Abstract
All humans carry a small fraction of archaic ancestry across the genome, the legacy of gene flow from Neanderthals, Denisovans, and other hominids into the ancestors of modern humans. While the effects of Neanderthal ancestry on human fitness and health have been explored more thoroughly, there are fewer examples of adaptive introgression of Denisovan variants. Here, we study the gene MUC19, for which some modern humans carry a Denisovan-like haplotype. MUC19 is a mucin, a glycoprotein that forms gels with various biological functions, from lubrication to immunity. We find the diagnostic variants for the Denisovan-like MUC19 haplotype at high frequencies in admixed Latin American individuals among global population, and at highest frequency in 23 ancient Indigenous American individuals, all predating population admixture with Europeans and Africans. We find that some Neanderthals––Vindija and Chagyrskaya––carry the Denisovan-like MUC19 haplotype, and that it was likely introgressed into human populations through Neanderthal introgression rather than Denisovan introgression. Finally, we find that the Denisovan-like MUC19 haplotype carries a higher copy number of a 30 base-pair variable number tandem repeat relative to the Human-like haplotype, and that copy numbers of this repeat are exceedingly high in American populations. Our results suggest that the Denisovan-like MUC19 haplotype served as the raw genetic material for positive selection as American populations adapted to novel environments during their movement from Beringia into North and then South America.
Introduction
It is widely accepted that most modern humans of non-African ancestry carry both Neanderthal and Denisovan genomic variants, and that these variants have been the targets of natural selection [Yang et al., 2012; Lohse and Frantz, 2014]. For the majority of genes found in modern humans, archaic variants appear to have been removed from the gene pool by purifying natural selection due to a lower historical effective population size in archaic populations [Sankararaman et al., 2016; Petr et al., 2019; Zhang et al., 2020]. However, a handful of archaic variants have risen to high frequency in modern humans through positive linked selection, which previous studies have concluded is a result of natural selection acting on introgressed variants [Racimo et al., 2015; Zhang et al., 2023]. In populations exposed to novel or changing environments, introgressed variants have been implicated as the genetic source for unique adaptations, and several Neanderthal variants that may have facilitated human adaptations have been identified [Mendez et al. 2012; Sankararaman et al., 2014; Vernot and Akey, 2014; Gittelman et al., 2016;Sams et al., 2016; Dannemann and Kelso, 2017; Marnetto, et al. 2017]. In contrast, the study of Denisovan introgression has identified far fewer candidate genes, outside of the lipid metabolism locus TBX15/WARS2 in Inuit from Greenland and Native Americans, and the case of high-altitude adaptation in Tibetans primarily driven by the EPAS1 gene [Huerta-Sánchez et al. 2014; Racimo et al. 2017; Zhang et al., 2021].
Interbreeding with Neanderthals and Denisovans may have therefore facilitated adaptation to the myriad of novel environments that modern humans encountered [Fan et al., 2016], and several studies have identified signatures of adaptive introgression in Eurasian and Oceanian populations. Indigenous American populations, however, present the greatest potential for studying local adaptation as they are the descendants of individuals who populated the American continent in a process that started approximately 25,000 years before present [Tamm et al., 2007]. In the 25,000 years since, these individuals would have encountered manifold novel environments, far different from the environment their ancestral population was adapted to in the Beringian steppe [Beck et al., 2018].
In a previous study, we computed the Population Branch Statistic (PBS, Yi et al., 2010) using SNPs within archaic introgressed tracts in admixed populations in the Americas to identify targets of adaptive introgression. We found the region surrounding MUC19 (a gene involved in immunity) harbors several variants with outlier PBS scores in Mexicans (MXL) from the 1000 Genomes Project (TGP), suggesting that the archaic allele was at high frequency in Mexicans [Witt et al., 2023]. Earlier studies had reported that this region has one of the largest densities of Denisovan alleles in Mexicans [Racimo et al., 2017], and MUC19 was also reported under positive selection in North American Indigenous populations [Reynolds et al., 2019].
In this study, we conducted a closer inspection of the MUC19 gene, a candidate for adaptive introgression, to 1) confirm that this region exhibits both signatures of introgression and positive selection in MXL, 2) determine whether other admixed populations have the introgressed segment and 3) the role of recent admixture in diluting this signal. Notably, we find a Denisovan-like haplotype segregating at high frequency in most populations in the American continent, and surprisingly that both the Vindija and Chagyrskaya Neanderthals also harbor the Denisovan-like haplotype, which may suggest introgression from Denisovans into the late Neanderthals. Our analysis also shows that MXL individuals exhibit differences in archaic allele frequency at coding sites in MUC19, and differences in copy number compared to other populations. These results point to a complex pattern of multiple introgression events, one of which may have played a unique role in the evolutionary history of Indigenous American populations.
Results
Signatures of adaptive introgression at MUC19 in admixed populations from the Americas.
