Abstract
Introduction
Urinary tract infection (UTI) is the second most common infectious disease affecting more than 150 million people globally annually. Uropathogenic E. coli (UPEC), the predominant cause of UTI, can occur as a biofilm associated with antimicrobial resistance (AMR). There is a data gap on global AMR patterns from low-income settings, including Tanzania. Data on antimicrobial susceptibility patterns in relation to biofilm formation will help in the proper selection of antibiotics and the fight against AMR.
Methods
This analytical cross-sectional study was conducted among consecutively selected outpatients (n = 344) from January to May 2022 at Morogoro Regional Referal Hospital. Mid-stream urine samples were collected aseptically from symptomatic patients. A significant UTI was defined when more than 105 colonies/ml of urine were recorded. Kirby Bauer’s disc diffusion method was used for antibiotics susceptibility patterns and a Congo Red Agar method was used to determine biofilm formation. Two-sided χ2 test or Fisher’s exact test, Cohen’s kappa coefficient and logistic regression were used for data analysis. A p-value < 0.05 was considered statistically significant.
Results
The prevalence of UTIs was 41% (141/344) and elders (>=60 years) had five times higher odds of having UTI as compared to adolescents (p < 0.001). E. coli was the most predominant bacteria (47%; 66/141), which displayed moderate susceptibility against ciprofloxacin (59.1%) and nitrofurantoin (57.6%). A total of 72 (51%) of all isolated bacteria were multi-drug resistant. All isolated bacteria demonstrated high resistance (> 85%) against ampicillin and co-trimoxazole. In this study, 51.5% (34/66) were biofilm-forming E. coli and demonstrated relatively higher antibiotic resistance as compared to non-biofilm forming bacteria (p < 0.05).
Conclusion
We report high antibiotic resistance against commonly used antibiotics. Slightly more than half of the isolated bacteria were biofilm forming E. coli. A need to strengthen stewardship programs is urgently advocated.
Keywords: UTI, Antimicrobial resistance, Biofilm forming bacteria
Background
Urinary tract infection (UTI) is a disease of public health importance affecting more than 150 million people [1] with a financial burden of about $6 billion worldwide each year [2]. It is one of the most common infectious diseases, second to upper respiratory tract infections [3]. More than 50% of all women and at least 12% of men experience UTI in their lifetime [4]. Data regarding the prevalence of UTI among children and pregnant women are largely available [5–7], probably because of their susceptibility to secondary complications. Nevertheless, data on UTI prevalence in the general population are scanty in Tanzania indicating the need to regularly monitor the burden of UTIs to inform policy decisions.
For the majority of UTIs, bacteria from the Enterobacteriaceae family are the most frequent culprits. Uropathogenic Escherichia coli (UPEC) are the most prevalent strains causing UTIs, accounting for about 80% of uncomplicated UTIs and about 95% of community and hospital-acquired infections [8, 9]. Bacteria colonize the urinary tract by either freely attaching to the epithelial surface (planktonic) reversibly [2], or forming biofilm [10] defined by the presence of bacterial aggregates embedded in a self-produced extracellular matrix [11].
The human immune system has trouble recognizing bacteria and getting rid of infections with biofilm. Additionally, antibiotics penetration to the deeper layers of the biofilm matrix is difficult and may expose bacteria to sub-therapeutic antimicrobial concentrations leading to resistance [12]. Consequently, biofilm-associated infections tolerate the standard 5–10-day antibiotics treatment [13, 14], indicating the need for multi-targeted or combination antibiotics therapies. Availability of data on the biofilm forming patterns in relation to antimicrobial resistance (AMR) may guide the designing of appropriate antibiotics policy guidelines and, eventually, the prevention of AMR. Few studies have reported the relationship between biofilm forming and AMR [15–17]. However, such data are scarce from Sub-Saharan Africa and Tanzania included [18, 19].
Data regarding antimicrobial susceptibility patterns are reported [20]. However, the patterns of AMR differ across geographical locations and there is data gap in many low-income settings [21]. The present study investigated the prevalence of UTI in the general population, biofilm forming ability of UPEC and AMR in clinical isolates at Morogoro referral regional hospital, Tanzania.
Methods
Study design and setting
This was an analytical cross-sectional study conducted at Morogoro referral regional hospital (MRRH). MRRH is located in the Morogoro region, about 190 km west of Dar es Salaam, the largest business city in Tanzania (Fig. 1). The study site choice was motivated by the scarcity of data regarding AMR patterns from the region, which is required for antimicrobial stewardship activities.
Fig. 1.
Study site map. The top right is the map of the Morogoro region. Bottom right is the map of Morogoro Urban. On the left is the map of Tanzania. The study site map was originally generated using ArcGIS software version 10.7.1 (Esri, California, USA; https://www.esri.uconn.edu/software/arcgis-student/)
Patient recruitment and data collection
The study targeted all patients attending outpatient clinics with any symptoms of dysuria, urination frequency, urgency, fever, hematuria and suprapubic pain. A standard pilot-tested questionnaire was used to collect patients’ socio-demographic and clinical characteristics. At enrollment, patient’s age, sex, level of education and occupation were recorded. Patients who met the inclusion criteria were conveniently recruited for this study. Outpatients with symptoms unrelated to UTI and those who were admitted were excluded.
