeloperoxidase; NETs, neutrophil extracellular traps; SD, standard deviation.
LHQK suppressed pyroptosis induced by LPS
Pyroptosis is another robust inflammatory response upon pathogen infection and plays a crucial pathological role in ALI development. Researchers also reported that NETs could promote macrophage pyroptosis in sepsis. 5 , 18 To assess whether LHQK affects the activation of pyroptosis, western blot analysis was employed to determine the protein level of pyroptosis‐related proteins, including caspase‐11, GSDMD, GSDMD N‐terminal domain, pro‐caspase‐1, cleaved‐caspase‐1, NLRP3, pro‐IL‐1β, and cleaved‐IL‐1β. Compared with the control group, LPS increased the protein level of caspase‐11, GSDMD N‐terminal domain, pro‐caspase‐1, cleaved‐caspase‐1, NLRP3 and cleaved‐IL‐1β in the lung of mice (Figure 4a–g). By contrast, LHQK considerably suppressed the activation of caspase‐11 and cleaved‐caspase‐1, cleavage of GSDMD N‐ terminal domain, and maturation of IL‐1β (Figure 4a–g), indicating an effective inhibition of pyroptosis by LHQK.
Figure 4.
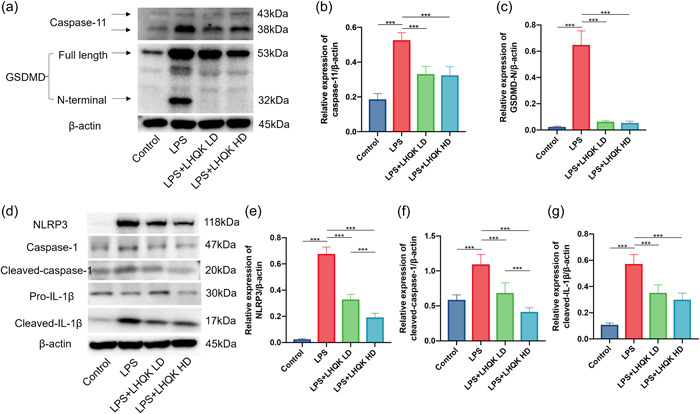
LHQK suppressed caspase‐11 and caspase‐1‐dependent pyroptosis induced by LPS. (a) Levels of caspase‐11, GSDMD, GSDMD N‐terminal domain, and β‐actin proteins in the lung tissues of mice were measured by western blot analysis. (b and c) Quantitative analysis of caspase‐11 and GSDMD‐N expression. (d) Levels of NLRP3, pro‐caspase‐1, cleaved‐caspase‐1, pro‐IL‐1β, cleaved‐IL‐1β, and β‐actin proteins in the lung tissues of mice were measured by western blot analysis. (e–g) Quantitative analysis of NLRP3, cleaved‐caspase‐1, and cleaved‐IL‐1β expression. N = 6–8 in each group. The data are presented as the mean ± SD, and statistical differences were assessed by one‐way ANOVA. ***p < 0.001. ANOVA, analysis of variance; IL, interleukin; LHQK, Lianhua Qingke; LPS, lipopolysaccharide; SD, standard deviation.
LHQK alleviated ALI through multiple pathways
To decipher molecular pathways affected by LHQK treatment in ALI/ARDS, RNA sequencing (RNA‐Seq) was performed using lung tissues from the LPS and LHQK‐treated groups (low dose, LD). Compared with the control group, 1898 genes were upregulated, and 1818 genes were downregulated in lung tissues of the LPS group, while 185 genes were upregulated, and 191 genes were downregulated in lung tissues of the LHQK‐treated group (LD) (Figure 5a,b). Venn plot showed that 75 of the genes downregulated by LPS were attenuated by LHQK, whereas 72 of the genes upregulated by LPS were downregulated by LHQK (Figure 5c,d). Several inflammatory related genes, including IL‐6, C‐C Motif Chemokine Ligand 4 (CCL4), interleukin 1 receptor antagonist (IL1RN), and adiponectin (ADIPOQ), upreguled by LPS were downregulated by LHQK. Furthermore, to interpret the mechanisms of LHQK against ALI from a systematic perspective, functionally grouped KEGG pathways and gene ontology enrichment analysis of the biological process (BP) were explored (Figure 5e). The key signaling pathways of LHQK in the treatment of ALI include HIF‐1 signaling pathway, hematopoietic cell lineage, and cytosolic DNA‐sensing pathway. In addition, negative regulation of MAP kinase activity, negative regulation of cytokine‐mediated signaling pathway, alpha‐beta T cell activation and differentiation involved in immune response, negative regulation of calcium ion transport, regulation of platelet‐derived growth factor receptor signaling pathway, and so forth, are the key BPs for the therapeutic effect of LHQK on ALI.
