Abstract
Diverse glutamatergic projection neurons (PNs) mediate myriad processing streams and output channels of the cerebral cortex. Yet, how different types of neural progenitors, such as radial glial cells (RGs) and intermediate progenitors (IPs), produce PN diversity and hierarchical organization remains unclear. A fundamental issue is whether RGs constitute a homogeneous, multipotent lineage capable of generating all major PN types through a temporally regulated developmental program, or whether RGs comprise multiple transcriptionally heterogenous pools, each fated to generate a subset of PNs. Beyond RGs, the role of IPs in PN diversification remains underexplored. Addressing these questions requires tracking PN developmental trajectories with cell type resolution – from transcription factor-defined RGs and IPs to their PN progeny, defined not only by laminar location, but projection patterns and gene expression. Advances in cell type resolution genetic fate mapping, axon tracing, and spatial transcriptomics may provide the technical capability for answering these fundamental questions.
Introduction
The cerebral cortex is comprised of dozens of functional areas mediating numerous information processing streams that form comprehensive representations of the internal and external world. Sensory, motor, and cognitive functions are integrated across preferentially connected areal subnetworks, which in turn influence subcortical brain regions via corticofugal output channels. At the cellular level, cortical processing streams and output channels are implemented by a diverse set of glutamatergic excitatory projection neurons (PNs). PNs are traditionally distinguished by their laminar cell body position, morphology, axonal projection patterns, and an array of molecular markers [1]. A renaissance in large-scale single cell RNA sequencing (scRNAseq) has recently contributed to a comprehensive and quantitative analysis of PN types and their organization. Recent progress has revealed the hierarchical organization of several major PN classes and subclasses, yielding over one hundred distinguishable transcriptomically defined PN subpopulations (t-types) in the mouse cortex [2–6]. This hierarchical organization of major PN classes and types appears to be largely conserved across mammalian species [3, 5–8]. Additionally, large-scale single cell reconstruction [9–11] and electrophysiologic studies [12, 13] have uncovered a variety of morphologically (m-types) and electrophysiologically defined (e-types) PN types, respectively; and techniques such as Patch-Seq [14] begin to provide correspondence between t-, m-, and e-types. Although fine grained “atomic” PN types remain to be reliably clarified and enumerated across studies, a multimodal consensus has emerged around the major classes, subclasses, and supertypes of cortical PNs [15]. Given the impressive progress in defining and cataloguing the multitude of PNs, a major question remains about how this spectacular diversity and its hierarchical organization are generated during cortical development [16, 17]. Answers to this question will not only illuminate the developmental genetic basis of cortical circuit organization but also shed light on the pathogenic mechanisms of neurodevelopment disorders.
All cortical PNs are generated from progenitors in the germinal zone lining the embryonic cerebral ventricles of the telencephalon [16]. Cortical development begins with the specification of a single layer of neuroepithelial cells lining the ventricles, which then divide symmetrically to amplify the stem cell pool that forms the ventricular zone (VZ). This is followed by differentiation of neuroepithelial cells into the radial glia cells (RGs) or apical progenitors, whose cell bodies reside in the VZ. Some RGs divide asymmetrically to both self-renew and produce one neuron by a process called direct neurogenesis (dNG). However, most RG asymmetric divisions generate intermediate progenitors (IPs), which typically migrate to the subventricular zone (SVZ) and divide symmetrically to produce two PNs; this process is called indirect neurogenesis (iNG). Newborn neurons migrate radially toward the pial surface using RGs as a scaffold to reach their destination in the cortical plate in an inside-out sequence. Thus, the spatial position of RGs within the VZ across anterior-posterior (A-P) and medial-lateral (M-L) axes of the cerebral ventricle wall determines the areal identity of their progeny PNs in the mature cortex, while PN birth order broadly determines their laminar location [16, 18].
Beyond this basic scheme of cortical neurogenesis, a fundamental unresolved issue is how cortical progenitors, especially RGs and IPs, give rise to the diversity of PN types and their hierarchical organization. Since the Boulder Committee postulated [19] that a single type of multipotent VZ progenitor gives rise to all cortical neuron types, several broad and competing models of cortical neural progenitor lineage organization and fate specification have been proposed and examined over subsequent decades [16, 20]. Despite the accumulation of multiple lines of experimental results using increasingly sophisticated methods, there has been enduring debates on the validity of these models. In this brief review, we highlight the mismatch in resolution between the fine-grained multi-modal analysis of PN types in mature cortex and the developmental studies that distinguish “PN types” largely based on their laminar location. We suggest that deciphering the progenitor basis of PN diversity requires methods to track their developmental trajectories with cell type resolution – from transcription factor-defined RGs to their PN progeny defined by multimodal features from laminar location to projection patterns and gene expression. We forecast how the integration of several emerging technologies may finally provide the much-needed cell type trajectory resolution for discovering the progenitor basis of PN diversity.