Urine sample collection and processing
Urine samples were collected according to the standard procedures as previously described [22]. For adult participants, the midstream urine method was used and the catch clean method was used for children. Participants were instructed to collect urine samples in two separate sterile containers. One bottle containing the sample was used for the dipstick urine test to detect leucocytes at the study health facility. The other container was immediately stored in the refrigerator (4 °C), where culture was done within 24 h after sample collection [22].
Isolation of microorganisms
Urine samples were plated on cysteine lactose electrolytes deficient (CLED) agar medium. A loop calibrated to deliver approximately 0.001 mL was used to inoculate urine in CLED agar plates and incubated aerobically at 37oC for 24 h. The growth of one type of organism of > 105 colony-forming units was considered bacteriuria [23, 24]. Clinical isolates were identified and confirmed biochemically using standard laboratory workflow [25, 26]. Confirmed bacteria isolates were suspended in nutrient broth, supplemented with 16% glycerol, and frozen at -80oC. The isolated bacteria samples were used for testing antimicrobial susceptibility and biofilm production.
Antimicrobial susceptibility testing
Antimicrobial susceptibility tests were performed according to standards of Clinical and Laboratory Standards Institute (CLSI) guidelines using Kirby ‘Bauer’s disc diffusion method [27]. Briefly, bacterial suspensions in physiological saline solution were spread platted on Mueller-Hinton Agar. Antimicrobial-impregnated disks, selected based on CLSI, were then placed on the culture medium surface. Commercially available antibiotic sensitivity discs that are widely used in Tanzania, including ampicillin (10 µg), amoxicillin/clavulanic acid (20/10µg), ciprofloxacin (5 µg), co-trimoxazole (trimethoprim/ sulfamethoxazole) (1.25 µg/23.75), ceftriaxone (30 µg), and nitrofurantoin (300µg) were tested. For P. aeruginosa, additional antibiotic sensitivity discs, including imipenem (10 µg), amikacin (30 µg) and gentamicin (10 µg), were also tested. After incubating the plates at 37°C for 18–24 hours, the diameter (nearest whole mm) of the inhibition zones [27] for each antibiotic was measured. The interpretation breakpoints were based on whether the bacterium was susceptible (S), intermediate (I), or resistant (R) to the tested drugs according to the CLSI recommendations [27]. Antibiograms were validated using standardized control strains of E. coli (ATCC 25922), S. aureus (ATCC 25923) and P. aeruginosa (ATCC 27853).
Determination of biofilm formation
To detect biofilm forming bacteria isolates, modified Congo red Agar method was used as previously described [28]. Briefly, bacteria strains were inoculated on blood base agar − 2 (BAB2) (40 gms/L BAB2, 10 gms/L glucose and 1000ml water), with 0.4 gms/L Congo red stain. At first, the Congo red stain was prepared as an aqueous solution and autoclaved (121 °C for 15 min) separately from the other medium constituents. The Congo red solution was added when the agar was cooled and incubated aerobically at 37 °C for 24 /48 hours. Black colonies with a dry crystalline consistency were interpreted as positive biofilm results. A red colored colony indicated negative biofilm results. E. coli ATCC 25,922 was used as positive control and S. aureus ATCC 25,932 as negative control for the CRA method.
Statistical analysis
Statistical package for Social Sciences (SPSS) software (IBM, Armonk, NY, USA) was used to analyze the data. Descriptive statistics (median, IQR, mean + SD, percentages) were used for patients’ socio-demographic characteristics. Comparison between proportions for categorical variables in two independent groups was performed using the two-sided χ2 test or Fisher’s exact test. Cohen’s kappa coefficient was used to test the agreement of positive and negative UTIs between urine deep stick and bacteria culture methods. Multi-drug resistance (MDR) was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories [29]. Binary logistic regression assessed the association between UTI and independent variables. A p-value < 0.05 was considered statistically significant.
Ethical consideration
The study was approved by the Institutional review board of the Muhimbili University of Health and Allied Sciences (MUHAS) before the commencement of the study (DA.282/298/01.C/920). Permission to conduct the study was requested from the Morogoro regional referral hospital’s medical officer in charge. Written consent was obtained from all participants before enrollment.
Results
Socio-demographic characteristics of participants
A total of 344 patients attending the outpatient clinic presenting with UTI symptoms were recruited for this study from January to May 2022. Participants had a median age of 25.5 (1–83) years; the majority (63%) were young adults between 19 and 59 years old. About three-quarters of the participants (76.4%) were women. More than half of the female adults were pregnant (60%). Almost half of the adult participants (52.1%) had formal employment and three quarter (66.7%) were living with a partner. The socio-demographic characteristics of participants are shown in Table 1.
Table 1.