Figure 5.
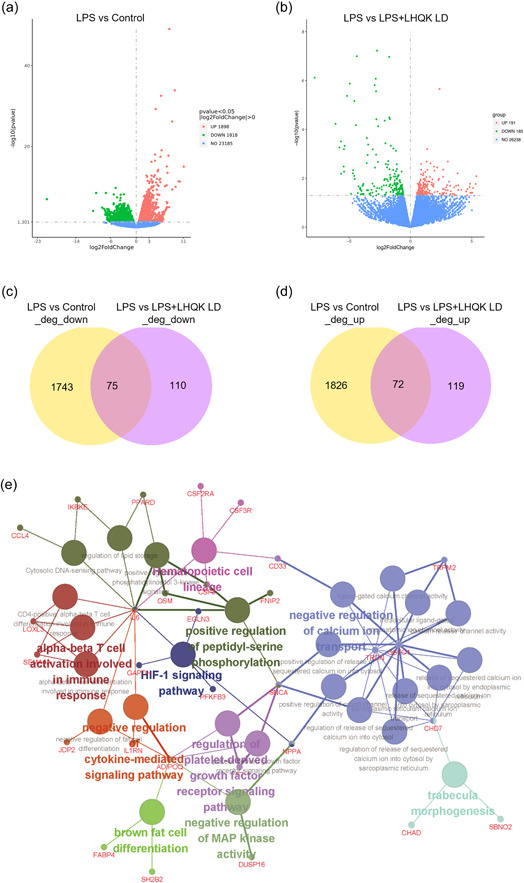
RNA sequencing and GO and KEGG pathway analysis. (a and b) Volcano maps. (c and d) Venn plots of DEGs. (e) BP and KEGG pathways enrichment analysis of DEGs reversed by LHQK (compared with ALI group). Large nodes represent BP and KEGG pathways. Small nodes represent DEGs mapped to BP and KEGG pathways. Nodes with the same color represent items with similar biological functions. BP, biological process; DEGs, differentially expressed genes; GO, Gene Ontology; LHQK, Lianhua Qingke; LPS, lipopolysaccharide; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Active compounds in LHQK showed potential regulatory effects on NETs and pyroptosis in LPS‐induced ALI
To explain the key material basis of LHQK with regulatory effects on regulating NETs and the pyroptosis, we collected the genes closely related to the NETs and pyroptosis and constructed the compound‐target (C‐T) networks for 14 compounds identified from LHQK by Wang et al. 24 (Figure 6a,b). According to the node importance parameter degree, active components, including emodin, chlorogenic acid, hesperidin, rhein, and chrysophanol (synonym: 1,8‐dihydroxy‐3‐methylanthraquinone), are predicted as key components with potential regulatory effects on NETs and pyroptosis. The key targets for active compounds of LHQK, such as PIK3CD, MMP2, MAPK1, MAPK3, AKT1, CXCL8, IL‐6, and TNF, are mainly key regulators involved in pulmonary diseases. 56 , 57 , 58 RNA‐Seq results showed that, among these key target genes, IL‐6 was also significantly downregulated by LHQK in ALI (Figure 5e).
Figure 6.