Multipotent progenitors with temporal competence to successively generate all PN types
Across the embryonic cerebral ventricle, morphogen gradients induce multiple opposing gradients of transcription factor (TF) expression in the VZ neuroepithelial cells and RGs, which shape the emergence of cortical areas [16, 18, 21]. A major unanswered question is whether beyond these TF gradients, neighboring RGs within the same VZ region are largely homogenous or are further differentiated into transcriptionally distinguishable pools with different fate potentials.
A widely influential model of progenitor organization posits that a single lineage of RGs generates all types of PNs and that the competence of a given RG to generate specific types becomes progressively limited over the course of development [16, 20]. In support of this model, classic transplantation experiments demonstrate that early-stage progenitors transplanted into the late-stage cortex are capable of producing all types across cortical layers, but late-stage progenitors transplanted into the early-stage cortex are competent only to produce superficial-layer types [22–25]. In addition, retroviral lineage tracing experiments show that single progenitors labelled early in corticogenesis are competent to produce neurons across all layers, whereas progenitors labelled later in corticogenesis mostly give rise to PNs residing in superficial layers [26–28]. Furthermore, in vitro studies using both primary dissociated cortical progenitors and embryonic stem cell-derived cortical progenitors demonstrate that they autonomously recapitulate the sequential generation of a number of PN types that are characteristic of corticogenesis in vivo [28, 29]. Perhaps a bedrock observation in support of this model comes from genetic fate mapping and clonal analysis in mice [30]. Mosaic Analysis with Double Markers (MADM) labels subpopulations of clonal progeny arising from individual RGs with single cell resolution, revealing that RGs sequentially produce distinct quanta of PN progeny as neurogenesis proceeds and that early born progeny of individual RG clones often spanned cortical lamina while later born progeny were restricted to upper layers. However, a major limitation of all these previous studies is the coarse binary distinction of PN types according to upper or lower layers, with minimal consideration of their projection targets or gene expression. For example, the IT class of PNs resides across layers, and thus previous studies would not have the resolution to identify a hypothetical RG subpopulation fated to produce only IT type PNs. Furthermore, these prior studies also treat all RGs as genetically equivalent and do not delineate between RG subsets based on differential gene expression patterns. For example, MADM analysis used a single Emx1-CreER transgenic line to probe the RG population [30], which may not represent endogenous Emx1 expression pattern as a knock-in line would provide, and also stopped short of testing the fate potential of RG pools defined by other TFs with potentially more restricted expression patterns.
The hypothesis of a homogenous RG population in the VZ that constitutes a uniform progenitor pool assumes that individual RGs share a largely homogenous gene expression profile, but this assumption has not been rigorously examined. Although a large set of TFs are expressed in the VZ, many showing gradients across the A-P and M-L axes, transcription profiles of individual RGs has only recently begun to be examined by scRNAseq. Several recent studies report relatively homogenous gene expression among cortical progenitors [4, 31, 32]. However, the interpretation of these results is complicated by at least two factors. First, the sequencing depth in these studies was relatively shallow, limiting the genetic resolution of their transcription profiles; thus subtler differential gene expression may have been missed or overlooked. Second, although global gene expression across RGs may be relatively uniform, yielding a single cluster from transcriptional clustering algorithms, the differential expression of even a single crucial TF could seed a distinct fate potential from very early neurogenic stages. Thus, there may still be critical fate specifying differences in gene expression among RGs that are simply insufficient to be detected by recent sequencing and algorithmic detection strategies.
In summary, decades of studies using available experimental tools have together established the foundational concept that RGs are a largely homogeneous, multipotent progenitor pool that progressively regulates their temporal competence during cortical neurogenesis to generate PNs that reside across cortical layers. On the other hand, none of these studies so far exclude the possibility of multiple distinct pools of RGs, each of limited multipotency but that still may demonstrate successive temporal competence during development to generate different sets of PN types that can be resolved by their projection and gene expression patterns.
Evidence for fate-restricted RGs
The possible presence of fate-restricted RGs was hinted at by the observation that a number of TFs that mark predominantly upper- (e.g. Cux2) or lower-layer (e.g. Fezf2) PNs are also expressed during neurogenesis in VZ progenitors [33–39]. These observations suggested that some molecular differences that distinguish lower- and upper-layer PNs might already coexist in subsets of RGs, and raised the possibility that neurogenic RGs might be comprised of heterogeneous populations. Evidence for RG heterogeneity also came from several genetic studies on the mechanisms governing the production of early-born lower-layer neurons versus late-born upper-layer neurons. For example, the bHLH TFs Ngn1 and Ngn2 are required for the specification of regional and laminar fates of PNs during lower-layer but not upper-layer neurogenesis [40]. Instead, specification of upper-layer PNs requires Pax6 and Tlx [40]. In addition, FOXP1 is expressed at high levels in RGs during early neurogenesis and bias the production of PNs toward deep layer fates [41]. These studies suggest that distinct molecular pathways may control the basic differentiation programs of RGs that differentially generate lower- and upper-layer excitatory neurons.