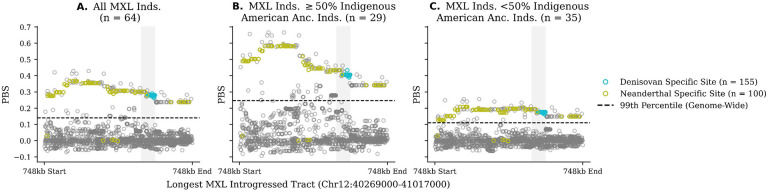
We compiled introgressed tracts that overlap by at least one base pair of the NCBI RefSeq coordinates for MUC19 (hg19, Chr12:40787196–40964559). Figure S1 shows the density of introgressed tracts for all non-African populations in the region, using introgression maps from Skov et al. [2018]. All non-African populations harbor the introgressed haplotype, but at much smaller frequencies than the admixed populations from the Americas. Given this, we then took a 748kb window containing the longest introgressed tract found in Mexicans (MXL, hg19, Chr12:40269000–41017000) to compute PBS scores for each SNP in that region (Figure S2). Figure 1 shows the results for MXL, a population with a large component of Indigenous American ancestry [Martin et al., 2017]. We find that, in MXL, there are many SNPs with statistically significant PBS values in that region (375/2043), all which present values above the 99th percentile of genome-wide PBS values (P-value: 9.413e-4; see Methods).
Figure 1.
Population Branch Statistic (PBS) values for SNPs within the 748kb (Chr12:40269000–41017000) longest introgressed tract for a population from Mexico (MXL) using a Han Chinese population (CHB) and a Central European population (CEU) as outgroups. Cyan points represent sites shared uniquely with the sequenced Denisovan, and olive points represent sites shared uniquely with Neanderthals, gray points represent sites that are not shared uniquely with either Neanderthals or Denisovans. The shaded region corresponds to the focal 72kb region with Denisovan affinities. The dotted line represents the 99th percentile of PBS scores for all SNPs genome-wide with respect to each comparison; points above the dotted line represent SNPs with significant PBS values. A) All individuals in the MXL populations, B) only MXL individuals with 50% or more genome-wide Indigenous American ancestry, C) only MXL individuals with less than 50% genome-wide Indigenous American ancestry. Python code to replicate this figure is available at: https://github.com/David-Peede/MUC19/blob/main/figure_nbs/figure_1_v_submission.ipynb.
To test if the signal for positive selection is being driven by the Indigenous American components of ancestry, we took a subset of 29 individuals in the MXL populations harboring more than 50% of Indigenous American ancestry genome-wide [Witt et al., 2023]. We recomputed PBS using the genomes of these individuals and found that PBS values for archaic variants were significantly elevated, suggesting that this region was likely targeted by selection before admixture with European and African populations (Table S1–S2). However, as this pattern can be confounded by heterosis [Zhang et al., 2020], we ran simulations but found that heterosis does not lead to the PBS values observed at this gene region (see Supplement).
Identification of the most likely donor of the introgressed haplotype at MUC19
We compared the distribution of introgressed tracts across all non-African populations (Figure S1). We focused our analyses on two regions. First the region with the highest density of introgressed tracts across individuals—a 72kb region (hg19, Chr12:40758000–40830000) within MUC19 and with some overlap with LRRK2 (see shaded gray region in Figure 1). As the inferred introgressed tracts in MXL all span beyond this region, we also consider a second region containing the longest introgressed tract found in MXL (same as the PBS section above), and encompassing the 72kb region. We compare the haplotypes in these two regions to the sequenced archaic humans to investigate the diversity of haplotypes at MUC19 and to identify the most likely archaic donor. We calculated the sequence divergence—the number of pairwise differences normalized by the effective sequence length—between all haplotypes in the TGP and the diplotypes for one Denisovan, and the three high-coverage Neanderthal individuals (Figure S3, Table S3).
The focal 72kb region exhibits a sharply bimodal distribution of sequence divergences between affinities to the sequenced Denisovan and the Altai Neanderthal, separating all haplotypes into two non-overlapping sets, with the exception of seven recombinant haplotypes (Figure 2, Figure S4). We infer that non-African populations harbor a Denisovan-like introgressed haplotype at the 72kb region, suggesting that the donor population was more closely related to the sequenced Denisovan than to the Altai Neanderthal. Interestingly, for the Vindija and Chagyrskaya Neanderthals, all haplotypes from the TGP grouped together at intermediate distances (Figure S3). Notably, this 72kb region is an extreme outlier for Denisovan-specific SNP density (Denisovan-specific derived SNPs: 84, P-value: <3.242e-5; Denisovan-specific ancestral SNPs: 65, P-value: <3.242e-5; Figure S5, Table S4) while having a relatively average Neanderthal-specific SNP density (Neanderthal-specific derived SNPs: 5, P-value: 0.59; Neanderthal-specific ancestral SNPs: 0, P-value: >0.99968; Figure S6, Table S5) compared to other similarly-sized regions across the genome.
Figure 2.
Sequence divergence for the MUC19 72kb region—number of pairwise differences between a modern human haplotype and an archaic diplotype normalized by the effective sequence length—between all individuals in the TGP for each global superpopulation by column: A) Africa (AFR), B) South Asia (SAS), C) East Asia (EAS), D) Europe (EUR), E) America (AMR). The first row of each column shows the distribution of sequence divergence from the sequenced Denisovan. The second row of each column represents the normalized distance from the sequenced Denisovan on the x-axis compared to the normalized distance from the Altai Neanderthal on the y-axis. Python code to replicate this figure is available at: https://github.com/David-Peede/MUC19/blob/main/figure_nbs/figure_2_v_submission.ipynb.