Socio-demographic characteristics of participants
| Category | Variable | Frequency n (%) |
|---|---|---|
| Age (n = 344) | Median age (range) | 25.5 (1–83) |
| Children (1–12 years) | 63 (18.3) | |
| Adolescents (13–18) | 31 (9.0) | |
| Young adults (19–59) | 215 (62.5) | |
| Elders (≥60) | 35 (10.2) | |
| Sex (n = 344) | Male | 81 (23.6) |
| Female | 263 (76.4) | |
| Education Level (n = 250) | No formal education | 7 (2.8) |
| Primary | 87 (34.8) | |
| Secondary | 96 (38.4) | |
| Above secondary | 60 (24.0) | |
| Occupation (n = 165) | Formal employment | 86 (52.1) |
| Self employed | 63 (38.2) | |
| Subsistence farmers | 16 (9.7) | |
|
Pregnancy status (adult women) (n = 130) |
YES | 78 (60%) |
| NO | 52 (40%) | |
| Marital status (n = 150) | Single | 50 (33.3) |
| Married | 100 (66.7) |
Prevalence and determinants of UTI
The prevalence of UTI among patients attending the outpatient clinic presenting with UTI symptoms was 34.3% (118/344) and 41% (141/344) by dipstick urine analysis and culture, respectively. There was low agreement between dipstick urine analysis and culture in detecting positive UTI (Kappa = 0.11). The sensitivity and specificity of dipstick urine analysis were 43% and 69%, respectively, compared to the urine culture method.
The relationship between baseline parameters and the prevalence of UTI was evaluated. There was no significant difference in UTI prevalence between different levels of education (p = 0.13), marital status (p = 0.37) and occupation (p = 0.77). When comparing pregnant with non-pregnant adult women, there was no statistically discernible difference in the prevalence of UTI (p = 0.40). On logistic regression, elderly patients (≥ 60 years) had five times higher odds of having UTI as compared to adolescents. Female participants had almost two times higher odds of having UTI as compared to male participants at the borderline p = 0.05 (Table 2).
Table 2.
Predictors of UTI
| Variable | n (%) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| OR (95% CI) | p-value | aOR (95% CI) | p-value | ||
| Age (years) | |||||
| Children (1–12) | 18 (28.6) | 2.0 (0.8–5.2) | 0.15 | 1.9 (0.7–4.9) | 0.19 |
| Young adults (19–59) | 82 (38.1) | 1.3 (0.6–2.9) | 0.48 | 1.5 (0.7–3.3) | 0.36 |
| Elders (≥60) | 16 (45.7) | 4.5 (1.2–16.4) | 0.023 | 4.8 (1.3–17.9) | 0.017 |
| Adolescents (13–18) | 5 (16.1) | 1 | 1 | ||
| Sex | |||||
| Female | 90 (34.2) | 1.6 (0.9–2.9) | 0.09 | 1.9 (1.0-3.6) | 0.05 |
| Male | 20 (24.7) | 1 | 1 | ||
Antimicrobial susceptibility patterns
UPEC was the most common isolated bacteria, accounting for 47% (66/141) of all the isolated species. P. aeruginosa, K. pneumoniae and P. mirabilis contributed 17% (24/141), 11.4% (16/141) and 14.2% (20/141) of all isolated species, respectively. Other species, which were non-identified Gram negetive bacilli, accounted for 9.9% (14/141).
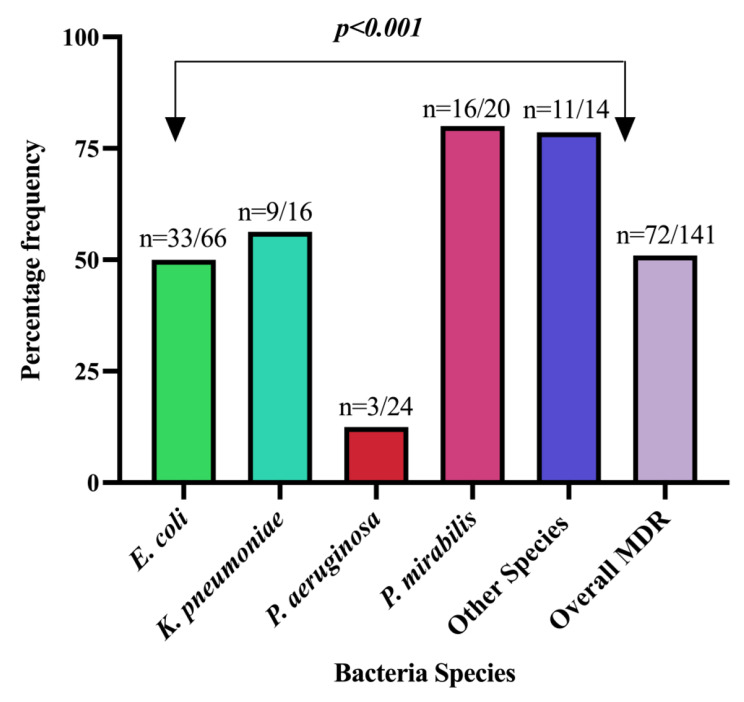
A total of 72 (51%) of all isolated bacteria demonstrated resistance to at least one agent in three or more antimicrobial categories (MDR) Fig. 2. For P. aeruginosa, MDR was assessed in three antimicrobial families namely aminoglycosides (amikacin and gentamicin) flouroquinolones (ciprofloxacin) and carbapenems (imipenem). Isolates of P. aeruginosa displayed low rates of MDR, which differed significantly compared to other isolated bacteria (Fig. 2).
