Abstract
Assembly of human T-cell leukemia virus type 1 (HTLV-1) particles is initiated by the trafficking of virally encoded Gag polyproteins to the inner leaflet of the plasma membrane (PM). Gag–PM interactions are mediated by the matrix (MA) domain, which contains a myristoyl group (myr) and a basic patch formed by lysine and arginine residues. For many retroviruses, Gag–PM interactions are mediated by phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]; however, previous studies suggested that HTLV-1 Gag–PM interactions and therefore virus assembly are less dependent on PI(4,5)P2. We have recently shown that PI(4,5)P2 binds directly to HTLV-1 unmyristoylated MA [myr(–)MA] and that myr(–)MA binding to membranes is significantly enhanced by inclusion of phosphatidylserine (PS) and PI(4,5)P2. Herein, we employed structural, biophysical, biochemical, and cell-based assays to identify residues involved in MA–membrane interactions. Our data revealed that the lysine-rich motif (Lys47, Lys48, and Lys51) constitutes the primary PI(4,5)P2–binding site. Furthermore, we show that arginine residues 3, 7, 14 and 17 located in the unstructured N-terminus are essential for MA binding to membranes containing PS and/or PI(4,5)P2. Both the lysine and arginine residues were confirmed to severely attenuate virus-like particle production, but only the lysine residues could be clearly correlated with reduced plasma membrane binding. These results support a mechanism by which HTLV-1 Gag targeting to the PM is mediated by a trio engagement of the myr group, Arg-rich and Lys-rich motifs. These findings advance our understanding of a key step in retroviral particle assembly.
Keywords: Human T-cell leukemia virus type 1 (HTLV-1), human immunodeficiency virus type 1 (HIV-1), Gag polyprotein, matrix (MA) protein, plasma membrane (PM)
Introduction
It is estimated that 10–20 million people worldwide are infected with human T-cell leukemia virus type 1 (HTLV-1), the first oncogenic human retrovirus to be discovered [1, 2]. HTLV-1 infection is strongly associated with a rapidly fatal form of adult T-cell leukemia/lymphoma (ATLL) and HTLV-1 associated myelopathy/tropical spastic paraparesis (HAM/TSP) [3–6]. The cumulative risk of developing either one of these diseases lies between 5–10% over the course of a lifetime [2]. HTLV-1 mainly infects CD4+ T-cells and subsequently propagates via mitotic and clonal expansion or cell-to-cell transmission, which contrasts with infection via free virus particles for human immunodeficiency virus type 1 (HIV-1) [7]. Due to the lower pathogenicity and divergent epidemiology of HTLV-1 compared to HIV-1, this oncogenic retrovirus may have received less attention. The lack of effective treatment for HTLV-1–associated diseases and the risk of future infection outbreaks necessitates the development of new therapeutics that target various phases of the virus replication cycle, including assembly.
Retroviruses encode a polyprotein called Gag which consists of matrix (MA), capsid (CA), nucleocapsid (NC), and short peptides. Post synthesis, Gag polyproteins for most retroviruses are targeted to the plasma membrane (PM) for assembly and virus release [8–21]. Gag–membrane interactions are mediated by the MA domain which contains an N-terminal myristoyl (myr) group and a highly basic region (HBR) [22–24]. For many retroviruses, Gag targeting to the PM was shown to be dependent on phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) [17, 19, 22, 25, 26], an essential signaling lipid in the inner leaflet of the PM [27]. For HIV-1, depletion of PI(4,5)P2 from the PM via overexpression of polyphosphoinositide 5-phosphatase IV (5ptaseIV), an enzyme that cleaves the phosphate group at the D5 position of PI(4,5)P2, led to retargeting of Gag from the PM to late endosomes [14]. Subsequent studies showed that 5ptaseIV also reduced the overall membrane binding of HIV-1 Gag [13]. Dependence of Gag assembly on PI(4,5)P2 has been recapitulated for other retroviruses including HIV-2 [19], Mason-Pfizer monkey virus [26, 28], murine leukemia virus [17], feline immunodeficiency virus [29], and avian sarcoma virus [22, 30, 31].
Although HTLV-1 Gag was shown to be localized at the PM and intracellular compartments in HeLa cells [32–35], Gag binding to the PM was less dependent on PI(4,5)P2 than for HIV-1 [36], suggesting that the interaction of HTLV-1 MA with PI(4,5)P2 is not a key determinant for HTLV-1 particle assembly [25, 36]. Chimeric switching of the MA domains of HIV-1 and HTLV-1 Gag proteins transferred PI(4,5)P2 dependence, demonstrating that lipid specificity is mediated by the MA domain of Gag [36]. Additionally, in vitro assays have shown that although PI(4,5)P2 enhanced HTLV-1 Gag binding to liposomes, Gag proteins bound efficiently to liposomes lacking PI(4,5)P2 but containing phosphatidylserine (PS) if a similar overall negative charge was maintained [36]. Subcellular localization studies of HTLV-1 Gag in HeLa cells using dual-color, z-scan fluorescence fluctuation spectroscopy and total internal reflection fluorescence microscopy revealed that HTLV-1 Gag puncta formation was observed at the PM even at lowest measurable concentrations of Gag. Unlike HIV-1 Gag, HTLV-1 Gag targeted to the PM as a monomer and lacked concentration dependent cytoplasmic Gag–Gag association prior to PM binding [37].
The HBR is highly conserved in most retroviral MA proteins and serves as the binding site for the polar head of acidic phospholipids in the inner leaflet of the PM [20, 38, 39]. It was shown that even a single amino acid substitution can strongly reduce MA binding to membranes containing PI(4,5)P2 and subsequently disrupt PM targeting and assembly of Gag [23, 31]. Myristoylation of MA is also a critical factor for efficient Gag binding to membranes [40–44]. A variety of structural and biophysical studies on retroviral MA binding to phospholipids and membrane mimetics such as bicelles, micelles, liposomes, and lipid nanodiscs provided invaluable insights on key molecular determinants of MA–mediated assembly [17, 19, 23, 24, 26, 28, 29, 31, 45–51].
In a recent study, we provided structural insights into the mechanism by which HTLV-1 Gag is targeted to the PM for assembly [51]. We characterized the interactions of the unmyristoylated MA protein (myr(–)MA) with lipids and liposomes and have shown that HTLV-1 MA contains a PI(4,5)P2 binding site and that PI(4,5)P2 and PS enhance myr(–)MA binding in a synergistic fashion. We also provided confocal microscopy evidence that formation of Gag puncta on the PM and therefore particle production is dependent on myristoylation [51]. In that study, the PI(4,5)P2 binding site was mapped to lysine- and arginine-rich motifs. However, it is still unclear which residues directly interact with PI(4,5)P2 and/or PS, how the interactions with PI(4,5)P2 modulate membrane binding, and whether specific residues in the HBR play key roles in membrane binding and therefore virus assembly and particle production.
In this report, we employed structural, biophysical, biochemical, and cell-based assays to identify residues involved in the interactions of HTLV-1 myr(–)MA and Gag with lipids and membranes. Our data revealed that the lysine-rich motif (Lys47, Lys48 and Lys51) constitutes the primary PI(4,5)P2–binding site. Furthermore, we show that arginine residues 3, 7, 14, and 17 located in the unstructured N-terminus are essential for MA binding to membranes containing PS and/or PI(4,5)P2. We also confirm that alanine substitutions of the lysine and arginine residues severely attenuate virus-like particle production, but only the lysine residues could be clearly correlated with reduced plasma membrane binding. These studies led us to propose a model in which HTLV-1 Gag targeting to the PM is mediated by a trio engagement of the Arg-rich and Lys-rich regions. Altogether, these findings advance our knowledge of a key step in retroviral particle assembly.
Results
Model of myr(–)MA99 bound to inositol 1,4,5-trisphosphate (IP3)
In a recent study, we characterized the interactions of the HTLV-1 myr(–)MA with lipids and liposomes and have shown that MA contains a PI(4,5)P2 binding site [51]. The PI(4,5)P2 binding site was mapped to a region involving multiple basic residues including a Lys-rich motif in helix II (Lys47, Lys48, and Lys51) and and Arg-rich motif in the unstructured N-terminus (Arg14 and Arg17). We also found that the acyl chains of PI(4,5)P2 do not appear to play a role in the interaction with myr(–)MA, and that interactions were mediated by the headgroup of PI(4,5)P2 [51]. A major aim of this study is to identify key residues involved in lipid and membrane binding. Due to technical challenges, we were unable to produce soluble, homogenous and monodisperse HTLV-1 myrMA for structural studies. Therefore, previous [51] and present studies were conducted with either a full-length myr(–)MA or with a myr(–)MA construct lacking the unstructured proline-rich C-terminus [myr(–)MA99] (Fig. 1).
Figure 1. HTLV-1 MA sequence and mutants.

(A) MA protein sequence with the lysine and arginine residues highlighted in blue. (B) Mutant constructs used in this study.
First, we attempted to determine the structure of the myr(–)MA99 in complex with a PI(4,5)P2 analog with truncated acyl chains (tr-PI(4,5)P2). However, we were unable to detect unambiguous intermolecular NOEs between the side chains of Lys and Arg residues and tr-PI(4,5)P2 due to signal broadening and possibly the large distance (> 5Å) between the inositol ring protons and side chains of Arg and Lys residues. Therefore, we performed docking analysis on the myr(–)MA99 protein with IP3, the polar headgroup of PI(4,5)P2. A model of the myr(–)MA99-IP3 complex was calculated using HADDOCK based on residues that exhibited significant 1H -15N chemical shift perturbations (CSPs) in the IP3 titration assay [51]. HADDOCK accounts for side-chain flexibility at the binding interface; however, it does not account for possible conformational changes in the protein backbone such as the flexible N-terminus, which may be required for the Arg-rich motif to interact with IP3. Therefore, all N-terminal Arg residues were excluded as active residues for docking purposes (see Materials & Methods for more details). Calculations yielded 195 structures in four clusters. Structures were clustered based on the positional root-mean-square deviation (RMSD) cut-off of 1 Å at the interaction interface. The first cluster contained 175 structures (Table S1), indicating a preferred binding orientation of IP3, where the phosphate groups on the 4th and 5th carbon are sandwiched between the side chains of Lys48 and Lys51, and Lys47 and Lys51, respectively (Fig. 2). Additionally, His44 appears to be involved in orienting the phosphate group on the 1st carbon. Altogether, these results provided additional support that PI(4,5)P2 binding to MA is mediated by interactions of the headgroup with the lysine-rich motif.
Figure 2. A model of HTLV-1 myr(–)MA99 bound to IP3.