To formally test for introgression and to identify the source population, we used the D+ statistic [Lopez-Fang et al., 2022; Peede et al., 2022]. We performed D+ (P1, P2; P3, Outgroup) tests with the following configurations: Yorubans (YRI) as P1; MXL as P2; and one of the four archaic genomes as P3 For comparisons involving the sequenced Denisovan as P3, we find a positive and significant D+ value for MXL suggesting more allele sharing with Denisovans compared to other global populations (Figure S7, Table S6). When we use other non-African TGP populations as P2, the only other population with a significant and positive D+ value is the Peruvian (PEL) population (Figure S7, Table S6). When the Altai Neanderthal is P3, we observe a negative and significant D+ value in MXL, suggesting the 72 kb region may not be of Neanderthal origin (Figure S8, Table S7). For all other comparisons of non-African populations as P2, involving a Neanderthal as P3, we observe non-significant D+ values (Figures S9–S10, Tables S8–S9). These D+ results confirm that the introgressed haplotypes in MXL—within the 72kb region—share more alleles with the sequenced Denisovan than with Neanderthals.
As the inferred introgressed tracts in MXL individuals are longer than the boundaries of the 72kb region, we also examined the region containing the longest introgressed tract in MXL—a 748kb region (Figure S2). Surprisingly, when we consider the longest introgressed tract, one that encompasses the 72kb region, we find that it is significantly closer exclusively to two Neanderthals than expected from the genomic background (Chagyrskaya sequence divergence: 0.0007, P-value: 0.006; Vindija sequence divergence: 0.0007, P-value: 0.007; Table S10). This result suggests that the donor of this longest introgressed tract likely came from a Neanderthal population that contains a region (72kb) that is more similar to the Denisovans (shaded gray region in Figure 1).
Denisovan-like introgression into Neanderthals and modern humans
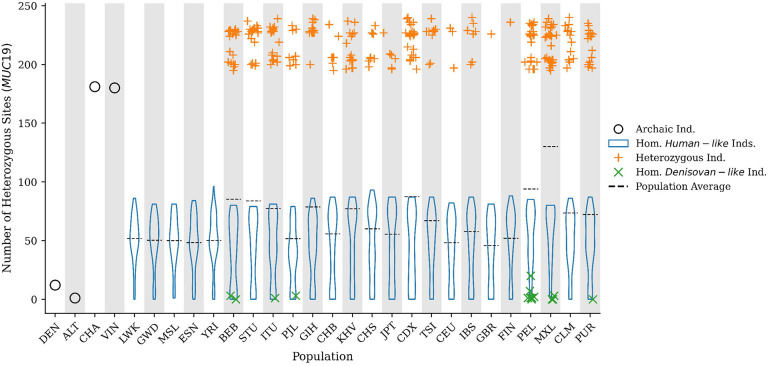
To understand why the Vindija and Chagyrskaya Neanderthals are not as distant to the Denisovan-like haplotype present in MXL (Figure S3), we computed the number of heterozygous sites per individual for the 72kb region. We find that individuals carrying exactly one Denisovan-like haplotype present significantly more heterozygous sites at MUC19 compared to the rest of their genome (average number of heterozygous sites: ~218, P-value: 3.242e-5; Figure S11, Table S11) which surpasses that of any African individual (orange crosses in Figure 3). Individuals carrying exactly two Denisovan-like haplotypes present significantly less heterozygous sites than expected at MUC19 relative to the rest of their genome (average number of heterozygous sites: ~3, P-value: 0.001; Figure S11, Table S11), while African individuals harbor an expected number of heterozygous sites (average number of heterozygous sites: ~50, P-value: 0.455; Figure S11, Table S11). Strikingly, we observe that the Vindija and Chagyrskaya Neanderthals also carry an elevated number of heterozygous sites (Vindija: 181, P-value: 0.0004; Chagyrskaya: 182, P-value: 0.0002) that is significantly higher than the Altai Neanderthal (heterozygous sites: 1, P-value: 0.799), the sequenced Denisovan (heterozygous sites: 12, P-value: 0.279), all African individuals, and comparable to modern humans carrying exactly one Denisovan-like haplotype (Figure 3, Figure S12, Table S12). This observation runs opposite to the genome-wide expectation for late-stage Neanderthals, as archaic humans have much lower heterozygosity than modern humans [Mafessoni et al., 2020]. The surprisingly high heterozygosity in the Vindija and Chagyrskaya Neanderthals suggest that they may also harbor one Denisovan-like haplotype.
Figure 3.