Fig. 2.
Proportion of MDR among the isolated bacteria species. Other species represent non-identified Gram negetive bacilli (p-value is based on the Chi-Square test)
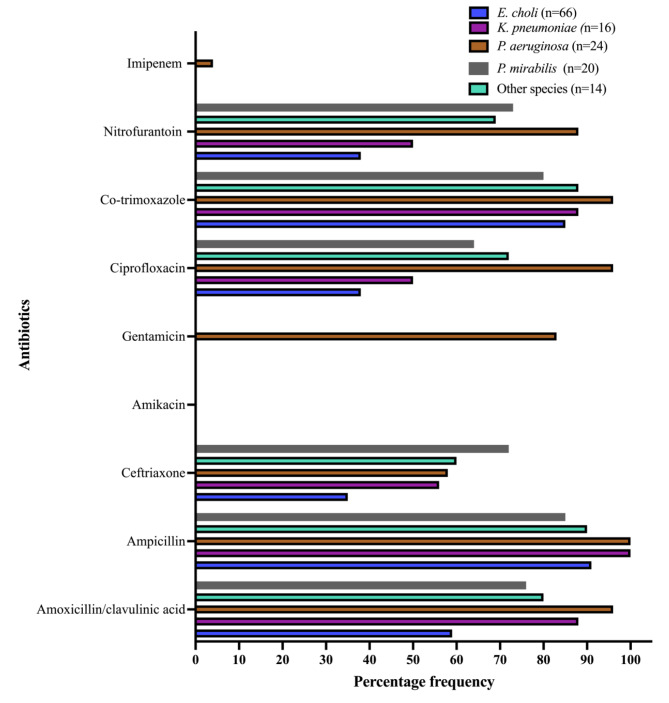
Isolated species demonstrated moderate to low susceptibility to the tested antibiotics. E. coli showed fairly susceptibility against ciprofloxacin (59.1%), nitrofurantoin (57.6%) and ceftriaxone (50%). P. aeruginosa displayed the highest susceptibility against amikacin (87.5%) and imipenem (83%) and lower susceptibility against gentamicin. K. pneumonia demonstrated fair susceptibility against ciprofloxacin (50%). Figure 3 presents the AMR patterns for the isolated species.
Fig. 3.
Antimicrobial resistance patterns among isolated bacteria (`n´ indicate the number of isolates under each bacteria specie)
Biofilm formation
The black colonies, with a dry crystalline consistency, were interpreted as positive biofilm results. In contrast, red colored colonies indicated negative biofilm results (Fig. 4). Of the 66 clinical isolates of E. coli, 34 (51.5%) formed biofilm on CRA.
Fig. 4.
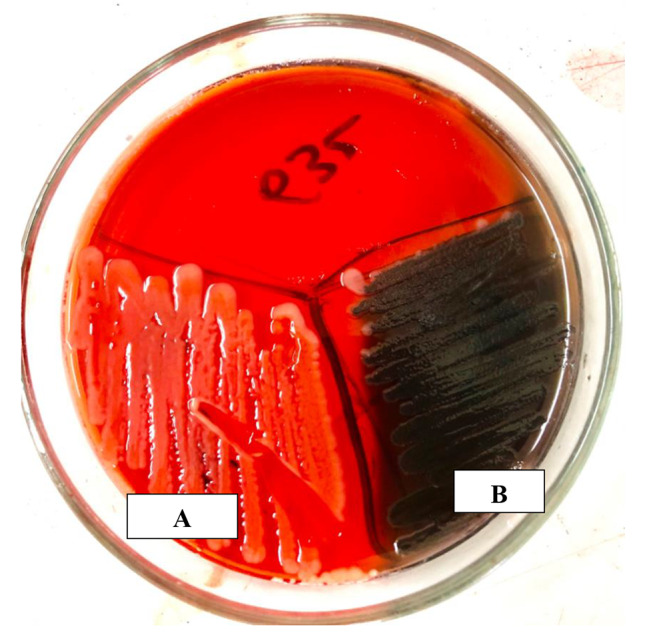
Congo Red Agar plates showing non-biofilm forming (A = red colored colonies) and biofilm forming (B = Dry crystalline Black colonies) UPEC isolates
More biofilm forming UPEC were MDR compared to UPEC non-biofilm forming isolates, although the difference was not statistically significant (p-value = 0.17). On the other hand, biofilm forming isolates demonstrated significantly higher resistance against co-trimoxazole and amoxycillin/clavulanic acid (p < 0.05), as shown in Table 3.
Table 3.
