Abstract
Pancreatic cancer (PC) is one of the most common malignancies. Surgical resection is a potential curative approach for PC, but most patients are unsuitable for operations when at the time of diagnosis. Even with surgery, some patients may still experience tumour metastasis during the operation or shortly after surgery, as precise prognosis evaluation is not always possible. If patients miss the opportunity for surgery and resort to chemotherapy, they may face the challenging issue of chemotherapy resistance. In recent years, liquid biopsy has shown promising prospects in disease diagnosis, treatment monitoring, and prognosis assessment. As a noninvasive detection method, liquid biopsy offers advantages over traditional diagnostic procedures, such as tissue biopsy, in terms of both cost-effectiveness and convenience. The information provided by liquid biopsy helps clinical practitioners understand the molecular mechanisms underlying tumour occurrence and development, enabling the formulation of more precise and personalized treatment decisions for each patient. This review introduces molecular biomarkers and detection methods in liquid biopsy for PC, including circulating tumour cells (CTCs), circulating tumour DNA (ctDNA), noncoding RNAs (ncRNAs), and extracellular vesicles (EVs) or exosomes. Additionally, we summarize the applications of liquid biopsy in the early diagnosis, treatment response, resistance assessment, and prognostic evaluation of PC.
Keywords: Circulating tumour cells, Circulating tumour DNA, Noncoding RNAs, Extracellular vesicles, Exosomes, Pancreatic cancer, Liquid biopsy
Introduction
Pancreatic cancer (PC) is one of the most common malignancies, and the number of PC cases has doubled over the past two decades. The incidence of PC varies significantly across regions and populations, with the highest rates observed in North America, Europe, and Australia [1–3]. Recent years have seen a rapid increase in deaths due to PC, which can be attributed to global population growth and age structure changes and is closely linked to social and economic development [4]. According to predictions, PC is expected to become the third leading cause of cancer-related deaths in the European Union [5]. By 2030, it is projected to overtake breast, prostate, and colorectal cancers and become the second leading cause of cancer-related deaths in the United States [6].
Pancreatic ductal adenocarcinoma (PDAC) is a primary histological subtype of PC, accounting for 90% of all cases [7, 8]. Surgical resection is one of the methods of a potential cure, but most PDAC patients are unsuitable for operations when they are diagnosed [9, 10]. Therefore, screening and diagnosis should be conducted as early as possible to ensure a positive outcome. The diagnosis of PDAC relies on endoscopic ultrasonography-guided fine needle aspiration (EUS-FNA), magnetic resonance imaging (MRI), and computed tomography (CT) [11–14]. However, there are some problems, such as invasiveness, high cost, and exposure of subjects to radiation [15, 16]. In addition, the molecular composition of tumours is complex and dynamic, and repeated endoscopy examinations create a significant burden on patients [17]. Although the potential role of diagnostic biomarkers of cancer is constantly evolving, reliable diagnostic biomarkers for PC are still lacking. For instance, carbohydrate antigen 19-9 (CA19-9) has been extensively studied as a biomarker for detecting PC [18]. However, due to the lack of specificity of CA19-9, it can be expressed in various liver and gallbladder diseases as well as other types of malignant tumours, and elevated levels can also occur in some benign obstructive diseases [19]. Fucosyltransferase 3 (also known as the Lewis gene) is the key enzyme involved in the biosynthesis of CA19-9. Approximately 5–10% of individuals are Lewis antigen-negative, which means they do not secrete or secrete very little CA19-9, and to some extent, this also hinders the diagnosis of PC [20]. Therefore, CA19-9 alone cannot offer a conclusive diagnosis and must be combined with different clinical presentations, imaging tests, and biomarkers [15, 21].
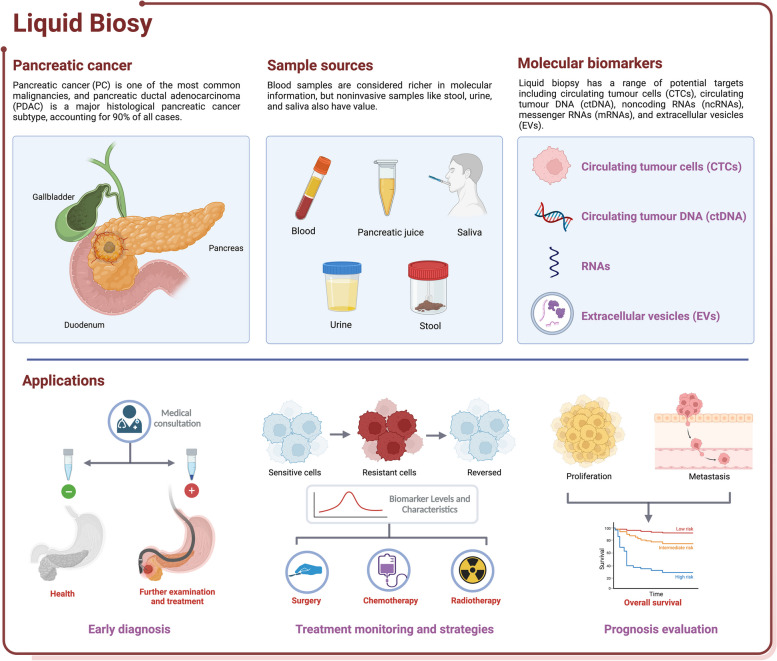
In recent years, liquid biopsy has garnered attention due to its advantages of lower invasiveness and the ability to continuously monitor cancer progression. While blood is considered the most critical biofluid for liquid biopsy (Fig. 1), other clinical samples, such as cerebrospinal fluid, saliva, ascites, pleural effusion, and urine, have also been used [22–26]. Different sample sources have unique characteristics, with the prevailing view suggesting that blood samples carry a richer molecular information profile. Although noninvasive samples such as stool, urine, and saliva may contain less biomarker information than blood, they can provide valuable information about the location of diseases. For instance, certain biomarkers in urine may be associated with kidney or bladder conditions [27], while stool biomarkers may be linked to digestive system disorders [28]. Currently, the potential targets of liquid biopsy are circulating tumour cells (CTCs), circulating tumour DNA (ctDNA), noncoding RNAs (ncRNAs), messenger RNAs (mRNAs), and extracellular vesicles (EVs), which can provide information about tumour genomics, transcriptomics, and proteomics.
Fig. 1.
Common samples, biomarkers, and clinical applications in liquid biopsy for pancreatic cancer. Blood is typically the most commonly used material in liquid biopsy, in addition to pancreatic juice, saliva, urine, and stool. Circulating tumour cells, circulating tumour DNA, noncoding RNAs, and extracellular vesicles are among the most common biomarkers. Liquid biopsy has a wide range of clinical applications, playing a crucial role in early diagnosis, treatment monitoring, and prognosis evaluation. Created with BioRender.com
Liquid biopsy exhibits high utility in the management of PC, with applications spanning early diagnosis, treatment strategies, drug resistance, recurrence monitoring, and prognosis assessment for PC patients. This review provides an overview of the biomarkers and detection methods utilized in liquid biopsy and their applications in the early diagnosis, treatment response, and prognosis evaluation of PC (Table 1). We also discuss the future trends of liquid biopsy and assess its limitations to improve current management strategies for patients.
Table 1.
The isolation and detection techniques in liquid biopsy
| Purpose | Methodology | Advantages | Disadvantages | Ref | |
|---|---|---|---|---|---|
| CTCs | isolation | density centrifugation | ease of operation | low detection rates of 24% to 40% in PDAC patients | [29] |
| CTCs filters | isolation of CTCs without capture agents | small-sized CTCs might be overlooked | [30] | ||
| flow cytometry | one of the most commonly used cell-sorting technologies for the analysis | lacked the ability to reveal sufficient morphological information to satisfy the standards set by pathologists for CTCs | [31] | ||
| microfluidic devices | the CTCs chip captures large numbers of viable CTCs in a single step | complex manufacturing process and high cost | [32] | ||
| dielectrophoresis | manipulate cells in accordance with their phenotype and membrane capacitance without the need for labeling or modification | Continuous optimization and fine-tuning of the electric field parameters are required | [33] | ||
| immuno-magnetic separation | excellent speed and efficiency for CTCs detection and characterization | EpCAM-based strategy fails to detect CTCs with low EpCAM expression | [34] | ||
| detection | high-resolution image scanning | an enrichment-free method to identify CTCs | limited applicability and processing speed | [35] | |
| mRNAs analysis and immunocytological staining | commonly used high-accuracy detection methods | complex operational procedures | [36] | ||
| mutational analysis | provide information on tumour evolution | a single mutation cannot fully represent the overall characteristics | [37] | ||
| single-cell next-generation sequencing | precise analysis of individual CTCs and revelation of tumour heterogeneity | complex sample handling | [38] | ||
| combined strategy | integrated platform combined deterministic lateral displacement, inertial focusing and magnetophoresis | high throughput and efficiency, isolating CTCs regardless of tumour surface epitopes | high manufacturing costs and complexity | [39] | |
| negative selection and 3D cell culture | compared to traditional negative selection, it greatly improves purity | cell proliferation cultured in a 3D environment is slower | [40] | ||
| CtDNA/ctRNA | mutation detection | ddPCR | high sensitivity | requires a larger peripheral blood volume | [41] |
| COLD-PCR | suitable for detecting rare and low-abundance mutations | needs to be combined with other detection methods | [42] | ||
| ARMS-PCR | high sensitivity, high accuracy, easy operation, and low cost | unable to achieve high-throughput and high-position detection | [43] | ||
| BEAMing | measurement of individual DNA molecules with high reliability and sensitivity | complex operational procedures | [44] | ||
| NGS | high throughput and high sensitivity | complex data analysis workflow and high cost | [45] | ||
| methylation detection | whole-genome bisulfite sequencing | high sensitivity and coverage of the entire genome | complex operational procedures and high cost | [46] | |
| DREAMing | simple, cost-effective, and has high sensitivity and specificity | only detects known loci | [47] | ||
| DISMIR | high sensitivity and relatively low detection cost | emphasizes the distribution of methylation across the entire genome rather than individual loci | [48] | ||
| NcRNAs | detection based on polymerase chain reaction | RT-qPCR, dPCR, and ddPCR | high sensitivity, simple and well-developed analysis program | controversy surrounding the standardization process | [49] |
| detection based on next generation sequencing | gene chips and RNA-seq | screening of ncRNAs can be accomplished | reduced specificity and elevated cost | [50] | |
| detection of expression and function | microarray | no amplification procedures are required | depends on known molecules | [51] | |
| nanostructured biochips | metallic nanoparticles, graphene oxide, quantum dots, and nanostructured polymers | high sensitivity allows it to detect molecules at very low concentrations | the fabrication process is more complex than traditional biochips | [52] | |
| EVs | isolation | ultracentrifugation | ease of operation and low cost | contamination and integrity are compromised | [53] |
| filtration | low cost and high efficiency | contamination and not suitable for all body fluids | [54] | ||
| size-exclusion chromatography | high particle integrity and less susceptible to contamination by soluble proteins | low particle yield and not suitable for all body fluids | [55] | ||
| membrane affinity | ease of operation and good commercial viability | low specificity in the enrichment of EVs subpopulations and susceptible to contamination | [56] | ||
| immunoaffinity capture | can separate specific subpopulations of particles | low yield | [57] | ||
| precipitation | high yield with good commercial viability | prone to contamination by soluble proteins | [58] | ||
| detection | traditional EVs detection methods, such as enzyme-linked immunosorbent assay, western blot, flow cytometry, and polymerase chain reaction | low cost | low efficiency and complex steps | [55, 59, 60] | |
| surface-enhanced Raman spectroscopy | a label-free, high-sensitivity technique | surface dependency | [61] | ||
| electron microscopy | reflect exosome structures | costly and sample processing can potentially modify the inherent properties | [62] | ||
| electro-chemical | high sensitivity and wide measurement range | high dependent | [63] | ||
| surface plasmon resonance | high sensitivity and real-time detection | high cost and surface dependency | [64] | ||
| colorimetric detection | ease of operation | susceptible to external interference | [65] | ||
| alternating current electrokinetic chips | high separation efficiency and sensitivity | complex design and manufacturing, limited to specific applications | [66] |
Biomarkers and detection methods
Circulating tumour cells
CTCs, which detach from the primary tumour, can enter the circulatory system and travel through the bloodstream. However, the majority of CTCs die in the peripheral blood within 1 to 2.5 hours due to mechanical forces or immune system attacks. Nevertheless, a small fraction of CTCs can survive and initiate distant metastasis [67, 68]. Numerous metastatic precursors within CTCs increase the risk of tumour metastasis and recurrence [69–71]. According to most perspectives, CTCs are believed to exhibit specific differences from primary tumours despite originating from primary tumours. This heterogeneity leads to their detachment from the primary tumour and acquisition of epithelial-mesenchymal transition (EMT) characteristics, facilitating intravascular infiltration and enhancing their potential for metastasis [72, 73].
The analysis process of CTCs mainly involves three stages: enrichment, detection, and characterization. Most enrichment methods are applied based on the surface phenotype or physical properties of CTCs. The CellSearch system, developed using an antibody targeting epithelial cell adhesion molecule (EpCAM), is the sole CTC detection technology approved by the U.S. Food and Drug Administration due to its ability to detect CTCs expressing EpCAM [29, 74]. However, this strategy cannot detect cells with low EpCAM expression due to the potential loss of epithelial antigens during the EMT process [34]. Furthermore, the abundance of CTCs varies across different types of cancer; the CellSearch system is more suitable for tumours with higher CTC abundance [75]. In microfluidic devices, affinity-based separation methods can also be employed. Designing microfluidic devices with varying materials, sizes, and structures to manipulate blood flow patterns creates additional opportunities for interacting CTCs and antibodies [32, 57, 76]. Pahattuge et al. [77] introduced a modular microfluidic system called SMART-Chip. They demonstrated that the SMART-Chip platform could significantly reduce the processing time by more than 50% when handling blood samples obtained from patients with PDAC and colorectal cancer compared to manual sample processing. Furthermore, microfabricated porous membranes can be employed to filtrate and isolate CTCs due to their size, which is larger than that of normal blood cells [78, 79].
CTCs are primarily detected using protein expression, immunocytochemistry, and nucleic acid methods. Flow cytometry allows for the quantitative assessment and characterization of protein expression in CTCs, offering the advantage of evaluating multiple biomarkers to characterize CTCs comprehensively. However, it exhibits lower sensitivity for detecting rare populations of CTCs [80]. Immunohistochemical staining and immunofluorescence are commonly employed techniques for detection and characterization purposes. Immunofluorescence, in particular, enables the visual confirmation of protein expression and localization by fluorescent markers. In the conventional cytofluorimetry approach, isolation is achieved through the utilization of specific antibodies that recognize markers selected on CTCs. This method utilizes monoclonal antibodies specifically targeting antigens expressed by CTCs, which results in the exclusion of CTCs that do not express such antigens but are present in the circulation. Consequently, this presents a challenge in obtaining or developing novel antibodies against specific targets [81]. The flexibility of immunofluorescence technology makes it a powerful tool for studying the protein expression of tumour cells. For instance, with the use of multimarker immunofluorescence panels, researchers can gain a more comprehensive understanding of the distribution and expression patterns of different CTC subtypes [82]. This not only aids in tumour classification and staging but also provides valuable insights for personalized therapy. In addition, CTCs can also be detected using techniques such as high-resolution image scanning, mutational analysis, and single-cell next-generation sequencing (scNGS). The molecular characteristics of CTCs were initially determined on enriched fractions, which provided limited information about tumour heterogeneity. In recent years, the rapidly advancing single-cell sequencing technology has become the preferred method for isolating individual CTCs and studying tumour heterogeneity. These technologies will facilitate the comprehensive characterization of CTCs at multiple omics and functional levels, enabling effective monitoring of the dynamic changes in tumour heterogeneity in individual cancer patients [83, 84].
Although the precise role of CTCs in tumour development remains elusive, they offer a valuable approach for obtaining comprehensive insights into tumours through liquid biopsy. In PC management, CTCs play a significant and beneficial role in patient diagnosis, prognostic evaluation, recurrence monitoring, and treatment decisions. In this regard, we have summarized the clinical applications of CTCs in various aspects of PC management in recent years (Table 2).
Table 2.