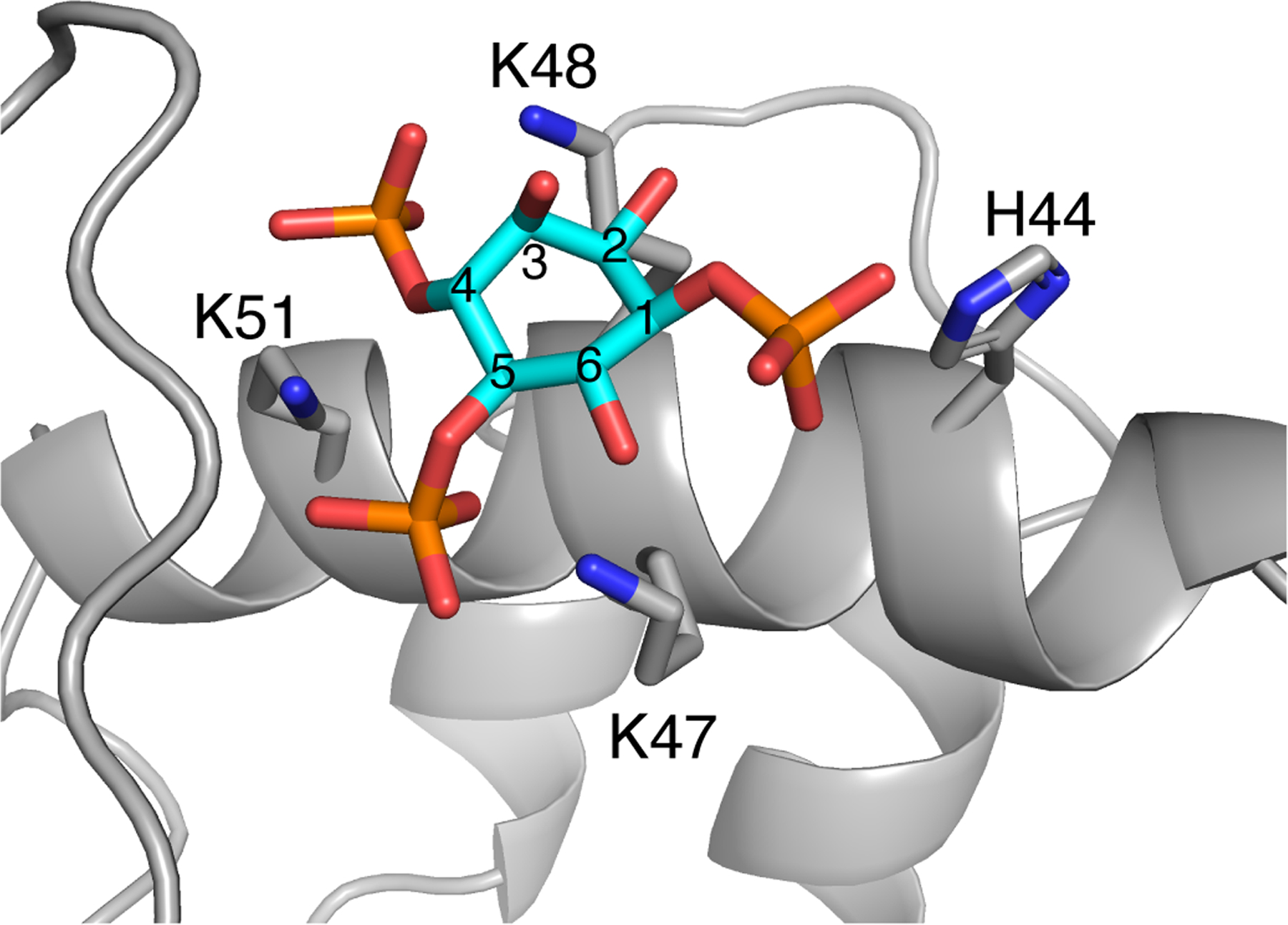
A representative structure from the first HADDOCK cluster of IP3 docked to myr(–)MA99. Residues capable of forming hydrogen bonds with IP3 are represented as sticks. Carbon atoms of the IP3 inositol ring are numbered (cyan).
Molecular dynamics simulations of myr(–)MA99 bound to IP3
Our recent NMR studies indicated that signals corresponding to Arg14 and Arg17 exhibited significant CSPs upon binding of IP3 [51]. To investigate the involvement of the N-terminal arginine residues in IP3 binding, we employed molecular dynamic (MD) simulations using the myr(–)MA99–IP3 model obtained from HADDOCK as starting coordinates. The trajectories of three simulations with identical starting parameters were calculated for a total of 300 ns each. Plotting of RMSD values for Cα atoms of the entire protein structure as a function of time showed that myr(–)MA99 did not undergo significant structural changes during the simulations as RMSD values remained below 1 nm for all three simulations (Figs. 3A and S1). The proximity of the Lys and Arg residues to IP3 over time was measured by calculating the distance of the Cζ of the N-terminal arginines and the Cε of lysines to the center of geometry (COG) of IP3, respectively (Fig. 3B,C). We observed a stabilization of the distance of all N-terminal arginines to IP3 within short timeframes, indicating that the arginine side chains form stable interactions with IP3 (Fig. 3B).
Figure 3. MD simulations of myr(–)MA99 bound to IP3.
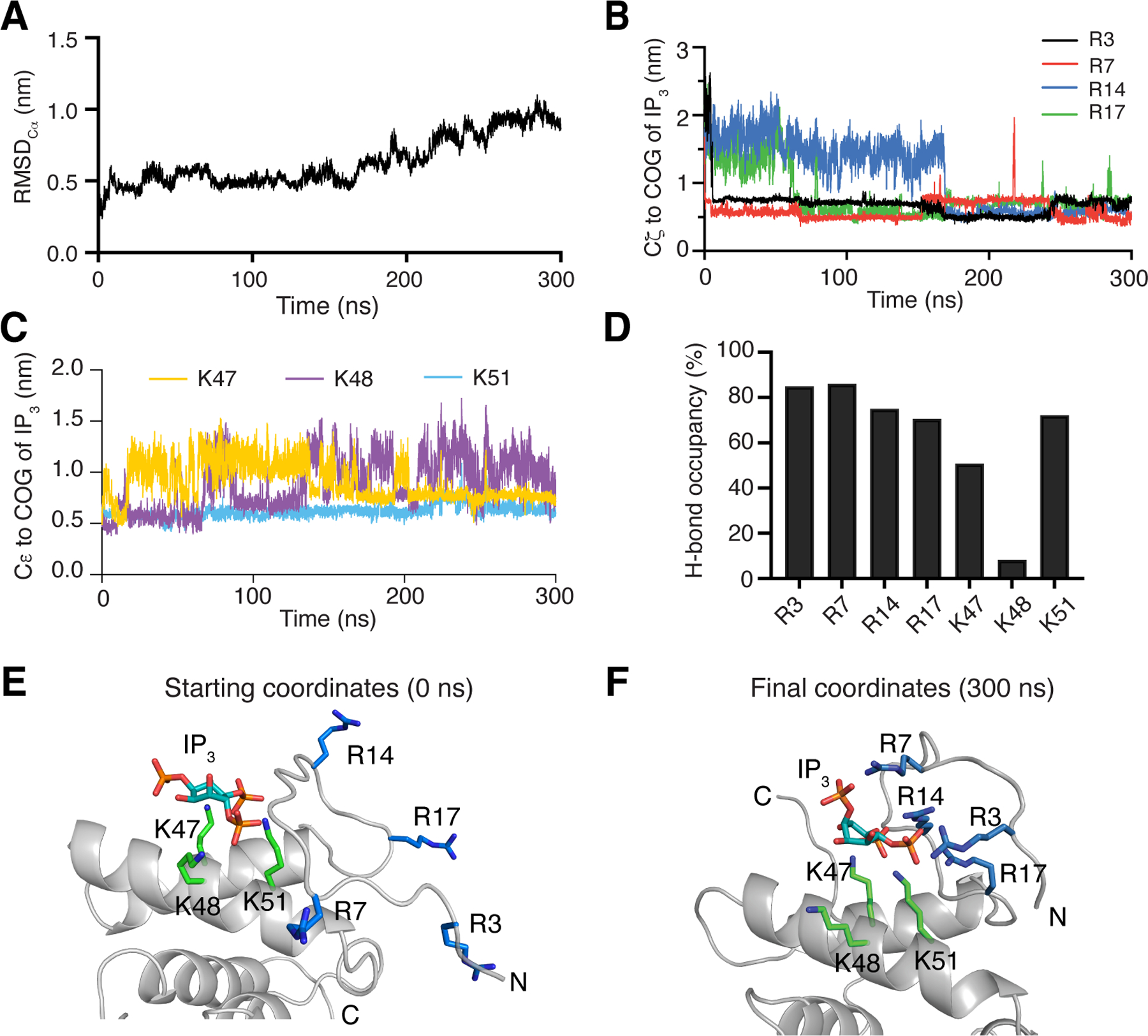
(A) RMSD values of Cα carbons of MA over time. (B) Distance of arginine side-chain Cζ carbons to the center of geometry (COG) of IP3 over time. (C) Distance of Cε lysine side-chain carbons to the COG of IP3. (D) H-bond occupancy of the equilibrated state (distance of all arginines to IP3 < 1 nm), analyzed using VMD. (E) Starting coordinates of the simulations. (F) Final coordinates after 300 ns of simulations. Arginine (blue) and lysine (green) residues are represented as sticks.
Interactions between IP3 and the side chains of Lys47, Lys48, and Lys51 were less stable, exhibiting more frequent dissociation from IP3 (Fig. 3C). Hydrogen bonds (H-bonds) between MA99 and IP3 were analyzed for the post-equilibrated state when the distance between arginine side chains to IP3 was consistently below 1 nm. We observed a high H-bond occupancy for the side chains of all N-terminal arginines (3, 7, 14, and 17) to IP3 (Fig. 3D). Distance and H-bond occupancy analysis for Lys47, Lys48, and Lys51 revealed that only two lysine residues form stable H-bonds with IP3 while the third remains largely disengaged throughout the simulation (Fig. 3D). However, we did not observe IP3 to be favoring a particular pair of lysines for this interaction, as both Lys47 and Lys48 could be engaged or disengaged, depending on which simulation was analyzed (Fig. S1). Similarly, we did not observe sequential arginine–IP3 interactions, as the order and timing of arginine–IP3 interactions varied among the simulations (Fig. S1). Therefore, the sequence of these events is likely of stochastic nature and no conclusions were drawn about the importance of certain residues over others, based on these simulations. However, all three simulations eventually converged to a point where all arginine residues interacted with IP3 which remained bound to the lysine rich motif (Fig. 3E–F). Due to the stronger H-bond capacity of the guanidinium group of arginine compared to the amino group of lysine, interactions between arginines and IP3, once established, were generally more stable than with Lys47, Lys48, and Lys51. Altogether, the MD simulations revealed that all arginine residues in the flexible N-terminus can interact with IP3 simultaneously.
Identification of MA residues critical for PI(4,5)P2 binding
Molecular docking and MD simulations provided a good initial model to guide the mutagenesis studies. To examine which residues in the Lys- and Arg-rich motifs are critical for PI(4,5)P2 binding, we constructed mutants with single, double or multiple alanine substitutions in the myr(–)MA protein (Fig. 1). With the exception of signals corresponding to residues in the vicinity of the mutation site, the HSQC spectra obtained for all myr(–)MA mutants are very similar to the spectrum of the wild-type (WT) protein (Fig. S2), indicating that amino acid substitutions had no adverse effect on the structure and folding of the protein. Next, we assessed binding of IP3 to myr(–)MA mutants using 2D 1H-15N HSQC titrations. For simplicity, the CSPs of the His44 signal is shown as an NMR marker (Fig. 4A) since this residue is located in proximity to the Lys-rich motif (Fig. 2). As shown, the His44 signal exhibited variable CSPs upon titration of WT and mutant myr(–)MA with IP3. CSP data were used to determine the dissociation constant (Kd) by fitting the CSPs as a function of IP3 concentration (Fig. 4B–C). Analysis of the NMR data revealed that MA K47A mutant exhibited the weakest binding of the three Lys-to-Ala mutants (~30-fold weaker than WT), whereas the effect of K48A and K51A mutations was rather moderate (Fig. 4). However, substitution of all three lysine residues (m3 mutant) led to severe attenuation of IP3 binding (Fig. 4), indicating that the Lys-rich motif is critical for PI(4,5)P2 binding.
Figure 4. NMR analysis of IP3 binding to WT and mutant myr(–)MA.