Drug resistance among biofilm forming and non-biofilm forming UPEC isolates
| Resistant to; | Biofilm forming E. coli n (%) N = 34 |
Non-biofilm forming E. coli n (%) N = 32 |
p-value |
|---|---|---|---|
| Amoxicillin/clavulanic acid | 22 (69) | 10 (31) | 0.013 |
| Ampicillin | 27 (54) | 23 (46) | 0.30* |
| Ceftriaxone | 12 (60) | 8 (40) | 0.18* |
| Ciprofloxacin | 15 (65) | 8 (35) | 0.09 |
| Co-trimoxazole | 27 (59) | 19 (41) | 0.01* |
| Nitrofurantoin | 13 (59) | 9 (41) | 0.11 |
| MDR | 20 (61) | 13 (39) | 0.17 |
*P-values are based on Fisher´s exact test
Discussion
UTI is a public health problem accounting for more than 15% of all the antibiotics prescriptions among outpatients [30]. Infections forming biofilm are associated with AMR and recurrent UTIs [31, 32] both of which are currently increasing globally [4]. Nevertheless, data regarding the characterization of biofilm-forming UTIs in relation to AMR rates in sub-Saharan Africa are scanty. We report, a substantial proportion of outpatient clinic patients with UTI symptoms had UTI, high AMR rate to commonly used antibiotics and 50% of UPEC isolates forming biofilm UTI associated with antibiotics resistance. The study included an outpatient population presenting with UTI symptoms regardless of sex and age to reflect the real-world population distribution. To the best of our knowledge, this is the first study to report biofilm-forming patterns of UPEC and their correlation with antibiotics resistance from Tanzania.
In this study, 41% of patients attending the outpatient clinic with any UTI symptoms were found to have culture-positive UTI. This prevalence is higher than the global prevalence, which ranges from 13 to 33% but is comparable to that of other resource-limited countries [33]. Our finding is also relatively lower than that reported among children with symptoms attending pediatric clinics [34] and among women attending outpatient clinics (63%) [35] in Dar es Salaam, Tanzania. On the other hand, our finding is higher than that reported among symptomatic women in Mwanza, Tanzania (18%) [36] and among asymptomatic pregnant women in Arusha (31.6%) northern part of Tanzania [37]. Our results indicate the diverse burden of UTI across different geographical locations, which might explain the differences in social determinants of UTI.
This study found a higher prevalence of UTI in women than men at the borderline (p = 0.05). Usually, women have shorter urethra, which is close to the perineum, than men; thus, they are more prone to UTI than men [38]. The fact that there were fewer male participants in the current study may account for the lack of significant differences. In contrast, older people had a considerably higher chance of getting a UTI than teenagers, which could be attributed to immunosenescence, an age-related immunological shift [39]. Other baseline characteristics did not show a significant effect on UTI.
Isolated species exhibited moderate to high resistance patterns against commonly used antibiotics. Half of all isolated bacteria species were resistant to at least one agent in more than two classes of antibiotics, referred to as multi-drug resistance (MDR). All isolated species showed high resistance rates against co-trimoxazole and ampicillin, similar to other reported findings from previous studies [40], suggesting that the drugs might have limited susceptibility against UTI and should be less considered in the treatment regimen. E. coli was the most predominant isolated species and displayed moderate resistance rates against ceftriaxone (34%), ciprofloxacin (37%) and nitrofurantoin (37%) which is comparable to several other previous studies [41, 42]. On the contrary, previous studies reported a higher resistance rate against ceftriaxone, ciprofloxacin and nitrofurantoin [36] [43]. Ciprofloxacin and ceftriaxone are the drugs of choice for UTI in Tanzania, suggesting that a substantial proportion of UTIs from the setting may be susceptible to the first-line antibiotics. However, isolated species exhibited high resistance against amoxicillin/clavulanic acid, the drug of choice for UTI in pregnant and adolescent women [44], similar to a previous study from Uganda [45].
P. aeruginosa demonstrated the highest sensitivity rates (> 80%) against amikacin and imipenem with low rates of MDR. The reason for this finding could be that these are reserved antibiotics and not commonly used in health facilities thus, they have low chances for resistance development. On the contrary, P. aeruginosa showed a higher rate of resistance (85–100%) among the commonly used antibiotics corroborating the findings of previous studies [46]. The finding implies that most UTIs associated with P. aeruginosa may not respond to commonly prescribed antibiotics. Our findings contribute to the knowledge about antibiotic resistance patterns peculiar to this geographical location.
About half of isolated E. coli were biofilm formers on Congo Red Agar (CRA). This finding suggest that biofilm forming UTIs are common in Tanzania similar to the findings of previous studies from other regions [15–17]. The susceptibility pattern showed that biofilm-forming E. coli were more MDR (60%) compared to non-biofilm formers (40%). Although statistical significance was not reached for the overall MDR, biofilm formers showed significantly higher resistance rates against ciprofloxacin and co-trimoxazole as compared to non-biofilm formers. Previous studies from other regions reported significant association between biofilm forming capability and AMR for the same antibiotics as the present study [15–17]. One reason to explain this might be the difficult penetration of antimicrobials to the deeper layers of the biofilm matrix which expose bacteria to sub-therapeutic antimicrobial concentrations leading to resistance [12]. This finding informs the policy makers on the importance of considering the impact of biofilm infections in policies against antimicrobial resistnance.