Clinical application of CTCs in PC in recent years
| Factors | Sensitivity | Specificity | AUC | Number of patients | Findings | Year of report | Ref | |
|---|---|---|---|---|---|---|---|---|
| Diagnosis | CTCs + CA19-9 | 0.95 | 80 | the combination of CTCs and CA19-9 improves the diagnosis of pancreatic cancer | 2022 | [85] | ||
| CTCs + Folate receptors + CA19-9 | 0.978 | 0.833 | 0.944 | 46 | folate receptors (+) CTCs, especially when combined with CA19‐9, have the potential as a biomarker for diagnosing pancreatic cancer | 2020 | [86] | |
| CTCs + EVs GPC1 | 1 | 0.8 | 22 | combining CTCs and GPC1-positive-exosome detection displayed 100% sensitivity and 80% specificity in PDAC | 2019 | [87] | ||
| CTCs | 0.75 | 0.964 | 0.867 | 72 | CTCs as a biomarker for diagnosis and staging in pancreatic cancer | 2016 | [88] | |
| CTCs + CA19-9 | 0.67 | 0.8 | 52 | CA19-9, combined with CTCs analysis, may represent an efficient method for diagnosing pancreatic adenocarcinoma in patients with of solid pancreatic tumours | 2017 | [89] | ||
| Prognosis | CTCs + KLF8 + vimentin | 40 | CTCs expressing Krüppel-like factor 8 and vimentin predict prognosis in pancreatic cancer | 2021 | [90] | |||
| CTCs | 0.8 | 100 | CTCs correctly identified patients with occult metastatic disease preoperatively | 2018 | [91] | |||
| Recurrence | CTCs + TIC markers | 60 | CTCs expressing markers of tumour-initiating cells predict survival and recurrence in PDAC | 2017 | [92] | |||
| CTCs + vimentin + cytokeratin | 50 | CTCs expressing vimentin and cytokeratin predict the recurrence of PDAC | 2016 | [93] | ||||
| CTCs | 36 | the presence or absence of CTCs in the blood of patients with PDAC could help predict the recurrence pattern after surgery | 2021 | [94] | ||||
| Therapy | CTCs + vimentin + CA19-9 | 100 | significantly reduced CTCs counts were observed after chemotherapy in subjects that responded to treatment | 2019 | [95] | |||
| CTCs | 0.82 | 0.85 | 0.871 | 200 | CTCs as a biomarker for 1-year recurrence for chemo-naive PDAC patients | 2018 | [96] |
Circulating tumour DNA
Cell-free DNA (cfDNA) is a crucial genetic component found in the bloodstream; its origin primarily stems from apoptotic, necrotic, and actively secreted fragments originating from healthy, inflamed, and tumour tissue. These fragments are typically approximately 150–180 base pairs in length [97–99]. CtDNA represents a distinctive subset of cfDNA released into the blood by CTCs. Compared to cfDNA, ctDNA is present in relatively lower amounts in the bloodstream, constituting only 1% (or even less than 0.01%) of cfDNA [98–100]. Most ctDNA fragments have lengths ranging from 160 to 200 base pairs, and they are less influenced by intratumoural heterogeneity compared to tumour tissues [97, 101, 102]. Additionally, ctDNA has a half-life of approximately 15 minutes to 2.5 hours, which means that it serves as a real-time tumour biomarker. In contrast, traditional blood protein biomarkers usually take weeks to manifest, and ctDNA can dynamically reflect the status of a tumour at a specific moment [103, 104]. Furthermore, ctDNA carries tumour-related genomic information, such as gene expression levels, mutations, the methylation status, and microsatellite instability. Compared to traditional biopsy markers, ctDNA is an ideal biomarker, especially for the real-time monitoring of treatment effectiveness and prognosis assessment.
CtDNA detection includes ctDNA preparation, library construction, analysis, and data alignment. One aspect of ctDNA detection focuses on genetic mutations. Single-base mutations have the potential to activate oncogenes, disrupting the balance between oncogenes and tumour suppressor genes, thereby instigating tumorigenesis. Another aspect involves DNA methylation, which plays a role in tumour initiation that is similar to that of DNA mutations [105, 106]. Mutation detection is a vital component of the analysis. Due to the extremely low abundance of ctDNA, employing highly sensitive techniques for detecting tumour mutations is crucial. Conventional approaches rely on polymerase chain reaction (PCR), but recent advancements in PCR and sequencing technologies have paved the way for alternative methods, including quantitative PCR (qPCR), digital PCR (dPCR), droplet digital PCR (ddPCR), and next-generation sequencing (NGS). qPCR allows real-time monitoring of DNA amplification with higher speed, reproducibility, and quantification. NGS platforms offer several advantages, including the ability to screen for unknown mutations and structural and copy-number variations. dPCR and ddPCR involve partitioning DNA samples into thousands or even millions of separate compartments or droplets, effectively reducing background noise associated with traditional methods and enabling the detection of tumour DNA at a variant allele frequency (VAF) below 0.1% [107–109]. In recent years, integrated detection strategies combining gene editing techniques, functional enzymes, and nanomaterials have been developed to effectively increase the net content of mutation fragments, thereby facilitating the identification of target gene mutations within ctDNA [106]. There are various methods for DNA methylation detection. Whole-genome bisulfite sequencing (WGBS-seq) is considered the gold standard for DNA methylation analysis. It can identify partially methylated regions in cancer cells. However, the sensitivity of this method may be compromised by DNA degradation [26, 110].
KRAS mutations are the most prevalent genetic alteration in PC. They are present in over 90% of patients and are considered an early driving factor in PDAC [108]. Castells et al. [111] demonstrated that the presence of KRAS mutations in plasma DNA served as a highly specific molecular marker for diagnosis and prognosis in a PDAC cohort of 44 patients. However, it is essential to emphasize that previous cfDNA sequencing results have not only identified mutations known to exist in tumours but have also uncovered a multitude of variations that are absent in tumour tissues [112]. In particular, some patients undergoing chemotherapy may harbour minimal residual lesions composed of drug-resistant cells. In such cases, the mutations detected in cfDNA in the bloodstream do not exclusively originate from tumour cells. CfDNA may also carry mutations from other sources, including those induced by the disease state or treatment. Undoubtedly, KRAS mutations are one of the vital indicators for evaluating PDAC, and their role has received widespread attention as a primary focus in many PC studies (Table 3). However, overall, the application of ctDNA and mutation analysis in PDAC still requires further strategies to thoroughly assess this detection method.
Table 3.
Clinical application of cfDNA/ctDNA in PC in recent years
| Testing methods | Markers | Number of patients | Stages | Findings | Year of report | Ref | |
|---|---|---|---|---|---|---|---|
| Diagnosis | ddPCR | KRAS | 105 | all stages PDAC | ddPCR increased the sensitivity and accuracy of EUS-FNA to 91.6% and 88.6% | 2022 | [113] |
| methylation-specific PCR | ADAMTS1, BNC1 | 39 | all stages PDAC | two-gene panel with highly promising sensitivity and specificity for detection of earliest stages of pancreatic cancer | 2019 | [114] | |
| methylation-specific PCR | BMP3, RASSF1A, BNC1, MESTv2, TFPI2, APC, SFRP1 and SFRP2 | 95 | all stages PDAC | patients with PDAC have a highly significant number of hypermethylated genes compared to patients with benign pancreatic diseases | 2016 | [115] | |
| Prognosis | ddPCR | KRAS | 108 | all stages PDAC | mutant-KRAS detection in the blood of PDAC patients is significantly associated with dismal prognosis for palliative and curative patients | 2023 | [116] |
| NGS | TP53, KRAS | 145 | all stages PDAC | cases that had detectable plasma ctDNA showed significantly shorter recurrence-free survival | 2022 | [117] | |
| BEAMing | KRAS | 61 | metastatic PDAC | RAS mutation analysis in cfDNA more accurately predicted the prognosis than tissue analysis | 2020 | [44] | |
| ddPCR | KRAS | 104 | metastatic PDAC | patients with metastases and detectable ctDNA had significantly shorter progression-free survival and overall survival times than patients without detectable ctDNA | 2019 | [118] | |
| Treatment | ddPCR | KRAS | 70 | metastatic PDAC | the change in magnitude of ctDNA during systemic treatment allows the prediction of treatment response and is associated with both overall survival and progression-free survival | 2022 | [119] |
| dPCR | KRAS | 47 | metastatic PDAC | KRAS mutation in ctDNA during chemotherapy could be a predictive factor for the disease progression of patients with PDAC | 2020 | [120] | |
| PCR-based-SafeSeqS assays | KRAS | 112 | resectable PDAC | a 24% decrease in the proportion of patients with detectable ctDNA following surgery | 2019 | [121] | |
| ddPCR | KRAS | 78 | localized, metastatic, and recurrent PDAC | no detection or disappearance of KRAS ctDNA within 6 months of treatment was significantly correlated with therapeutic responses to first-line chemotherapy | 2019 | [41] |
Noncoding RNAs
NcRNAs were once perceived to have a limited impact on tumour initiation and progression due to their inability to encode proteins. However, emerging evidence has highlighted the essential regulatory functions of ncRNAs. In addition to their capacity to modulate gene and protein expression, ncRNAs actively participate in diverse tumorigenic processes, including EMT, autophagy, and apoptosis [122–124]. NcRNAs can be classified into two main categories based on their lengths: small noncoding RNAs (sncRNAs), with a length of less than 200 nucleotides, and long noncoding RNAs (lncRNAs), with a length exceeding 200 nucleotides [125, 126]. In addition, circRNAs, which are RNA molecules with a circular structure, have been recognized for their significant regulatory roles in gene expression, cell proliferation, cell differentiation, and disease development in recent years. SncRNAs encompass several subtypes, including microRNAs (miRNAs), small nucleolar RNAs, small nuclear RNAs, piwi-interacting RNAs, and tRNA-derived small RNAs [127]. Among them, miRNAs are the most extensively studied factors in cancer research, and liquid biopsy identifies miRNAs actively secreted by CTCs and tumour cells themselves [128, 129]. MiRNAs can influence genes, with thousands of miRNAs regulating approximately 60% of the genes. Their principal function involves binding to recognition sites in the 3' untranslated region, thereby reducing mRNA stability and suppressing gene expression [130, 131].
LncRNAs play a regulatory role in protein and miRNA functions and expression levels and contribute to chromatin remodelling [130, 132]. Some lncRNAs are considered valuable biomarkers for PDAC because they mediate various processes involved in tumour cell progression, making them applicable for liquid biopsy in PDAC [122]. The detection methods for ncRNAs are similar to those for cfDNA and ctDNA. Standard techniques include qPCR, dPCR, ddPCR, gene chips, and NGS. Over the past few decades, many miRNA detection methods have been developed. The most commonly used methods include qPCR, hybridization chain reaction, rolling circle amplification, strand displacement amplification, and the use of media, such as graphene oxide and gold nanoparticles, to transfer probes into cells [133–136]. Zhao et al. [137] established a novel miRNA and circRNA detection approach based on an enhanced fluorescent signal. This method exhibits a significantly improved detection sensitivity and can be applied to both miRNA and circRNA detection. Additionally, Dittmar et al. [138] successfully applied the Abcam Fireplex™ (a novel limited-volume assay platform) to identify differential plasma miRNAs between early-stage PC cases and controls. Based on hydrogel particles, this platform enables the detection of up to 68 miRNAs in 20 μL of plasma per sample in a 96-well plate format without extensive isolation and purification steps. Likewise, improved detection methods can save lncRNA extraction and purification steps. Lou et al. [139] proposed a rapid colorimetric method to detect lncRNA HOTTIP in diluted serum; one significant benefit of this assay is its ability to be performed on diluted serum, eliminating the requirement for RNA extraction and purification. This detection method holds tremendous potential in PDAC clinical screening. In conclusion, these new technologies significantly differ from traditional detection methods. Although further advancements are required for their clinical application and efficacy validation, these technologies have opened up new avenues for detecting ncRNAs.
Extracellular vesicles
EVs are essential heterogeneous subcellular structures involved in intercellular communication and are composed of a phospholipid bilayer membrane with membrane proteins and glycoproteins [140]. EVs contain many bioactive molecules, including mRNAs, miRNAs, nucleic acids, lipids, proteins, transcription factors, and growth factors [140–142]. Viable cells actively secrete EVs and exhibit a ubiquitous presence in various bodily fluids. These vesicles are pivotal in enabling essential cellular communication within specific pathological and physiological contexts. Remarkably, tumour cells are also capable of releasing EVs, which actively participate in various mechanisms encompassing the initiation and progression of cancer, immune regulation, and neuronal communication [143]. According to the position statement released by the International Society for Extracellular Vesicles in 2018 [144], vesicles with a size of < 100 nm or < 200 nm are referred to as "small EVs," while those larger than 200 nm are termed "medium/large EVs." Based on their size and cellular origin, EVs can be classified into three main subtypes: exosomes, microvesicles, and apoptotic bodies. Exosomes have received considerable attention and have been extensively investigated among these subtypes. Initially considered cellular debris, similar to ncRNAs, exosomes have emerged as important molecules due to their involvement in diverse biological processes, including molecular transport, intercellular communication, and immune responses [145, 146]. The size of exosomes falls within the range of 30–160 nm [145, 147], classifying them as small EVs. Exosomes present distinctive advantages in the field of liquid biopsy. Compared to ctDNA released from apoptotic or necrotic cells, exosomes released by viable cells provide a more representative depiction of active tumour cell information. Additionally, exosomes exhibit better stability than ctDNA, which is attributed to their protective lipid bilayer [148–150]. With ongoing technological advancements and improved analytical capabilities, exosomes have the potential to become one of the most important alternative tools for liquid biopsy, and we have summarized the clinical applications of exosomal cargo or ncRNAs in various aspects of PC in recent years (Table 4).
Table 4.
Clinical application of exosomal cargo or ncRNAs in PC in recent years
| Exosomal cargo or ncRNAs | Type | Source | Number of patients | Findings | Year of report | Ref | |
|---|---|---|---|---|---|---|---|
| Diagnosis | miR-1246 and miR-4644 | miRNA | saliva | 12 | the relative expression ratios of miR-1246 and miR-4644 were significantly higher in the cancer group than these ratios in the control group | 2016 | [159] |
| miR-191, miR-21 and miR-451a | miRNA | serum | 61 | miR-191, -21, and -451a enclosed in serum exosomes, significantly up-regulated in pancreatic cancer, were sensitive diagnostic markers | 2018 | [160] | |
| miR-155 | miRNA | pancreatic juice | 35 | ex-miR-21 and ex-miR-155 levels discriminated PDAC patients from chronic pancreatitis patients with area under the curve values of 0.90 and 0.89, respectively | 2019 | [161] | |
| glypican-1 | protein | serum | 190 | ROC curves indicated that GPC1 + circulating exosomes revealed a perfect classifier with an AUC of 1.0, a sensitivity of 100%, and a specificity of 100% | 2015 | [102] | |
| Therapy | chitinase 3-like-1 and fibronectin | protein | macrophage | N/A | CHI3L1 and FN1 induce resistance of PDAC cells to GEM, and inhibitor treatment has proven effective | 2021 | [162] |
| miR-20a-5p | miRNA | plasma | 73 | the relative expression of miR-20a-5p in gemcitabine-resistant plasma of PC patients was significantly lower than in nonresistant patients | 2019 | [163] | |
| Recurrence | miR-451a | miRNA | plasma | 56 | miR-451a showed the highest upregulation in the stage II patients who showed recurrence after surgery | 2018 | [164] |
| miR-4525, miR-451a and miR-21 | miRNA | portal vein blood | 55 | high expression of miR-4525, miR-451a, and miR-21 in portal vein blood was associated with recurrence with higher sensitivity and specificity than that in peripheral blood | 2019 | [165] | |
| Prognosis | miR-3607-3p | miRNA | NK cells | 40 | miR-3607-3p is down-regulated in pancreatic cancer and decreased miR-3607-3p level predicts poor prognosis in pancreatic cancer patients | 2019 | [166] |
| IARS | circRNA | peripheral blood | 79 | circRNA IARS is highly expressed in pancreatic cancer, positively correlated with tumour metastasis, and negatively correlated with patient survival time | 2018 | [167] | |
| UPK1A-AS1 | lncRNA | serum | 75 | the patients with higher UPK1A-AS1 expression had a shorter PFS than those with lower UPK1A-AS1 expression | 2022 | [168] |
The isolation and characterization of EVs present certain challenges. One challenge arises from the low abundance of EVs in biological samples, necessitating highly sensitive techniques for their isolation and purification. Moreover, EVs are prone to contamination from non-EV proteins, lipoproteins, and high-density lipoproteins [151, 152]. Currently, major methods for EV isolation include the following: density-based (ultracentrifugation), size-based (filtration and size-exclusion chromatography), affinity-based (membrane affinity and immunoaffinity capture), precipitation (polyethylene glycol precipitation), and microfluidic technologies. Each method for extracellular vesicle isolation has advantages and limitations. The choice of method should be based on a comprehensive consideration of factors such as the intended purpose, sample characteristics, and experimental conditions. For example, ultracentrifugation, considered the gold standard for EV isolation, is time-consuming and may lead to potential damage to EVs [152]. Filtration is a simple and convenient method. However, it presents challenges in effectively removing impurities from the filter membrane and can still result in the deformation and lysis of EVs [153]. Novel techniques based on microfluidics have been developed to isolate EVs; size-based microfluidics use nanowire and micropillar structures to separate EVs with diameters in a certain range from smaller cellular debris, proteins, and other particles [154, 155]. Compared to conventional approaches, the consolidation of EV isolation and disease detection on a unified platform enhances the clinical viability of EV detection. Microfluidic technology offers significant advantages in this regard, as it allows for preserving EV morphology while minimizing contamination from proteins and other nanoparticles. Additionally, microfluidics exhibits characteristics such as portability, rapidity, low cost, and ease of operation, which are crucial for noninvasive disease detection [152, 155]. Zheng et al. [156] developed an alternating drop-shaped micropillar array to assist in capturing tumour-derived exosomes by Tim4-modified magnetic beads to improve the efficiency of exosome capture. This approach enables the effective extraction of tumour-derived exosomes and greatly enhances their purity. Combined analysis of different types of biomarkers on exosomal membrane surface proteins improves the accuracy of cancer diagnosis. Emerging EV isolation techniques have allowed EVs to rival other important biomarkers, such as CTCs and ctDNA. However, further refinement is still required to optimize this technology for practical clinical applications [154].