The first direct evidence for fate-restricted cortical progenitors was reported using genetic fate mapping from Cux2-Cre and Cux2-CreER mouse lines [42]. Cux2+ RGs can be identified even before the onset of neurogenesis and are intermingled with Cux2− RGs along the ventricular zone. Moreover, temporal lineage-tracing revealed that Cux2+ RGs generate PNs that predominantly reside in upper layers, whereas lower-layer neurons tend to arise from Cux2− RGs. Furthermore, Cux2+ RGs are hypothesized to remain primarily proliferative rather than neurogenic during early, lower-layer neurogenesis, and then transition to produce significant numbers of upper-layer neurons at later stages [42]. These results suggest that at least one lineage of RGs may be restricted in its fate potential even before the onset of neurogenesis. Unfortunately, these results were contested due to posited differences in transgene expression among different mouse strains, genetic drift from different breeding strategies, and discordance between Cre and reporter expression specifically in deep layers [43, 44]. Another factor that may have contributed to the confusion and debate is that, similar to earlier studies, an emphasis on defining PNs solely by their binary distinction as upper- versus of lower-layer PNs may have obscured transcriptional and projection-based features that define PNs from a putative Cux2+ lineage.
Perhaps the most compelling evidence for fate-restricted progenitors came from a recent study that distinguished two RG pools based on differential TF expression and distinguished their PN progeny based on axon projection patterns [45]. The key technical innovation is a genetic strategy that enabled fate mapping of RGs based on their differential expression of two TFs, LHX2 and FEZF2, in the same animal. Lhx2 and Fezf2 play important roles in cortical patterning and fate specification [46–48]. Combinatorial fate mapping using Lhx2-CreER and Fezf2-Flp gene knock-in lines unequivocally demonstrate the coexistence of highly intermixed Lhx2+/Fezf2− and Lhx2+/Fezf2+ RG subpopulations from the early period of neurogenesis. Strikingly, while the PN progeny from each RG pool are distributed across cortical layers, they show a categorical distinction in projection patterns: whereas Lhx2+/Fezf2−- derived PNs project across the corpus callosum but not to subcortical regions, Lhx2+/Fezf2+- derived PNs show the opposite pattern, projecting corticofugal axons to subcortical regions but not to the contralateral cortex. These results provide unequivocal evidence for the presence of separate TF-defined RG lineages that give rise to distinct projection-defined PN types. Such fate-restricted RGs are likely also multipotent and deploy their temporal competence to generate projection-defined PNs across layers. An intriguing result is that Lhx2+/Fezf2+ progeny also include a set of upper-layer non-callosal PNs, suggesting that “non-callosal” projections might define a prominent IT subclass that includes both lower and upper layer PNs. A major question is whether these two RG pools and the previously reported Cux2+ RG pool represent minor and unusual exceptions to the model of homogeneous, multipotent progenitors or whether they represent the tip of the iceberg for uncovering the presence of multiple fate-restricted RG pools. In addition, the relationship between the Lhx2/Fezf2-defined and the potential Cux2-defined RGs remains to be elucidated. A systematic fate mapping of TF-defined RGs and their mature PN progeny, as defined by their transcriptome, morphology, and projection patterns should help address these questions. The reliable targeting of TF-defined RGs and the developmental trajectory of their PN progeny will further provide experimental access to explore the underlying molecular genetic profiles and mechanisms.
Direct and indirect neurogenesis differentially contribute to PN diversity
Although apical RGs all reside in the VZ and can mediate direct neurogenesis (dNG), the large majority of PNs in rodents and especially in primates are produced from indirect neurogenesis (iNG) through intermediate progenitors (IPs) that reside largely in the SVZ. During evolution, RG-mediated dNG originated before the emergence of vertebrates, while IP-mediated iNG is thought to have originated in the last common ancestor of amniotes and further diversified along the amniote lineage [49–51]. In mammals, RGs are ubiquitous along the neural tube and generate neurons for the entire central nervous system, whereas IPs are restricted largely to the telencephalon that develops into a major part of the forebrain, particularly the cerebral cortex [52]. Compared to rodents, primates demonstrate an even greater expansion of IP subtypes in terms of both their transcriptomes and morphologies [53, 54], which may in turn have led to further expansion and diversity of cortical PN subtypes in non-human primates and humans. While iNG clearly contributes to the amplification of PNs, particularly upper-layer PNs [55–57], whether and how iNG contributes to PN diversification is still largely unknown.
A recent study designed a mouse genetic fate-mapping method to differentially visualize dNG and iNG in the same animal [58]. Within the neocortex, while dNG generates all major IT, PT, and CT classes, iNG differentially amplifies and diversifies PN types within each class, with disproportionally large contribution to the IT class. Importantly, dNG and iNG derived PN subtypes across as well as within genetically defined major subpopulations are extensively intermixed and show distinct projection patterns, indicating that they assemble fine mosaics of lineage-specified and evolutionarily-rooted cortical subnetworks. These results reveal the differential contribution of RG-dNG and IP-iNG to the diversification of PN types and suggest a ground-level lineage framework for understanding cortical development and evolution. When combined with the TF-based RG fate mapping methods outlined above, the developmental script for cortical PN diversification may become clear.