The observed high antibiotic resistance patterns might be explained by the community´s inappropriate and overuse of antibiotics [47]. Miserably, AMR causes a stall in achieving the global sustainable development goal number 3.3, which aims at reducing the burden of infectious diseases by the end of 2030 [48]. The findings of this study underscores the need for strenghthening antimicrobial stewardship in health facilities [49].
UTI is treated empirically in Tanzania and other resource-limited countries. In the majority of primary healthcare facilities, urine analysis is based on dipstick urine analysis, which in this study was found to have a lower sensitivity (45%) than urine culture. This suggests that a method used in the standard of care for diagnosing UTIs may miss a considerable portion of individuals who have severe bacteriuria. The findings underscore the need for integrating urine culture and sensitivity to properly manage UTI in our health facilities.
Conclusion and recommendations
We report a high prevalence of culture-positiveUTI among outpatients presenting with UTI symptoms from the study population. High prevalence rates of antibiotic resistance were also observed in clinical bacterial isolates. Slightly more than half of the isolated E. coli bacteria were biofilm forming, demonstrating higher antibiotic resistance to co-trimoxazole and amoxicillin/clavulanic acid than non-biofilm forming counterparts. A need to strengthen AMR stewardship programs in health facilities is urgently advocated in order to halt the spread of AMR.
Acknowledgements
We convey our sincere thanks to all participants who took part in this study. We also thank the Pharmaceutical Microbiology Laboratory (MUHAS) staff for their support during experiments.
Abbreviations
- CLSI
Clinical and laboratory standards institute
- MDR
Multi drug resistance
- SOPs
Standard Operating procedures
- UPEC
Uropathogenic E. coli
- MUHAS
Muhimbili University of Health and Allied Sciences
Author contributions
EMM designed the study performed the data collection and wrote the frist draft of the manuscript; EMM, JAM, RZS and KDM participated in data analysis. All authors read, edited and approved the final manuscript.
Funding
This work was funded by the Muhimbili University of Health and Allied Sciences through seed grants supported by the Swedish International Development Cooperation Agency– (Sida). The funders had no role in study design, data collection and analysis, the decision to publish, or the preparation of the manuscript.
Data Availability
All data generated or analyzed during this study are included in this manuscript.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study obtained approval from the Research and Ethics Committee of the Muhimbili University of Health and Allied Sciences. We confirm that all methods were performed in accordance with relevant guidelines and regulations. All participants gave written informed consent before recruitment.
Consent for publication
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stamm WE, Norrby SR. Urinary tract infections: Disease Panorama and Challenges. J Infect Dis Suppl. 2001;183:1–S4. doi: 10.1086/318850. [DOI] [PubMed] [Google Scholar]
- 2.Mann R, Mediati DG, Duggin IG, Harry EJ, Bottomley AL. Metabolic adaptations of Uropathogenic E. coli in the urinary tract. Front Cell Infect Microbiol. 2017;7:241–1. doi: 10.3389/fcimb.2017.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klumpp D, Rycyk M, Chen M, Thumbikat P, Sengupta S, Schaeffer A. Uropathogenic Escherichia coli induces extrinsic and intrinsic cascades to initiate Urothelial apoptosis. Infect Immun. 2006;74:5106–13. doi: 10.1128/IAI.00376-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brumbaugh AR, Smith SN, Mobley HL. Immunization with the yersiniabactin receptor, FyuA, protects against pyelonephritis in a murine model of urinary tract infection. Infect Immun. 2013;81(9):3309–16. doi: 10.1128/IAI.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sangeda R, Paul F, Mtweve D. Prevalence of urinary tract infections and antibiogram of uropathogens isolated from children under five attending Bagamoyo District Hospital in Tanzania: a cross-sectional study F1000Research 2021, 10(449).