Once EVs are extracted, they need to be measured and examined. Taking exosomes as an example, common analysis methods for detecting specific proteins include western blot, enzyme-linked immunosorbent assay, and flow cytometry techniques. However, these methods often require expensive equipment or relatively long analysis times [61, 157, 158]. Currently, there have been numerous reports on other EV detection and characterization methods, such as electrochemical, colorimetric detection surface-enhanced Raman spectroscopy (SERS), surface plasmon resonance (SPR), and biosensors based on nanomaterials [61, 63–65]. Li et al. [61] developed a direct and sensitive strategy for detecting exosomes in serum samples using hierarchical SERS substrate and detection probes. The superposition of hotspots between the hierarchical SERS substrate and SERS probes, combined with the proximity of SERS probes achieved through magnetic bead aggregation, resulted in a dual enhancement of the Raman signal. This not only enhances the sensitivity of exosome detection but also holds significant potential for early PC diagnosis. EVs have demonstrated tremendous potential and promising diagnostic, prognostic, and therapeutic prospects. The emergence of these novel detection methods and tools has contributed to the early diagnosis and therapeutic assessment of PC. However, it will take time to integrate them into clinical workflows and disease management strategies. While continuous technological innovation and improving isolation yields are crucial, clinical translation should be given equal attention.
Clinical applications in the management of PC
Early diagnosis
Currently, the main diagnostic methods for PC are EUS-FNA, MRI, and CT. However, most patients do not experience symptoms until the tumour obstructs the bile duct or invades surrounding nerves [34, 169]. Nonetheless, early detection is possible. Yachida et al. [170] proposed a diagnostic window of at least 10 years from the initial formation of a pancreatic tumour to the onset of symptoms, which provides a critical opportunity for early detection of PC through liquid biopsy. Early diagnosis can improve the success and survival rates of treatment and effectively reduce the side effects and cost of treatment.
CTCs are detectable at all stages of PDAC, including the precancerous lesion stage. However, CTCs can detach from the primary tumour and infiltrate the bloodstream, a crucial pathway for metastasis in advanced stages. Consequently, CTCs are seldom present in the early stages of the tumour, and detecting PDAC in the precancerous and early stages is challenging due to the limited sensitivity of CTC detection techniques [29]. These limitations currently impede the implementation of CTC-based early detection and screening of PDAC in the population. Nonetheless, the high specificity of CTCs renders them a valuable auxiliary diagnostic tool, as they are almost undetectable in healthy individuals [108]. A study conducted by Ankeny et al. [88] encompassed a cohort of nearly half of the early-stage PDAC patients (43.1% early-stage I/II), and CTCs demonstrated a sensitivity of 75.0% and specificity of 96.4% in the diagnosis of PDAC, with significant differences in CTC counts between PDAC and nonadenocarcinoma diseases. Compared to CTCs, which are often detectable in the late stages of cancer, circulating epithelial cells (CECs) may have the potential to identify patients earlier in the disease process because haematogenous dissemination may occur before tumour formation [171]. Rhim et al. [172] developed a sensitive method for labelling and tracking pancreatic epithelial cells in a mouse model of PC and detected pancreatic-origin CECs in the precancerous stage. After two years, the research team conducted a prospective blinded trial [173], employing a previously utilized detection platform for CTCs in patients with prostate cancer. The results indicated that among the 19 control individuals, only three individuals (15.8%) exhibited detectable CECs, with a maximum count of 3 CECs/ml. Conversely, in the cohort of 9 PDAC patients, 77.8% (7 individuals) exhibited detectable CECs, while among the 20 patients with pancreatic cystic lesions, 40% (8 individuals) demonstrated detectable CECs. Several other clinical studies have also confirmed the detectability of CECs in patients with benign, precancerous, and malignant pancreatic lesions, especially in those with highly atypical precancerous lesions [174, 175]. The application of single CECs or CECs combined with other molecular markers has demonstrated value in various types of tumours [176, 177]. Additionally, pancreatic-origin CECs may play a crucial role in tumour metastasis and provide initial evidence for the diagnostic value in PDAC, albeit requiring further validation with larger patient cohorts.
Similarly, ctDNA is derived from apoptotic and necrotic tumour cells, which are characteristic of advanced disease [178, 179]. KRAS mutations are commonly used as the primary target in ctDNA analysis. However, a study has demonstrated that single ctDNA analysis for KRAS mutations in plasma samples exhibits poor sensitivity, accuracy, and area under the curve (AUC) (35.2%, 51.0%, and 0.683, respectively) [113]. However, despite the identification of specific mutations in PC, these mutations can be shared with other cells and do not align exclusively with tumour cells [112]. This implies that in the analysis, these mutations cannot be regarded as the sole indicators exclusively associated with the tumour. Therefore, it is necessary to approach the presence and significance of these mutations in a more comprehensive and cautious manner. To provide a more comprehensive assessment of the value of ctDNA in liquid biopsy, many researchers are exploring strategies to combine ctDNA with other biomarkers. Cohen et al. [180] conducted a case‒control study involving 221 surgically resectable PC patients and 182 healthy individuals, and this study integrated the analysis of ctDNA mutations with protein markers. The key contribution of this study is the demonstration that genetic alterations can be detected with elevated protein markers. Importantly, the combination of ctDNA and protein markers performed better than any individual marker in screening tests. In another study [89] focusing on 68 patients with solid pancreatic tumours (58 malignant, 10 benign), a combination analysis involving CA19-9, ctDNA, and CTCs achieved a sensitivity of 78% and specificity of 91% for the diagnosis of PC. Furthermore, a registered clinical study in the United States (NCT03334708) is currently underway, with an anticipated enrolment of 700 participants. This study aims to develop blood-based biomarkers, including ctDNAs, for the early diagnosis and assessment of treatment response in PC. In addition to analysing mutational characteristics in PC, recent research has focused on integrating methylation markers as potential indicators for early diagnosis. For example, the methylation status of ADAMTS1 and BNC1 in cfDNA has demonstrated excellent diagnostic performance in detecting early-stage PDAC (sensitivity: 97.4%, specificity: 91.6%, AUC: 0.95) [114]. Although further optimization may be required for practical clinical applications, methylation markers offer a promising and noninvasive diagnostic strategy for identifying PDAC.
Despite the considerable interest in miRNAs as potential biomarkers, their widespread utilization for the clinical diagnosis of PC is currently lacking. Among the commonly investigated miRNAs, miR-196a, miR-196b, miR-885-5p, miR-122-5p, miR-210, and miR-21 have been extensively studied in this context. However, their application as validated diagnostic markers in routine clinical practice has yet to be established [181–186]. Dittmar et al. [138] employed an innovative hydrogel particle-based miRNA assay platform to analyse fluid samples with limited volume. They successfully identified molecular biomarkers (miR-34a-5p, miR-130a-3p, and miR-222-3p) that demonstrate suitability for stage II PDAC cases. Notably, when combined with CA19-9, the integration of these biomarkers resulted in an increased AUC from 0.89 (using CA19-9 alone) to 0.92, 0.94, and 0.92, respectively. It is important to emphasize the significant value of individual miRNAs in PC diagnosis, as they exhibit a favourable balance between effectiveness and cost-effectiveness. Some diagnostic models incorporating many biomarkers exhibit high diagnostic performance, but their complex composition and low reproducibility present challenges for practical clinical application. A comprehensive review of the literature conducted by Wnuk et al. [187] investigated the clinical value of circulating plasma miRNAs in PDAC. The analysis of 55 circulating miRNAs revealed that 66.10% exhibited superior diagnostic value compared to CA19-9, whereas only 23.73% of miRNAs performed worse. In all cases where miRNAs exhibited inferior diagnostic value compared to CA19-9, combinatorial strategies effectively enhanced the diagnostic performance of these miRNAs. The current reports on lncRNAs and circRNAs as early diagnostic markers for PDAC are limited. This is primarily due to the untapped potential of these molecules as PDAC biomarkers, particularly in the case of circRNAs, which is still in its early stages of research [188]. Most studies on lncRNAs and circRNAs have predominantly focused on their roles as therapeutic targets and prognostic markers for PDAC [189–192]. These studies have provided insights into the involvement of lncRNAs and circRNAs in the initiation and progression of PDAC, suggesting their potential as future diagnostic markers. However, further advancements in bioinformatics methods and functional characterization techniques are needed to fully explore their diagnostic capabilities.
Given the abundant biological information carried by EVs, the molecular contents of these vesicles can mirror crucial phenotypic traits of their parent cells. Consequently, there has been a recent surge in interest among researchers to investigate the potential utility of EVs as biomarkers for early-stage diagnostic purposes [193]. In a study conducted by Yu et al. [194], a comprehensive case‒control analysis was carried out, encompassing a cohort of 501 participants, among whom 284 were diagnosed with PDAC. This investigation aimed to assess the differential extracellular vesicle long RNA (exLR) levels in PDAC patients, individuals with chronic pancreatitis, and healthy control individuals. They developed a robust signature comprising eight distinct exLR molecules that demonstrate exceptional accuracy in discerning stage I/II cancer cases. The merged AUC of the signature reached an impressive value of 0.949. Glypican-1 (GPC1), a cell surface heparan sulfate proteoglycan, is specifically detected on PDAC cell-derived exosomes and not detectable in nontumor cells. Even without MRI results and noticeable pancreatic lesions, mutant KRAS mRNAs can be detected in circulating GPC1 + exosomes in the serum. This implies that GPC1 + exosomes may serve as biomarkers for detecting premalignant lesions [102, 195]. It is crucial to emphasize that the full-fledged implementation of extracellular vesicles in early diagnosis necessitates the standardization of their isolation and detection techniques, alongside the conduction of more extensive clinical investigations. Presently, an ongoing clinical study in the United States (NCT05625529) is focused on liquid biopsy and the utilization of extracellular vesicles for the early detection of PC.
Monitoring treatment response and resistance
Although surgery is the preferred treatment for PC patients, less than 20% of patients diagnosed with the disease can undergo surgical treatment due to the limitations of early detection methods [196]. Gemcitabine (GEM) is commonly used as a first-line treatment for PC, particularly in advanced-stage patients, and is considered highly effective. However, chemoresistance is a common issue that significantly limits the effectiveness of this treatment. Developing personalized treatment strategies with the help of liquid biopsy technologies before treatment is one potential solution to address this problem.
There is still some controversy regarding whether CTCs can predict treatment response. Most studies have found that a decrease in CTC counts signifies a favourable treatment response [73, 95, 96]. However, some research suggests that there may not be a significant difference in CTC counts between blood samples collected before and after chemotherapy, possibly due to variations in CTC identification and treatment strategies [29, 93, 197]. In addition to quantitative analyses, the molecular characteristics of CTCs are also frequently used to assess a patient's treatment response [198]. Some CTC measurement techniques enable genetic profiling of CTCs, allowing the detection of key gene mutations, such as those in KRAS, HER2, and TP53 [199–201]. Furthermore, programmed death ligand 1 (PD-L1) staining methods can be employed to evaluate the status of CTCs in patients receiving monoclonal antibody therapy, with PD-L1-negative CTC patients often achieving better treatment outcomes [202]. In most cases, CTCs express chemokine receptors, with CXC-motif chemokine receptor 4 (CXCR4) being the most commonly expressed receptor. Continuous monitoring of CXCR4 during treatment serves as a predictive biomarker, providing information to identify which patients are likely to benefit from treatment or develop resistance [203, 204]. Regarding drug sensitivity, Wu et al. [205] conducted a study wherein they collected CTCs from patients diagnosed with PDAC and expanded them ex vivo into organoids. The sensitivity of these organoids to nine drugs (GEM, 5-fluorouracil, erlotinib, irinotecan, olaparib, oxaliplatin, paclitaxel, palbociclib, and trametinib) was examined. A significant correlation was observed between the drug sensitivity of CTCs and clinical outcomes. This indicates that the drug sensitivity of CTCs holds the potential to predict therapeutic outcomes in PDAC, thus enabling the avoidance of ineffective treatments. CECs have been proposed as a potential tool to predict how patients will respond to antiangiogenic cancer therapies. However, it is important to recognize that their diverse phenotypes may exhibit different dynamics during the course of treatment. Given the unique characteristics of CECs and their crucial role in liquid biopsies, this avenue of research holds promise and warrants further exploration. A clinical trial focused on late-stage pancreatic cancer patients monitored CEC levels during neoadjuvant therapy and observed an overall increase in CECs in response to combination therapy that was attributed to chemotherapy-induced vascular damage exacerbating CEC release [206]. Furthermore, research concerning surgery, which is a common treatment method, has indicated that CEC levels typically decrease after tumour resection. This decline may result from the disruption of PDAC-derived growth factor recruitment of endothelial cells after tumour removal, subsequently reducing CEC levels [207].
The longitudinal assessment of ctDNA enables dynamic monitoring of disease trajectory, including treatment monitoring and the detection of minimal residual disease, and serves as an alternative biomarker for overall disease burden [208]. Tao et al. [209] conducted a study to examine the role of ctDNA in monitoring treatment response in a cohort of 17 PDAC patients who were treated with the FOLFIRINOX regimen (fluorouracil, irinotecan, and oxaliplatin). Among the 12 patients who responded to chemotherapy, 11 exhibited a reduction in the mutant allele fraction (MAF) of cfDNA. In contrast, the remaining 5 patients who developed chemotherapy resistance showed an increase in the ctDNA MAF during disease progression. These findings suggest that the levels of ctDNA partly reflect the tumour burden. In another study, Groot et al. [210] identified a substantial decrease in the probability of detecting ctDNA in the bloodstream of patients who underwent neoadjuvant chemotherapy compared to those who did not receive any preoperative chemotherapy (21% vs. 69%; p < 0.001). Although the practice of longitudinal ctDNA monitoring in PDAC cases remains limited, these studies underscore the potential of ctDNA as a crucial monitoring biomarker during the therapeutic course.
The emergence of chemoresistance presents formidable obstacles for nonsurgical candidates, thereby exacerbating their clinical predicament. Several miRNAs are considered key regulatory elements involved in acquiring chemoresistance in PDAC. Lu et al. [163] demonstrated that the expression of plasma miR-20a-5p in PDAC patients who exhibited resistance to GEM was markedly diminished compared to that in nonresistant patients (p < 0.01). The authors proposed that miR-20a-5p potentially regulates the expression of the RRM2 protein, thereby exerting an influence on the sensitivity of tumour cells to GEM. MiRNA levels have the potential to serve as informative indicators regarding disease progression, whether assessed before treatment initiation or during the treatment course. In a study by van der Sijde et al. [211], the elevated expression levels of serum miR-373-3p before FOLFIRINOX therapy was identified as a predictive factor for disease progression. Correspondingly, the reduced expression levels of miR-194-5p following a single cycle of FOLFIRINOX treatment indicated disease deterioration. LncRNA holds considerable importance in guiding the therapeutic approach for PC. Zhang et al. [168] demonstrated that UPK1A-AS1 expression significantly facilitates chemoresistance to oxaliplatin in PDAC. Elevated UPK1A-AS1 expression is directly associated with an unfavourable chemotherapy response and shorter progression-free survival in patients with advanced PDAC. These findings underscore the potential of ncRNA as a valuable tool for monitoring treatment response and evaluating tumour drug resistance in PDAC.