Number of heterozygous sites per individual across the MUC19 72kb introgressed region for the Denisovan (DEN), Altai Neanderthal (ALT), Chagyrskaya Neanderthal (CHA), Vindija Neanderthal (VIN), and the TGP modern human populations. Black dots represent archaic individuals, blue violin plots represents the distributions of homozygous individuals carrying two copies of the Human-like haplotype, orange crosses represents heterozygous individuals carrying one copy of each haplotype, green xs represent homozygous individuals carrying two copies of the Denisovan-like haplotype, and the dashed lines represent the population average. Python code to replicate this figure is available at: https://github.com/David-Peede/MUC19/blob/main/figure_nbs/figure_3_v_submission.ipynb.
Due to the inability to phase Ancient DNA data, to investigate if the Chagyrskaya and Vindija Neanderthals carry a Denisovan-like haplotype, we developed an approach referred to as Pseudo-Ancestry Painting (PAP, see Methods) to assign the two alleles at a heterozygous sites to two source populations. Specifically, if we use a MXL individual who is homozygous for the Denisovan-like haplotype and a YRI individual who carries no Denisovan-like haplotypes, we find that ~93% of heterozygous sites in the 72kb region (Chagyrskaya: 169/181; Vindija: 168/180) in the Neanderthals are fixed differences between the MXL and YRI individuals. This further supports that both Chagyrskaya and Vindija Neanderthals have the Denisovan-like haplotype that is present in the MXL individuals. We found that using a MXL and a YRI individual maximizes the number of heterozygous sites (in the Chagyrskaya and Vindija Neanderthals) whose alleles are present in the two sources (Figure S13, Table S13).
Additionally, we performed four tests for gene flow between the archaic individuals using the D+ statistic for the 72kb regions that provide evidence that the Chagyrskaya and Vindija Neanderthals harbor one copy of the Denisovan-like haplotype at the 72kb MUC19 region. For the first two comparisons, where the Altai Neanderthal is P1, either the Chagyrskaya and Vindija Neanderthals are P2, and the sequenced Denisovan is P3, we observe significant and positive D+ values, supporting gene flow between the Denisovan and the Chagyrskaya and Vindija Neanderthals (Figure S14, Table S14). For the other two comparisons, where the Chagyrskaya Neanderthal is P1, the Vindija Neanderthal is P2, and P3 is either the Altai Neanderthal or Denisovan, we observed D+ values that do not significantly differ from zero (Figure S15, Table S15).
In sum, our analysis suggests that non-Africans have a mosaic region of archaic ancestry, with a small Denisovan haplotype (72kb) embedded in a large Neanderthal haplotype (748kb), that was inherited through Neanderthals, who themselves acquired Denisovan ancestry from an earlier introgression event. This is consistent with the literature, where Denisovan introgression into Neanderthals was rather common [Slon et al., 2018; Peter, 2020].
Introgression introduced missense mutations at MUC19
Inspecting the 72kb introgressed region to check the location of archaic alleles reveals that the Denisovan-like haplotype in humans carries six non-synonymous sites where the archaic allele codes for a different amino acid relative to the Human-like haplotype. We then quantified the allele frequencies for these six derived Denisovan-specific missense mutations in present-day populations and in 23 ancient Indigenous American genomes that predate European colonization and the African slave trade (Figure 4, Figure S16, Table S16).
Figure 4.
Heatmap depicting the frequency of six Denisovan-specific derived missense mutations in the Denisovan-like haplotype in Latin American populations, as well as three Neanderthals, and a panel of ancient Indigenous American individuals. The top of each column denotes the position (in hg19) of the six Denisovan-specific derived missense mutations and the amino acid change (ancestral amino acid → derived amino acid). Top panel: amino acid genotype for the missense mutations in the four archaic individuals and fixed in modern Africans. Lower panel: allele frequencies in the Latin American populations depicted individually—Puerto Rico (PUR), Colombia (CLM), Peru (PEL), and Mexico (MXL). The “n” represents the number of chromosomes in each sample. For the Ancient Americans, the proportion of derived missense mutations relative to the number of chromosomes that pass quality control are given for each site. Python code to replicate this figure is available at: https://github.com/David-Peede/MUC19/blob/main/figure_nbs/figure_5_v_submission.ipynb.
In modern Admixed American populations, we find that the Denisovan-specific missense mutations are segregating at high frequencies with respect to the expectation for archaic ancestry (Figure 4, Table S17). We find the Denisovan-specific missense mutations at varying frequencies, between 0.005 and 0.167 in European, East Asian, and Southeast Asian populations (Table S17). We additionally confirmed the presence of the Denisovan-like MUC19 haplotype amongst the 15 Papuan individuals (Denisovan-like haplotype frequency: 0.1) present in the Simons Genome Diversity Project (SGDP), a population with a much larger component of Denisovan ancestry, and that the Papuans with the Denisovan-like haplotype have all six Denisovan-specific missense mutations (Table S17).
In the 23 ancient pre-European colonization American individuals, we find that each of the six Denisovan-specific missense mutations are segregating at higher frequencies than in any Admixed American population. When we quantify the frequency of these mutations in 22 Indigenous Americans from the SGDP, we find that all six Denisovan-specific missense are segregating at a frequency of ~0.364, which is similar to the ancient Americans and higher than any Admixed American population in the TGP (Table S17–19). Taken together these results suggest that admixture events have diluted the Denisovan ancestry observed at the 72kb MUC19 region.