- 6.Chandra Bala Sekharan KRK, Evod Ernest Kuwingwa and Devarajan Dinesh Kumar Evaluation of the prevalence of urinary tract infection in females aged 6–50 years at Kinondoni District, Tanzania. Sci Internat. 2017;5:42–6. doi: 10.17311/sciintl.2017.42.46. [DOI] [Google Scholar]
- 7.Msaki BP, Mshana SE, Hokororo A, Mazigo HD, Morona D. Prevalence and predictors of urinary tract infection and severe malaria among febrile children attending Makongoro health centre in Mwanza city, North-Western Tanzania. Arch Public Health. 2012;70(1):4. doi: 10.1186/0778-7367-70-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foxman B. The epidemiology of urinary tract infection. Nat Rev Urol. 2010;7(12):653–60. doi: 10.1038/nrurol.2010.190. [DOI] [PubMed] [Google Scholar]
- 9.Dhakal B, Kulesus RR, Mulvey M. Mechanisms and consequences of bladder cell invasion by uropathogenic Escherichia coli. Eur J Clin Invest. 2008;38(Suppl 2):2–11. doi: 10.1111/j.1365-2362.2008.01986.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhao F, Yang H, Bi D, Khaledi A, Qiao M. A systematic review and meta-analysis of antibiotic resistance patterns, and the correlation between biofilm formation with virulence factors in uropathogenic E. coli isolated from urinary tract infections. Microb Pathog. 2020;144:104196. doi: 10.1016/j.micpath.2020.104196. [DOI] [PubMed] [Google Scholar]
- 11.Hall-Stoodley L, Stoodley P, Kathju S, Høiby N, Moser C, Costerton JW, Moter A, Bjarnsholt T. Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol Med Microbiol. 2012;65(2):127–45. doi: 10.1111/j.1574-695X.2012.00968.x. [DOI] [PubMed] [Google Scholar]
- 12.Soto SM, Smithson A, Horcajada JP, Martinez JA, Mensa JP, Vila J. Implication of biofilm formation in the persistence of urinary tract infection caused by uropathogenic Escherichia coli. Clin Microbiol Infect. 2006;12(10):1034–6. doi: 10.1111/j.1469-0691.2006.01543.x. [DOI] [PubMed] [Google Scholar]
- 13.Mulvey MA, Schilling JD, Hultgren SJ. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun. 2001;69(7):4572–9. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schilling JD, Lorenz RG, Hultgren SJ. Effect of trimethoprim-sulfamethoxazole on recurrent bacteriuria and bacterial persistence in mice infected with uropathogenic Escherichia coli. Infect Immun. 2002;70(12):7042–9. doi: 10.1128/IAI.70.12.7042-7049.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shrestha LB, Bhattarai NR, Khanal B. Antibiotic resistance and biofilm formation among coagulase-negative staphylococci isolated from clinical samples at a tertiary care hospital of eastern Nepal. Antimicrob Resist Infect Control. 2017;6:89. doi: 10.1186/s13756-017-0251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah C, Baral R, Bartaula B, Shrestha LB. Virulence factors of uropathogenic Escherichia coli (UPEC) and correlation with antimicrobial resistance. BMC Microbiol. 2019;19(1):204. doi: 10.1186/s12866-019-1587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karigoudar RM, Karigoudar MH, Wavare SM, Mangalgi SS. Detection of biofilm among uropathogenic Escherichia coli and its correlation with antibiotic resistance pattern. J Lab Physicians. 2019;11(1):17–22. doi: 10.4103/JLP.JLP_98_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Neill J. (2016). Tackling Drug-Resistant Infections Globally - Final Report and Recommendations: https://amrreview.org/Publications. Accessed on January 06 2023.
- 19.World Health Organization. Global action plan on antimicrobial resistance. (2015). https://www.who.int/antimicrobial-resistance/publications/global-action-plan/en/. Accessed on December 28 2022.
- 20.Tadesse BT, Ashley EA, Ongarello S, Havumaki J, Wijegoonewardena M, González IJ, Dittrich S. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis. 2017;17(1):616. doi: 10.1186/s12879-017-2713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Global burden of Bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–55. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baron EJ. Specimen Collection, Transport, and Processing: Bacteriology: Manual of Clinical Microbiology, Eleventh Edition. edn.: American Society of Microbiology; 2015.
- 23.Smelov V, Naber K, Bjerklund Johansen TE. Improved classification of urinary tract infection: future considerations. Eur Urol Suppl. 2016;15(4):71–80. doi: 10.1016/j.eursup.2016.04.002. [DOI] [Google Scholar]
- 24.Tandogdu Z, Wagenlehner FME. Global epidemiology of urinary tract infections. Curr Opin Infect Dis. 2016;29(1):73–9. doi: 10.1097/QCO.0000000000000228. [DOI] [PubMed] [Google Scholar]
- 25.Wauters G, Vaneechoutte M. Approaches to the identification of AerobicGram-Negative Bacteria. Manual of Clinical Microbiology, Eleventh Edition. edn.: American Society of Microbiology; 2015.
- 26.General Approaches to Identification of Aerobic Gram-Positive Cocci. In: Manual of Clinical Microbiology. edn. ; 2015: 350–353.
- 27.CLSI. Performance Standards for Antimicrobial susceptibility testing. Volume 31st editionst, 100CLSI supplement MS ed. Clinical and Laboratory Standards Institute; 2021.