Summary and perspective
The diversity of PNs contributes to the awesome computational power of cortical circuits underlying a wide range of cortical functions. Recent single cell transcriptomic, anatomical, and physiological studies have revealed over a hundred PN types and their hierarchical organization. Understanding the developmental origin of this PN diversity will shed light on the biological basis of PN classification and organization, and also provide the starting point for studying the developmental assembly and plasticity of cortical circuits. The difficulties of studying the developmental specification of PN types include their multi-modal definitions and substantial diversity in the mature cortex, the complexity of progenitor organization in the embryonic telencephalon, and the extensive and complex process linking progenitor fate to PN postmitotic differentiation. The key challenge is to systematically track this developmental trajectory with cell type resolution – from transcriptionally-defined progenitor pools and their lineage progression to PN types defined by multi-modal features, such as projection patterns and transcriptomic profiles.
Decades of studies have leveraged, but also are limited by, available methods to track the relationship between cortical progenitors and their PN progeny. Classic transplantation studies are limited to tissue level resolution and distinguished progenitors and PNs mainly according to their spatial locations in VZ versus SVZ of the embryonic ventricle wall or in upper versus lower cortical layers, respectively. Subsequent viral lineage tracing and genetic fate mapping studies have achieved cellular and even clonal resolution yet are limited by their lack of cell type resolution - they neither distinguish progenitors by gene expression nor distinguish PNs by projection and transcriptome. Recent single-cell RNAseq analysis across developmental stages provide pseudo-time lineage trajectories of transcriptionally-defined cell populations [4, 31, 32], and the incorporation of cell-heritable viral barcodes provide additional information for tracing cell lineage relationships [59, 60]. However, the resolution of cell types and their temporal relationship in these studies is limited by sequencing depth and the use of statistical clustering algorithms across developmental stages; thus, these inferred lineage relationships need to be validated by ground truth datasets. Recent combinatorial genetic fate mapping [45, 58] begins to provide ground truth lineage relationships between TF-defined RGs and projection-defined PNs. However, such studies are currently constrained by the availability of driver lines and limited by scalability and throughput.
Considering the strength and limitations of these emerging approaches, we suggest that an integration of combinatorial genetic fate mapping and single-cell genomics and spatial transcriptomics in mice may facilitate major progress in deciphering the progenitor basis of PN diversity. For example, targeted scRNAseq of fate-mapped PNs from TF-defined progenitors across developmental stages will provide ground truth transcriptome trajectories with bona fide lineage labels. Spatial transcriptomics of such fate-mapped samples will be more scalable without the need to purify lineage-labeled PNs and will preserve their spatial location and distribution patterns. Targeted single-cell ATACseq and chromatin mapping will further reveal transcriptional cis-regulatory elements and their target genes in lineage-labeled PN types. Such datasets with biological validity can be used to improve computational algorithms that can then be applied to large scale datasets across species. Together, these integrated studies may help elucidate the epigenomic landscapes, transcriptional trajectories, and ultimately the developmental genetic programs embedded in progenitors that shape the diversity of PN types and their organization. This knowledge will also facilitate understanding the pathogenic mechanisms of neurodevelopmental disorders.