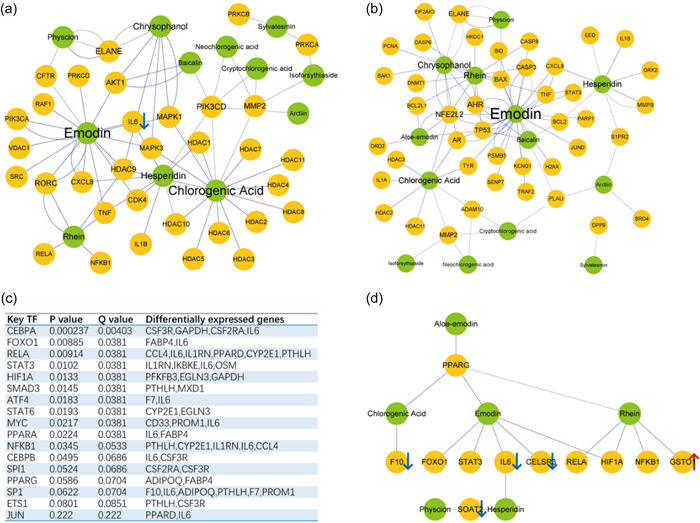
The compound‐target (C‐T) networks. (a) The (C‐T) networks for genes are closely related to the neutrophil extracellular traps (NETs) process. (b) The (C‐T) networks for genes are closely related to the pyroptosis. (c) Key transcription factors (TFs) predicted by the TRRUST database. (d) The (C‐T) networks for TFs and DEGs are regulated by LHQK. If a compound targets a protein/gene, there is a link between them. The green and yellow nodes represent the compound and target, respectively. Node size is proportional to its degree. The blue edge represents the known interactions, and the gray edge represents the predicted interactions. The red upward arrow indicates that the gene is upregulated by LHQK. The blue down arrow indicates that the gene is downregulated by LHQK (compared with the ALI group). DEGs, differentially expressed genes; LHQK, Lianhua Qingke.
Additionally, TRRUST version 2 49 was used to predict the possible TFs targets that cause differential gene changes from RNA‐Seq data analysis (p < 0.05 as the threshold). A total of 17 transcription regulators were obtained, of which 12 were significantly different (p < 0.05) (Figure 6c). Furthermore, C‐T networks were constructed for these TFs and differentially expressed genes (DEGs) regulated by LHQK. The results showed that six of the 12 significantly different TFs, including FOXO1, RELA, STAT3, HIF1A, NFKB1, and PPARG, were shown to be regulated by active compounds such as emodin, chlorogenic acid, hesperidin, and rhein (Figure 6d). Of note, three of the target TFs(RELA, STAT3, and NFKB1) were shown in the list of genes closely related to NETs (Figure 6a) or pyroptosis (Figure 6b).
DISCUSSION
ALI and ARDS are serious clinically common diseases characterized by acute onset and pulmonary inflammation arising and caused by various pathological factors, frequently resulting in significant morbidity and mortality. 1 , 2 Despite extensive efforts, 59 , 60 , 61 , 62 there is still no specific and effective treatment for ALI/ARDS. With the exemplified success of artemisinin from ancient Chinese medicine books in the fight against malaria, the understanding and application of TCM have received extensive attention globally. TCM is effective and reliable in treating many respiratory diseases, including ALI induced by respiratory infections and refractory chronic lung diseases, particularly during early‐phase interventions. 63 LHQK has been recommended for acute tracheobronchitis, cough, expectoration with a dry throat, and chest distress. Previous studies have proved the therapeutic effects of LHQK on ALI induced by bacteria and smoking exposure, and pneumonia caused by HCoVs (HCoV‐229E and SARS‐CoV‐2) in an animal model and human patients. 23 , 24 , 25 , 26 , 64 , 65 Clinical studies shown the effects of LHQK on relieving cough and expectoration in symptomatic COVID‐19 cases. 25 , 64 , 65 Wang et al. demonstrated LHQK ameliorated pneumonia caused by HCoVs in mice by coordinating the Th‐mediated immune responses to reduce levels of pro‐inflammatory cytokines. 24 However, the exact underlying mechanisms of LHQK on ALI/ARDS have not been fully elucidated. In this study, we demonstrated that LHQK is a novel therapeutic strategy for ALI/ARDS due to its inhibition of NETs formation and pyroptosis.
The therapeutic efficacy of LHQK in a mouse model of LPS‐induced ALI was evaluated to study the effect and underlying mechanisms of LHQK on ALI/ARDS. Lung histopathological analysis with H&E staining and ELISA detection demonstrated that LHQK could effectively attenuate LPS‐induced lung injury and inhibit the release of inflammatory cytokines, such as IL‐6, TNF‐α, and IL‐1β. Furthermore, LHQK could reduce LPS‐induced mucus secretion. Overall, our findings proved the protective effect of LHQK against ALI.