EVs play a pivotal role in acquiring GEM resistance in PDAC, particularly under conditions of prolonged drug exposure. Mikamori et al. [212] elucidated that extended GEM treatment upregulates the expression of miR-155 within PDAC cells. Moreover, they have substantiated a positive correlation between miR-155 expression levels and the secretion of extracellular vesicles, which promote GEM resistance in clinical samples. Nevertheless, it is imperative to acknowledge that the development of GEM resistance in organisms is not invariably irreversible. In a separate investigation [162], the authors unveiled the capability of macrophage-derived extracellular vesicular factors, namely, chitinase 3-like-1 (CHI3L1) and fibronectin (FN1), to induce resistance to GEM in PDAC cells. Treatment interventions involving the administration of CHI3L1 and FN1 inhibitors, canertinib and pifithrin, respectively, have demonstrated partial restoration of GEM resistance. These findings suggest the potential utilization of these inhibitors as adjunctive therapeutic modalities in managing GEM-resistant PDAC patients.
Evaluation of prognosis, metastasis, and recurrence
Accurate prognostic assessment is crucial for determining appropriate treatment strategies in patients with surgically resectable PC. It should be emphasized that ideal biomarkers are generally considered to possess both high sensitivity and high specificity. However, very few biomarkers can perfectly exhibit both high sensitivity and high specificity, even within excellent multimarker panels [213]. It is typically necessary to find a reasonable balance between these two aspects to meet varying clinical needs and objectives. In diagnosis, diagnostic tests with high specificity are typically used to determine whether a patient has a specific disease. However, highly sensitive circulating biomarkers are more useful in prognosis assessment, as they can detect the ongoing progression or recurrence of a disease, enabling health care professionals to implement timely therapeutic measures.
CTCs and ctDNA also play a significant role in prognosis assessment, providing information that can reveal the tendency for tumour metastasis and recurrence, as well as the overall survival (OS) of PC patients. This viewpoint has been confirmed by several studies [91, 94, 214–217]. Although the CTC count is commonly used as the determining criterion in studies, distinguishing CTC subgroups can also reflect the tumour status to some extent. Certain CTC subgroups with specific phenotypes, for example, indicate the tendency of the tumour for metastasis [218, 219]. Semaan et al. [33] identified and characterized 4 CTC subpopulations that can be used for the clinical stratification of PC, providing a valuable perspective for applying liquid biopsy technologies in prognostic prediction. Some studies suggest that CTC distribution shows spatial heterogeneity and that portal venous blood may be a better option for assessing PDAC prognosis than peripheral venous blood [220]. The CTCs count from intraoperative portal venous blood has been linked to poor prognosis in resectable PDAC patients [221]. Some studies have focused on the factors expressed on the surface of CTCs. In a study of 55 PDAC patients, Nitschke et al. [222] discovered that CTC positivity (≥ 3 CTCs) was significantly linked to shorter recurrence-free survival (p = 0.002). Moreover, they proposed the expression of RARRES1 on CTCs as a novel biomarker for treatment failure and early recurrence. During follow-up observations, some PDAC patients experience tumour recurrence after surgery. Interestingly, as the tumour recurs, the number of CECs increases once again. This phenomenon may be linked to the ongoing release of CECs by recurrent tumours, and these cells may play a role in tumour growth and invasion [207]. Many studies suggest that pretreatment levels of CECs are not associated with treatment response, specific disease stages, histological adverse prognostic features, or overall survival [174, 223–225]. However, monitoring specific subsets of CECs with overlapping phenotypes reveals a unique potential for prognosis prediction in PDAC, which may help in more accurately predicting disease progression and survival for patients [206].
Research on ctDNA primarily focuses on three KRAS mutations (G12D, G12V, and G12R) [226]; however, G12R has a lower detection rate than the other two mutations [227, 228]. Ako et al. [227] studied postoperative recurrence and overall survival in PDAC by analysing two KRAS mutations (G12D and G12V); they discovered that patients with both KRAS mutations had significantly lower disease-free survival. Interestingly, in a separate study, Guo et al. [216] specifically examined the influence of the G12D mutation on the prognosis of PDAC. Among 26 patients with PDAC, those with the KRAS G12D mutation had notably reduced overall survival (12.1 vs. 24.9 months, p < 0.001) and recurrence-free survival (6.3 vs. 17.4 months, p < 0.001) compared to those without the mutation. Notably, patients with the KRAS G12D mutation exhibited a distinct early recurrence trend and poorer clinical outcomes.
In previous studies, several reports have confirmed the predictive role of ncRNAs in PC. Most of these studies constructed multifactor prognostic risk models and performed survival analyses with these models [189, 229, 230]. Kandimalla and colleagues [231] developed a risk model for PDAC that includes nine miRNAs (miR-192-5p, 194-5p, 194-3p, 215-5p, 375-3p, 552-3p, and 1251-5p). They used this model to predict the survival outcomes of PDAC patients and evaluated the feasibility of applying the model to liquid biopsy. The results showed that PDAC patients with high-risk scores had a significantly shorter 5-year OS rate than those with low-risk scores (8.6% vs. 48.4%; HR = 2.85 [95% CI 1.41–5.76]; p = 0.02). This study helps identify high-risk patients and predict the prognosis of PDAC patients in clinical settings. However, such studies typically need to be validated in larger populations, and an excessive number of factors in the model hinder its application in clinical practice.
In terms of EVs, Reese et al. [232] demonstrated the diagnostic and prognostic potential of serum exosomal miR-200 in PDAC in the context of extracellular vesicles; this study showed that low expression of miR-200b in EpCAM-positive serum exosomes and miR-200c in total serum exosomes were favourable for the prognosis of PDAC. Clinical studies have shown that various substances can be detected in purified exosomes from the plasma of tumour patients. Tumour cells overexpressing PD-L1 can evade immune system surveillance and invade neighbouring tissues. A study [233] found that patients with systemic advanced PDAC had high levels of exosomal PD-L1. Nevertheless, the statistical analysis did not show a significant correlation between the level of exosomal PD-L1 and survival outcomes. In addition to the PD-L1/PD-1 axis, other immune checkpoints have also been found to be involved with exosomes. The CD155/TIGIT axis, for instance, plays a role in tumour immune evasion, similar to the PD-1/PD-L1 axis. High expression of CD155 and TIGIT has been associated with adverse prognosis in many tumours [234–236]. Furthermore, PDAC cells have been shown to express high levels of survivin. Exosomes containing survivin can induce tumour resistance and provide an additional advantage for tumour progression. Research has indicated that KRAS-dependent cancer cells produce exosomes rich in survivin, promoting cancer cell survival and resistance and ultimately leading to poor prognosis [237]. Some studies have attempted to integrate different biomarkers, which seem to have greater advantages. Yang et al. [238] discovered that combining multiple factors offers more advantages than using a single biomarker. They developed a five-factor biomarker panel for blood samples that includes EV-CK18 mRNA, EV-CD63 mRNA, EV-miR-409, cfDNA concentrations, and CA19-9. This biomarker panel exhibited not only excellent diagnostic capabilities for PDAC (with an accuracy of 92%, sensitivity of 88%, specificity of 95%, and AUC of 0.95) but also outperformed traditional imaging methods in detecting the occult metastasis of PDAC (with an accuracy of 84%, sensitivity of 78%, specificity of 88%, and AUC of 0.85).
Limitations and prospects
In the face of the threat posed by PC, surgery appears to be the only curative method, but only for patients without detected metastasis. However, due to the limitations of early diagnosis techniques, 80% of patients miss the opportunity for surgery and resort to chemotherapy. Nevertheless, chemoresistance remains an unavoidable problem. Even with surgery, some patients may still experience tumour metastasis during the operation or shortly after surgery, as precise prognosis evaluation is not always possible. Currently, liquid biopsy in clinical practice aims to address the following three issues: 1) achieving an early and accurate diagnosis of PC to provide patients with more treatment options; 2) accurately assessing metastasis and recurrence risks before surgery to avoid unnecessary treatments; and 3) selecting appropriate chemotherapy drugs and accurately evaluating treatment response for patients who are not eligible for surgical resection.
In recent years, liquid biopsy has shown promising prospects in disease diagnosis, treatment monitoring, and prognosis assessment [239–241]. As a noninvasive detection method, liquid biopsy is more advantageous than traditional tissue biopsy in terms of both economic and convenience aspects, especially when tissue samples cannot be obtained. Although cells, molecules, and biomarkers originating from tumours can potentially promote tumour metastasis, we can leverage detection technologies in this process to enhance the precision of disease diagnosis (Fig. 2). Disease progression is dynamic, highlighting the need for liquid biopsy to enable multiple sampling and dynamic observation of disease development. Based on liquid biopsy results, physicians can make more informed clinical decisions and improve disease treatment strategies, which is particularly urgent for diseases with high mortality rates, such as PC.
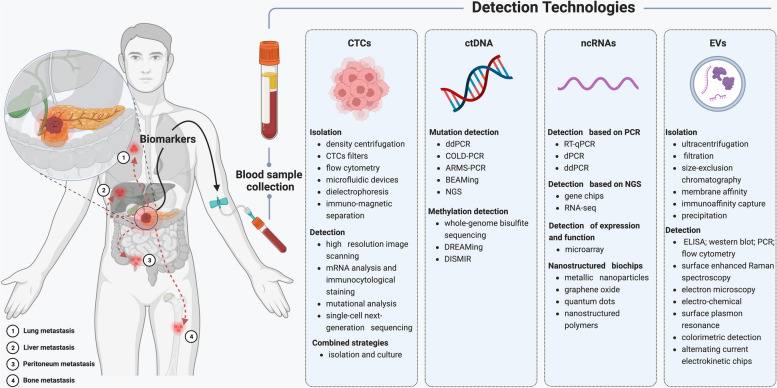
Fig. 2.
Common metastasis pathways of pancreatic cancer and detection techniques of liquid biopsy. Tumour cells, molecules, and biomarkers originating from the tumour itself are considered significant factors in the metastasis of pancreatic cancer. The liver is the most frequent site of metastasis in pancreatic cancer, followed by the lungs and peritoneum. Currently, various detection techniques are employed to identify different biomarkers, aiming to acquire more tumour characteristics. Created with BioRender.com
We should acknowledge that liquid biopsy still has certain limitations. First, liquid biopsy is still in its developmental stage with relatively low technological maturity; the complex detection process and inefficient equipment hinder its wider adoption. For example, the low concentrations of CTCs and ctDNA lead to reduced sensitivity, while contamination during sample extraction and processing may result in false-positive and false-negative outcomes. Different sample processing methods also result in low sensitivity and specificity of liquid biopsy; therefore, improving the detection level is the key to further developing and applying liquid biopsy. Second, liquid biopsy is a new detection technology that lacks standardized operating procedures and uniform data analysis methods, which can affect its accuracy and reliability to some extent. It is necessary to develop standardized analytical processes alongside technological improvements to enhance its clinical application value. Additionally, the occurrence and development of diseases involve the interaction of multiple organs, cells, and various biological molecules. Thus, an isolated liquid biopsy can only reflect the level of specific molecules or biomarkers and cannot comprehensively reflect the complex features of diseases. Therefore, it is necessary to consider integrating other examination indicators, such as CT, MRI, and ultrasound. Finally, most research results cannot be directly applied in clinical practice and require rigorous prospective trials in larger populations. Strict validation and evaluation should be conducted before clinical application. In summary, we believe that the future of liquid biopsy development should prioritize the development of new technologies and analysis platforms while simultaneously improving the operating and analytical procedures. In addition, large-scale clinical trials should be actively promoted.
Conclusion
In conclusion, the application of liquid biopsy in the clinical management of PC aligns with the concept of precision medicine. Biological samples obtained through noninvasive procedures can provide detailed information about various aspects of the tumour, which aids in monitoring tumour development and evaluating treatment responses. Moreover, this information assists clinical physicians in understanding the molecular mechanisms of tumour occurrence and development and providing more accurate and personalized treatment decisions for each patient. There are also some limitations, including low-sensitivity detection techniques, nonstandardized analysis workflows, and small sample sizes; these limitations are significant barriers of liquid biopsy. It cannot be denied that with the continuous advancement of technological methods and large-scale clinical trials, many biomarkers have begun to demonstrate their value, indicating broad prospects for their application. Liquid biopsy will become an indispensable technology for tumour diagnosis and treatment.
Acknowledgements
We would like to extend our sincere gratitude to Professor Ying Cheng and the reviewers for their invaluable and constructive comments provided during the manuscript revision process. The graphics in this article were supported and licensed by BioRender.
Abbreviations
- PC
Pancreatic cancer
- PDAC
Pancreatic ductal adenocarcinoma
- EUS-FNA
Endoscopic ultrasonography-guided fine needle aspiration
- MRI
Magnetic resonance imaging
- CT
Computed tomography
- CA19-9
Carbohydrate antigen 19-9
- CTCs
Circulating tumour cells
- ctDNA
Circulating tumour DNA
- ncRNAs
Noncoding RNAs
- mRNAs
Messenger RNAs
- EVs
Extracellular vesicles
- EMT
Epithelial-mesenchymal transition
- EpCAM
Epithelial cell adhesion molecule
- scNGS
Single-cell next-generation sequencing
- cfDNA
Cell-free DNA
- PCR
Polymerase chain reaction
- qPCR
Quantitative PCR
- dPCR
Digital PCR
- ddPCR
Droplet digital PCR
- NGS
Next-generation sequencing
- VAF
Variant allele frequency
- WGBS-seq
Whole-genome bisulfite sequencing
- sncRNAs
Small noncoding RNAs
- lncRNAs
Long noncoding RNAs
- miRNAs
MicroRNAs
- SERS
Surface-enhanced Raman spectroscopy
- SPR
Surface plasmon resonance
- CECs
Circulating epithelial cells
- AUC
Area under the curve
- exLR
Extracellular vesicle long RNA
- GPC1
Glypican-1
- GEM
Gemcitabine
- PD-L1
Programmed death ligand 1
- CXCR4
CXC-motif chemokine receptor 4
- MAF
Mutant allele fraction
- CHI3L1
Chitinase 3-like-1
- FN1
Fibronectin
- OS
Overall survival
Authors’ contributions
ZB and PQ designed the study, WKC and ZB collected the related papers and drafted the manuscript, PQ and WX generated figures and critically reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qi Pan, Email: qpan@cmu.edu.cn.
Bei Zhao, Email: zhaobei95@163.com.