To estimate the effect on the protein of these missense substitutions, we relied on Grantham scores [Grantham, 1974]—a quantification of protein evolution based on predicted chemical qualities. Notably, one of the Denisovan-specific missense mutations found at position 40821871 (rs17467284) results in an amino acid change with score 102, classified as moderately radical in Li et al. [1985], and falling within an exon conserved across vertebrates (PhyloP score 5.15, Figure S17, [Pollard et al., 2010]. Furthermore, this missense mutation falls between two Von Willebrand factor D domains, which play an important role in the formation of mucin polymers and gel-like matrices [Javitt et al., 2020]. Our results suggest that this missense mutation is a strong candidate for impacting its translated protein, which may affect the polymerization properties of MUC19 and the viscosity of the mucin matrix.
Admixed individuals exhibit an elevated number of variable number tandem repeats at MUC19
MUC19 is structurally similar to other mucins, containing a variable number tandem repeat with a 30 base-pair repeat motif (Figure S18), which is 46.4kb away from the core 72kb haplotype, but within the larger 748kb introgressed region. To test if the Human-like and Denisovan-like MUC19 haplotypes differ in the number of repeats, we calculated the number of repeat copies of the 30 base-pair motif in the TGP individuals (see Methods).
Surprisingly, individuals in American populations carrying the Denisovan-like MUC19 haplotype present an elevated number of repeats relative to most other individuals in the TGP (Figure 5). We found that non-American populations range from an average of 345 to 355 repeats (Table S20). In contrast, admixed individuals from the Americas have on average 417 copies. Furthermore, we find that on average individuals harboring at least one Denisovan-like haplotype have a significantly elevated number of repeat copies, than individuals harboring two Human-like haplotypes (Denisovan-like mean: 446, Human-like mean: 349, Welch’s T-test P-value: 6.423e-28). We find that Admixed American populations have a significantly greater proportion of individuals with repeat copies greater than the non-African mean, than all other non-African populations (Fisher’s exact test, odds ratio: 2.2, P-value: 4.976e-11; Table S21).
Figure 5.
Number of repeat copies of a 30 base-pair motif in the TGP individuals at MUC19. Blue violin plots represent the distributions of homozygous individuals carrying two copies of the Human-like haplotype, orange crosses represents heterozygous individuals carrying one copy of each haplotype, green Xs represent homozygous individuals carrying two copies of the Denisovan-like haplotype, and the dashed lines represent the population average. Repeat copies appeared similar in length to the reference human genome (287.5 copies) in Denisovan and Neanderthal archaic genomes. Python code to replicate this figure is available at: https://github.com/David-Peede/MUC19/blob/main/figure_nbs/figure_6_v_submission.ipynb.
Importantly, these findings were corroborated through analysis of a subset of these samples using long-read sequence data from the Human Pangenome Reference Consortium (HPRC) and Human Genome Structural Variant Consortium (HGSVC), that revealed an extra four copies of a 3,171 bp segment of the MUC19 tandem repeat exclusively in American samples. This structural variant effectively doubles the size of the ~12kb coding exon that harbors the tandem repeat (Figures S19–S20, Tables S22–S23). This suggests that functional differences between the Human-like and Denisovan-like may lie in the elevated number of the 30 base-pair motifs carried in the Denisovan-like haplotype, and that the positive selection detected in American populations may be acting on haplotypes carrying elevated copy numbers of the 30 base-pair motif.
Discussion
The study of adaptive archaic introgression is in its infancy, but has already illuminated candidate genomic regions that affect the health and overall fitness of global populations. In this study, we pinpointed several aspects of the gene MUC19 that highlight its importance as a candidate to study adaptive introgression: the haplotype that spans this gene in modern humans is likely of Denisovan-like origin; the haplotype introduced six missense mutations that are at high frequency in both Indigenous and Admixed American populations; individuals with the archaic haplotype carry a higher copy number of proline, threonine, and serine (PTS) tandem repeats relative to the non-archaic haplotype, and their functional differences may help explain how mainland Indigenous Americans adapted to their environments, which remains under-explored. To our knowledge, this study provides the first example of natural selection acting on archaic alleles at coding sites and the first example of natural selection acting on VNTRs.
Here, we find the Denisovan-like haplotype of MUC19 segregating at high frequency in most American populations, including representative populations of North, Central, and South America. Our results are consistent with the Denisovan-like haplotype being responsible for the signals of positive selection at the MUC19 locus in admixed American populations, suggesting that this haplotype is a candidate for adaptation in these populations. The high frequency of the Denisovan-like MUC19 in modern admixed American populations is encouraging, supporting a large fitness role for this haplotype.