NETs are beneficial antibacterial defense structures released by neutrophils to bind and eliminate pathogens. 7 , 54 However, the exaggerated release of NETs may contribute to sustained inflammation and immunothrombosis and be correlated with morbidity and mortality of ARDS. 5 , 9 , 66 Recently, Zou et al. 67 also reported that NETs formation was strongly correlated with acute‐phase reactants in patients with COVID‐19, and they found that serum from individuals with COVID‐19 could trigger NETs release from control neutrophils in vitro. Therefore, maintaining a critical balance of NETs is vital to prevent lung injury triggered by bacterial or viral infection. Since NETs could be indirectly triggered by LPS and contribute to ALI, 54 , 68 we evaluated the LPS‐induced NETs formation with or without LHQK treatment. The elevated levels of cf‐DNA, CitH3‐DNA, and the specific markers of NETs (MPO and CitH3) by LPS were significantly inhibited by LHQK, indicating that LHQK could effectively attenuate lung injury and reduce lung inflammation by inhibiting LPS‐induced NETs formation.
Pyroptosis, a form of programmed cell death, may also be augmented by NETs and contribute to the development of ALI. 5 , 12 , 18 , 69 Accumulating data proved that macrophages enhanced inflammatory responses during ARDS by undergoing pyroptosis when challenged by infectious and noninfectious stimuli. 70 , 71 Moreover, SARS‐CoV‐2 can activate inflammasomes directly or indirectly, further leading to pyroptosis. 72 Thus, targeting the process of pyroptosis may serve as a logical therapeutic strategy for ALI/ARDS. To address the role of LHQK in pyroptosis, a series of pyroptosis‐related biomolecules were evaluated by western blot. We found that LPS increased the protein level of NLRP3, a canonical inflammasome, and activated caspase‐1 and caspase‐11, thus cleaving pro‐IL‐1β and GSDMD into mature IL‐1β and GSDMD‐N, respectively. GSDMD‐N could translocate to the cell membrane to form membrane pores, releasing mature IL‐1β and pyroptosis. All the increased protein levels triggered by LPS were attenuated after LHQK treatment, suggesting a therapeutic effect of LHQK for ALI/ARDS through inhibiting pyroptosis.
Furthermore, RNA‐Seq data analysis, additional bioinformatic analysis, and network pharmacological analysis indicated that LHQK or its active compounds may play protective functions in LPS‐induced ALI/ARDS through modulating genes and TFs directly and indirectly related to NETs and pyroptosis. In the RNA‐Seq data, we also found additional targets of LHQK, including genes related to inflammatory regulation and genes involved in specific cellular functions, such as mucus clearance and antioxidants, which may be due to the multiple protective functions of TCM. These findings provide a hint for the future study of LHQK. However, further in‐depth studies are still required to validate the specific active components of LHQK that function in different pathways of ALI/ARDS.
AUTHOR CONTRIBUTIONS
Wenjun Peng and Wensi Zhu: Data curation and formal analysis; software and writing—original draft. Lin Tong and Ainiwaer Rouzi: Data curation; software and formal analysis. Hui Qi: Bioinformatic analysis; writing—review and editing, and visualization. Yuanyuan Wu, Linxiao Han, Ludan He, Yu Yan, Ting Pan, Jie Liu and Qin Wang: Data curation and software. Zhenhua Jia: Resources and funding acquisition. Yuanlin Song, Qiaoliang Zhu and Jian Zhou: Conceptualization; methodology; funding acquisition and supervision.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
All animal experiments were conducted in accordance with the approval of Experimental Animal Ethical Committee of Fudan University (Animal experiment proposal approved on 20201021).