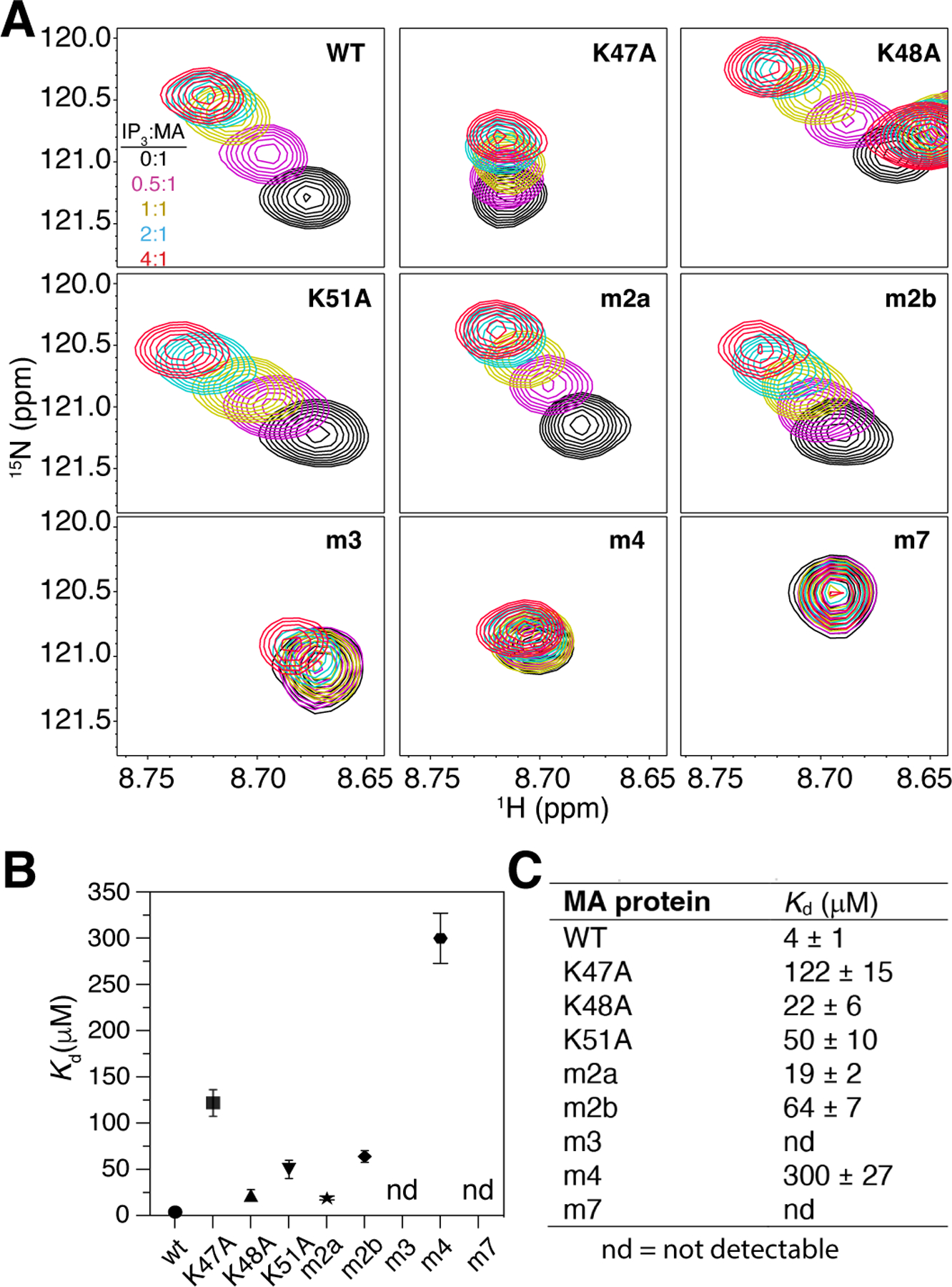
(A) Overlay of 2D 1H-15N HSQC spectra showing the His44 signal upon titration of WT and mutant myr(–)MA with IP3 [100 μM, 35 °C; IP3:MA = 0:1 (black), 0.5:1 (magenta), 1:1 (yellow), 2:1 (cyan), 4:1 (red)] in 20 mM MES (pH 6) and 2 mM TCEP. (B) Graphical and (C) numerical representation of Kd values of WT and mutant myr(–)MA binding to IP3.
We have recently shown that NMR signals corresponding to Arg14 and Arg17 exhibited CSPs upon tr-PI(4,5)P2 titration [51]. MD simulations also suggested that the arginine residues in the N-terminus can potentially interact with IP3. To assess the role of the Arg-rich motif in PI(4,5)P2 binding, we conducted similar 2D NMR titrations on myr(–)MA Arg-to-Ala mutants with IP3. Our NMR data showed that mutation of Arg3 and Arg7 (m2a) and Arg14 and Arg17 (m2b) led to 5- and 16-fold reduction in IP3 binding, respectively (Fig. 4), indicating that Arg14 and Arg17 are more important for IP3 binding. Interestingly, mutation of all four arginine residues (m4) led to a dramatic decrease in affinity (75-fold; Fig. 4), demonstrating that the N-terminal arginines play an important role in IP3 binding, consistent with our MD simulations. Finally, we show that substitution of all seven residues in the Lys- and Arg-rich motifs (m7 mutant) completely abrogated binding of IP3 (Fig. 4). In summary, these findings indicated that the Lys-rich motif is the main docking site for IP3 and that the flexibility of the N-terminal loop allows arginine residues, especially Arg14 and Arg17, to interact with IP3 and therefore enhance binding.
Interaction of myr(–)MA mutants with liposomes
MD simulations and IP3 binding studies suggested that the flexibility of the rather long and unstructured N-terminus allows for potential and variable interactions of all arginine residues with PI(4,5)P2. However, it is not clear whether such interactions are specific to PI(4,5)P2 or can occur with other acidic lipids such as PS. We next sought to delineate the roles of Arg-rich and Lys-rich motifs in the context of a membrane mimetic and whether myr(–)MA interacts distinctly with PI(4,5)P2 and PS. We employed a sensitive 1H NMR–based assay (Fig. 5A) to characterize binding of myr(–)MA mutants to large unilamellar vesicles (LUVs) containing native lipids. This approach was utilized to characterize interaction of WT HTLV-1, HIV-1 and ASV MA proteins to LUVs [23, 31, 51]. The assay allows for measurement of the unbound protein fraction in solution under equilibrium conditions with the LUV–bound fraction, which can provide quantitative binding measurements such as Kd values and important information on the synergy of membrane components and cooperativity of binding [51–53]. Using this assay on the WT myr(–)MA protein, we have recently shown that PI(4,5)P2 and PS do not compete for the same binding site but instead bind to distinct sites, or at least by different mechanisms, enhancing myr(–)MA binding in a synergistic fashion [51].
Figure 5. Interaction of WT and mutant myr(–)MA with LUVs.
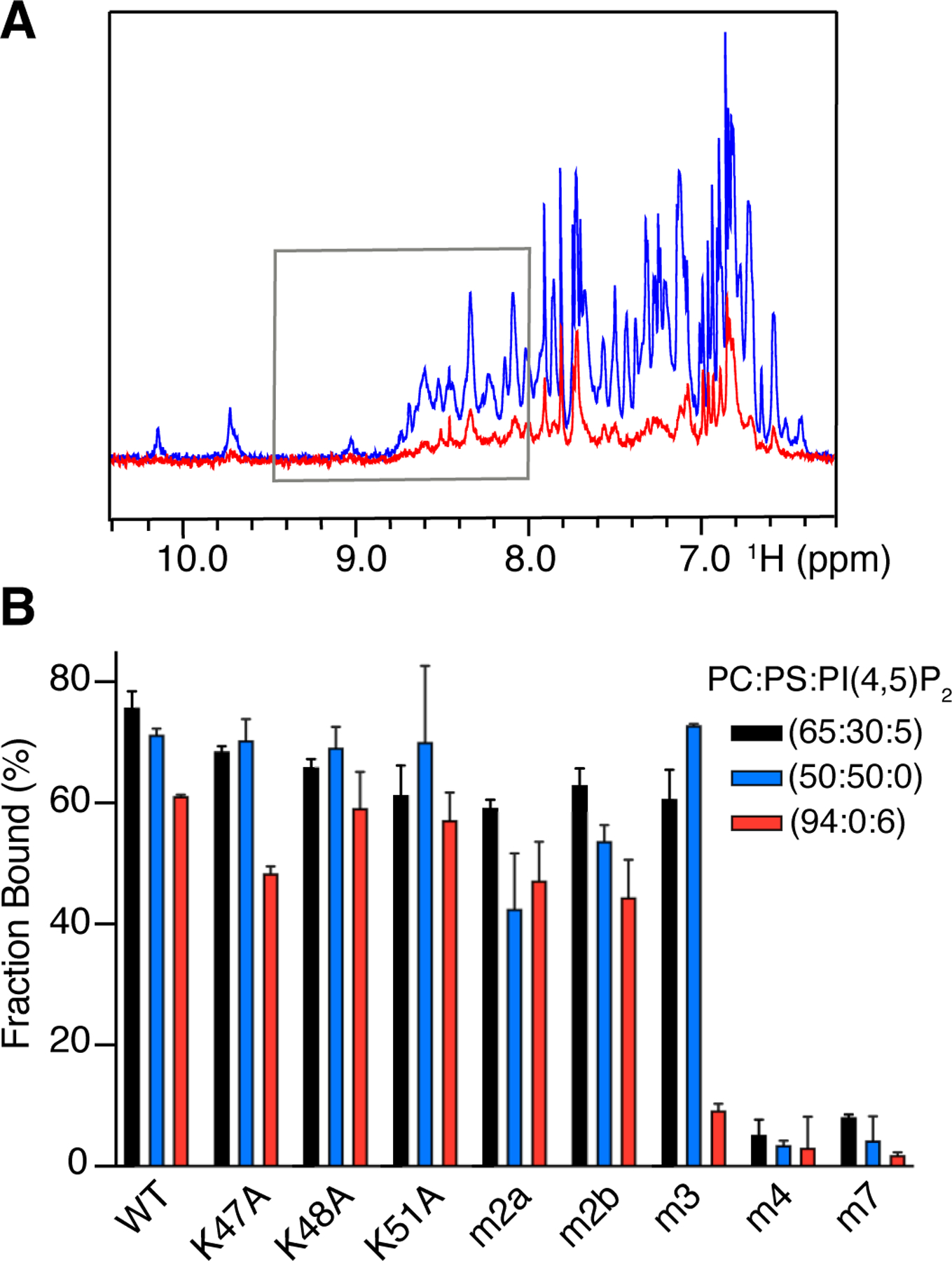
(A) Overlay of 1H NMR spectra of free myr(–)MA (50 µM) and in the presence of 250 ug of LUVs with 60 mol. % PC, 30 mol. % PS, and 5 mol. % PI(4,5)P2 (red). Gray box marks the area of spectra integration. (B) Bar graph of WT and mutant myr(–)MA binding to LUVs with varying composition of PC:PS:PI(4,5)P2 [black (65:30:5), blue (50:50:0), and red (94:0:6)]. Error bars were calculated from three replicates.