Figure 1. Diversity of Glutamatergic PNs and models of PN diversification in the developing neocortex.
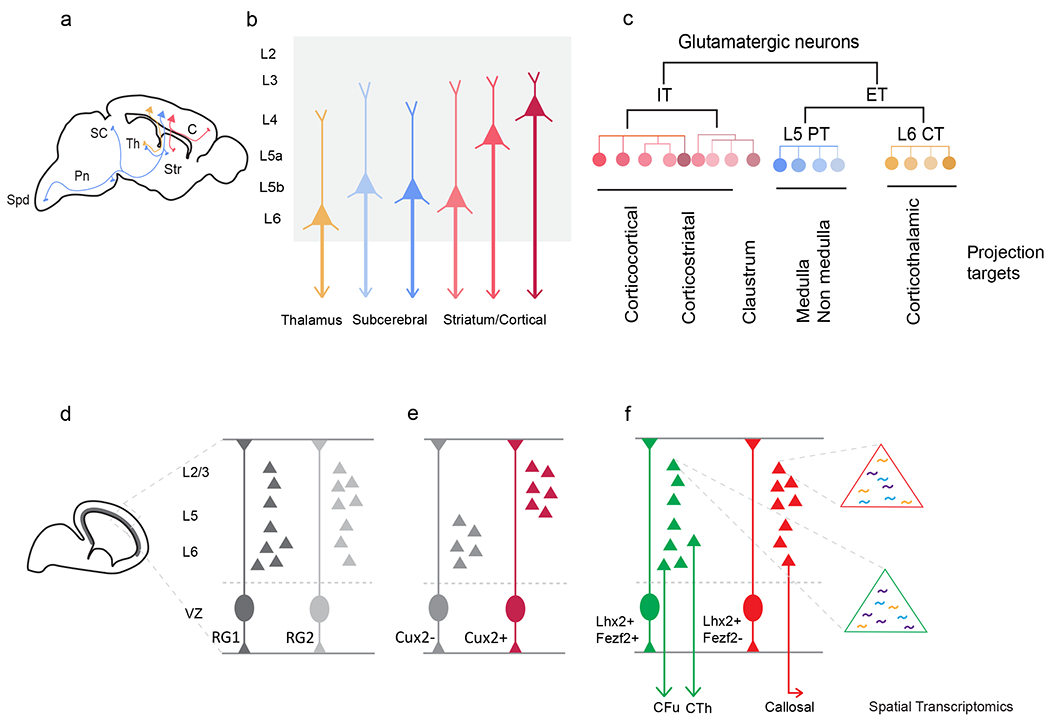
a. Sagittal schematic showing major PN classes and their projection targets to intratelencephalic regions (IT, Red), subcerebral targets (PT, Blue), and thalamus (CT, Yellow). C, cortex; SC, superior colliculus; Str, striatum; Th, thalamus; Pn, pons; Spd, spinal cord. b. Schematic representation of major PN types based on their laminar location and projection targets. c. Hierarchical organization of PN based on transcriptomic profiles and projection targets in the neocortex. ET, Extratelencephalic; PT, Pyramidal tract; CT, Corticothalamic. d. Sagittal schematic of embryonic mouse brain at E12.5 showing ventricular zone (Gray) where RG progenitors reside and differentiate. Model of uniform multipotent progenitors, where a single lineage of RGs generates all types of PNs and the competence of RG to generate specific types becomes progressively restricted over the course of development. E. Model of fate-restricted progenitors, where distinct lineages of RGs co-exist and are specified to generated different types of PNs. Cux2− RGs are competent to generate layer 5-6 corticofugal PN while Cux2+ RGs generate L2-4 corticocortical PN. F. Fate-mapping based on TF (Fezf2 and Lhx2)-defined RGs derived PN population suggests the presence of fate-restricted progenitors [Ref.45]. PNs generated from RGLhx2+Fezf2+ are extratelencephalic (Green) while RGLhx2+Fezf2− derived PN project to intratelencephalic regions (Red). Schematic representation of the use of spatial transcriptomics to reveal the molecular identity of fate-mapped PNs from TF-defined progenitors (ET, Green; IT, Red). CFu, Corticofugal; CTh, Corticothalamic.
Figure 2: Differential contribution of dNG and iNG to cortical PN diversity.
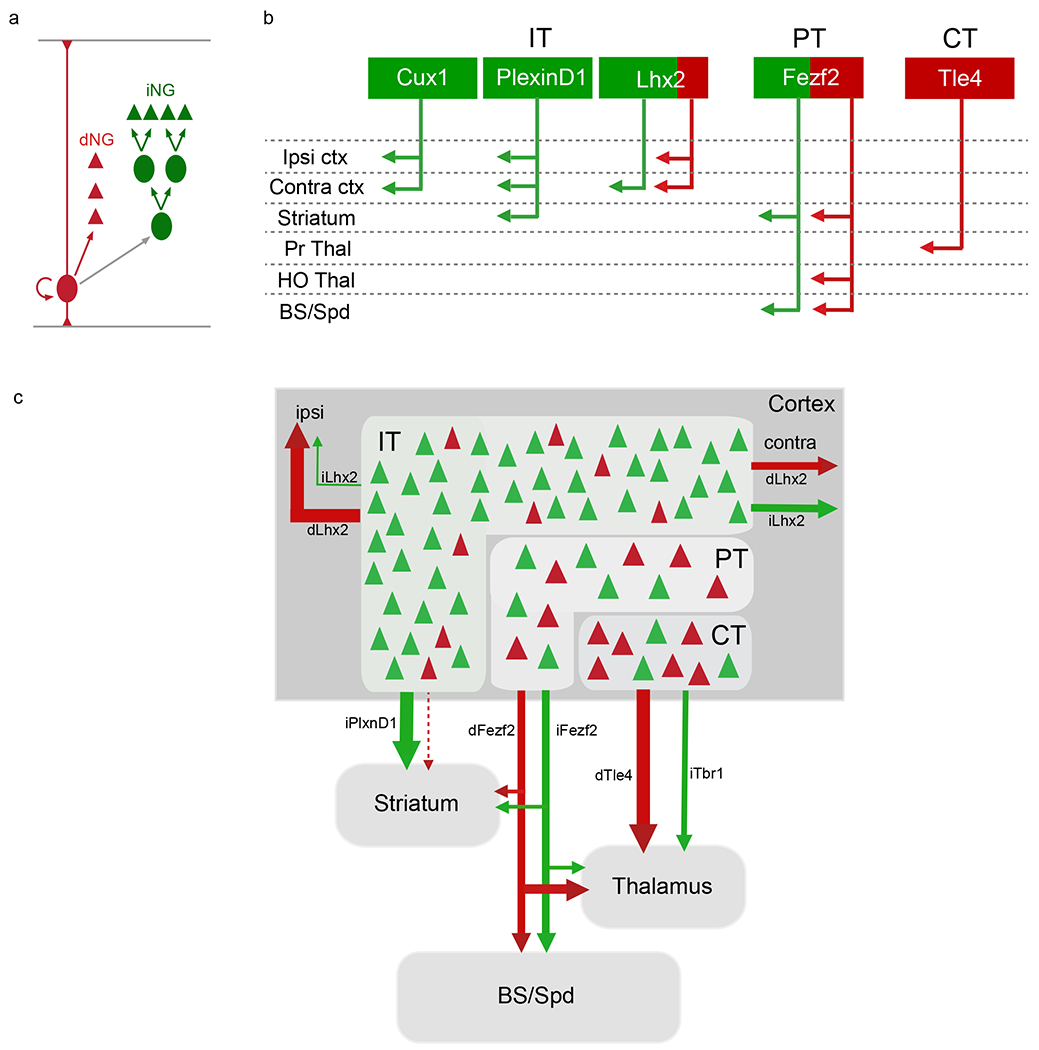
a. Within the cerebral ventricular zone, RGs mediate direct neurogenesis (dNG, red) and via IPs, indirect neurogenesis (iNG, green) to produce PNs (triangles). An intersection-subtraction genetic fate-mapping scheme allows simultaneous visualization of dNG and iNG [Ref.58]. b. Distinct genetic and projection defined PN types generated by dNG (red) and iNG (green) across IT, PT and CT subcategories. The schematic summarizes inducible PN driver lines used for this analysis as shown in the corresponding boxes with distinct projections to different cortical targets [Ref.45, 58]. c. dNG (red) generates PNs across layers in the IT, PT and CT categories, whereas iNG (green) differentially amplifies and diversifies genetically defined PN types within each PN class. dNG- and iNG-derived PN subcategories are highly intermixed and show distinct projection patterns both across and within genetically defined subpopulations; thus dNG and iNG assemble a lineage-based fine mosaic of cortical projections and possibly subnetworks.
Highlights.
the hierarchical organization of PN types is jointly defined by their transcriptome, anatomy, and physiology
lineage tracing needs to distinguish TF-defined RGs as well as projection- and transcriptionally-defined PNs
emerging evidence of fate-restricted RGs is not mutually exclusive with models of multipotency and temporal competence