ACKNOWLEDGMENTS
We would like to acknowledge Dr. Zhonghua Zhao for his help in immunofluorescence staining of lung sections, and all members of Jian Zhou's lab for their valuable discussions. This work was supported by Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine [ZYYCXTD‐D‐202206], National Natural Science Foundation of China [82070045, 82270040], National Key Research and Development Program of China [2022YFA0806200], Shanghai Municipal Science and Technology Major Project [ZD2021CY001], Shanghai Municipal Key Clinical Specialty [shslczdzk02201], Science and Technology Commission of Shanghai Municipality [20DZ2261200, 21DZ2200600] and Shanghai Jinshan Municipal Health Commission [GWV‐10.1‐XK26, JSZK2019A01].
Peng W, Qi H, Zhu W, Tong L, Rouzi A, Wu Y, Han L, He L, Yan Y, Pan T, Liu J, Wang Q, Jia Z, Song Y, Zhu Q, Zhou J. Lianhua Qingke ameliorates lipopolysaccharide‐induced lung injury by inhibiting neutrophil extracellular traps formation and pyroptosis. Pulm Circ. 2023;13:e12295. 10.1002/pul2.12295
Wenjun Peng, Hui Qi, Wensi Zhu, Lin Tong, and Ainiwaer Rouzi contributed equally to this work.
Contributor Information
Yuanlin Song, Email: song.yuanlin@zs-hospital.sh.cn.
Qiaoliang Zhu, Email: zhuqiaoliang111@126.com.
Jian Zhou, Email: zhou.jian@fudan.edu.cn.
DATA AVAILABILITY STATEMENT
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Phua J, Badia JR, Adhikari NKJ, Friedrich JO, Fowler RA, Singh JM, Scales DC, Stather DR, Li A, Jones A, Gattas DJ, Hallett D, Tomlinson G, Stewart TE, Ferguson ND. Has mortality from acute respiratory distress syndrome decreased over time?: a systematic review. Am J Respir Crit Care Med. 2009;179(3):220–227. [DOI] [PubMed] [Google Scholar]
- 2. Máca J, Jor O, Holub M, Sklienka P, Burša F, Burda M, Janout V, Ševčík P. Past and present ARDS mortality rates: a systematic review. Respir Care. 2017;62(1):113–122. [DOI] [PubMed] [Google Scholar]
- 3. Mercat A, Richard JCM, Vielle B, Jaber S, Osman D, Diehl JL, Lefrant JY, Prat G, Richecoeur J, Nieszkowska A, Gervais C, Baudot J, Bouadma L, Brochard L, Expiratory Pressure (Express) Study Group f. Positive end‐expiratory pressure setting in adults with acute lung injury and acute respiratory distress syndrome: a randomized controlled trial. JAMA. 2008;299(6):646–655. [DOI] [PubMed] [Google Scholar]
- 4. Bonaventura A, Liberale L, Carbone F, Vecchié A, Diaz‐Cañestro C, Camici G, Montecucco F, Dallegri F. The pathophysiological role of neutrophil extracellular traps in inflammatory diseases. Thromb Haemost. 2018;118(1):006–027. [DOI] [PubMed] [Google Scholar]
- 5. Li H, Li Y, Song C, Hu Y, Dai M, Liu B, Pan P. Neutrophil extracellular traps augmented alveolar macrophage pyroptosis via AIM2 inflammasome activation in LPS‐Induced ALI/ARDS. J Inflamm Res. 2021;14:4839–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–1022. [DOI] [PubMed] [Google Scholar]
- 7. Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, Feramisco J, Nizet V. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol. 2006;16(4):396–400. [DOI] [PubMed] [Google Scholar]
- 8. Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 2018;18(2):134–147. [DOI] [PubMed] [Google Scholar]
- 9. Lefrançais E, Mallavia B, Zhuo H, Calfee CS, Looney MR. Maladaptive role of neutrophil extracellular traps in pathogen‐induced lung injury. JCI Insight. 2018;3(3):e98178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wallach D, Kang TB, Dillon CP, Green DR. Programmed necrosis in inflammation: toward identification of the effector molecules. Science. 2016;352(6281):aaf2154. [DOI] [PubMed] [Google Scholar]
- 11. Liu Y, Zhou J, Luo Y, Li J, Shang L, Zhou F, Yang S. Honokiol alleviates LPS‐induced acute lung injury by inhibiting NLRP3 inflammasome‐mediated pyroptosis via Nrf2 activation in vitro and in vivo. Chin Med. 2021;16(1):127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu B, Wang Z, He R, Xiong R, Li G, Zhang L, Fu T, Li C, Li N, Geng Q. Buformin alleviates sepsis‐induced acute lung injury via inhibiting NLRP3‐mediated pyroptosis through an AMPK‐dependent pathway. Clin Sci. 2022;136(4):273–289. [DOI] [PubMed] [Google Scholar]