A larger implication of our findings is that archaic ancestry could have been a useful source of standing genetic variation as the early Indigenous American populations adapted to new environments, with genes like MUC19 and other mucins possibly mediating important fitness effects [Xu et al., 2016]. The variation in the copy number of PTS tandem repeats present in global populations dovetails with this idea. Yet, in American populations, particular haplotypes carrying the most extreme copy numbers were selected and are now very frequent, indicating an adaptive role driven by environmental pressures particular to the Americas. Another implication for living American individuals is that if archaic variants such as the Denisovan-like MUC19 haplotype are adaptive, the depletion of archaic ancestry in the Indigenous component of Latin American genomes by European and African population admixture could carry negative fitness effects. This is a recurring idea in personalized medicine serving admixed individuals, where epistasis between genome elements from diverging ancestries can lead to negative health effects such as, for example, increased incidences of autoimmune conditions [Martin et al., 2017]. This is a complex topic, however, as admixture can also carry positive fitness effects, such as breaking up deleterious gene interactions that are otherwise fixed in a population.
Another interesting aspect of MUC19 is the evolutionary history of the introgressed region. We find that while the 72kb-region that spans MUC19 is most similar to the sequenced Denisovan, it is contained within a much larger introgressed haplotype. The size of the longest introgressed haplotype in MXL is 748kb, and is most likely of Neanderthal origin. While the Altai Neanderthal does not harbor the Denisovan-like haplotype in the 72kb region, the other two late Neanderthals (Vindija and Chagyrskaya) do. Our results suggest that late Neanderthals experienced introgression from a Denisovan-like population before Neanderthals introgressed into modern humans. Another striking feature of the introgressed haplotype in the 72kb region is that the sequenced Denisovan is highly divergent from both the Altai Neanderthal and modern Africans, which is one of the signatures expected under a model of super-divergent introgression. Simulating under a demographic model with parameters inferred in Hubisz et al., [2020] cannot explain the summary statistics observed for this region (see Supplement). This in turn may suggest that these two highly divergent haplotypes were maintained in archaic populations via balancing selection [Viscardi et al., 2018]. Balancing selection in the form of heterozygous advantage may also explain why the Denisovan-like haplotype is found at high frequencies in American populations yet did not reach fixation in any sampled population. More generally, the evolutionary history of this regions suggests a complex history that involves recurrent introgression and natural selection, and it parallels findings from other regions of the genome (Neanderthal mtDNA, Y chromosome, and KNL1 spindle gene [Posth et al., 2017; Petr et al., 2019;Peyrégne et al., 2022]).
Perhaps the largest knowledge gap in why the Denisovan-like haplotype of MUC19 would be under positive selection is determining its underlying function. Mucins are secreted glycoproteins responsible for the gel-like properties and the viscosity of the mucus [Pajic et al., 2022]. MUC19 is associated with gland function in humans, which can be important components of immune response, in particular its role in mucus formation in tracheal submucosal glands in preventing pathogenic infections [Chen et al., 2004; Zhu et al., 2011;Reynolds et al., 2019]. The differences in copy numbers of the 30bp PTS tandem repeat domains carried by individuals harboring the Human-like and Denisovan-like haplotypes certainly suggests MUC19 variants differ in function as a consequence of different molecular binding affinities between variants. This is the case in other mucins, such as MUC7, where variants carrying different numbers of PTS repeats exhibit different microbe-binding properties [Xu et al., 2016; Xu et al., 2017]. If the two variants of MUC19 also present differential binding properties, this would lend support towards why positive selection would increase the frequency of the Denisovan-like haplotype in American populations. As there is limited medical literature associating variation in MUC19 with human fitness, further experimental validation investigating how VNTRs or the Denisovan-specific missense mutations affect function, is necessary to understand the role the Denisovan-like haplotype may exert on the translated MUC19 protein, and how it modifies its function during the formation of mucin polymers.
Finally, beyond improving our understanding of how archaic variants facilitated adaptation in novel environments, our findings also highlight the importance of studying archaic introgression in understudied populations, such as admixed Latin Americans [Villanea and Witt, 2022]. Genetic variation in American populations is less well-characterized than other global populations, due to the difficulties in deconvoluting Indigenous ancestries from European and African ancestries, and to a lesser extent Southeast Asian ancestry, following 500 years of European colonization [Martin et al., 2017]. This knowledge gap is exacerbated by the high cost of performing genomic studies, building infrastructure, and generating scientific capacity in Latin America—but it is a worthwhile investment—as our study shows that leveraging these populations can lead to the identification of exciting candidate loci that can expand our understanding of adaptation from archaic standing variation.
Our study also shows that harnessing methods developed in evolutionary biology are useful for identifying candidate variants underlying a biological function. Future functional and evolutionary studies of the MUC19 region will not only provide insights into the specific mechanisms of how the variation at this gene confers a selective advantage, but may also be informative about specific evolutionary events that occurred in the history of humans.
Methods
Data Processing
Modern human genome data
Sequence data for the MUC19 locus were obtained from a publicly available global reference panel, the 1,000 Genomes Project Phase III (TGP), which contains a diverse set of individuals from multiple populations [1000 Genomes Project Consortium, 2015]. The autosomal variant sites from the integrated callset VCF files for the TGP were downloaded from http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502. Data for MUC19 in the Papuan and present day Indigenous American individuals was obtained from the Simons Genome Diversity Project (SGDP) [Wong et al., 2020]. The autosomal variant sites VCF files for the SGDP were downloaded from https://sharehost.hms.harvard.edu/genetics/reich_lab/sgdp/phased_data2021. It should be noted here, that the analyses in the section “Copy number polymorphism of a 30bp tandem repeat motif between the Human-like and a Denisovan-like haplotypes” were conducted on the TGP data aligned to the hg38 reference assembly, while all other analyses completed using the hg19 reference which was soft masked for repetitive regions.