Herein, we assessed binding of myr(–)MA mutants to LUVs containing variable compositions of PI(4,5)P2 and PS. Titration of LUVs containing PI(4,5)P2 to single lysine myr(–)MA mutants resulted in only a moderate reduction of the LUV–bound protein fraction for the three lipid compositions tested (Fig. 5B). Compared to the WT myr(–)MA protein, noticeable differences were only observed for the K47A mutant (~10% less protein bound to LUVs; Fig. 5B). Mutation of all three lysines (m3) inhibited binding of PI(4,5)P2 LUVs dramatically (Fig. 5B). These results are consistent with the data obtained for the mutant proteins in the IP3 NMR titration assay (Fig. 4). Next, to test whether PI(4,5)P2 binding is confined to the Lys-rich motif we employed the NMR assay with LUVs made with 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC) and PS. Strikingly, mutations of lysine residues (single or triple) had little to no effect on binding to LUVs containing 50% PS (Fig. 5B), suggesting that lysine residues are not key to PS binding and that binding of myr(–)MA to PS is perhaps mediated by the N-terminal arginines. Based on these findings, we hypothesized that the Lys- and Arg-rich motifs may play distinct roles in binding to membrane binding. To test this hypothesis, we assessed binding of double arginine mutants (m2a and m2b) to LUVs containing either PS or PI(4,5)P2 and observed a decrease in affinity (Fig. 5B). The m2a and m2b mutants bind to LUVs containing PI(4,5)P2 or PS with similar affinities (Fig. 5B). However, mutation of all four arginines (m4) resulted in a complete loss of binding to LUVs containing either of PS, PI(4,5)P2, or PS:PI(4,5)P2 (Fig. 5B). Likewise, mutation of all lysine and arginine residues (m7) abrogated binding to all tested LUVs. The lack of m4 binding to any of the LUVs indicates that the N-terminal arginines are essential for binding to PI(4,5)P2 and PS. In contrast, the role of the lysine-rich motif appears to be confined to mediating interactions with PI(4,5)P2. However, without the contribution of the N-terminal arginines, the affinity to PI(4,5)P2 is not sufficient to facilitate noticeable binding to LUVs containing only PI(4,5)P2. Altogether, these data support functionally distinct membrane interacting surfaces on HTLV-1 MA, a specific PI(4,5)P2–binding site (Lys-rich motif), and a PS–binding site (Arg-rich motif) that also significantly enhances the affinity to PI(4,5)P2. This is the first example in which a retroviral MA protein binding to membranes is mediated by two distinct motifs that bind to distinct lipids on the membrane.
Virus-like particle (VLP) production of HTLV-1 Gag mutants
To investigate the contributions of the PI(4,5)P2 (Lys-rich) and PS (Arg-rich) binding motifs to the interaction with the PM, we performed several assays examining membrane binding in cultured cells. First, PM binding ability was assessed indirectly by examining VLP production of HTLV-1 Gag mutants described in Fig. 1. As interaction with the PM is requisite for particle production, the reduction in VLP production may suggest a decreased ability to interact with the PM. VLPs were produced by transfecting 293T cells with plasmids expressing HTLV-1 Gag (WT or mutant) and envelope (Env) at a 5:1 ratio. VLPs were collected from the cell culture supernatant and detected by using an immunoblot with an anti-HTLV-1 p24 antibody. Particle production efficiency was determined by normalizing the p24 detected in VLPs to p24 levels detected from cell lysates that were normalized by GAPDH expression level. Differences in particle production compared to WT were determined by using the one-way ANOVA multiple comparisons test. HTLV-1 Gag G2A, which lacks myristylation (and efficient membrane binding), resulted in a severe reduction in particle production (~ 9% of WT; Fig. 6), consistent with previous studies [34, 51, 54]. Alanine substitutions of the Lys- or Arg-rich motifs virtually eliminated particle production, as the m3 and m4 Gag mutants exhibited less than 1% of particle production compared to WT Gag (Fig. 6). This result agrees with a previous study that reported no particle production for HTLV-1 Gag mutants with lysine-to-isoleucine substitutions at positions 47, 48, and 51 [34]. The severe reduction in particle production for the m3 Gag mutant supports the conclusion that the PI(4,5)P2 binding site significantly contributes to PM binding, a step that is crucial for HTLV-1 particle production. Mutation of the Arg-rich motif resulted in a HTLV-1 Gag protein variant (m4) that was poorly expressed in cells (Fig. 6A). Therefore, the reduction in m4 Gag particle production may be due to reduced levels of Gag expression (and/or reduced Gag stability) rather than that of reduced membrane binding.
Figure 6. Particle production analysis.
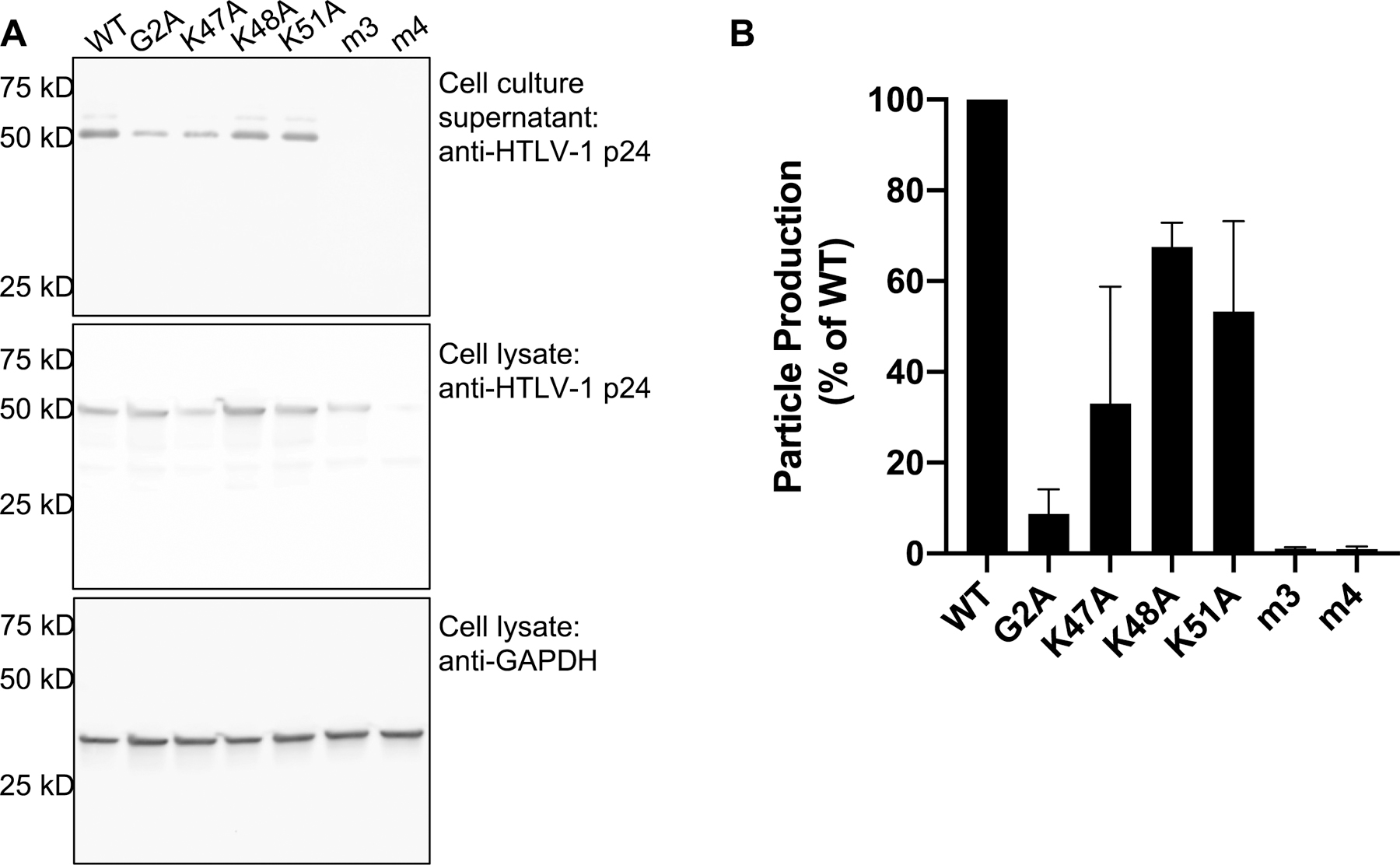
(A) Representative immunoblot of cell lysate and cell culture supernatants from 293T cells transfected with WT and mutant HTLV-1 Gag. Gag was detected from cell culture supernatants and cell lysates by using an anti-Gag p24 antibody. GAPDH was detected in cell lysates by using an anti-GAPDH antibody. (B) Particle production efficiency was determined by analyzing immunoblot band intensities of Gag from cell culture supernatant normalized to the Gag band intensity from cell lysates (normalized to the GAPDH band intensity from cell lysates). The average particle production efficiency is expressed as the percent of WT particle production. Error bars represent the standard deviation of three independent biological replicates.
To further investigate the role of the PI(4,5)P2 binding motif, we analyzed particle production of single amino acid substitutions (i.e., K47A, K48A, or K51A). All single amino acid substitutions resulted in a decrease in particle production, but less severe than the HTLV-1 Gag m3 mutant. Among the single mutants, the K47A substitution led to the most significant phenotype, reducing particle production to 33% of WT (Fig. 6). Particle production for the K48A and K51A mutants was reduced to 68% and 53% of WT, respectively (Fig. 6). These results are consistent with the NMR findings obtained for the binding of MA single mutants to IP3, where K47A exhibited the most significant change in binding affinity (Fig. 4). Except for the K48A mutant which reduced particle production to 7% of WT, a previous study reported similar observations in particle production with MA mutants in the context of a full-length molecular clone [34]. The discrepancy in the K48A mutant data is probably due to different viral constructs (i.e., VLP versus molecular clone) or cell type differences. The intermediate reduction in particle production attributed to the individual MA residues (i.e., Lys47, Lys48, and Lys51) implies that while each residue contributes to PI(4,5)P2 and PM binding, there is no single residue that is dominant in mediating interaction with the PM.
Contributions of MA basic residues in HTLV-1 Gag binding to the PM
To directly investigate the role of the Lys- and Arg-rich motifs in the interaction of HTLV-1 Gag with the PM, we examined Gag–PM binding in cell culture by using three assays. First, we assessed the subcellular localization of HTLV-1 Gag by transfection of HeLa cells with a 3:1 ratio of unlabeled Gag along with Gag having a C-terminal EYFP tag and analyzed Gag distribution in cells using z-stacks collected by confocal microscopy. Lack of Gag binding to the PM can eliminate Gag puncta formation given that Gag translocation to the PM is requisite for Gag multimerization at the PM [37, 55]. The HTLV-1 Gag G2A construct showed reduced membrane binding, leading to a diffuse cytoplasmic expression pattern (Fig. 7A). This diffuse cytoplasmic localization of the HTLV-1 Gag G2A mutant was previously reported in several cell types [34, 51]. We observed that single amino acid substitutions of lysine residues resulted in punctate localization of Gag, comparable to WT, with no serious defect in membrane binding (Fig. 7A, C). Substitution of all three lysine residues (i.e., m3) or all four N-terminal arginine residues (i.e., m4) did not impact Gag puncta formation (Fig. 7A). There was no apparent difference in m4 Gag protein expression compared to that of WT and to the other mutants, as determined by confocal microscopy. Therefore, the reduction in particle production observed with the m3 mutant as well as the individual lysine amino acid substitutions (Fig. 6) is not strongly correlated with Gag puncta formation.
Figure 7. Gag localization in cells and membrane binding.