References
- 1.Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016;22:9694–9705. doi: 10.3748/wjg.v22.i44.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raimondi S, Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an overview. Nat Rev Gastroenterol Hepatol. 2009;6:699–708. doi: 10.1038/nrgastro.2009.177. [DOI] [PubMed] [Google Scholar]
- 3.Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol. 2021;18:493–502. doi: 10.1038/s41575-021-00457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong MCS, Jiang JY, Liang M, Fang Y, Yeung MS, Sung JJY. Global temporal patterns of pancreatic cancer and association with socioeconomic development. Sci Rep. 2017;7:3165. doi: 10.1038/s41598-017-02997-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferlay J, Partensky C, Bray F. More deaths from pancreatic cancer than breast cancer in the EU by 2017. Acta Oncol. 2016;55:1158–1160. doi: 10.1080/0284186X.2016.1197419. [DOI] [PubMed] [Google Scholar]
- 6.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 7.Cid-Arregui A, Juarez V. Perspectives in the treatment of pancreatic adenocarcinoma. World J Gastroenterol. 2015;21:9297–9316. doi: 10.3748/wjg.v21.i31.9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lux A, Kahlert C, Grützmann R, Pilarsky C. c-Met and PD-L1 on Circulating Exosomes as Diagnostic and Prognostic Markers for Pancreatic Cancer. Int J Mol Sci. 2019;20(13):3305. doi: 10.3390/ijms20133305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neuzillet C, Tijeras-Raballand A, Bourget P, Cros J, Couvelard A, Sauvanet A, Vullierme MP, Tournigand C, Hammel P. State of the art and future directions of pancreatic ductal adenocarcinoma therapy. Pharmacol Ther. 2015;155:80–104. doi: 10.1016/j.pharmthera.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Martin-Perez E, Domínguez-Muñoz JE, Botella-Romero F, Cerezo L, Matute Teresa F, Serrano T, Vera R. Multidisciplinary consensus statement on the clinical management of patients with pancreatic cancer. Clin Transl Oncol. 2020;22:1963–1975. doi: 10.1007/s12094-020-02350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semaan A, Bernard V, Lee JJ, Wong JW, Huang J, Swartzlander DB, Stephens BM, Monberg ME, Weston BR, Bhutani MS, et al. Defining the Comprehensive Genomic Landscapes of Pancreatic Ductal Adenocarcinoma Using Real-World Endoscopic Aspiration Samples. Clin Cancer Res. 2021;27:1082–1093. doi: 10.1158/1078-0432.CCR-20-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashida R, Kitano M. Endoscopic ultrasound-guided tissue acquisition for pancreatic ductal adenocarcinoma in the era of precision medicine. Dig Endosc. 2022;34:1329–1339. doi: 10.1111/den.14344. [DOI] [PubMed] [Google Scholar]
- 13.Barat M, Marchese U, Pellat A, Dohan A, Coriat R, Hoeffel C, Fishman EK, Cassinotto C, Chu L, Soyer P. Imaging of Pancreatic Ductal Adenocarcinoma: An Update on Recent Advances. Can Assoc Radiol J. 2023;74:351–361. doi: 10.1177/08465371221124927. [DOI] [PubMed] [Google Scholar]
- 14.Anta JA, Martínez-Ballestero I, Eiroa D, García J, Rodríguez-Comas J. Artificial intelligence for the detection of pancreatic lesions. Int J Comput Assist Radiol Surg. 2022;17:1855–1865. doi: 10.1007/s11548-022-02706-z. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Sanagapalli S, Stoita A. Challenges in diagnosis of pancreatic cancer. World J Gastroenterol. 2018;24:2047–2060. doi: 10.3748/wjg.v24.i19.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou H, Zhu L, Song J, Wang G, Li P, Li W, Luo P, Sun X, Wu J, Liu Y, et al. Liquid biopsy at the frontier of detection, prognosis and progression monitoring in colorectal cancer. Mol Cancer. 2022;21:86. doi: 10.1186/s12943-022-01556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misek DE, Patwa TH, Lubman DM, Simeone DM. Early detection and biomarkers in pancreatic cancer. J Natl Compr Canc Netw. 2007;5:1034–1041. doi: 10.6004/jnccn.2007.0086. [DOI] [PubMed] [Google Scholar]
- 19.Mann DV, Edwards R, Ho S, Lau WY, Glazer G. Elevated tumour marker CA19-9: clinical interpretation and influence of obstructive jaundice. Eur J Surg Oncol. 2000;26:474–479. doi: 10.1053/ejso.1999.0925. [DOI] [PubMed] [Google Scholar]
- 20.Guo M, Luo G, Lu R, Shi W, Cheng H, Lu Y, Jin K, Yang C, Wang Z, Long J, et al. Distribution of Lewis and Secretor polymorphisms and corresponding CA19-9 antigen expression in a Chinese population. FEBS Open Bio. 2017;7:1660–1671. doi: 10.1002/2211-5463.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marrelli D, Caruso S, Pedrazzani C, Neri A, Fernandes E, Marini M, Pinto E, Roviello F. CA19-9 serum levels in obstructive jaundice: clinical value in benign and malignant conditions. Am J Surg. 2009;198:333–339. doi: 10.1016/j.amjsurg.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 22.Erben V, Poschet G, Schrotz-King P, Brenner H. Comparing Metabolomics Profiles in Various Types of Liquid Biopsies among Screening Participants with and without Advanced Colorectal Neoplasms. Diagnostics (Basel). 2021;11(3):561. doi: 10.3390/diagnostics11030561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Ji H, Yang W, Zhang M, Guo Y, Li B, Wang J, Chen R, Chen Y, Wang X. Liquid biopsy using ascitic fluid and pleural effusion supernatants for genomic profiling in gastrointestinal and lung cancers. BMC Cancer. 2022;22:1020. doi: 10.1186/s12885-022-09922-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, Sausen M, James N, Rettig EM, Guo T, Pickering CR, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015;7:293ra104. doi: 10.1126/scitranslmed.aaa8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Mattos-Arruda L, Mayor R, Ng CKY, Weigelt B, Martínez-Ricarte F, Torrejon D, Oliveira M, Arias A, Raventos C, Tang J, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol. 2022;15:131. doi: 10.1186/s13045-022-01351-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshi M, Murthy V, Takahashi H, Huyser M, Okano M, Tokumaru Y, Rashid OM, Matsuyama R, Endo I, Takabe K. Urine as a Source of Liquid Biopsy for Cancer. Cancers (Basel). 2021;13(11):2652. doi: 10.3390/cancers13112652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zygulska AL, Pierzchalski P. Novel Diagnostic Biomarkers in Colorectal Cancer. Int J Mol Sci. 2022;23(2):852. doi: 10.3390/ijms23020852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeo D, Bastian A, Strauss H, Saxena P, Grimison P, Rasko JEJ. Exploring the Clinical Utility of Pancreatic Cancer Circulating Tumor Cells. Int J Mol Sci. 2022;23(3):1671. doi: 10.3390/ijms23031671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khoja L, Backen A, Sloane R, Menasce L, Ryder D, Krebs M, Board R, Clack G, Hughes A, Blackhall F, et al. A pilot study to explore circulating tumour cells in pancreatic cancer as a novel biomarker. Br J Cancer. 2012;106:508–516. doi: 10.1038/bjc.2011.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao L, Su L, Yuan C, Ma Z, Zhang L, Bo S, Niu Y, Lu S, Xiu D. Postoperative metastasis prediction based on portal vein circulating tumor cells detected by flow cytometry in periampullary or pancreatic cancer. Cancer Manag Res. 2019;11:7405–7425. doi: 10.2147/CMAR.S210332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semaan A, Bernard V, Kim DU, Lee JJ, Huang J, Kamyabi N, Stephens BM, Qiao W, Varadhachary GR, Katz MH, et al. Characterisation of circulating tumour cell phenotypes identifies a partial-EMT sub-population for clinical stratification of pancreatic cancer. Br J Cancer. 2021;124:1970–1977. doi: 10.1038/s41416-021-01350-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martini V, Timme-Bronsert S, Fichtner-Feigl S, Hoeppner J, Kulemann B. Circulating Tumor Cells in Pancreatic Cancer: Current Perspectives. Cancers (Basel). 2019;11(11):1659. doi: 10.3390/cancers11111659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeo D, Kao S, Gupta R, Wahlroos S, Bastian A, Strauss H, Klemm V, Shrestha P, Ramirez AB, Costandy L, et al. Accurate isolation and detection of circulating tumor cells using enrichment-free multiparametric high resolution imaging. Front Oncol. 2023;13:1141228. doi: 10.3389/fonc.2023.1141228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sergeant G, Roskams T, van Pelt J, Houtmeyers F, Aerts R, Topal B. Perioperative cancer cell dissemination detected with a real-time RT-PCR assay for EpCAM is not associated with worse prognosis in pancreatic ductal adenocarcinoma. BMC Cancer. 2011;11:47. doi: 10.1186/1471-2407-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulemann B, Rösch S, Seifert S, Timme S, Bronsert P, Seifert G, Martini V, Kuvendjiska J, Glatz T, Hussung S, et al. Pancreatic cancer: Circulating Tumor Cells and Primary Tumors show Heterogeneous KRAS Mutations. Sci Rep. 2017;7:4510. doi: 10.1038/s41598-017-04601-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu J, Gemenetzis G, Kinny-Köster B, Habib JR, Groot VP, Teinor J, Yin L, Pu N, Hasanain A, van Oosten F, et al. Pancreatic circulating tumor cell detection by targeted single-cell next-generation sequencing. Cancer Lett. 2020;493:245–253. doi: 10.1016/j.canlet.2020.08.043. [DOI] [PubMed] [Google Scholar]
- 39.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao C-J, Hsieh C-H, Wang H-M, Chou W-P, Chiu T-K, Chang J-H, Chao AC, Wu M-H. Isolation of label-free and viable circulating tumour cells (CTCs) from blood samples of cancer patients through a two-step process: negative selection-type immunomagnetic beads and spheroid cell culture-based cell isolation. RSC Adv. 2017;7:29339–29349. doi: 10.1039/C7RA03663A. [DOI] [Google Scholar]
- 41.Watanabe F, Suzuki K, Tamaki S, Abe I, Endo Y, Takayama Y, Ishikawa H, Kakizawa N, Saito M, Futsuhara K, et al. Longitudinal monitoring of KRAS-mutated circulating tumor DNA enables the prediction of prognosis and therapeutic responses in patients with pancreatic cancer. PLoS ONE. 2019;14:e0227366. doi: 10.1371/journal.pone.0227366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mauger F, How-Kit A, Tost J. COLD-PCR Technologies in the Area of Personalized Medicine: Methodology and Applications. Mol Diagn Ther. 2017;21:269–283. doi: 10.1007/s40291-016-0254-8. [DOI] [PubMed] [Google Scholar]
- 43.Semrad T, Barzi A, Lenz HJ, Hutchins IM, Kim EJ, Gong IY, Tanaka M, Beckett L, Holland W, Burich RA, et al. Pharmacodynamic separation of gemcitabine and erlotinib in locally advanced or metastatic pancreatic cancer: therapeutic and biomarker results. Int J Clin Oncol. 2015;20:518–524. doi: 10.1007/s10147-014-0730-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toledano-Fonseca M, Cano MT, Inga E, Rodríguez-Alonso R, Gómez-España MA, Guil-Luna S, Mena-Osuna R, de la Haba-Rodríguez JR, Rodríguez-Ariza A, Aranda E. Circulating Cell-Free DNA-Based Liquid Biopsy Markers for the Non-Invasive Prognosis and Monitoring of Metastatic Pancreatic Cancer. Cancers (Basel). 2020;12(7):1754. doi: 10.3390/cancers12071754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng H, Liu C, Jiang J, Luo G, Lu Y, Jin K, Guo M, Zhang Z, Xu J, Liu L, et al. Analysis of ctDNA to predict prognosis and monitor treatment responses in metastatic pancreatic cancer patients. Int J Cancer. 2017;140:2344–2350. doi: 10.1002/ijc.30650. [DOI] [PubMed] [Google Scholar]
- 46.Davis AA, Iams WT, Chan D, Oh MS, Lentz RW, Peterman N, Robertson A, Shah A, Srivas R, Wilson TJ, et al. Early Assessment of Molecular Progression and Response by Whole-genome Circulating Tumor DNA in Advanced Solid Tumors. Mol Cancer Ther. 2020;19:1486–1496. doi: 10.1158/1535-7163.MCT-19-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pisanic TR, 2nd, Athamanolap P, Poh W, Chen C, Hulbert A, Brock MV, Herman JG, Wang TH. DREAMing: a simple and ultrasensitive method for assessing intratumor epigenetic heterogeneity directly from liquid biopsies. Nucleic Acids Res. 2015;43:e154. doi: 10.1093/nar/gkv795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Wei L, Zhang X, Zhang W, Wang H, Zhong B, Xie Z, Lv H, Wang X. DISMIR: Deep learning-based noninvasive cancer detection by integrating DNA sequence and methylation information of individual cell-free DNA reads. Brief Bioinform. 2021;22(6):bbab250. [DOI] [PMC free article] [PubMed]
- 49.Badowski C, He B, Garmire LX. Blood-derived lncRNAs as biomarkers for cancer diagnosis: the Good, the Bad and the Beauty. NPJ Precis Oncol. 2022;6:40. doi: 10.1038/s41698-022-00283-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller S, Raulefs S, Bruns P, Afonso-Grunz F, Plötner A, Thermann R, Jäger C, Schlitter AM, Kong B, Regel I, et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14:94. doi: 10.1186/s12943-015-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu XL, Liu DJ, Yan TT, Yang JY, Yang MW, Li J, Huo YM, Liu W, Zhang JF, Hong J, et al. Analysis of long non-coding RNA expression profiles in pancreatic ductal adenocarcinoma. Sci Rep. 2016;6:33535. doi: 10.1038/srep33535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martino S, Tammaro C, Misso G, Falco M, Scrima M, Bocchetti M, Rea I, De Stefano L, Caraglia M. microRNA Detection via Nanostructured Biochips for Early Cancer Diagnostics. Int J Mol Sci. 2023;24(9):7762. doi: 10.3390/ijms24097762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Luo G, Zhang K, Cao J, Huang C, Jiang T, Liu B, Su L, Qiu Z. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kγ to Promote Pancreatic Cancer Metastasis. Cancer Res. 2018;78:4586–4598. doi: 10.1158/0008-5472.CAN-17-3841. [DOI] [PubMed] [Google Scholar]
- 54.Xu YF, Xu X, Bhandari K, Gin A, Rao CV, Morris KT, Hannafon BN, Ding WQ. Isolation of extra-cellular vesicles in the context of pancreatic adenocarcinomas: Addition of one stringent filtration step improves recovery of specific microRNAs. PLoS ONE. 2021;16:e0259563. doi: 10.1371/journal.pone.0259563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buenafe AC, Dorrell C, Reddy AP, Klimek J, Marks DL. Proteomic analysis distinguishes extracellular vesicles produced by cancerous versus healthy pancreatic organoids. Sci Rep. 2022;12:3556. doi: 10.1038/s41598-022-07451-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stranska R, Gysbrechts L, Wouters J, Vermeersch P, Bloch K, Dierickx D, Andrei G, Snoeck R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J Transl Med. 2018;16:1. doi: 10.1186/s12967-017-1374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen K, Amontree J, Varillas J, Zhang J, George TJ, Fan ZH. Incorporation of lateral microfiltration with immunoaffinity for enhancing the capture efficiency of rare cells. Sci Rep. 2020;10:14210. doi: 10.1038/s41598-020-71041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ludwig AK, De Miroschedji K, Doeppner TR, Börger V, Ruesing J, Rebmann V, Durst S, Jansen S, Bremer M, Behrmann E, et al. Precipitation with polyethylene glycol followed by washing and pelleting by ultracentrifugation enriches extracellular vesicles from tissue culture supernatants in small and large scales. J Extracell Vesicles. 2018;7:1528109. doi: 10.1080/20013078.2018.1528109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao J, Guo M, Song Y, Liu S, Liao R, Zhang Y, Zhang Y, Yang Q, Gu Y, Huang X. Serum exosomal and serum glypican-1 are associated with early recurrence of pancreatic ductal adenocarcinoma. Front Oncol. 2022;12:992929. doi: 10.3389/fonc.2022.992929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New Technologies for Analysis of Extracellular Vesicles. Chem Rev. 2018;118:1917–1950. doi: 10.1021/acs.chemrev.7b00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J, Li Y, Chen S, Duan W, Kong X, Wang Y, Zhou L, Li P, Zhang C, Du L, Wang C. Highly Sensitive Exosome Detection for Early Diagnosis of Pancreatic Cancer Using Immunoassay Based on Hierarchical Surface-Enhanced Raman Scattering Substrate. Small Methods. 2022;6:e2200154. doi: 10.1002/smtd.202200154. [DOI] [PubMed] [Google Scholar]