Archaic human genome data
The autosomal all-sites VCF files for the four high-coverage archaic genomes were downloaded from https://www.eva.mpg.de/genetics/genome-projects and the ancestral allele calls in fasta format for the hg19 assembly using the Enredo, Pecan, Ortheus (EPO) pipeline was download from http://ftp.ensembl.org/pub/release-74/fasta/ancestral_alleles [Paten et al. 2008a, Paten et al. 2008b, Herrero et al. 2016]. The autosomal VCF files for the archaic genomes were initially merged using BCFtools v1.13, and after the initial merging the resulting VCF files were filtered to only include sites that were mono-allelic or bi-allelic with an ancestral allele call present and where at least one archaic had a MQ >= 25 and GQ >= 40—archaics that did not meet this threshold were coded as missing data.
Combined data set
The autosomal VCF files for each modern human dataset (i.e., TGP and SGDP) and archaic genomes were initially merged using BCFtools v1.13, after the initial merging the resulting VCF files were filtered to only include sites with mono-allelic or bi-allelic SNPs with an ancestral allele call present and where at least one archaic had a MQ >= 25 and GQ >= 40—archaics that did not meet this threshold were coded as missing data. Since the TGP genotypes were imputed and only include information for variable sites—unlike the archaic data which contains information for all sites—any site that was originally absent in the TGP data but present in archaic data we assumed to be homozygous for the reference allele in the TGP data [Huerta-Sánchez et al 2014]. After the final dataset was curated we annotated coding sites using SnpEff v5.1 [Cingolani et al., 2012].
Pre-Contact Indigenous American Genomes
Genomic data for MUC19 in ancient individuals was generated by combining high coverage (>1X) pre-European contact genomes from the literature, including nine individuals from California, one from Ontario [Scheib et al., 2018], four from Peru [Lindo et al., 2018] four from Patagonia [de la Fuente et al., 2018], one from Alaska [Moreno-Mayar et al., 2018], one from Montana [Rasmussen et al., 2014], and three from Central Mexico [Villa-Islas et al., 2023].
Sequence reads were downloaded in fastq format and converted to bam format using bwa v7.17 [Li and Durbin, 2009]. Reads were then sorted, duplicates were removed, and all non-autosomal chromosomes were removed using Samtools v1.9 [Li et al., 2009]. Using ANGSD v0.92 we further filtered out reads that had a quality score less than 30 and then determined the read depth of the alleles present at the six sites (hg19 Chr12: 40808672, 40808726, 40815060, 40821795, 40821847, 40821871) where the Denisovan is the only archaic who is fixed for the derived allele missense mutation, which is absent in the Altai Neanderthal. Then for each ancient individual, we determined the genotype at each of the six Denisovan-specific missense sites which we had sequencing information for by first considering any allele that had a read depth of two or greater, and then ensured that site was mono-allelic or bi-allelic for only the derived and/or ancestral allele(s).
Positive Selection
We utilized the population branch statistic (PBS) to assess if the Denisovan-like haplotype has been subjected to positive selection in Admixed American populations. PBS uses the logarithmic transformation of pairwise estimates of FST to measure the branch length in the target population since its divergence from the two control populations [Yi et al., 2010]. We chose MXL as target populations—as they harbor the largest number of Denisovan-like haplotypes and are the least-admixed populations [Martin et al., 2017]—and CEU and CHB as our control populations. To assess if the PBS values SNPs in the 748kb longest introgressed tract region are elevated in Indigenous American individuals we conducted three separate PBS analyses on the MXL target population: 1) with all 64 MXL individuals, 2) only for the 29 MXL individuals with 50% or more Indigenous American ancestry as defined by [Martin et al., 2017], and 3) only for the 35 MXL individuals with less than 50% Indigenous American ancestry as defined by [Martin et al., 2017]. To account for differences in sample sizes we used Hudson’s estimator of FST as it has been shown to not only be a conservative estimator, but is also robust to differences in sample sizes [Bhatia et al., 2013]. We chose this experimental design to assess if the signal of positive selection is elevated in individuals with high proportions of Indigenous American ancestry—as genomic data is scarce for Indigenous American populations—and to assess if recent admixture with European and African populations dilutes the signal of positive selection. For each analysis we used an outlier approach to identify if a given SNP exhibits signatures consistent with positive selection by setting the significance threshold at the genome-wide 99th PBS percentile.
To calculate the P-value for the 748kb longest introgressed tract region we computed the observed mean PBS(MXL:CHB:CEU) value for the longest introgressed tract region and compare the observed mean to a distribution of mean PBS values from the genomic background of 748kb non-overlapping windows with comparable effective sequence length density—i.e., an effective sequence length of at least 400kb amongst the combined dataset—and determined proportion of windows from the genomic background with a mean PBS value greater than or equal to what we observed at the 748kb region.