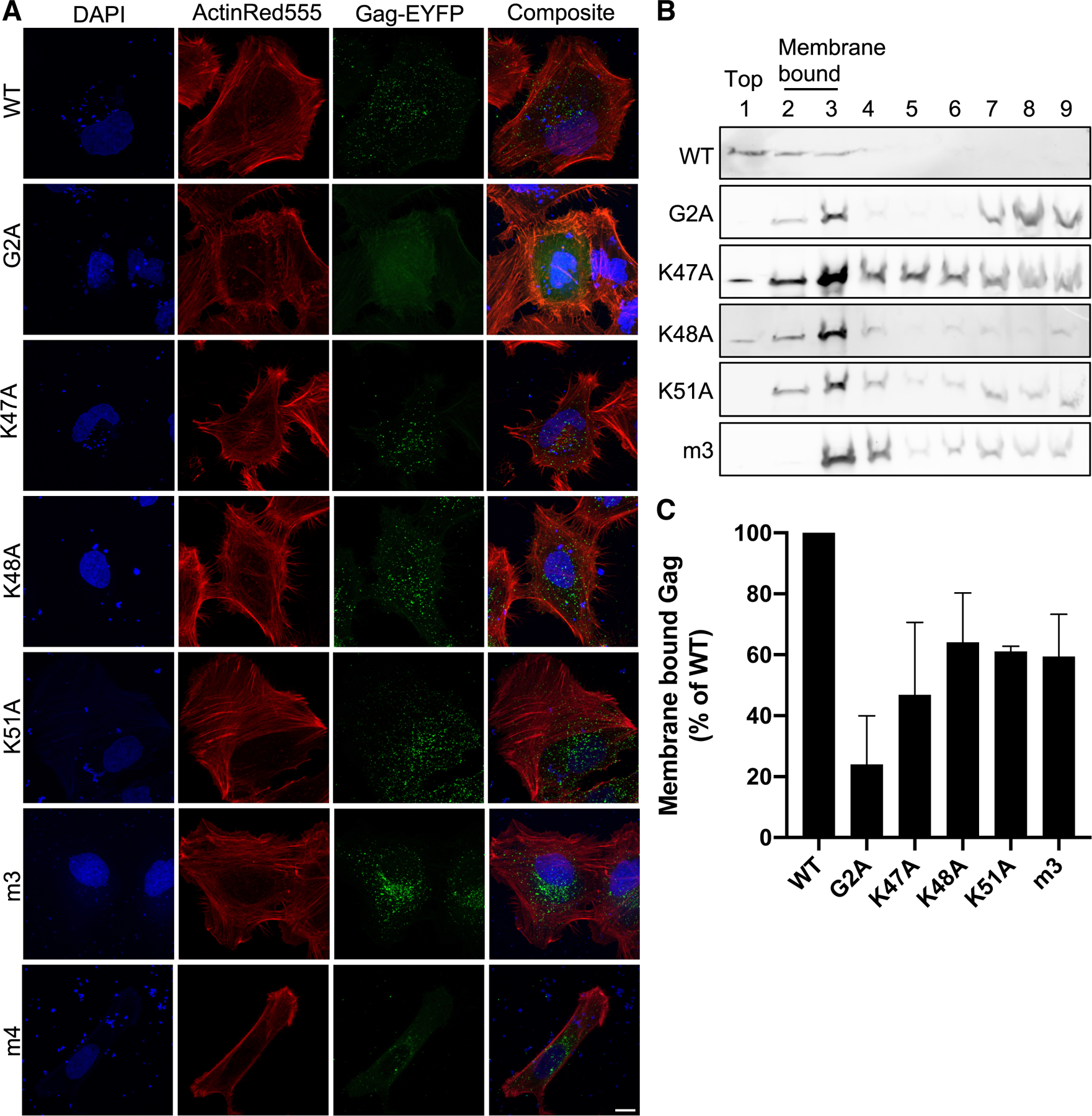
(A) Subcellular localization of Gag. HeLa cells were transfected with HTLV-1 Gag-EYFP. Gag (green), nuclei (blue; stained with DAPI), and actin (red; stained with ActinRed555) were analyzed in fixed cells by confocal microscopy. Shown are z-projections of a representative z-stack. Scale bar = 10 µm. (B) Immunoblot analysis. Hela cell lysates were fractionated in sucrose gradients. Gag was detected by using an anti-HTLV-1 p24 antibody. Membrane bound Gag is present in fractions 2 and 3 and unbound Gag in fractions 7–9. Fraction 1 = top fraction; fraction 9 = bottom fraction. Immunoblot is representative of three independent experiments. (C) Membrane-associated Gag. The relative percentage of membrane-associated Gag for each mutant was determined by the sum of Gag detected in fractions 2 and 3 out of the sum of Gag detected in all fractions normalized to WT Gag. Error bars represent the standard deviation from three independent replicates.
Second, we used equilibrium flotation centrifugation to further investigate Gag–membrane interactions. This assay separates membrane-bound Gag from cytoplasmic Gag. Cell lysates from HeLa cells transiently expressing HTLV-1 Gag and Env were placed at the bottom of a sucrose gradient, where ultracentrifugation results in membrane-bound Gag migrating up to the interface between the 10 and 65% sucrose layers [42, 56, 57]. Following ultracentrifugation, fractions were assessed for Gag–membrane interactions by immunoblot analysis (Fig 7B). Fractions 2 and 3 were at the interface of the 10 and 65% sucrose layers and would contain membrane bound proteins. Calculating the percent of total Gag in fractions 2 and 3 allows for the determination of the membrane binding ability of Gag. Binding of the G2A construct was reduced to 24% of WT (p = 0.002; one-way ANOVA) (Fig. 7C). Single alanine substitutions for Lys47, Lys48, and Lys51 reduced Gag membrane binding by 47% (p = 0.0033), 64% (p = 0.0414), and 61% (p = 0.0266) of WT, respectively (Fig. 7C). The m3 mutant exhibited reduced Gag membrane binding to 59% (p = 0.0208) of WT Gag (Fig. 7C). This observation implies the lysine-rich motif is required for PM binding. The approximate 40% reduction in membrane-bound Gag suggests involvement of other residues in the interaction. The low level of m4 expression in cells prohibited efficient analysis of Gag–membrane binding by equilibrium flotation centrifugation.
Third, we investigated the interaction of Gag with the PM by using two-photon z-scan fluorescence microscopy [55], which enables direct and quantitative monitoring of Gag–PM binding in living cells. Briefly, a z-scan was acquired by moving the two-photon excitation volume vertically through a cell expressing HTLV-1 Gag-EYFP (Fig. 8A). The resulting fluorescence intensity profile versus height through the cell encodes the vertical distribution of fluorescent Gag along the scan trajectory. Intensity traces acquired from z-scans of the WT and each Gag mutant were fit to a model of the cell that allows fluorescence to originate from the bottom PM, the cytoplasm, and the top PM. This model also incorporates the effect of the calibrated shape of the two-photon excitation volume on the shape of the intensity traces. The fit decomposes the intensity trace into its components originating from the cytoplasm and from the top or bottom PM (Fig. 8B). Amplitudes of the decomposed intensity traces determine the membrane intensity fraction m (Materials and Methods).
Figure 8. Cellular distribution of WT and Gag mutants by z-scan fluorescence imaging.
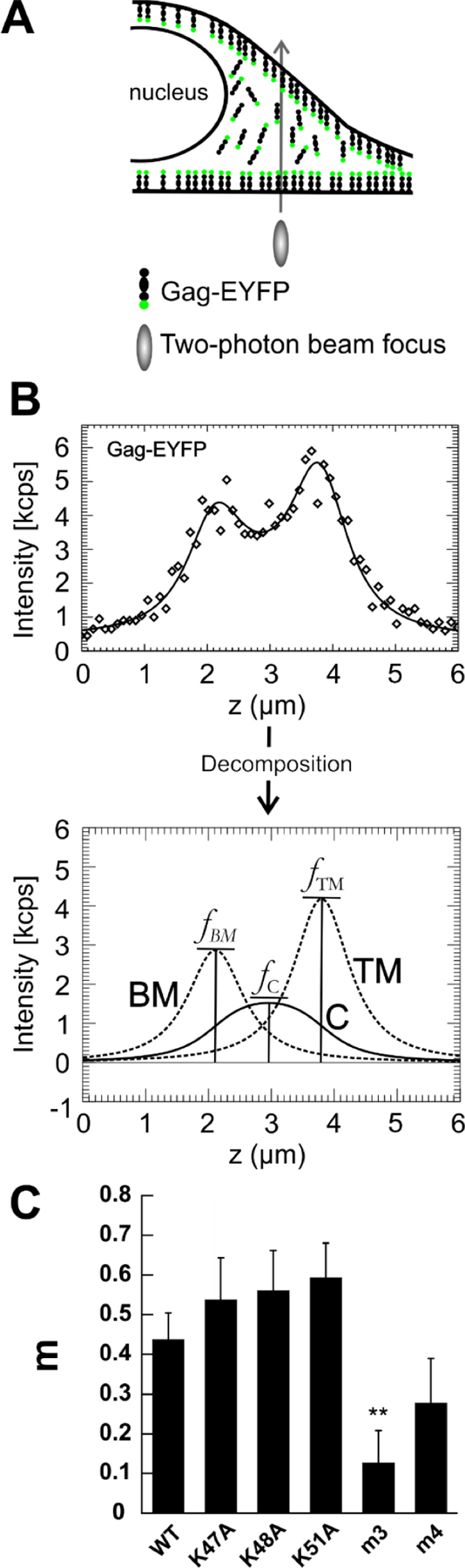
(A) Diagram of the cross-section of an adherent cell depicting the cellular distribution of Gag. Shown is membrane-associated and cytosolic Gag-EYFP, as well as the two-photon focus and its vertical movement through the cell along the z-axis. (B) Fluorescence z-scan intensity trace and decomposition. The intensity trace (diamonds) of the total fluorescence measured along the z-axis is fit (solid line) to a model which decomposes it into traces representing the signal from the cytoplasm (C), the top plasma membrane (TM) and the bottom plasma membrane (BM), whose amplitudes , , and are used to calculate membrane intensity fractions (m). (C) Membrane intensity fractions. Shown are the weighted average membrane intensity fractions m of WT and Gag mutants. Error bars correspond to the dispersion corrected SEM [67] (n = 20 – 38). The m3 mutant showed a statistically different m relative to WT Gag (p=0.004). All other mutants had values statistically indistinguishable from WT Gag.
The WT HTLV-1 Gag protein has been previously reported to bind to the PM without providing m estimates [37]. Our z-scan study determined an m value of ~0.4 for WT Gag at nanomolar cytosolic concentrations (Figs. 8C and S3). The average m values of the single mutants (K47A, K48A and K51A) were statistically indistinguishable from that of WT (mWT = 0.44 ± 0.07, mK47A = 0.5 ± 0.1, mK48A = 0.56 ± 0.10, mK51A = 0.59 ± 0.09), indicating that single mutations of Lys47, Lys48 or Lys51 did not impact Gag-membrane interactions. In contrast, the m3 mutant was significantly different (mm3 = 0.13 ± 0.08, p = 0.004). While the m4 mutant showed on average a lower m value relative to WT, this reduction was not statistically significant. Taken together, these results highlight the importance of the myr group and the basic amino acid motifs in membrane targeting.
Discussion
The HTLV-1 Gag polyprotein was shown to be localized at the inner leaflet of the PM and intracellular compartments in HeLa cells [32–35]. For many retroviruses, Gag targeting to the PM is mediated by PI(4,5)P2 [17, 19, 22, 25, 26]. For HTLV-1, the finding that Gag binding is less dependent on PI(4,5)P2 led to the suggestion that the MA domain may not contain a well-defined PI(4,5)P2 binding site and that the interaction of MA with PI(4,5)P2 is not a key determinant for HTLV-1 particle assembly [25, 36]. In a recent study, we have shown that the HTLV-1 MA protein binds directly to PI(4,5)P2 and that a large basic surface formed by Lys- and Arg-rich motifs plays a role in PI(4,5)P2 and membrane binding [51]. We have shown that binding of myr(–)MA to PI(4,5)P2 is mediated by the headgroup, demonstrating the electrostatic nature of the interaction. We have also shown that the presence of PS in membranes enhanced the affinity of myr(–)MA to PI(4,5)P2, suggesting that PI(4,5)P2 and PS do not compete for the same binding site [51]. Instead, we hypothesized that PI(4,5)P2 and PS bind to distinct sites, thereby enhancing myr(–)MA binding in a synergistic fashion. Confocal microscopy data have shown that Gag is localized to the inner leaflet of the PM of the infected cell, while the G2A Gag mutant lacking myristoylation is diffuse and cytoplasmic [51]. These findings provided significant insights into the mechanism by which HTLV-1 Gag protein binds to the inner leaflet of the PM for assembly and release of immature particles. However, several mechanistic details of Gag assembly were unresolved. Among these are the specific roles of the Lys- and Arg-rich motifs in HTLV-1 MA binding to membranes, and whether Gag–membrane binding is mediated by engagement of PI(4,5)P2 and PS to distinct basic motifs on MA.