- 13. Cheng KT, Xiong S, Ye Z, Hong Z, Di A, Tsang KM, Gao X, An S, Mittal M, Vogel SM, Miao EA, Rehman J, Malik AB. Caspase‐11‐mediated endothelial pyroptosis underlies endotoxemia‐induced lung injury. J Clin Invest. 2017;127(11):4124–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang J, Zhao Y, Shao F. Non‐canonical activation of inflammatory caspases by cytosolic LPS in innate immunity. Curr Opin Immunol. 2015;32:78–83. [DOI] [PubMed] [Google Scholar]
- 15. Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660–665. [DOI] [PubMed] [Google Scholar]
- 16. Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. Inflammasome‐activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535(7610):153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kovacs SB, Miao EA. Gasdermins: effectors of pyroptosis. Trends Cell Biol. 2017;27(9):673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen L, Zhao Y, Lai D, Zhang P, Yang Y, Li Y, Fei K, Jiang G, Fan J. Neutrophil extracellular traps promote macrophage pyroptosis in sepsis. Cell Death Dis. 2018;9(6):597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Y, Liu Q, Xie H, Yin S, Wu L, Yu X, Fan L, Lin L. Is Chinese Medicine injection applicable for treating acute lung injury and acute respiratory distress syndrome? A systematic review and meta‐analysis of randomized controlled trials. Chin J Integr Med. 2020;26(11):857–866. [DOI] [PubMed] [Google Scholar]
- 20. Gao P, Zhao Z, Zhang C, Wang C, Long K, Guo L, Li B. The therapeutic effects of traditional Chinese Medicine Fusu agent in LPS‐induced acute lung injury model rats. DDDT. 2018;12:3867–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li GH, Sun XJ, Deng YH, Lai JY, Liu Q. Meta‐analysis and trial sequential analysis of Qingjin Huatan Decoction for treating community‐acquired pneumonia in elderly. Zhongguo Zhong Zhi = Chin J Chin Materia Med. 2020;45(11):2658–2667. [DOI] [PubMed] [Google Scholar]
- 22. Zheng W, Huang X, Lai Y, Liu X, Jiang Y, Zhan S. Glycyrrhizic acid for COVID‐19: findings of targeting pivotal inflammatory pathways triggered by SARS‐CoV‐2. Front Pharmacol. 2021;12:631206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deng L, Xia W, Liu X, et al. Protective effect and mechanism of Lianhua Qingke tablets on acute bronchitis in rats. Central South Pharmacy. 2020;18(6):919–923. [Google Scholar]
- 24. Wang M, Li W, Cui W, Hao Y, Mi Y, Wang H, Hou Y, Jia Z. The therapeutic promises of Lianhua Qingke in the mice model of coronavirus pneumonia (HCoV‐229E and SARS‐CoV‐2). Chin Med. 2021;16(1):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang L, Wu L, Xu X, Yuan Y, Jiang R, Yan X, Zhang X, Gao Y, Shang H, Lian B, Hu J, Mei J, Wu S, Liu Q. Efficacy and safety of Lianhua Qingke tablets in the treatment of mild and Common‐Type COVID‐19: a randomized, controlled, multicenter clinical study. Evid‐Based Compl Altern Med. 2022;2022:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jin L, Xu Y, Yuan H. Effects of four types of integrated Chinese and Western medicines for the treatment of COVID‐19 in China: a network meta‐analysis. Rev Assoc Med Bras. 2020;66(6):771–777. [DOI] [PubMed] [Google Scholar]
- 27. Zhen G, Jing J, Fengsen L. Traditional Chinese Medicine classic herbal formula Xiaoqinglong decoction for acute exacerbation of chronic obstructive pulmonary disease: a systematic review protocol. Medicine. 2018;97(52):e13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hsieh CF, Lo C, Liu CH, Lin S, Yen HR, Lin TY, Horng JT. Mechanism by which ma‐xing‐shi‐gan‐tang inhibits the entry of influenza virus. J Ethnopharmacol. 2012;143(1):57–67. [DOI] [PubMed] [Google Scholar]