- 62.Tatischeff I, Larquet E, Falcón-Pérez JM, Turpin PY, Kruglik SG. Fast characterisation of cell-derived extracellular vesicles by nanoparticles tracking analysis, cryo-electron microscopy, and Raman tweezers microspectroscopy. J Extracell Vesicles. 2012;1. 10.3402/jev.v1i0.19179. [DOI] [PMC free article] [PubMed]
- 63.Li S, Ma Q. Electrochemical nano-sensing interface for exosomes analysis and cancer diagnosis. Biosens Bioelectron. 2022;214:114554. doi: 10.1016/j.bios.2022.114554. [DOI] [PubMed] [Google Scholar]
- 64.Bellassai N, D'Agata R, Jungbluth V, Spoto G. Surface Plasmon Resonance for Biomarker Detection: Advances in Non-invasive Cancer Diagnosis. Front Chem. 2019;7:570. doi: 10.3389/fchem.2019.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu D, Li Y, Wang M, Gu J, Xu W, Cai H, Fang X, Zhang X. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer. 2022;21:56. doi: 10.1186/s12943-022-01509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis JM, Vyas AD, Qiu Y, Messer KS, White R, Heller MJ. Integrated Analysis of Exosomal Protein Biomarkers on Alternating Current Electrokinetic Chips Enables Rapid Detection of Pancreatic Cancer in Patient Blood. ACS Nano. 2018;12:3311–3320. doi: 10.1021/acsnano.7b08199. [DOI] [PubMed] [Google Scholar]
- 67.Kowalik A, Kowalewska M, Góźdź S. Current approaches for avoiding the limitations of circulating tumor cells detection methods-implications for diagnosis and treatment of patients with solid tumors. Transl Res. 2017;185:58–84.e15. doi: 10.1016/j.trsl.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 68.Agashe R, Kurzrock R. Circulating Tumor Cells: From the Laboratory to the Cancer Clinic. Cancers (Basel). 2020;12(9):2361. doi: 10.3390/cancers12092361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Castro-Giner F, Aceto N. Tracking cancer progression: from circulating tumor cells to metastasis. Genome Med. 2020;12:31. doi: 10.1186/s13073-020-00728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tayoun T, Faugeroux V, Oulhen M, Aberlenc A, Pawlikowska P, Farace F. CTC-Derived Models: A Window into the Seeding Capacity of Circulating Tumor Cells (CTCs) Cells. 2019;8(10):1145. doi: 10.3390/cells8101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aceto N, Toner M, Maheswaran S, Haber DA. En Route to Metastasis: Circulating Tumor Cell Clusters and Epithelial-to-Mesenchymal Transition. Trends Cancer. 2015;1:44–52. doi: 10.1016/j.trecan.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 72.Mavroudis D. Circulating cancer cells. Ann Oncol. 2010;21(Suppl 7):vii95–100. doi: 10.1093/annonc/mdq378. [DOI] [PubMed] [Google Scholar]
- 73.Lin D, Shen L, Luo M, Zhang K, Li J, Yang Q, Zhu F, Zhou D, Zheng S, Chen Y, Zhou J. Circulating tumor cells: biology and clinical significance. Signal Transduct Target Ther. 2021;6:404. doi: 10.1038/s41392-021-00817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dirix L, Buys A, Oeyen S, Peeters D, Liègeois V, Prové A, Rondas D, Vervoort L, Mariën V, Laere SV, Vermeulen P. Circulating tumor cell detection: A prospective comparison between Cell Search® and RareCyte® platforms in patients with progressive metastatic breast cancer. Breast Cancer Res Treat. 2022;193:437–444. doi: 10.1007/s10549-022-06585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gruijs M, Zeelen C, Hellingman T, Smit J, Borm FJ, Kazemier G, Dickhoff C, Bahce I, de Langen J, Smit EF, et al. Detection of Circulating Tumor Cells Using the Attune NxT. Int J Mol Sci. 2022;24(1):21. doi: 10.3390/ijms24010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rajta I, Huszánk R, Szabó AT, Nagy GU, Szilasi S, Fürjes P, Holczer E, Fekete Z, Járvás G, Szigeti M, et al. Tilted pillar array fabrication by the combination of proton beam writing and soft lithography for microfluidic cell capture: Part 1 Design and feasibility. Electrophoresis. 2016;37:498–503. doi: 10.1002/elps.201500254. [DOI] [PubMed] [Google Scholar]
- 77.Pahattuge TN, Freed IM, Hupert ML, Vaidyanathan S, Childers K, Witek MA, Weerakoon-Ratnayake K, Park D, Kasi A, Al-Kasspooles MF, et al. System Modularity Chip for Analysis of Rare Targets (SMART-Chip): Liquid Biopsy Samples. ACS Sens. 2021;6:1831–1839. doi: 10.1021/acssensors.0c02728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, Fleisher M, Stadler W, Datar RH, Tai YC, Cote RJ. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hosokawa M, Yoshikawa T, Negishi R, Yoshino T, Koh Y, Kenmotsu H, Naito T, Takahashi T, Yamamoto N, Kikuhara Y, et al. Microcavity array system for size-based enrichment of circulating tumor cells from the blood of patients with small-cell lung cancer. Anal Chem. 2013;85:5692–5698. doi: 10.1021/ac400167x. [DOI] [PubMed] [Google Scholar]
- 80.Lowes LE, Allan AL. Circulating Tumor Cells and Implications of the Epithelial-to-Mesenchymal Transition. Adv Clin Chem. 2018;83:121–181. doi: 10.1016/bs.acc.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 81.Couto-Cunha A, Jerónimo C, Henrique R. Circulating Tumor Cells as Biomarkers for Renal Cell Carcinoma: Ready for Prime Time? Cancers (Basel). 2022;15(1):287. doi: 10.3390/cancers15010287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee DH, Yoon W, Lee A, Han Y, Byun Y, Kang JS, Kim H, Kwon W, Suh YA, Choi Y, et al. Multi-biomarker panel prediction model for diagnosis of pancreatic cancer. J Hepatobiliary Pancreat Sci. 2023;30:122–132. doi: 10.1002/jhbp.986. [DOI] [PubMed] [Google Scholar]
- 83.Gao Y, Ni X, Guo H, Su Z, Ba Y, Tong Z, Guo Z, Yao X, Chen X, Yin J, et al. Single-cell sequencing deciphers a convergent evolution of copy number alterations from primary to circulating tumor cells. Genome Res. 2017;27:1312–1322. doi: 10.1101/gr.216788.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Keller L, Pantel K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat Rev Cancer. 2019;19:553–567. doi: 10.1038/s41568-019-0180-2. [DOI] [PubMed] [Google Scholar]
- 85.Chen J, Wang H, Zhou L, Liu Z, Tan X. A combination of circulating tumor cells and CA199 improves the diagnosis of pancreatic cancer. J Clin Lab Anal. 2022;36:e24341. doi: 10.1002/jcla.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng H, He W, Yang J, Ye Q, Cheng L, Pan Y, Mao L, Chu X, Lu C, Li G, et al. Ligand-targeted polymerase chain reaction for the detection of folate receptor-positive circulating tumour cells as a potential diagnostic biomarker for pancreatic cancer. Cell Prolif. 2020;53:e12880. doi: 10.1111/cpr.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buscail E, Alix-Panabières C, Quincy P, Cauvin T, Chauvet A, Degrandi O, Caumont C, Verdon S, Lamrissi I, Moranvillier I, et al. High Clinical Value of Liquid Biopsy to Detect Circulating Tumor Cells and Tumor Exosomes in Pancreatic Ductal Adenocarcinoma Patients Eligible for Up-Front Surgery. Cancers (Basel). 2019;11(11):1656. doi: 10.3390/cancers11111656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ankeny JS, Court CM, Hou S, Li Q, Song M, Wu D, Chen JF, Lee T, Lin M, Sho S, et al. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br J Cancer. 2016;114:1367–1375. doi: 10.1038/bjc.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sefrioui D, Blanchard F, Toure E, Beaussire L, Dolfus C, Perdrix A, Paresy M, Antonietti M, Iwanicki-Caron I, et al. Diagnostic value of CA19.9, circulating tumour DNA and circulating tumour cells in patients with solid pancreatic tumours. Br J Cancer. 2017;117:1017–1025. doi: 10.1038/bjc.2017.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu P, Liu HY, Liu FC, Gu FM, Yuan SX, Huang J, Pan ZY, Wang WJ. Circulating Tumor Cells Expressing Krüppel-Like Factor 8 and Vimentin as Predictors of Poor Prognosis in Pancreatic Cancer Patients. Cancer Control. 2021;28:10732748211027163. doi: 10.1177/10732748211027163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Court CM, Ankeny JS, Sho S, Winograd P, Hou S, Song M, Wainberg ZA, Girgis MD, Graeber TG, Agopian VG, et al. Circulating Tumor Cells Predict Occult Metastatic Disease and Prognosis in Pancreatic Cancer. Ann Surg Oncol. 2018;25:1000–1008. doi: 10.1245/s10434-017-6290-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Poruk KE, Blackford AL, Weiss MJ, Cameron JL, He J, Goggins M, Rasheed ZA, Wolfgang CL, Wood LD. Circulating Tumor Cells Expressing Markers of Tumor-Initiating Cells Predict Poor Survival and Cancer Recurrence in Patients with Pancreatic Ductal Adenocarcinoma. Clin Cancer Res. 2017;23:2681–2690. doi: 10.1158/1078-0432.CCR-16-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Poruk KE, Valero V, 3rd, Saunders T, Blackford AL, Griffin JF, Poling J, Hruban RH, Anders RA, Herman J, Zheng L, et al. Circulating Tumor Cell Phenotype Predicts Recurrence and Survival in Pancreatic Adenocarcinoma. Ann Surg. 2016;264:1073–1081. doi: 10.1097/SLA.0000000000001600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park Y, Jun HR, Choi HW, Hwang DW, Lee JH, Song KB, Lee W, Kwon J, Ha SH, Jun E, Kim SC. Circulating tumour cells as an indicator of early and systemic recurrence after surgical resection in pancreatic ductal adenocarcinoma. Sci Rep. 2021;11:1644. doi: 10.1038/s41598-020-80383-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wei T, Zhang X, Zhang Q, Yang J, Chen Q, Wang J, Li X, Chen J, Ma T, Li G, et al. Vimentin-positive circulating tumor cells as a biomarker for diagnosis and treatment monitoring in patients with pancreatic cancer. Cancer Lett. 2019;452:237–243. doi: 10.1016/j.canlet.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 96.Gemenetzis G, Groot VP, Yu J, Ding D, Teinor JA, Javed AA, Wood LD, Burkhart RA, Cameron JL, Makary MA, et al. Circulating Tumor Cells Dynamics in Pancreatic Adenocarcinoma Correlate With Disease Status: Results of the Prospective CLUSTER Study. Ann Surg. 2018;268:408–420. doi: 10.1097/SLA.0000000000002925. [DOI] [PubMed] [Google Scholar]
- 97.Dhayat SA, Yang Z. Impact of circulating tumor DNA in hepatocellular and pancreatic carcinomas. J Cancer Res Clin Oncol. 2020;146:1625–1645. doi: 10.1007/s00432-020-03219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yong E. Cancer biomarkers: Written in blood. Nature. 2014;511:524–526. doi: 10.1038/511524a. [DOI] [PubMed] [Google Scholar]
- 99.Li YZ, Kong SN, Liu YP, Yang Y, Zhang HM. Can Liquid Biopsy Based on ctDNA/cfDNA Replace Tissue Biopsy for the Precision Treatment of EGFR-Mutated NSCLC? J Clin Med. 2023;12(4):1438. doi: 10.3390/jcm12041438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Campos-Carrillo A, Weitzel JN, Sahoo P, Rockne R, Mokhnatkin JV, Murtaza M, Gray SW, Goetz L, Goel A, Schork N, Slavin TP. Circulating tumor DNA as an early cancer detection tool. Pharmacol Ther. 2020;207:107458. doi: 10.1016/j.pharmthera.2019.107458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sausen M, Parpart S, Diaz LA., Jr Circulating tumor DNA moves further into the spotlight. Genome Med. 2014;6:35. doi: 10.1186/gm552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee VH, Kwong DL, Leung TW, Choi CW, Lai V, Ng L, Lam KO, Ng SC, Sze CK, Tong CC, et al. Prognostication of serial post-intensity-modulated radiation therapy undetectable plasma EBV DNA for nasopharyngeal carcinoma. Oncotarget. 2017;8:5292–5308. doi: 10.18632/oncotarget.14137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grønbaek K, Hother C, Jones PA. Epigenetic changes in cancer. APMIS. 2007;115:1039–1059. doi: 10.1111/j.1600-0463.2007.apm_636.xml.x. [DOI] [PubMed] [Google Scholar]
- 106.Wen X, Pu H, Liu Q, Guo Z, Luo D. Circulating Tumor DNA-A Novel Biomarker of Tumor Progression and Its Favorable Detection Techniques. Cancers (Basel). 2022;14(24):6025. doi: 10.3390/cancers14246025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Raufi AG, May MS, Hadfield MJ, Seyhan AA, El-Deiry WS. Advances in Liquid Biopsy Technology and Implications for Pancreatic Cancer. Int J Mol Sci. 2023;24(4):4238. doi: 10.3390/ijms24044238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heredia-Soto V, Rodríguez-Salas N, Feliu J. Liquid Biopsy in Pancreatic Cancer: Are We Ready to Apply It in the Clinical Practice?. Cancers (Basel). 2021;13(8):1986. doi: 10.3390/cancers13081986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Castells A, Puig P, Móra J, Boadas J, Boix L, Urgell E, Solé M, Capellà G, Lluís F, Fernández-Cruz L, et al. K-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: diagnostic utility and prognostic significance. J Clin Oncol. 1999;17:578–584. doi: 10.1200/JCO.1999.17.2.578. [DOI] [PubMed] [Google Scholar]
- 112.Razavi P, Li BT, Brown DN, Jung B, Hubbell E, Shen R, Abida W, Juluru K, De Bruijn I, Hou C, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25:1928–1937. doi: 10.1038/s41591-019-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang R, Zhao Y, Wang Y, Zhao Z, Chen Q, Duan Y, Xiong S, Luan Z, Wang J, Cheng B. Diagnostic and Prognostic Values of KRAS Mutations on EUS-FNA Specimens and Circulating Tumor DNA in Patients With Pancreatic Cancer. Clin Transl Gastroenterol. 2022;13:e00487. doi: 10.14309/ctg.0000000000000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Eissa MAL, Lerner L, Abdelfatah E, Shankar N, Canner JK, Hasan NM, Yaghoobi V, Huang B, Kerner Z, Takaesu F, et al. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin Epigenetics. 2019;11:59. doi: 10.1186/s13148-019-0650-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Henriksen SD, Madsen PH, Larsen AC, Johansen MB, Drewes AM, Pedersen IS, Krarup H, Thorlacius-Ussing O. Cell-free DNA promoter hypermethylation in plasma as a diagnostic marker for pancreatic adenocarcinoma. Clin Epigenetics. 2016;8:117. doi: 10.1186/s13148-016-0286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nitschke C, Markmann B, Walter P, Badbaran A, Tölle M, Kropidlowski J, Belloum Y, Goetz MR, Bardenhagen J, Stern L, et al. Peripheral and Portal Venous KRAS ctDNA Detection as Independent Prognostic Markers of Early Tumor Recurrence in Pancreatic Ductal Adenocarcinoma. Clin Chem. 2023;69:295–307. doi: 10.1093/clinchem/hvac214. [DOI] [PubMed] [Google Scholar]
- 117.Watanabe K, Nakamura T, Kimura Y, Motoya M, Kojima S, Kuraya T, Murakami T, Kaneko T, Shinohara Y, Kitayama Y, et al. Tumor-Informed Approach Improved ctDNA Detection Rate in Resected Pancreatic Cancer. Int J Mol Sci. 2022;23(19):11521. doi: 10.3390/ijms231911521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bernard V, Kim DU, San Lucas FA, Castillo J, Allenson K, Mulu FC, Stephens BM, Huang J, Semaan A, Guerrero PA, et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology. 2019;156:108–118.e104. doi: 10.1053/j.gastro.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kirchweger P, Kupferthaler A, Burghofer J, Webersinke G, Jukic E, Schwendinger S, Wundsam H, Biebl M, Petzer A, Rumpold H. Prediction of response to systemic treatment by kinetics of circulating tumor DNA in metastatic pancreatic cancer. Front Oncol. 2022;12:902177. doi: 10.3389/fonc.2022.902177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sugimori M, Sugimori K, Tsuchiya H, Suzuki Y, Tsuyuki S, Kaneta Y, Hirotani A, Sanga K, Tozuka Y, Komiyama S, et al. Quantitative monitoring of circulating tumor DNA in patients with advanced pancreatic cancer undergoing chemotherapy. Cancer Sci. 2020;111:266–278. doi: 10.1111/cas.14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee B, Lipton L, Cohen J, Tie J, Javed AA, Li L, Goldstein D, Burge M, Cooray P, Nagrial A, et al. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann Oncol. 2019;30:1472–1478. doi: 10.1093/annonc/mdz200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Takahashi K, Taniue K, Ono Y, Fujiya M, Mizukami Y, Okumura T. Long Non-Coding RNAs in Epithelial-Mesenchymal Transition of Pancreatic Cancer. Front Mol Biosci. 2021;8:717890. doi: 10.3389/fmolb.2021.717890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Donati S, Aurilia C, Palmini G, Falsetti I, Iantomasi T, Brandi ML. Autophagy-Related ncRNAs in Pancreatic Cancer. Pharmaceuticals (Basel). 2022;15(12):1547. doi: 10.3390/ph15121547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang M, Dang P, Liu Y, Qiao B, Sun Z. Noncoding RNAs in pyroptosis and cancer progression: Effect, mechanism, and clinical application. Front Immunol. 2022;13:982040. doi: 10.3389/fimmu.2022.982040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huynh NP, Anderson BA, Guilak F, McAlinden A. Emerging roles for long noncoding RNAs in skeletal biology and disease. Connect Tissue Res. 2017;58:116–141. doi: 10.1080/03008207.2016.1194406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Aurilia C, Donati S, Palmini G, Miglietta F, Falsetti I, Iantomasi T, Brandi ML. Are Non-Coding RNAs Useful Biomarkers in Parathyroid Tumorigenesis? Int J Mol Sci. 2021;22(19):10465. doi: 10.3390/ijms221910465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smolkova B, Kataki A, Earl J, Ruz-Caracuel I, Cihova M, Urbanova M, Buocikova V, Tamargo S, Rovite V, Niedra H, et al. Liquid biopsy and preclinical tools for advancing diagnosis and treatment of patients with pancreatic neuroendocrine neoplasms. Crit Rev Oncol Hematol. 2022;180:103865. doi: 10.1016/j.critrevonc.2022.103865. [DOI] [PubMed] [Google Scholar]