In this report, we have shown that the lysine-rich motif in HTLV-1 MA constitutes the primary PI(4,5)P2 binding site, and that the arginine-rich motif is essential for MA binding to membranes containing PS and/or PI(4,5)P2. Substitution of residues in the Lys- and Arg-rich regions reduced binding to membranes and severely attenuated VLP production, indicating that these sites are critical for virus assembly. Analysis of molecular docking of the myr(–)MA protein with IP3 suggested Lys47, Lys48, and Lys51 are involved in IP3 binding. While we were able to establish that Lys47 is more important than Lys48 and Lys51 for IP3 binding, single alanine substitutions only had a moderate effect on binding of myr(–)MA to IP3 or to PI(4,5)P2–containing LUVs. Only substitution of all three lysines had a detrimental effect on binding to IP3 and LUVs containing PI(4,5)P2. This result is consistent with the MD simulations conducted on the myr(–)MA99–IP3 complex, which revealed that at least two lysines are needed for IP3 binding (Figs. 3 and S1). We have also shown that binding of IP3 to myr(–)MA is further enhanced by interactions with the Arg-rich motif, especially Arg14 and Arg17. This appears to be possible due to the unusually long, unstructured N-terminus of MA which allows for the conformational flexibility to facilitate these interactions. This feature appears to be unique for HTLV-1 MA as it was not observed for other retroviral MA proteins.
MD simulations have shown that arginine residues can potentially interact with IP3, which is reflected by the significant decrease in the binding affinity of m2b and m4 mutants. These arginines also seem to play a role in binding of myr(–)MA to PI(4,5)P2–containing LUVs, as the m4 mutant was deficient in binding to LUVs containing PI(4,5)P2. We have recently shown that MA binding to membranes is driven by electrostatic interactions as incorporation of PS yielded a similar fraction of protein bound as to PI(4,5)P2–enriched LUVs, if total negative charge was maintained [51]. Additionally, the observation that PI(4,5)P2 and PS do not compete for the same binding site and that they bind to myr(–)MA binding in a synergistic fashion led to the hypothesis that the arginine residues are involved in PS binding. Indeed, disruption of the PI(4,5)P2 binding site by substitution of the lysine residues (m3) only abrogated binding to LUVs containing PI(4,5)P2 as the LUVs containing PS interacted with myr(–)MA efficiently (Fig. 5B). This result indicated that MA interactions with PS are mediated by the Arg-rich motif. It is reasonable to conclude, based on our data, that in the absence of PS, arginines can interact with PI(4,5)P2 even though the Lys-rich motif is considered the primary binding site. However, in the presence of both lipids, arginines may preferentially interact with PS. Taken together, these findings support a mechanism by which HTLV-1 Gag targeting to the PM is mediated by a trio engagement of the myr group, structured Lys-rich, and unstructured Arg-rich motifs (Fig. 9).
Figure 9. Model of HTLV-1 myrMA bound to membrane.
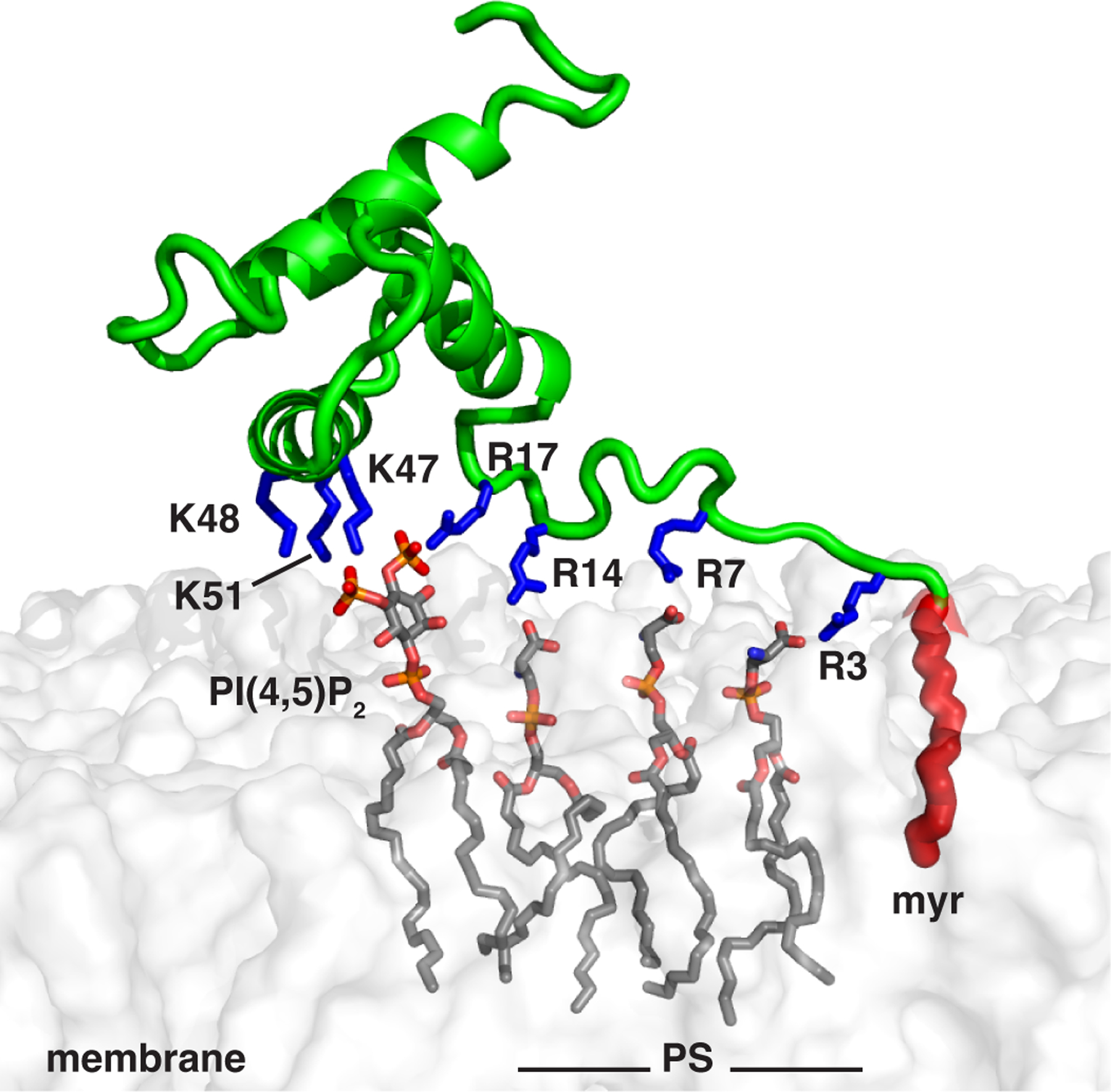
myrMA–membrane interactions are mediated by the myr group (red) and electrostatic interactions between the Lys- and Arg-rich motifs with negatively charged PM lipids (PS and PI(4,5)P2). Membrane was constructed using CHARMM-GUI membrane builder [69].
Our cell-based data support the conclusion that efficient HTLV-1 Gag targeting to the PM is mediated by the myr group and Lys-rich motif. The G2A and m3 mutants implicate these residues in PM targeting and particle production. Equilibrium flotation centrifugation and fluorescence z-scan analyses further demonstrated the importance of the myr group and the Lys-rich motifs in membrane targeting of the G2A and m3 Gag mutants, respectively. Reduced membrane binding was variable for the single mutants in the equilibrium flotation centrifugation and z-scan analysis. The equilibrium flotation centrifugation assay was performed on populations of cells with various Gag expression levels, whereas fluorescence z-scan analysis was performed on individual cells at low expression levels of Gag. The role of the Arg-rich motif cannot be fully interpreted from our cell culture assays, as reduced m4 Gag protein expression (as determined by immunoblot analysis) does not allow for a rigorous analysis of particle production or membrane binding by equilibrium flotation centrifugation. It is notable that detection of fluorescently-labeled m4 Gag did not reveal differences in expression relative to WT Gag and other mutants. This could be due to the method of detection or by the relative sensitivity of detection of Gag-EYFP by both confocal microscopy and by fluorescent z-scan analysis. In the specific case of the fluorescently labeled m4 Gag variant, the role of the Arg-rich motif in PM binding was not corroborated. Gag subcellular localization by confocal microscopy may not readily reveal a PM binding defect, as Gag puncta can form intracellularly, particularly at higher expression levels. A Gag mutant having substitutions at Lys or Arg residues can still be myristoylated, which would still allow for Gag puncta formation in the absence of efficient particle production. Fluorescent z-scan analysis shows a decrease in m4 binding to membranes, but it is not statistically significant.
As discussed above, our data indicated that the interaction of HTLV-1 myr(–)MA with PI(4,5)P2 is confined to a Lys-rich motif, and that substitutions in this region do not interfere with PS binding. However, elimination of these lysine residues was detrimental for VLP production, therefore PI(4,5)P2 appears to be a major determinant for HTLV-1 particle production. At first glance, this result appears to not be in complete agreement with previous studies that suggested PI(4,5)P2 may be less important for HTLV-1 than for HIV-1 [36]. One possible explanation is that 5ptaseIV, which hydrolyzes the 5’-phosphate group of PI(4,5)P2, does not completely eliminate the negative charge of PI(4,5)P2 in the PM. It is likely that residual charge of PI(4)P is sufficient to mediate HTLV-1 Gag binding to the PM and subsequent VLP release, albeit at reduced levels [36]. We cannot rule out additional roles of the Lys- and Arg-rich motifs in virus production, independent of PI(4,5)P2. For example, previous studies have shown that two MA mutations (R17L and K48I) did not alter the binding of HTLV-1 Gag to membranes but caused profound defects of particle release and infectivity [34]. Further studies to investigate the interplay of PI(4,5)P2 vs. PS and Lys- vs. Arg-rich motifs in HTLV-1 Gag binding to the PM and virus assembly are warranted.
In summary, our data support a novel mechanism by which HTLV-1 Gag targeting to the PM is mediated by the engagement of the myr group and the Arg- and Lys-rich motifs. These observations advance our understanding of a key step in deltaretroviral particle assembly and highlight important fundamental differences in the HTLV-1 particle assembly pathway from other retroviruses, including lentiviruses such as HIV-1. This argues for the importance of comparative analysis in shedding light into the diversity of mechanistic differences among retroviruses regarding virus-host cell interactions and viral replication. Elucidation of the molecular determinants of MA–membrane interactions may help in the development of new antiviral therapeutic agents that inhibit Gag assembly on the PM and ultimately virus production.
Experimental procedures
Sample Preparation
Plasmid construction.
A molecular clone harboring the MA gene and encoding for amino acids 1–130 was used [51]. The MA mutant constructs were generated using a QuickChange Lightning site-directed mutagenesis kit (Agilent Technologies). Forward and reverse primers (Integrated DNA Technologies) extended 15 base-pairs on either side of the mutation codon. DNA sequences were verified at the Heflin Genomics Core at the University of Alabama at Birmingham.