direct and indirect neurogenesis differentially contribute to PN diversity
Acknowledgement
This work was supported in part by NIMH grants 1DP1MH129954-01 and 5U19MH114821-03 to Z.J.H. J.B.R was supported by an NINDS grant 5K12NS098482-05 and a Duke University Strong Start Award. D.H. was supported by the Human Frontier Science Program long-term fellowship LT000075/2014-L and NARSAD Young Investigator grant no. 26327. We apologize to the authors whose work was not cited because of space limitation.
Footnotes
Conflict of interest – None of the authors have any conflict of interest to disclose.
References
- 1.Harris KD and Shepherd GM, The neocortical circuit: themes and variations. Nat Neurosci, 2015. 18(2): p. 170–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tasic B, et al. , Shared and distinct transcriptomic cell types across neocortical areas. Nature, 2018. 563(7729): p. 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodge RD, et al. , Conserved cell types with divergent features in human versus mouse cortex. Nature, 2019. 573(7772): p. 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Bella DJ, et al. , Molecular logic of cellular diversification in the mouse cerebral cortex. Nature, 2021. 595(7868): p. 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study provides a comprehensive atlas of single cell RNA and ATAC sequencing from mouse cortex at multiple developmental stages from early corticogenesis to early postnatal ages. From this data, they infer a dendrogram of neuronal lineage development and predict key genes that may mediate divergence of neuronal fate down each lineage branch.
- 5.Network BICC, A multimodal cell census and atlas of the mammalian primary motor cortex. Nature, 2021. 598(7879): p. 86–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang R, et al. , Conservation and divergence of cortical cell organization in human and mouse revealed by MERFISH. Science, 2022. 377(6601): p. 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nowakowski TJ, et al. , Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science, 2017. 358(6368): p. 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakken TE, et al. , Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature, 2021. 598(7879): p. 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munoz-Castaneda R, et al. , Cellular anatomy of the mouse primary motor cortex. Nature, 2021. 598(7879): p. 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng H, et al. , Morphological diversity of single neurons in molecularly defined cell types. Nature, 2021. 598(7879): p. 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao L, et al. , Single-neuron projectome of mouse prefrontal cortex. Nat Neurosci, 2022. 25(4): p. 515–529. [DOI] [PubMed] [Google Scholar]
- 12.Scala F, et al. , Phenotypic variation of transcriptomic cell types in mouse motor cortex. Nature, 2021. 598(7879): p. 144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee BR, et al. , Scaled, high fidelity electrophysiological, morphological, and transcriptomic cell characterization. Elife, 2021. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berg J, et al. , Human neocortical expansion involves glutamatergic neuron diversification. Nature, 2021. 598(7879): p. 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study describes Patch-seq, a method that combines electrophysiological recording, biocytin labeling and morphological analysis, and single cell RNA sequencing in human cortex. This powerful pipeline provides a strategy to begin to correlate transcriptionally-defined t-types, electrophysiologically-defined e-types, and morphologically-defined m-types of cortical pyramidal neurons.