- 128.Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 129.Galoș D, Gorzo A, Balacescu O, Sur D. Clinical Applications of Liquid Biopsy in Colorectal Cancer Screening: Current Challenges and Future Perspectives. Cells. 2022;11(21):3493. doi: 10.3390/cells11213493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zandieh MA, Farahani MH, Rajabi R, Avval ST, Karimi K, Rahmanian P, Razzazan M, Javanshir S, Mirzaei S, Paskeh MDA, et al. Epigenetic regulation of autophagy by non-coding RNAs in gastrointestinal tumors: Biological functions and therapeutic perspectives. Pharmacol Res. 2023;187:106582. doi: 10.1016/j.phrs.2022.106582. [DOI] [PubMed] [Google Scholar]
- 131.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 132.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-I. [DOI] [PubMed] [Google Scholar]
- 133.Mi Z, Zhongqiang C, Caiyun J, Yanan L, Jianhua W, Liang L. Circular RNA detection methods: A minireview. Talanta. 2022;238:123066. doi: 10.1016/j.talanta.2021.123066. [DOI] [PubMed] [Google Scholar]
- 134.Deng R, Zhang K, Li J. Isothermal Amplification for MicroRNA Detection: From the Test Tube to the Cell. Acc Chem Res. 2017;50:1059–1068. doi: 10.1021/acs.accounts.7b00040. [DOI] [PubMed] [Google Scholar]
- 135.Zhao Y, Chen F, Li Q, Wang L, Fan C. Isothermal Amplification of Nucleic Acids. Chem Rev. 2015;115:12491–12545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- 136.Masud MK, Umer M, Hossain MSA, Yamauchi Y, Nguyen NT, Shiddiky MJA. Nanoarchitecture Frameworks for Electrochemical miRNA Detection. Trends Biochem Sci. 2019;44:433–452. doi: 10.1016/j.tibs.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 137.Zhao G, Yan X, Zhang Y, Deng J, Liang X. Sensitive detection of MiRNA and CircRNA through DSN enzyme cooperating NEase assisted dual signal amplification. Anal Biochem. 2022;654:114744. doi: 10.1016/j.ab.2022.114744. [DOI] [PubMed] [Google Scholar]
- 138.Dittmar RL, Liu S, Tai MC, Rajapakshe K, Huang Y, Longton G, DeCapite C, Hurd MW, Paris PL, Kirkwood KS, et al. Plasma miRNA Biomarkers in Limited Volume Samples for Detection of Early-stage Pancreatic Cancer. Cancer Prev Res (Phila) 2021;14:729–740. doi: 10.1158/1940-6207.CAPR-20-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lou UK, Wong CH, Chen Y. A simple and rapid colorimetric detection of serum lncRNA biomarkers for diagnosis of pancreatic cancer. RSC Adv. 2020;10:8087–8092. doi: 10.1039/C9RA07858D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nicoletti A, Negri M, Paratore M, Vitale F, Ainora ME, Nista EC, Gasbarrini A, Zocco MA. Zileri Dal Verme L: Diagnostic and Prognostic Role of Extracellular Vesicles in Pancreatic Cancer: Current Evidence and Future Perspectives. Int J Mol Sci. 2023;24(1):885. doi: 10.3390/ijms24010885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li SR, Man QW, Gao X, Lin H, Wang J, Su FC, Wang HQ, Bu LL, Liu B, Chen G. Tissue-derived extracellular vesicles in cancers and non-cancer diseases: Present and future. J Extracell Vesicles. 2021;10:e12175. doi: 10.1002/jev2.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Doyle LM, Wang MZ. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells. 2019;8(7):727. doi: 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Qian F, Huang Z, Zhong H, Lei Q, Ai Y, Xie Z, Zhang T, Jiang B, Zhu W, Sheng Y, et al. Analysis and Biomedical Applications of Functional Cargo in Extracellular Vesicles. ACS Nano. 2022;16:19980–20001. doi: 10.1021/acsnano.2c11298. [DOI] [PubMed] [Google Scholar]
- 144.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dai J, Su Y, Zhong S, Cong L, Liu B, Yang J, Tao Y, He Z, Chen C, Jiang Y. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct Target Ther. 2020;5:145. doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. doi: 10.1016/S0021-9258(18)48095-7. [DOI] [PubMed] [Google Scholar]
- 147.Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, Shao Y, Zheng S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther. 2020;5:144. doi: 10.1038/s41392-020-00258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, Breakefield XO, Skog JK. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32:466–477. doi: 10.1016/j.annonc.2021.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yu J, Ostowari A, Gonda A, Mashayekhi K, Dayyani F, Hughes CCW, Senthil M. Exosomes as a Source of Biomarkers for Gastrointestinal Cancers. Cancers (Basel). 2023;15(4):1263. doi: 10.3390/cancers15041263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 152.Irmer B, Chandrabalan S, Maas L, Bleckmann A, Menck K. Extracellular Vesicles in Liquid Biopsies as Biomarkers for Solid Tumors. Cancers (Basel). 2023;15(4):1307. doi: 10.3390/cancers15041307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zeringer E, Barta T, Li M, Vlassov AV. Strategies for isolation of exosomes. Cold Spring Harb Protoc. 2015;2015:319–323. doi: 10.1101/pdb.top074476. [DOI] [PubMed] [Google Scholar]
- 154.Yang F, Liao X, Tian Y, Li G. Exosome separation using microfluidic systems: size-based, immunoaffinity-based and dynamic methodologies. Biotechnol J. 2017;12(4). 10.1002/biot.201600699. [DOI] [PubMed]
- 155.Liu J, Chen Y, Pei F, Zeng C, Yao Y, Liao W, Zhao Z. Extracellular Vesicles in Liquid Biopsies: Potential for Disease Diagnosis. Biomed Res Int. 2021;2021:6611244. doi: 10.1155/2021/6611244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zheng L, Wang H, Zuo P, Liu Y, Xu H, Ye BC. Rapid On-Chip Isolation of Cancer-Associated Exosomes and Combined Analysis of Exosomes and Exosomal Proteins. Anal Chem. 2022;94:7703–7712. doi: 10.1021/acs.analchem.2c01187. [DOI] [PubMed] [Google Scholar]
- 157.Armakolas A, Kotsari M, Koskinas J. Liquid Biopsies, Novel Approaches and Future Directions. Cancers (Basel). 2023;15(5):1579. doi: 10.3390/cancers15051579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lee M, Kim J, Jang M, Park C, Lee JH, Lee T. Introduction of Nanomaterials to Biosensors for Exosome Detection: Case Study for Cancer Analysis. Biosensors (Basel). 2022;12(8):648. doi: 10.3390/bios12080648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Machida T, Tomofuji T, Maruyama T, Yoneda T, Ekuni D, Azuma T, Miyai H, Mizuno H, Kato H, Tsutsumi K, et al. miR-1246 and miR-4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol Rep. 2016;36:2375–2381. doi: 10.3892/or.2016.5021. [DOI] [PubMed] [Google Scholar]
- 160.Goto T, Fujiya M, Konishi H, Sasajima J, Fujibayashi S, Hayashi A, Utsumi T, Sato H, Iwama T, Ijiri M, et al. An elevated expression of serum exosomal microRNA-191, - 21, -451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer. 2018;18:116. doi: 10.1186/s12885-018-4006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nakamura S, Sadakari Y, Ohtsuka T, Okayama T, Nakashima Y, Gotoh Y, Saeki K, Mori Y, Nakata K, Miyasaka Y, et al. Pancreatic Juice Exosomal MicroRNAs as Biomarkers for Detection of Pancreatic Ductal Adenocarcinoma. Ann Surg Oncol. 2019;26:2104–2111. doi: 10.1245/s10434-019-07269-z. [DOI] [PubMed] [Google Scholar]
- 162.Xavier CPR, Castro I, Caires HR, Ferreira D, Cavadas B, Pereira L, Santos LL, Oliveira MJ, Vasconcelos MH. Chitinase 3-like-1 and fibronectin in the cargo of extracellular vesicles shed by human macrophages influence pancreatic cancer cellular response to gemcitabine. Cancer Lett. 2021;501:210–223. doi: 10.1016/j.canlet.2020.11.013. [DOI] [PubMed] [Google Scholar]
- 163.Lu H, Lu S, Yang D, Zhang L, Ye J, Li M, Hu W. MiR-20a-5p regulates gemcitabine chemosensitivity by targeting RRM2 in pancreatic cancer cells and serves as a predictor for gemcitabine-based chemotherapy. Biosci Rep. 2019;39(5):BSR20181374. [DOI] [PMC free article] [PubMed]
- 164.Takahasi K, Iinuma H, Wada K, Minezaki S, Kawamura S, Kainuma M, Ikeda Y, Shibuya M, Miura F, Sano K. Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J Hepatobiliary Pancreat Sci. 2018;25:155–161. doi: 10.1002/jhbp.524. [DOI] [PubMed] [Google Scholar]
- 165.Kawamura S, Iinuma H, Wada K, Takahashi K, Minezaki S, Kainuma M, Shibuya M, Miura F, Sano K. Exosome-encapsulated microRNA-4525, microRNA-451a and microRNA-21 in portal vein blood is a high-sensitive liquid biomarker for the selection of high-risk pancreatic ductal adenocarcinoma patients. J Hepatobiliary Pancreat Sci. 2019;26:63–72. doi: 10.1002/jhbp.601. [DOI] [PubMed] [Google Scholar]
- 166.Sun H, Shi K, Qi K, Kong H, Zhang J, Dai S, Ye W, Deng T, He Q, Zhou M. Natural Killer Cell-Derived Exosomal miR-3607-3p Inhibits Pancreatic Cancer Progression by Targeting IL-26. Front Immunol. 2019;10:2819. doi: 10.3389/fimmu.2019.02819. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 167.Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X, Li X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang X, Zheng S, Hu C, Li G, Lin H, Xia R, Ye Y, He R, Li Z, Lin Q, et al. Cancer-associated fibroblast-induced lncRNA UPK1A-AS1 confers platinum resistance in pancreatic cancer via efficient double-strand break repair. Oncogene. 2022;41:2372–2389. doi: 10.1038/s41388-022-02253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chari ST. Detecting early pancreatic cancer: problems and prospects. Semin Oncol. 2007;34:284–294. doi: 10.1053/j.seminoncol.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Choi JI, Jang SI, Hong J, Kim CH, Kwon SS, Park JS, Lim JB. Cancer-initiating cells in human pancreatic cancer organoids are maintained by interactions with endothelial cells. Cancer Lett. 2021;498:42–53. doi: 10.1016/j.canlet.2020.10.012. [DOI] [PubMed] [Google Scholar]
- 172.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rhim AD, Thege FI, Santana SM, Lannin TB, Saha TN, Tsai S, Maggs LR, Kochman ML, Ginsberg GG, Lieb JG, et al. Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology. 2014;146:647–651. doi: 10.1053/j.gastro.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cauley CE, Pitman MB, Zhou J, Perkins J, Kuleman B, Liss AS, Fernandez-Del Castillo C, Warshaw AL, Lillemoe KD, Thayer SP. Circulating Epithelial Cells in Patients with Pancreatic Lesions: Clinical and Pathologic Findings. J Am Coll Surg. 2015;221:699–707. doi: 10.1016/j.jamcollsurg.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kuvendjiska J, Müller F, Bronsert P, Timme-Bronsert S, Fichtner-Feigl S, Kulemann B. Circulating Epithelial Cells in Patients with Intraductal Papillary Mucinous Neoplasm of the Pancreas. Life (Basel). 2023;13(7):1570. doi: 10.3390/life13071570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Roa-Colomo A, López Garrido MÁ, Molina-Vallejo P, Rojas A, Sanchez MG, Aranda-García V, Salmeron J, Romero-Gomez M, Muntane J, Padillo J, et al. Hepatocellular carcinoma risk-stratification based on ASGR1 in circulating epithelial cells for cancer interception. Front Mol Biosci. 2022;9:1074277. doi: 10.3389/fmolb.2022.1074277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lin HC, Liou MJ, Hsu HL, Hsieh JC, Chen YA, Tseng CP, Lin JD. Combined analysis of circulating epithelial cells and serum thyroglobulin for distinguishing disease status of the patients with papillary thyroid carcinoma. Oncotarget. 2016;7:17242–17253. doi: 10.18632/oncotarget.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. [DOI] [PMC free article] [PubMed]
- 180.Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P, Schmidt CM, Yip-Schneider MT, Allen PJ, Schattner M, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci U S A. 2017;114:10202–10207. doi: 10.1073/pnas.1704961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu G, Shao C, Li A, Zhang X, Guo X, Li J. Diagnostic Value of Plasma miR-181b, miR-196a, and miR-210 Combination in Pancreatic Cancer. Gastroenterol Res Pract. 2020;2020:6073150. doi: 10.1155/2020/6073150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bartsch DK, Gercke N, Strauch K, Wieboldt R, Matthäi E, Wagner V, Rospleszcz S, Schäfer A, Franke FS, Mintziras I, et al. The Combination of MiRNA-196b, LCN2, and TIMP1 is a Potential Set of Circulating Biomarkers for Screening Individuals at Risk for Familial Pancreatic Cancer. J Clin Med. 2018;7(10):295. [DOI] [PMC free article] [PubMed]
- 183.Franklin O, Jonsson P, Billing O, Lundberg E, Öhlund D, Nyström H, Lundin C, Antti H, Sund M. Plasma Micro-RNA Alterations Appear Late in Pancreatic Cancer. Ann Surg. 2018;267:775–781. doi: 10.1097/SLA.0000000000002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhou X, Lu Z, Wang T, Huang Z, Zhu W, Miao Y. Plasma miRNAs in diagnosis and prognosis of pancreatic cancer: A miRNA expression analysis. Gene. 2018;673:181–193. doi: 10.1016/j.gene.2018.06.037. [DOI] [PubMed] [Google Scholar]
- 185.Vieira NF, Serafini LN, Novais PC, Neto FSL, Cirino MLA, Kemp R, Ardengh JC, Saggioro FP, Gaspar AF, Sankarankutty AK, et al. The role of circulating miRNAs and CA19-9 in pancreatic cancer diagnosis. Oncotarget. 2021;12:1638–1650. doi: 10.18632/oncotarget.28038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yuan W, Tang W, Xie Y, Wang S, Chen Y, Qi J, Qiao Y, Ma J. New combined microRNA and protein plasmatic biomarker panel for pancreatic cancer. Oncotarget. 2016;7:80033–80045. doi: 10.18632/oncotarget.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wnuk J, Strzelczyk JK, Gisterek I. Clinical Value of Circulating miRNA in Diagnosis, Prognosis, Screening and Monitoring Therapy of Pancreatic Ductal Adenocarcinoma-A Review of the Literature. Int J Mol Sci. 2023;24(6):5113. [DOI] [PMC free article] [PubMed]
- 188.Sharma GG, Okada Y, Von Hoff D, Goel A. Non-coding RNA biomarkers in pancreatic ductal adenocarcinoma. Semin Cancer Biol. 2021;75:153–168. doi: 10.1016/j.semcancer.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xu G, Ji Y, Wang L, Xu H, Shen C, Ye H, Yang X. M6A-Related Long Non-Coding RNA Displays Utility in Predicting Prognosis, Portraying the Tumor Immune Microenvironment and Guiding Immunotherapy in Pancreatic Ductal Adenocarcinoma. Vaccines (Basel). 2023;11(3):499. doi: 10.3390/vaccines11030499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wu Q, Chen L, Miao D, Jin Y, Zhu Z. Prognostic signature based on m6A-related lncRNAs to predict overall survival in pancreatic ductal adenocarcinoma. Sci Rep. 2022;12:3079. doi: 10.1038/s41598-022-07112-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wu H, Wang B, Wang L, Xue Y. circular RNAs 0000515 and 0011385 as potential biomarkers for disease monitoring and determining prognosis in pancreatic ductal adenocarcinoma. Oncol Lett. 2022;23:56. doi: 10.3892/ol.2021.13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Xu Y, Yao Y, Gao P, Cui Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun. 2019;509:138–142. doi: 10.1016/j.bbrc.2018.12.088. [DOI] [PubMed] [Google Scholar]