Protein expression and purification.
WT and mutant myr(–)MA, and myr(–)MA99 proteins were overexpressed and purified as described [51]. Purity of protein samples was confirmed by SDS-page gels and verified by NMR.
Unilamellar Vesicles (LUVs)
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (PS), porcine brain PI(4,5)P2 (Avanti Polar Lipids) were used as received. Lipids were mixed in appropriate ratios and solvent was evaporated under a stream of air, followed by lyophilization. Dried lipids were then resuspended in a buffer containing 50 mM sodium phosphates (pH 7.4) and 100 mM NaCl by repeated brief vortexing and allowed to rehydrate for 45 min at room temperature. Lipid suspension was then passed 30 times through a 100 nm pore filter in an extruder (Avanti Polar Lipids). LUVs were stored at 4 °C and used within 24 h. Final total lipid concentration in LUV stocks was 10 mg/mL.
Lipid NMR titrations
NMR data were collected at 35 ºC on a Bruker Avance III (600 MHz 1H) equipped with a cryogenic triple-resonance probe, processed with NMRPIPE [58] and analyzed with NMRVIEW [59]. 1H-15N HSQC NMR titrations with IP3 were conducted with 50–100 μM samples of 15N-labeled WT or mutant myr(–)MA in 20 mM MES (pH 6.0) and 2 mM TCEP. A stock solution of IP3 was prepared in water at 10 mM. CSPs were calculated as , where and are 1H and 15N chemical shift changes, respectively. Dissociation constants were calculated by non-linear least-square fitting algorithm in gnuplot software (http://www.gnuplot.info) using the equation:
where is chemical shift difference between complex and free protein, total concentration of lipid, and [P]0 total concentration of protein.
LUV NMR titrations
Samples for NMR titration contained 25 μM WT or mutant myr(–)MA in 50 mM sodium phosphates (pH 7.4), 100 mM NaCl, 2 mM TCEP, 250 μg LUVs with varying PC, PS and/or PI(4,5)P2 concentrations, and 5% D2O (vol/vol) in a total volume of 500 μL. 1H NMR spectra with excitation sculpting water suppression were recorded for each sample and integral intensity measured in the region 9.5–8.0 ppm. The amount of protein bound to LUVs was determined as the difference between integrals of samples with and without LUVs.
HADDOCK docking
The structure of the myr(–)MA99–I(1,4,5)P3 complex was calculated using HADDOCK (v. 2.4 web server) [60, 61]. The structure of IP3 (6 negative charges) was built and energy-minimized in Avogadro (75) using universal force field (76). Docking of IP3 was calculated for the structure of myr(–)MA99 (PDB ID 7M1W). Active residues in helix II of myr(–)MA99 were selected based on mutagenesis data and residues displaying larger than average standard deviation of + 1 for all CSPs (His44, Lys47, Lys48, and Lys51). Residues in 6.5 Å proximity to active residues with greater than 40% side-chain solvent accessibility were defined as passive residues. The N-terminal arginines were excluded as active residues despite displaying significant CSPs due to the limited ability of HADDOCK to handle protein flexibility. Calculations were performed with recommended settings for protein–ligand docking.
Molecular dynamics simulations
A representative protein structure from the first HADDOCK cluster of myr(–)MA99 docked to IP3 was used as starting coordinates for all MD simulations. Ligand topology was prepared using CGenFF [62, 63] and a charmm2gmx script obtained from the MacKerell laboratory (University of Maryland). All MD simulations were performed on the Cheaha HPC system with Gromacs [64] using the CHARMM36 force field. MD simulations were performed in a dodecahedral periodic box filled with TIP3P water in the absence of NaCl to approximate conditions used in our experiments. Solvent was substituted for a single chlorine ion at a random position to neutralize the protein charge. Steepest descent minimization of the solvent water was performed with positional restraints for protein and lipid. This was followed by temperature and pressure equilibration for 100 ps with protein and ligand restrained. Trajectories of three identical equilibrated systems were calculated for 300 ns each. Electrostatic interactions were calculated by the particle-mesh Ewald method and Lennard-Jones cutoffs were set at 1.0 nm. Trajectories were visualized and hydrogen bonds analyzed using VMD [65]. RMSD values and distances were calculated using Gromacs.
Particle production assay
Cells were transfected by using the PEI transfection reagent (Sigma-Aldrich) at a 1:3 weight to volume ratio. HTLV-1 Gag was produced from pN3 HTLV-1 WT Gag plasmid, pN3 HTLV-1 Gag G2A [37]. Site-directed MA domain mutants were created by using Gibson Assembly (New England Biolabs) with primers containing the following mutations, K47A, K48A, K51A, m2a, m2b, m3 and m4. HTLV-1 Env was produced from an Env expression plasmid, pCMV-ENV, which was graciously provided by Kathryn Jones and Marie-Dokhelar [66]. To produce VLPs, Gag and Env expression plasmids were transfected into cells at 5:1, respectively. Forty-eight hours post-transfection, VLPs were concentrated from cell culture supernatants by filtering through a 0.22 µm filter and concentrating by ultracentrifugation by using a 50.2 Ti rotor at 150,000 x g for 90 min through a 5 mL 8% OptiPrep (Sigma-Aldrich) cushion. VLP pellets were resuspended in PBS + 0.1% Triton X-100 and concentrated 100X of the initial volume. 293T cells (ATCC) were harvested, washed with PBS and lysed in PBS + 0.1% Triton X-100. Cell lysates and cell culture supernatants were normalized for protein concentration by using the BCA assay according to the manufacturers’ instructions (Thermo Fisher Scientific). Each sample (30 µg) was run on a 14% SDS-PAGE gel and transferred to a nitrocellulose membrane (Bio-Rad). Gag was detected by using a 1:1000 dilution of a mouse monoclonal antibody against HTLV-1 p24 (6G9; Santa Cruz Biotechnology) and a 1:2500 dilution of a secondary antibody goat anti-mouse IgG StarBright Blue 700 (Bio-Rad). Cellular GAPDH was detected by using a 1:3000 dilution of an anti-GAPDH hFAB Rhodamine antibody (Bio-Rad). Immunoblots were imaged by using a ChemiDoc Touch system and analyzed with ImageLab software (Bio-Rad).
Confocal microscopy analysis of Gag subcellular distribution
HeLa cells were co-transfected with pN3 HTLV-1 Gag and pN3 Gag-EYFP at a 3:1 ratio, respectively [66] and with pCMV-ENV, at a 5:1 total (Gag:Env) as described above. Sixteen-hours post-transfection, cells were fixed with 4% paraformaldehyde and permeabilized. The cytoskeleton was visualized by using the ActinRed 555 ReadyProbes reagent (Life Technologies) and cell nuclei were visualized by using the NucBlue Fixed Cell ReadyProbes reagent (Life Technologies) following the protocols provided by the manufacturer. Z-stack images of cells were collected by a Zeiss LSM700 confocal laser scanning microscope with a Plan-Apochromat 63x/1.4 aperture (NA) oil objective.
Equilibrium flotation centrifugation
Six hours post transfection, HeLa cells were washed with PBS and resuspended in 200 µL of 10 mM Tris-HCl (pH 7.5) with 4 mM EDTA. Cells were sonicated by using a Qsonica Q500 sonicator for 2, 10 second pulses at 20% amplitude. Cellular debris was pelleted and removed by centrifugation at 2000 x g for 3 min. Cell lysates were mixed in 1.05 mL of 86.9% (wt/vol) sucrose in 10 mM Tris-HCl (pH 7.5) with 1 mM EDTA (TE) buffer at the bottom of a 5 mL tube (Beckman Coulter) to result in a final concentration of 73% (wt/vol). Next, 2.74 mL of 65% sucrose in TE buffer was layered above, followed by 1.27 mL of a 10% sucrose solution in TE buffer. Samples were centrifuged for 16 h at 100,000 x g in a SW55Ti rotor (Beckman Coulter). Nine equal fractions were collected, going from the top to the bottom. HTLV-1 p24 in each fraction was detected by using immunoblot analysis as described in the particle production assay.
Fluorescence z-scan imaging
Dual-color z-scan data was collected and analyzed as described [67]. U2OS cells were maintained in Dulbecco’s modified Eagle’s serum with 10% fetal bovine serum. Cells were plated into chambered 8-well slides (Cellvis, C8-1.5H-N) and transfected with either WT (pN3 HTLV-1 Gag-EYFP) or mutant Gag expression plasmid (85 ng per well) and an mCherry expression plasmid (15 ng per well) by using GenJet (SignaGen Laboratories) 12–24 h prior to z-scan imaging. mCherry served as a cytoplasmic marker. Measurements were conducted at room temperature in Dulbecco’s phosphate-buffered saline (DPBS). Data were collected on multiple cells at varying cytoplasmic concentrations of Gag-EYFP (≤ 80 nM), in locations of the cells without Gag puncta. Instrumentation is as described [67]. The instrument point spread function (PSF) was modeled as the modified squared Gaussian-Lorentzian PSF [68]. Gag-EYFP intensity traces were modeled as a delta-slab-delta geometry and the mCherry reference channel was modeled as a single slab geometry [67]. The fit identifies the amplitudes , , and of the decomposed fluorescence intensity contributions from the cytoplasm, bottom PM, and top PM, respectively (Fig. 8B). The membrane fluorescence intensity fraction (m) for the top and bottom PM is determined by
where is the amplitude or , respectively. Weighted average m values were determined by considering the SEM of repeated z-scan traces. Since no statistically significant differences in m for the TM and BM were observed, the weighted average of all measured m values was reported.
Supplementary Material
Acknowledgments
This work was supported by grants 9 R01 AI150901–10 from the National Institutes of Health (NIH) to JSS; R01 GM098550 (to LMM); T32 AI083196 and F31 AI147805 (to HMH), T90 DE022732 and F30 DE031829 (to NAW), TL1R002493 (to RA); and T90 DE022732 (to IA). The High-Field NMR facility at the University of Alabama at Birmingham was established through NIH grant 1S10RR026478 and is currently supported through the O’Neal Comprehensive Cancer center (NCI grant P30 CA013148).
Abbreviations:
- HTLV-1
human T-cell leukemia virus type 1
- myrMA
myristoylated matrix
- myr(–)MA
unmyristoylated matrix
- VLP
virus-like particle
- HBR
highly basic region
- PM
plasma membrane
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- MD
molecular dynamics MD
- NMR
nuclear magnetic resonance
- HSQC
heteronuclear single quantum coherence
- CSP
chemical shift perturbation
- IP3
inositol 1,4,5-trisphosphate
- PC
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- PS
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine
- LUV
large unilamellar vesicle
Footnotes
Disclaimer: “The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.”
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Availability.
The raw data described in the manuscript can be shared upon request by directly contacting saad@uab.edu.