- 28.Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol. 1989;42(8):872–4. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 30.Annual Epidemiological . Report 2014 - Antimicrobial Resistance and Healthcare-associated infections. Stockholm, Sweden: European Centre for Disease Prevention and Control; 2015. [Google Scholar]
- 31.McLellan LK, Hunstad DA. Urinary tract infection: Pathogenesis and Outlook. Trends Mol Med. 2016;22(11):946–57. doi: 10.1016/j.molmed.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiles TJ, Kulesus RR, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol. 2008;85(1):11–9. doi: 10.1016/j.yexmp.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Öztürk R, Murt A. Epidemiology of urological infections: a global burden. World J Urol. 2020;38(11):2669–79. doi: 10.1007/s00345-019-03071-4. [DOI] [PubMed] [Google Scholar]
- 34.Sangeda RZ, Paul F, Mtweve DM. Prevalence of urinary tract infections and antibiogram of uropathogens isolated from children under five attending Bagamoyo District Hospital in Tanzania: a cross-sectional study. F1000Research. 2021;10:449. doi: 10.12688/f1000research.52652.1. [DOI] [Google Scholar]
- 35.Sekharan CB, Kumari KR, Kuwingwa EE, Devarajan Dinesh Kumar. 2017. Evaluation of the Prevalence of Urinary Tract Infection in Females Aged 6–50 Years at Kinondoni District, Tanzania. Science Int. 5: 42–46. 2015.
- 36.Masinde A, Gumodoka B, Kilonzo A, Mshana SE. Prevalence of urinary tract infection among pregnant women at Bugando Medical Centre, Mwanza, Tanzania. Tanzan J Health Res. 2009;11(3):154–9. doi: 10.4314/thrb.v11i3.47704. [DOI] [PubMed] [Google Scholar]
- 37.Chandra Bala Sekharan DDK. Koneru Ratna Kumari, Cecilia Alphonce Joachim: determination of prevalence of urinary tract infection among the pregnant women with Lower Abdominal Pain. UKJPB. 2016;5(2):50–5. [Google Scholar]
- 38.Johansen TE, Botto H, Cek M, Grabe M, Tenke P, Wagenlehner FM, Naber KG. Critical review of current definitions of urinary tract infections and proposal of an EAU/ESIU classification system. Int J Antimicrob Agents. 2011;38:64–70. doi: 10.1016/j.ijantimicag.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Baarz BR, Rink L. Rebalancing the unbalanced aged immune system - a special focus on zinc. Ageing Res Rev. 2022;74:101541. doi: 10.1016/j.arr.2021.101541. [DOI] [PubMed] [Google Scholar]
- 40.Rizwan M, Akhtar M, Najmi AK, Singh K. Escherichia coli and Klebsiella pneumoniae Sensitivity/Resistance pattern towards Antimicrobial Agents in primary and simple urinary tract infection patients visiting University Hospital of Jamia Hamdard New Delhi. Drug Res. 2018;68(7):415–20. doi: 10.1055/a-0576-0079. [DOI] [PubMed] [Google Scholar]
- 41.Kadigi DMMF, Moyo S, Matee MI. Etiology and Antimicrobial susceptibility patterns of bacterial agents causing urinary tract infection in children under five years, dar es salaam. J Biotech. 2020, 2(1).
- 42.Bryce A, Hay AD, Lane IF, Thornton HV, Wootton M, Costelloe C. Global prevalence of antibiotic resistance in paediatric urinary tract infections caused by Escherichia coli and association with routine use of antibiotics in primary care: systematic review and meta-analysis. BMJ. 2016;352:i939. doi: 10.1136/bmj.i939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmed M, Moremi N, Mirambo MM, Hokororo A, Mushi MF, Seni J, Kamugisha E, Mshana SE. Multi-resistant gram negative enteric bacteria causing urinary tract infection among malnourished underfives admitted at a tertiary hospital, northwestern, Tanzania. Ital J Pediatr. 2015;41(1):44. doi: 10.1186/s13052-015-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ministry of Health. Standard Treatment Guidelines & National Essential Medicines List Tanzania Mainland. https://www.medbox.org/pdf/5e148832db60a2044c2d5cdb. Accessed on November 25, 2022.
- 45.Johnson B, Stephen BM, Joseph N, Asiphas O, Musa K, Taseera K. Prevalence and bacteriology of culture-positive urinary tract infection among pregnant women with suspected urinary tract infection at Mbarara regional referral hospital, South-Western Uganda. BMC Pregnancy Childbirth. 2021;21(1):159. doi: 10.1186/s12884-021-03641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X, Niu S, Zhang L. Antimicrobial susceptibilities and clinical characterization of Pseudomonas aeruginosa isolates from urinary tract infections. Urol Int. 2014;93(4):464–9. doi: 10.1159/000358493. [DOI] [PubMed] [Google Scholar]
- 47.Eulambius M, Osward N. Evaluation of Ceftriaxone utilization among selected hospitals in Dar-es-Salaam, Tanzania. JDDT. 2019; 9(6):7 – 1. 2018.
- 48.Ekesa B, Blomme G, Garming H. Dietary diversity and nutritional status of pre-school children from Musa-dependent households in Gitega (Burundi) and Butembo (Democratic Republic of Congo). Afr J Food Agric Nutr Dev. 2011, 11(4).
- 49.Sangeda R, et al. Assessment of implementation of Antimicrobial Resistance Surveillance and Antimicrobial Stewardship Programs in Tanzanian Health Facilities a Year after Launch of the National Action Plan. Front Public Health. 2020;8:554092. doi: 10.3389/fpubh.2020.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this manuscript.