- 29. Peng W, Chang M, Wu Y, Zhu W, Tong L, Zhang G, Wang Q, Liu J, Zhu X, Cheng T, Li Y, Chen X, Weng D, Liu S, Zhang H, Su Y, Zhou J, Li H, Song Y. Lyophilized powder of mesenchymal stem cell supernatant attenuates acute lung injury through the IL‐6‐p‐STAT3‐p63‐JAG2 pathway. Stem Cell Res Ther. 2021;12(1):216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Caudrillier A, Kessenbrock K, Gilliss BM, Nguyen JX, Marques MB, Monestier M, Toy P, Werb Z, Looney MR. Platelets induce neutrophil extracellular traps in transfusion‐related acute lung injury. J Clin Invest. 2012;122(7):2661–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 2010;28(5):511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA‐Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12(3):R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y, Zhang S, Li F, et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48(D1):D1031–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrián‐Uhalte E, Davies M, Dedman N, Karlsson A, Magariños MP, Overington JP, Papadatos G, Smit I, Leach AR. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45(D1):D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Davis AP, Wiegers TC, Johnson RJ, Sciaky D, Wiegers J, Mattingly CJ. Comparative toxicogenomics database (CTD): update 2023. Nucleic Acids Res. 2023;51(D1):D1257–D1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: augmenting protein‐chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44(D1):D380–D384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nature Biotechnol. 2007;25(2):197–206. [DOI] [PubMed] [Google Scholar]
- 42. Yao ZJ, Dong J, Che YJ, Zhu MF, Wen M, Wang NN, Wang S, Lu AP, Cao DS. TargetNet: a web service for predicting potential drug‐target interaction profiling via multi‐target Sar models. J Comput Aided Mol Des. 2016;30(5):413–424. [DOI] [PubMed] [Google Scholar]
- 43. Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42(Web Server issue):W32–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bosc N, Atkinson F, Felix E, Gaulton A, Hersey A, Leach AR. Large scale comparison of QSAR and conformal prediction methods and their applications in drug discovery. J Cheminf. 2019;11(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Z, Guo F, Wang Y, Li C, Zhang X, Li H, Diao L, Gu J, Wang W, Li D, He F. BATMAN‐TCM: a bioinformatics analysis tool for molecular mechANism of Traditional Chinese Medicine. Sci Rep. 2016;6:21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro‐Watanabe M. KEGG for taxonomy‐based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587–D592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, Sirota‐Madi A, Olender T, Golan Y, Stelzer G, Harel A, Lancet D. GeneCards version 3: the human gene integrator. Database. 2010;2010:baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S, Kang B, Jeong D, Kim Y, Jeon HN, Jung H, Nam S, Chung M, Kim JH, Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46(D1):D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24(2):282–284. [DOI] [PubMed] [Google Scholar]
- 52. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug‐in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Matthay MA, Ware LB, Zimmerman GA. The acute respiratory distress syndrome. J Clin Invest. 2012;122(8):2731–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. [DOI] [PubMed] [Google Scholar]
- 55. Khan MA, Farahvash A, Douda DN, Licht JC, Grasemann H, Sweezey N, Palaniyar N. JNK activation turns on LPS‐ and Gram‐negative bacteria‐induced NADPH oxidase‐dependent suicidal NETosis. Sci Rep. 2017;7(1):3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mukaida N. Pathophysiological roles of interleukin‐8/CXCL8 in pulmonary diseases. Am J Physiol‐Lung Cell Mol Physiol. 2003;284(4):L566–L577. [DOI] [PubMed] [Google Scholar]