References
- [1].Gessain A, Cassar O. Epidemiological Aspects and World Distribution of HTLV-1 Infection. Front Microbiol 2012;3:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bangham CRM. Human T Cell Leukemia Virus Type 1: Persistence and Pathogenesis. Annu Rev Immunol 2018;36:43–71. [DOI] [PubMed] [Google Scholar]
- [3].Osame M, Matsumoto M, Usuku K, Izumo S, Ijichi N, Amitani H, et al. Chronic progressive myelopathy associted with elevated antibodies to human T-lymphotropic virus type I and adult T-cell leukemia-like cells. Annals of Neurology 1987;21:117–22. [DOI] [PubMed] [Google Scholar]
- [4].Gessain A, Barin F, Vernant JC, Gout O, Maurs L, Calender A, et al. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet 1985;2:407–10. [DOI] [PubMed] [Google Scholar]
- [5].Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type-C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci 1980;75:7415–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus (ATLV) from cell lines of human adult T-cell leukemia and its implication in the diseases. Proc Natl Acad Sci 1982;79:2031–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gross C, Wiesmann V, Millen S, Kalmer M, Wittenberg T, Gettemans J, et al. The Tax-Inducible Actin-Bundling Protein Fascin Is Crucial for Release and Cell-to-Cell Transmission of Human T-Cell Leukemia Virus Type 1 (HTLV-1). PLoS Pathog 2016;12:e1005916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Finzi A, Orthwein A, Mercier J, Cohen EA. Productive Human Immunodeficiency Virus Type 1 Assembly Takes Place at the Plasma Membrane. J Virol 2007;81:7476–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gousset K, Ablan SD, Coren LV, Ono A, Soheilian F, Nagashima K, et al. Real-time visualization of HIV-1 GAG trafficking in infected macrophages. PLoS Pathog 2008;4:e1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Joshi A, Ablan SD, Soheilian F, Nagashima K, Freed EO. Evidence that productive human immunodeficiency virus type 1 assembly can occur in an intracellular compartment J Virol 2009;83:5375–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jouvenet N, Neil SJD, Bess C, Johnson MC, Virgen CA, Simon SM, et al. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol 2006;4:e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Welsch S, Keppler OT, Habermann A, Allespach I, Krijnse-Locker J, Kräusslich H-G. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog 2007;3:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag membrane binding. J Virol 2008;82:2405–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci U S A 2004;101:14889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ghanam RH, Samal AB, Fernandez TF, Saad JS. Role of the HIV-1 matrix protein in Gag intracellular trafficking and targeting to the plasma membrane for virus assembly. Front Microbiol 2012;3:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vlach J, Saad JS. Structural and molecular determinants of HIV-1 Gag binding to the plasma membrane. Front Microbiol 2015;6:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hamard-Peron E, Juillard F, Saad JS, Roy C, Roingeard P, Summers MF, et al. Targeting of murine leukemia virus gag to the plasma membrane is mediated by PI(4,5)P2/PS and a polybasic region in the matrix. J Virol 2010;84:503–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Prchal J, Kroupa T, Ruml T, Hrabal R. Interaction of Mason-Pfizer monkey virus matrix protein with plasma membrane. Front Microbiol 2014;4:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Saad JS, Ablan SD, Ghanam RH, Kim A, Andrews K, Nagashima K, et al. Structure of the myristylated HIV-2 MA protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J Mol Biol 2008;382:434–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Freed EO. HIV-1 assembly, release and maturation. Nat Rev Microbiol 2015;13:484–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol 2008;18:203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Watanabe SM, Medina GN, Eastep GN, Ghanam RH, Vlach J, Saad JS, et al. The matrix domain of the Gag protein from avian sarcoma virus contains a PI(4,5)P2-binding site that targets Gag to the cell periphery. J Biol Chem 2018;293:18841–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mercredi PY, Bucca N, Loeliger B, Gaines CR, Mehta M, Bhargava P, et al. Structural and Molecular Determinants of Membrane Binding by the HIV-1 Matrix Protein. J Mol Biol 2016;428:1637–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vlach J, Saad JS. Trio engagement via plasma membrane phospholipids and the myristoyl moiety governs HIV-1 matrix binding to bilayers. Proc Natl Acad Sci U S A 2013;110:3525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Inlora J, Collins DR, Trubin ME, Chung JY, Ono A. Membrane binding and subcellular localization of retroviral Gag proteins are differentially regulated by MA interactions with phosphatidylinositol-(4,5)-bisphosphate and RNA. mBio 2014;5:e02202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Prchal J, Srb P, Hunter E, Ruml T, Hrabal R. The Structure of Myristoylated Mason-Pfizer Monkey Virus Matrix Protein and the Role of Phosphatidylinositol-(4,5)-Bisphosphate in Its Membrane Binding. J Mol Biol 2012;423:427–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mandal K Review of PIP2 in Cellular Signaling, Functions and Diseases. Int J Mol Sci 2020;21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stansell E, Apkarian R, Haubova S, Diehl WE, Tytler EM, Hunter E. Basic residues in the Mason-Pfizer monkey virus gag matrix domain regulate intracellular trafficking and capsid-membrane interactions. J Virol 2007;81:8977–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brown LA, Cox C, Baptiste J, Summers H, Button R, Bahlow K, et al. NMR structure of the myristylated feline immunodeficiency virus matrix protein. Viruses 2015;7:2210–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nadaraia-Hoke S, Bann DV, Lochmann TL, Gudleski-O’Regan N, Parent LJ. Alterations in the MA and NC domains modulate phosphoinositide-dependent plasma membrane localization of the Rous sarcoma virus Gag protein. J Virol 2013;87:3609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Vlach J, Eastep GN, Ghanam RH, Watanabe SM, Carter CA, Saad JS. Structural basis for targeting avian sarcoma virus Gag polyprotein to the plasma membrane for virus assembly. J Biol Chem 2018;293:18828–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Heidecker G, Lloyd PA, Soheilian F, Nagashima K, Derse D. The role of WWP1-Gag interaction and Gag ubiquitination in assembly and release of human T-cell leukemia virus type 1. J Virol 2007;81:9769–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Le Blanc I, Blot V, Bouchaert I, Salamero J, Goud B, Rosenberg AR, et al. Intracellular distribution of human T-cell leukemia virus type 1 Gag proteins is independent of interaction with intracellular membranes. J Virol 2002;76:905–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Le Blanc I, Rosenberg AR, Dokhelar MC. Multiple functions for the basic amino acids of the human T-cell leukemia virus type 1 matrix protein in viral transmission. J Virol 1999;73:1860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang H, Machesky NJ, Mansky LM. Both the PPPY and PTAP motifs are involved in human T-cell leukemia virus type 1 particle release. J Virol 2004;78:1503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Inlora J, Chukkapalli V, Derse D, Ono A. Gag localization and virus-like particle release mediated by the matrix domain of human T-lymphotropic virus type 1 Gag are less dependent on phosphatidylinositol-(4,5)-bisphosphate than those mediated by the matrix domain of HIV-1 Gag. J Virol 2011;85:3802–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fogarty KH, Berk S, Grigsby IF, Chen Y, Mansky LM, Mueller JD. Interrelationship between cytoplasmic retroviral Gag concentration and Gag-membrane association. J Mol Biol 2014;426:1611–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Thornhill D, Olety B, Ono A. Relationships between MA-RNA binding in cells and suppression of HIV-1 Gag mislocalization to intracellular membranes. J Virol 2019. [DOI] [PMC free article] [PubMed]
- [39].Olety B, Ono A. Roles played by acidic lipids in HIV-1 Gag membrane binding. Virus Res 2014;193:108–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A 1990;87:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhou W, Resh MD. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol 1996;70:8540–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol 1997;71:6582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nguyen DH, Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol 2000;74:3264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Provitera P, El-Maghrabi R, Scarlata S. The effect of HIV-1 Gag myristoylation on membrane binding. J Mol Biol 2006;119:23–32. [DOI] [PubMed] [Google Scholar]
- [45].Saad JS, Loeliger E, Luncsford P, Liriano M, Tai J, Kim A, et al. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J Mol Biol 2007;366:574–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A 2006;103:11364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Anraku K, Fukuda R, Takamune N, Misumi S, Okamoto Y, Otsuka M, et al. Highly sensitive analysis of the interaction between HIV-1 Gag and phosphoinositide derivatives based on surface plasmon resonance. Biochemistry 2010;49:5109–16. [DOI] [PubMed] [Google Scholar]
- [48].Shkriabai N, Datta SA, Zhao Z, Hess S, Rein A, Kvaratskhelia M. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry 2006;45:4077–83. [DOI] [PubMed] [Google Scholar]
- [49].Fernandes F, Chen K, Ehrlich LS, Jin J, Chen MH, Medina GN, et al. Phosphoinositides direct equine infectious anemia virus gag trafficking and release. Traffic 2011;12:438–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Murphy RE, Samal AB, Vlach J, Mas V, Prevelige PE, Saad JS. Structural and biophysical characterizations of HIV-1 matrix trimer binding to lipid nanodiscs shed light on virus assembly. J Biol Chem 2019;294:18600–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Herrmann D, Zhou LW, Hanson HM, Willkomm NA, Mansky LM, Saad JS. Structural Insights into the Mechanism of Human T-cell Leukemia Virus Type 1 Gag Targeting to the Plasma Membrane for Assembly. J Mol Biol 2021;433:167161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bodner CR, Dobson CM, Bax A. Multiple tight phospholipid-binding modes of alpha-synuclein revealed by solution NMR spectroscopy. J Mol Biol 2009;390:775–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ceccon A, D’Onofrio M, Zanzoni S, Longo DL, Aime S, Molinari H, et al. NMR investigation of the equilibrium partitioning of a water-soluble bile salt protein carrier to phospholipid vesicles. Proteins 2013;81:1776–91. [DOI] [PubMed] [Google Scholar]
- [54].Hayakawa T, Miyazaki T, Misumi Y, Kobayashi M, Fujisawa Y. Myristoylation-dependent membrane targeting and release of the HTLV-I gag precursor, Pr53gag, in yeast. Genes & Development 1992;119:273–7. [DOI] [PubMed] [Google Scholar]
- [55].Fogarty KH, Chen Y, Grigsby IF, Macdonald PJ, Smith EM, Johnson JL, et al. Characterization of cytoplasmic Gag-gag interactions by dual-color z-scan fluorescence fluctuation spectroscopy. Biophys J 2011;100:1587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bergmann JE, Fusco PJ. The M protein of vesicular stomatitis virus associates specifically with the basolateral membranes of polarized epithelial cells independently of the G protein. J Cell Biol 1988;107:1707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ono A, Freed EO. Binding of Human Immunodeficiency Virus Type 1 gag to membrane: Role of the matrix amino terminus. J Virol 1999;73:4136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 1995;6:277–93. [DOI] [PubMed] [Google Scholar]
- [59].Johnson BA, Blevins RA. NMRview: a Computer Program for the Visualization and Analysis of NMR Data. J Biomol NMR 1994;4:603–14. [DOI] [PubMed] [Google Scholar]
- [60].Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc 2003;125:1731–7. [DOI] [PubMed] [Google Scholar]
- [61].van Zundert GCP, Rodrigues J, Trellet M, Schmitz C, Kastritis PL, Karaca E, et al. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J Mol Biol 2016;428:720–5. [DOI] [PubMed] [Google Scholar]
- [62].Yu W, He X, Vanommeslaeghe K, MacKerell AD, Jr. Extension of the CHARMM General Force Field to sulfonyl-containing compounds and its utility in biomolecular simulations. J Comput Chem 2012;33:2451–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 2010;31:671–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013;29:845–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph 1996;14:33–8, 27–8. [DOI] [PubMed] [Google Scholar]
- [66].Delamarre L, Rosenberg AR, Pique C, Pham D, Dokhelar MC. A novel human T-leukemia virus type 1 cell-to-cell transmission assay permits definition of SU glycoprotein amino acids important for infectivity. J Virol 1997;71:259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Angert I, Karuka SR, Hennen J, Chen Y, Albanesi JP, Mansky LM, et al. Sensitive Detection of Protein Binding to the Plasma Membrane with Dual-Color Z-Scan Fluorescence. Biophys J 2020;118:281–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Macdonald PJ, Chen Y, Wang X, Chen Y, Mueller JD. Brightness analysis by Z-scan fluorescence fluctuation spectroscopy for the study of protein interactions within living cells. Biophys J 2010;99:979–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 2008;29:1859–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data described in the manuscript can be shared upon request by directly contacting saad@uab.edu.