- 193.Zhang W, Campbell DH, Walsh BJ, Packer NH, Liu D, Wang Y. Cancer-derived small extracellular vesicles: emerging biomarkers and therapies for pancreatic ductal adenocarcinoma diagnosis/prognosis and treatment. J Nanobiotechnology. 2022;20:446. doi: 10.1186/s12951-022-01641-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yu S, Li Y, Liao Z, Wang Z, Wang Z, Li Y, Qian L, Zhao J, Zong H, Kang B, et al. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut. 2020;69:540–550. doi: 10.1136/gutjnl-2019-318860. [DOI] [PubMed] [Google Scholar]
- 195.Hsu SK, Jadhao M, Liao WT, Chang WT, Lin IL, Chiu CC. The Role of Exosomes in Pancreatic Ductal Adenocarcinoma Progression and Their Potential as Biomarkers. Cancers (Basel). 2023;15(6):1776. doi: 10.3390/cancers15061776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zeng S, Pöttler M, Lan B, Grützmann R, Pilarsky C, Yang H. Chemoresistance in Pancreatic Cancer. Int J Mol Sci. 2019;20(18):4504. doi: 10.3390/ijms20184504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kim H, Heo CM, Oh J, Chung HH, Lee EM, Park J, Lee SH, Lee KH, Lee KT, Lee JK, et al. Clinical significance of circulating tumor cells after chemotherapy in unresectable pancreatic ductal adenocarcinoma. Transl Oncol. 2022;16:101321. doi: 10.1016/j.tranon.2021.101321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ju S, Chen C, Zhang J, Xu L, Zhang X, Li Z, Chen Y, Zhou J, Ji F, Wang L. Detection of circulating tumor cells: opportunities and challenges. Biomark Res. 2022;10:58. doi: 10.1186/s40364-022-00403-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pan X, Zhang X. Utility of circulating tumor cells and DNA in the management of advanced colorectal cancer. Future Oncol. 2020;16:1289–1299. doi: 10.2217/fon-2020-0073. [DOI] [PubMed] [Google Scholar]
- 200.Beije N, Onstenk W, Kraan J, Sieuwerts AM, Hamberg P, Dirix LY, Brouwer A, de Jongh FE, Jager A, Seynaeve CM, et al. Prognostic Impact of HER2 and ER Status of Circulating Tumor Cells in Metastatic Breast Cancer Patients with a HER2-Negative Primary Tumor. Neoplasia. 2016;18:647–653. doi: 10.1016/j.neo.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Garrido-Navas MC, García-Díaz A, Molina-Vallejo MP, González-Martínez C, Alcaide Lucena M, Cañas-García I, Bayarri C, Delgado JR, González E, Lorente JA, Serrano MJ. The Polemic Diagnostic Role of TP53 Mutations in Liquid Biopsies from Breast, Colon and Lung Cancers. Cancers (Basel). 2020;12(11):3343. doi: 10.3390/cancers12113343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nicolazzo C, Raimondi C, Mancini M, Caponnetto S, Gradilone A, Gandini O, Mastromartino M, Del Bene G, Prete A, Longo F, et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci Rep. 2016;6:31726. doi: 10.1038/srep31726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Fusi A, Liu Z, Kümmerlen V, Nonnemacher A, Jeske J, Keilholz U. Expression of chemokine receptors on circulating tumor cells in patients with solid tumors. J Transl Med. 2012;10:52. doi: 10.1186/1479-5876-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gardner KP, Tsai S, Aldakkak M, Gironda S, Adams DL. CXCR4 expression in tumor associated cells in blood is prognostic for progression and survival in pancreatic cancer. PLoS ONE. 2022;17:e0264763. doi: 10.1371/journal.pone.0264763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wu YH, Hung YP, Chiu NC, Lee RC, Li CP, Chao Y, Shyr YM, Wang SE, Chen SC, Lin SH, et al. Correlation between drug sensitivity profiles of circulating tumour cell-derived organoids and clinical treatment response in patients with pancreatic ductal adenocarcinoma. Eur J Cancer. 2022;166:208–218. doi: 10.1016/j.ejca.2022.01.030. [DOI] [PubMed] [Google Scholar]
- 206.Starlinger P, Brugger P, Reiter C, Schauer D, Sommerfeldt S, Tamandl D, Kuehrer I, Schoppmann SF, Gnant M, Brostjan C. Discrimination between circulating endothelial cells and blood cell populations with overlapping phenotype reveals distinct regulation and predictive potential in cancer therapy. Neoplasia. 2011;13:980–990. doi: 10.1593/neo.11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sabbaghian MS, Rothberger G, Alongi AP, Gagner JP, Goldberg JD, Rolnitzky L, Chiriboga L, Hajdu CH, Zagzag D, Basch R, Shamamian P. Levels of elevated circulating endothelial cell decline after tumor resection in patients with pancreatic ductal adenocarcinoma. Anticancer Res. 2010;30:2911–2917. [PubMed] [Google Scholar]
- 208.Dall'Olio FG, Marabelle A, Caramella C, Garcia C, Aldea M, Chaput N, Robert C, Besse B. Tumour burden and efficacy of immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2022;19:75–90. doi: 10.1038/s41571-021-00564-3. [DOI] [PubMed] [Google Scholar]
- 209.Wei T, Zhang Q, Li X, Su W, Li G, Ma T, Gao S, Lou J, Que R, Zheng L, et al. Monitoring Tumor Burden in Response to FOLFIRINOX Chemotherapy Via Profiling Circulating Cell-Free DNA in Pancreatic Cancer. Mol Cancer Ther. 2019;18:196–203. doi: 10.1158/1535-7163.MCT-17-1298. [DOI] [PubMed] [Google Scholar]
- 210.Groot VP, Mosier S, Javed AA, Teinor JA, Gemenetzis G, Ding D, Haley LM, Yu J, Burkhart RA, Hasanain A, et al. Circulating Tumor DNA as a Clinical Test in Resected Pancreatic Cancer. Clin Cancer Res. 2019;25:4973–4984. doi: 10.1158/1078-0432.CCR-19-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.van der Sijde F, Homs MYV, van Bekkum ML, van den Bosch TPP, Bosscha K, Besselink MG, Bonsing BA, de Groot JWB, Karsten TM, Groot Koerkamp B, et al. Serum miR-373–3p and miR-194–5p Are Associated with Early Tumor Progression during FOLFIRINOX Treatment in Pancreatic Cancer Patients: A Prospective Multicenter Study. Int J Mol Sci. 2021;22(20):10902. doi: 10.3390/ijms222010902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mikamori M, Yamada D, Eguchi H, Hasegawa S, Kishimoto T, Tomimaru Y, Asaoka T, Noda T, Wada H, Kawamoto K, et al. MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Sci Rep. 2017;7:42339. doi: 10.1038/srep42339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Amaral MJ, Oliveira RC, Donato P, Tralhão JG. Pancreatic Cancer Biomarkers: Oncogenic Mutations, Tissue and Liquid Biopsies, and Radiomics-A Review. Dig Dis Sci. 2023;68:2811–2823. doi: 10.1007/s10620-023-07904-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hugenschmidt H, Labori KJ, Borgen E, Brunborg C, Schirmer CB, Seeberg LT, Naume B, Wiedswang G. Preoperative CTC-Detection by Cell Search(®) Is Associated with Early Distant Metastasis and Impaired Survival in Resected Pancreatic Cancer. Cancers (Basel). 2021;13(3):485. doi: 10.3390/cancers13030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sun Y, Wu G, Cheng KS, Chen A, Neoh KH, Chen S, Tang Z, Lee PF, Dai M, Han RPS. CTC phenotyping for a preoperative assessment of tumor metastasis and overall survival of pancreatic ductal adenocarcinoma patients. EBioMedicine. 2019;46:133–149. doi: 10.1016/j.ebiom.2019.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Guo S, Shi X, Shen J, Gao S, Wang H, Shen S, Pan Y, Li B, Xu X, Shao Z, Jin G. Preoperative detection of KRAS G12D mutation in ctDNA is a powerful predictor for early recurrence of resectable PDAC patients. Br J Cancer. 2020;122:857–867. doi: 10.1038/s41416-019-0704-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kirchweger P, Kupferthaler A, Burghofer J, Webersinke G, Jukic E, Schwendinger S, Weitzendorfer M, Petzer A, Függer R, Rumpold H, Wundsam H. Circulating tumor DNA correlates with tumor burden and predicts outcome in pancreatic cancer irrespective of tumor stage. Eur J Surg Oncol. 2022;48:1046–1053. doi: 10.1016/j.ejso.2021.11.138. [DOI] [PubMed] [Google Scholar]
- 218.DiPardo BJ, Winograd P, Court CM, Tomlinson JS. Pancreatic cancer circulating tumor cells: applications for personalized oncology. Expert Rev Mol Diagn. 2018;18:809–820. doi: 10.1080/14737159.2018.1511429. [DOI] [PubMed] [Google Scholar]
- 219.Kamyabi N, Huang J, Lee JJ, Bernard V, Semaan A, Stephens B, Hurd MW, Vanapalli SA, Maitra A, Guerrero PA. A microfluidic device for label-free isolation of tumor cell clusters from unprocessed blood samples. Biomicrofluidics. 2019;13:044111. doi: 10.1063/1.5111888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pan Y, Li D, Yang J, Wang N, Xiao E, Tao L, Ding X, Sun P, Li D. Portal Venous Circulating Tumor Cells Undergoing Epithelial-Mesenchymal Transition Exhibit Distinct Clinical Significance in Pancreatic Ductal Adenocarcinoma. Front Oncol. 2021;11:757307. doi: 10.3389/fonc.2021.757307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Song BG, Kwon W, Kim H, Lee EM, Han YM, Kim H, Byun Y, Lee KB, Lee KH, Lee KT, et al. Detection of Circulating Tumor Cells in Resectable Pancreatic Ductal Adenocarcinoma: A Prospective Evaluation as a Prognostic Marker. Front Oncol. 2020;10:616440. doi: 10.3389/fonc.2020.616440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Nitschke C, Markmann B, Tölle M, Kropidlowski J, Belloum Y, Goetz MR, Schlüter H, Kwiatkowski M, Sinn M, Izbicki J, et al. Characterization of RARRES1 Expression on Circulating Tumor Cells as Unfavorable Prognostic Marker in Resected Pancreatic Ductal Adenocarcinoma Patients. Cancers (Basel). 2022;14(18):4405. doi: 10.3390/cancers14184405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mancuso P, Colleoni M, Calleri A, Orlando L, Maisonneuve P, Pruneri G, Agliano A, Goldhirsch A, Shaked Y, Kerbel RS, Bertolini F. Circulating endothelial-cell kinetics and viability predict survival in breast cancer patients receiving metronomic chemotherapy. Blood. 2006;108:452–459. doi: 10.1182/blood-2005-11-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bidard FC, Mathiot C, Degeorges A, Etienne-Grimaldi MC, Delva R, Pivot X, Veyret C, Bergougnoux L, de Cremoux P, Milano G, Pierga JY. Clinical value of circulating endothelial cells and circulating tumor cells in metastatic breast cancer patients treated first line with bevacizumab and chemotherapy. Ann Oncol. 2010;21:1765–1771. doi: 10.1093/annonc/mdq052. [DOI] [PubMed] [Google Scholar]
- 225.Strijbos MH, Gratama JW, Schmitz PI, Rao C, Onstenk W, Doyle GV, Miller MC, de Wit R, Terstappen LW, Sleijfer S. Circulating endothelial cells, circulating tumour cells, tissue factor, endothelin-1 and overall survival in prostate cancer patients treated with docetaxel. Eur J Cancer. 2010;46:2027–2035. doi: 10.1016/j.ejca.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 226.Miglio U, Oldani A, Mezzapelle R, Veggiani C, Paganotti A, Garavoglia M, Boldorini R. KRAS mutational analysis in ductal adenocarcinoma of the pancreas and its clinical significance. Pathol Res Pract. 2014;210:307–311. doi: 10.1016/j.prp.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 227.Ako S, Kato H, Nouso K, Kinugasa H, Terasawa H, Matushita H, Takada S, Saragai Y, Mizukawa S, Muro S, et al. Plasma KRAS mutations predict the early recurrence after surgical resection of pancreatic cancer. Cancer Biol Ther. 2021;22:564–570. doi: 10.1080/15384047.2021.1980312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hadano N, Murakami Y, Uemura K, Hashimoto Y, Kondo N, Nakagawa N, Sueda T, Hiyama E. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br J Cancer. 2016;115:59–65. doi: 10.1038/bjc.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wang Y, Ji Y, Xu Q, Huang W. Prognostic N6-methyladenosine (m6A)-related lncRNA patterns to aid therapy in pancreatic ductal adenocarcinoma. Front Genet. 2022;13:866340. doi: 10.3389/fgene.2022.866340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yang H, Zhang W, Ding J, Hu J, Sun Y, Peng W, Chu Y, Xie L, Mei Z, Shao Z, Xiao Y. A novel genomic instability-derived lncRNA signature to predict prognosis and immune characteristics of pancreatic ductal adenocarcinoma. Front Immunol. 2022;13:970588. doi: 10.3389/fimmu.2022.970588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kandimalla R, Shimura T, Mallik S, Sonohara F, Tsai S, Evans DB, Kim SC, Baba H, Kodera Y, Von Hoff D, et al. Identification of Serum miRNA Signature and Establishment of a Nomogram for Risk Stratification in Patients With Pancreatic Ductal Adenocarcinoma. Ann Surg. 2022;275:e229–e237. doi: 10.1097/SLA.0000000000003945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Reese M, Flammang I, Yang Z, Dhayat SA. Potential of Exosomal microRNA-200b as Liquid Biopsy Marker in Pancreatic Ductal Adenocarcinoma. Cancers (Basel). 2020;12(1):197. doi: 10.3390/cancers12010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Park SJ, Park JY, Shin K, Hong TH, Lee M, Kim Y, Kim IH. Clinical significance of serum-derived exosomal PD-L1 expression in patients with advanced pancreatic cancer. BMC Cancer. 2023;23:389. doi: 10.1186/s12885-023-10811-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Freed-Pastor WA, Lambert LJ, Ely ZA, Pattada NB, Bhutkar A, Eng G, Mercer KL, Garcia AP, Lin L, Rideout WM, 3rd, et al. The CD155/TIGIT axis promotes and maintains immune evasion in neoantigen-expressing pancreatic cancer. Cancer Cell. 2021;39:1342–1360.e1314. doi: 10.1016/j.ccell.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Murakami D, Matsuda K, Iwamoto H, Mitani Y, Mizumoto Y, Nakamura Y, Matsuzaki I, Iwamoto R, Takahashi Y, Kojima F, et al. Prognostic value of CD155/TIGIT expression in patients with colorectal cancer. PLoS ONE. 2022;17:e0265908. doi: 10.1371/journal.pone.0265908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Wang D, Gu Y, Yan X, Huo C, Wang G, Zhao Y, Teng M, Li Y. Role of CD155/TIGIT in Digestive Cancers: Promising Cancer Target for Immunotherapy. Front Oncol. 2022;12:844260. doi: 10.3389/fonc.2022.844260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Chang WH, Nguyen TT, Hsu CH, Bryant KL, Kim HJ, Ying H, Erickson JW, Der CJ, Cerione RA, Antonyak MA. KRAS-dependent cancer cells promote survival by producing exosomes enriched in Survivin. Cancer Lett. 2021;517:66–77. doi: 10.1016/j.canlet.2021.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yang Z, LaRiviere MJ, Ko J, Till JE, Christensen T, Yee SS, Black TA, Tien K, Lin A, Shen H, et al. A Multianalyte Panel Consisting of Extracellular Vesicle miRNAs and mRNAs, cfDNA, and CA19-9 Shows Utility for Diagnosis and Staging of Pancreatic Ductal Adenocarcinoma. Clin Cancer Res. 2020;26:3248–3258. doi: 10.1158/1078-0432.CCR-19-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Satoh K. Molecular Approaches Using Body Fluid for the Early Detection of Pancreatic Cancer. Diagnostics (Basel). 2021;11(2):375. doi: 10.3390/diagnostics11020375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Malara N, Kovacs G, Bussu F, Ferrazzo T, Garo V, Raso C, Cornacchione P, Iezzi R, Tagliaferri L. Liquid Biopsy-Guided Interventional Oncology: A Proof of Concept with a Special Focus on Radiotherapy and Radiology. Cancers (Basel). 2022;14(19):4676. doi: 10.3390/cancers14194676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Vidal L, Pando E, Blanco L, Fabregat-Franco C, Castet F, Sierra A, Macarulla T, Balsells J, Charco R, Vivancos A. Liquid biopsy after resection of pancreatic adenocarcinoma and its relation to oncological outcomes. Systematic review and meta-analysis. Cancer Treat Rev. 2023;120:102604. doi: 10.1016/j.ctrv.2023.102604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.