- 57. Jena A, Mishra S, Deepak P, Kumar‐M P, Sharma A, Patel YI, Kennedy NA, Kim AHJ, Sharma V, Sebastian S. Response to SARS‐CoV‐2 vaccination in immune mediated inflammatory diseases: systematic review and meta‐analysis. Autoimmun Rev. 2022;21(1):102927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang C, Zhou J, Wang J, Li S, Fukunaga A, Yodoi J, Tian H. Progress in the mechanism and targeted drug therapy for COPD. Signal Transduct Target Ther. 2020;5(1):248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang Z, Yu T, Hou Y, Zhou W, Ding Y, Nie H. Mesenchymal stem cell therapy for ALI/ARDS: therapeutic potential and challenges. Curr Pharm Des. 2022;28(27):2234–2240. [DOI] [PubMed] [Google Scholar]
- 60. Lin X, Dean DA. Gene therapy for ALI/ARDS. Crit Care Clin. 2011;27(3):705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhu YG, Qu JM, Zhang J, Jiang HN, Xu JF. Novel interventional approaches for ALI/ARDS: cell‐based gene therapy. Mediators Inflamm. 2011;2011:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu C, Xiao K, Xie L. Advances in the use of exosomes for the treatment of ALI/ARDS. Front Immunol. 2022;13:971189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang J, Wu Q, Ding L, Song S, Li Y, Shi L, Wang T, Zhao D, Wang Z, Li X. Therapeutic effects and molecular mechanisms of bioactive compounds against respiratory diseases: Traditional Chinese Medicine theory and high‐frequency use. Front Pharmacol. 2021;12:734450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ge L, Zhu H, Wang Q, Li M, Cai J, Chen Y, Chen Y, Ding B, Fang B, Fei Y, Feng J, Guo X, Jiang R, Jiang Y, Li G, Li X, Li X, Liang Q, Liu J, Liu J, Liu Q, Liu S, Lu Y, Miao Q, Qi W, Shang H, Shi L, Tan X, Tang X, Wang X, Wang X, Xia W, Yang K, Yang L, Ye Y, Zhou Q, Zhang H, Zhang J, Zhang Z, Zhang Z, Zou X, Li J, Wu D. Integrating Chinese and Western Medicine for COVID‐19: a living evidence‐based guideline (version 1). J Evid‐Based Med. 2021;14(4):313–332. [DOI] [PubMed] [Google Scholar]
- 65. Liang SB, Fang M, Liang CH, Lan HD, Shen C, Yan LJ, Hu XY, Han M, Robinson N, Liu JP. Therapeutic effects and safety of oral Chinese patent medicine for COVID‐19: a rapid systematic review and meta‐analysis of randomized controlled trials. Complement Ther Med. 2021;60:102744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Middleton EA, He XY, Denorme F, Campbell RA, Ng D, Salvatore SP, Mostyka M, Baxter‐Stoltzfus A, Borczuk AC, Loda M, Cody MJ, Manne BK, Portier I, Harris ES, Petrey AC, Beswick EJ, Caulin AF, Iovino A, Abegglen LM, Weyrich AS, Rondina MT, Egeblad M, Schiffman JD, Yost CC. Neutrophil extracellular traps contribute to immunothrombosis in COVID‐19 acute respiratory distress syndrome. Blood. 2020;136(10):1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zuo Y, Yalavarthi S, Shi H, Gockman K, Zuo M, Madison JA, Blair CN, Weber A, Barnes BJ, Egeblad M, Woods RJ, Kanthi Y, Knight JS. Neutrophil extracellular traps in COVID‐19. JCI Insight. 2020;5(11):e138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu S, Su X, Pan P, Zhang L, Hu Y, Tan H, Wu D, Liu B, Li H, Li H, Li Y, Dai M, Li Y, Hu C, Tsung A. Neutrophil extracellular traps are indirectly triggered by lipopolysaccharide and contribute to acute lung injury. Sci Rep. 2016;6:37252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jiang R, Xu J, Zhang Y, Zhu X, Liu J, Tan Y. Ligustrazine alleviate acute lung injury through suppressing pyroptosis and apoptosis of alveolar macrophages. Front Pharmacol. 2021;12:680512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fan EKY, Fan J. Regulation of alveolar macrophage death in acute lung inflammation. Respir Res. 2018;19(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sauler M, Bazan IS, Lee PJ. Cell death in the lung: the apoptosis‐necroptosis axis. Annu Rev Physiol. 2019;81:375–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vora SM, Lieberman J, Wu H. Inflammasome activation at the crux of severe COVID‐19. Nat Rev Immunol. 2021;21(11):694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.