- 15.Yao Z, et al. , A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell, 2021. 184(12): p. 3222–3241 e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greig LC, et al. , Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci, 2013. 14(11): p. 755–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lodato S and Arlotta P, Generating neuronal diversity in the mammalian cerebral cortex. Annu Rev Cell Dev Biol, 2015. 31: p. 699–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadwell CR, et al. , Development and Arealization of the Cerebral Cortex. Neuron, 2019. 103(6): p. 980–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Embryonic vertebrate central nervous system: revised terminology. The Boulder Committee. Anat Rec, 1970. 166(2): p. 257–61. [DOI] [PubMed] [Google Scholar]
- 20.Franco SJ and Muller U, Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron, 2013. 77(1): p. 19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodworth MB, et al. , SnapShot: cortical development. Cell, 2012. 151(4): p. 918–918 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McConnell SK, Fates of visual cortical neurons in the ferret after isochronic and heterochronic transplantation. J Neurosci, 1988. 8(3): p. 945–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McConnell SK and Kaznowski CE, Cell cycle dependence of laminar determination in developing neocortex. Science, 1991. 254(5029): p. 282–5. [DOI] [PubMed] [Google Scholar]
- 24.Frantz GD and McConnell SK, Restriction of late cerebral cortical progenitors to an upper-layer fate. Neuron, 1996. 17(1): p. 55–61. [DOI] [PubMed] [Google Scholar]
- 25.Desai AR and McConnell SK, Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development, 2000. 127(13): p. 2863–72. [DOI] [PubMed] [Google Scholar]
- 26.Luskin MB, Pearlman AL, and Sanes JR, Cell lineage in the cerebral cortex of the mouse studied in vivo and in vitro with a recombinant retrovirus. Neuron, 1988. 1(8): p. 635–47. [DOI] [PubMed] [Google Scholar]
- 27.Reid CB, Liang I, and Walsh C, Systematic widespread clonal organization in cerebral cortex. Neuron, 1995. 15(2): p. 299–310. [DOI] [PubMed] [Google Scholar]
- 28.Shen Q, et al. , The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci, 2006. 9(6): p. 743–51. [DOI] [PubMed] [Google Scholar]
- 29.Gaspard N, et al. , An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature, 2008. 455(7211): p. 351–7. [DOI] [PubMed] [Google Scholar]
- 30.Gao P, et al. , Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell, 2014. 159(4): p. 775–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Telley L, et al. , Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science, 2019. 364(6440). [DOI] [PubMed] [Google Scholar]; *The authors use FlashTag to label “isochronic cohorts” of ventricle-adjacent excitatory progenitors, followed by single cell RNA sequencing at various intervals, to uncover developmental genetic programs of temporally linked neuronal progenitors and their early progeny. They find that while early born neurons predictably express deep layer markers, later born neurons first express some deep layer markers before upregulating canonical upper layer markers. Further, early neuronal gene expression progresses from intrinsic proteins to a more extracellularly receptive array of effectors.
- 32.Ruan X, et al. , Progenitor cell diversity in the developing mouse neocortex. Proc Natl Acad Sci U S A, 2021. 118(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirata T, et al. , Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Dev Dyn, 2004. 230(3): p. 546–56. [DOI] [PubMed] [Google Scholar]
- 34.Arlotta P, et al. , Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron, 2005. 45(2): p. 207–21. [DOI] [PubMed] [Google Scholar]
- 35.Molyneaux BJ, et al. , Fezl is required for the birth and specification of corticospinal motor neurons. Neuron, 2005. 47(6): p. 817–31. [DOI] [PubMed] [Google Scholar]
- 36.Chen JG, et al. , Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci U S A, 2005. 102(49): p. 17792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nieto M, et al. , Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol, 2004. 479(2): p. 168–80. [DOI] [PubMed] [Google Scholar]
- 38.Zimmer C, et al. , Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex, 2004. 14(12): p. 1408–20. [DOI] [PubMed] [Google Scholar]
- 39.Molyneaux BJ, et al. , Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci, 2009. 29(39): p. 12343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuurmans C, et al. , Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J, 2004. 23(14): p. 2892–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson CA, et al. , Foxp1 Regulates Neural Stem Cell Self-Renewal and Bias Toward Deep Layer Cortical Fates. Cell Rep, 2020. 30(6): p. 1964–1981 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study demonstrates that Foxp1 is expressed in radial glia and plays a crucial role in the generation of early-born, deep layer pyramidal neurons until its downregulation at the transition to upper layer neurogenesis. Prolonged overexpression of Foxp1 extends the maintenance of a neurogenic radial glia population.
- 42.Franco SJ, et al. , Fate-restricted neural progenitors in the mammalian cerebral cortex. Science, 2012. 337(6095): p. 746–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gil-Sanz C, et al. , Lineage Tracing Using Cux2-Cre and Cux2-CreERT2 Mice. Neuron, 2015. 86(4): p. 1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckler MJ, et al. , Cux2-positive radial glial cells generate diverse subtypes of neocortical projection neurons and macroglia. Neuron, 2015. 86(4): p. 1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matho KS, et al. , Genetic dissection of the glutamatergic neuron system in cerebral cortex. Nature, 2021. 598(7879): p. 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]; *In addition to generating a comprehensive toolbox of knock-in mouse lines that provide genetic access to a number of key cortical excitatory neuronal populations, the authors demonstrate through a sophisticated intersectional fate mapping strategy that Lhx2+/Fezf2− and Lhx2+/Fezf2+ radial glia are intermixed in the early ventricular zone. At mature cortical stages, both lineages span cortical layers, but Lhx2+/Fezf2+ progeny are nearly exclusively non-callosal. This provides additional evidence for the existence of fate restricted radial glia subpopulations that are slated to produce only a subset of excitatory pyramidal neurons defined by their axonal projections.
- 46.Chou SJ and Tole S, Lhx2, an evolutionarily conserved, multifunctional regulator of forebrain development. Brain Res, 2019. 1705: p. 1–14. [DOI] [PubMed] [Google Scholar]
- 47.Lodato S, et al. , Gene co-regulation by Fezf2 selects neurotransmitter identity and connectivity of corticospinal neurons. Nat Neurosci, 2014. 17(8): p. 1046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsyporin J, et al. , Transcriptional repression by FEZF2 restricts alternative identities of cortical projection neurons. Cell Rep, 2021. 35(12): p. 109269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cardenas A, et al. , Evolution of Cortical Neurogenesis in Amniotes Controlled by Robo Signaling Levels. Cell, 2018. 174(3): p. 590–606 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cardenas A and Borrell V, Molecular and cellular evolution of corticogenesis in amniotes. Cell Mol Life Sci, 2020. 77(8): p. 1435–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villalba A, Gotz M, and Borrell V, The regulation of cortical neurogenesis. Curr Top Dev Biol, 2021. 142: p. 1–66. [DOI] [PubMed] [Google Scholar]
- 52.Haubensak W, et al. , Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A, 2004. 101(9): p. 3196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, et al. , Transcriptional priming as a conserved mechanism of lineage diversification in the developing mouse and human neocortex. Sci Adv, 2020. 6(45). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pebworth MP, et al. , Human intermediate progenitor diversity during cortical development. Proc Natl Acad Sci U S A, 2021. 118(26). [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study provides the first extensive characterization of transcriptomic and morphological subtypes of intermediate progenitors in the developing human cortex. Interestingly, distinct transcriptomic subtypes correlate with phases of neurogenesis but do not appear to correlate with intermediate progenitor morphologic subtypes.
- 55.Kriegstein A, Noctor S, and Martinez-Cerdeno V, Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci, 2006. 7(11): p. 883–90. [DOI] [PubMed] [Google Scholar]
- 56.Vasistha NA, et al. , Cortical and Clonal Contribution of Tbr2 Expressing Progenitors in the Developing Mouse Brain. Cereb Cortex, 2015. 25(10): p. 3290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mihalas AB, et al. , Intermediate Progenitor Cohorts Differentially Generate Cortical Layers and Require Tbr2 for Timely Acquisition of Neuronal Subtype Identity. Cell Rep, 2016. 16(1): p. 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huilgol D LJ, Galbavy W, Wang B-S, He M, Suryanarayana SM, Huang ZJ, Direct and indirect neurogenesis generate a mosaic of distinct glutamatergic projection neuron types and cortical subnetworks, in bioRXiv. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Using a sophisticated intersectional genetic labeling strategy, the authors identify neuronal progeny that arise from direct neurogenesis from radial glia or indirect neurogenesis from intermediate progenitors. They show that direct and indirect neurogenesis differentially contribute to the major cortical excitatory neuronal classes. Intratelencephalic neurons, the most evolutionarily advanced and diverse excitatory cortical population, are generated almost exclusively through indirect neurogenesis.
- 59.Bandler RC, et al. , Single-cell delineation of lineage and genetic identity in the mouse brain. Nature, 2022. 601(7893): p. 404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]; *The authors use viral barcoding of neuronal progenitors at early embryonic stages and subsequent single cell RNA sequencing at postnatal ages to track the lineage relationships of clonally related progeny. They demonstrate that early-labeled progenitors generate larger numbers of progeny across a greater number of neuronal and glial transcriptional classes than later-labeled progenitors. A major finding from this study is that GABAergic neuronal precursors of the ventral telencephalon demonstrate marked lineage divergence.
- 60.Ratz M, et al. , Clonal relations in the mouse brain revealed by single-cell and spatial transcriptomics. Nat Neurosci, 2022. 25(3): p. 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study utilized viral delivery of oligonucleotide barcodes to neuronal progenitors in the mouse forebrain at embryonic stages followed by dissociation and single cell RNA sequencing at postnatal ages. They describe high clonal fate restriction of both hippocampal neurons and microglia. Regarding cortical neurons, they describe a nearly equal split between clones that generate progeny across cell types and those that generate progeny restricted to only one cell type, though they caution that with a low sampling rate of clonal progeny this is not conclusive evidence for fate restricted progenitors. They also note excitatory progenitors capable of generating progeny that reside across cortical layers.