Identification of the MUC19 Introgressed Region
To identify the genome coordinates of the introgressed region for the modern human MUC19, we downloaded the inferred introgressed tracts from Skov et al. [2018] for chromosome 12, and only retained inferred tracts that had a posterior probability greater than or equal to 0.8, and only retained inferred tracts that overlapped with at least one base pair of the MUC19 NCBI RefSeq coordinates for the hg19 assembly (Chr12:40787196–40964559). We then identified two distinct regions, the region containing the longest archaic tract in any MXL individual (Chr12:40269000–41017000, in NA19789), and a focal 72kb region (Chr12:40758000–40830000) which has the highest density of inferred introgressed tracts amongst non-African populations in the TGP (see Figure S1–S2).
Archaic SNP Density
To assess if TGP individuals at our focal 72kb MUC19 region harbor more archaic SNPs than expected we computed the number of Denisovan-specific and Neanderthal-specific SNPs. For a SNP to first be considered an archaic allele, we require that an allele must be rare in the African superpopulation (i.e., at a frequency less than 0.01) and present in at least one non-African individual. For the SNP to be considered Denisovan-specific we further required the archaic allele to be fixed in the sequenced Denisovan and not fixed in any of the three high-coverage Neanderthals. Similarly, for a SNP to be considered Neanderthal-specific we further required the archaic allele to be fixed in at least one Neanderthal and not fixed in the sequenced Denisovan. To assess if our 72kb MUC19 region has a higher derived and ancestral allele archaic SNP density than expected, we compared the observed archaic SNP density to a distribution of archaic SNP densities from the genomic background of 72kb non-overlapping windows with comparable effective sequence length density—i.e., an sequence length of at least 40kb amongst the combined dataset. To calculate P-values, we determined the proportion of windows from the genomic background with an archaic SNP density greater than or equal to what we observed at the 72kb MUC19 region. After correcting for two multiple comparisons (i.e., one per allelic state) for both Denisovan-specific and Neanderthal-specific SNPs using the Bonferroni correction, a P-value less than 0.025 is considered significant.
Sequence Divergence
To assess the extent of divergence between MUC19 haplotypes harbored by the various individuals in this study, we calculated sequence divergence which corresponds to number of pairwise differences between chromosomes normalized by the effective sequence length—i.e., the total number of sites that passed quality control. We use the term haplotype to refer to a single chromosome from a phased modern human individual and diplotype to refer to the two chromosomes of an archaic individual for which phasing is not possible.
Identifying the Donor of the Longest Introgressed Tract found in MXL
To determine the most likely archaic source of the 748kb longest introgressed tract (Chr12:40787196–40964559) found in an MXL individual (i.e., NA19789), inferred from Skov et al. [2018] we computed the sequence divergence between each of the NA19789’s chromosomes and the four archaic diplotypes. To identify the archaic donor for the 748kb longest introgressed tract, we compared the observed sequence divergence to a distribution of sequence divergence from the genomic background of 748kb non-overlapping windows with comparable effective sequence length density—i.e., an effective sequence length of at least 400kb amongst the combined dataset. To calculate P-values, we determined the proportion of windows from the genomic background with a sequence divergence less than or equal to what we observed at the 748kb region. After correcting for four multiple comparisons per chromosome using the Bonferroni correction, a P-value less than 0.0125 is considered significant.
Modern Human Haplotype-Archaic Diplotype Divergence
To determine the haplotype identity for the 72kb MUC19 region in TGP individuals, we calculated the sequence divergence for all pairwise possibilities between each TGP chromosome and each archaic diplotype. We then characterized a TGP haplotype as being Denisovan-like if the sequence divergence to the sequenced Denisovan was less than 0.00156 (or 80.5 pairwise differences between a single TGP chromosome and the two archaic chromosomes), corresponding to the valley of the bimodal distribution in Figure 2. Seven TGP chromosomes exhibited intermediary sequence divergence levels with respect to the sequenced Denisovan between 0.00157 and 0.00252 and were determined to be recombinant haplotypes (see Figure S4) and all TGP chromosomes with a sequence divergence larger than 0.00252 include all African chromosomes and were considered to be Human-like haplotypes. Additionally, we computed sequence divergence between all chromosomes for the 15 Papuan individuals from the SGDP dataset and each archaic diplotype and using the same sequence divergence threshold as was done for the TGP dataset identified three Denisovan-like haplotypes and 27 Human-like haplotypes amongst the Papuans.
African and Denisovan-like Individuals-Archaic Diplotype Divergence
To understand if African individuals and individuals carrying the Denisovan-like haplotypes are on average more or less divergent than expected with the sequenced Denisovan we calculated the average sequence divergence for all African individuals (n = 504), heterozygous individuals (n = 255) who carry one copy of the Denisovan-like haplotype, and homozygous individuals (n = 16) who carry two copies of the Denisovan-like haplotype for the focal 72kb MUC19 region. To assess significance, we compared the observed average sequence divergence to a distribution of average sequence divergence from the genomic background of 72kb windows with comparable effective sequence length density—i.e., an effective sequence length of at least 40kb. To calculate P-values for the African and heterozygous individuals, we determine the proportion of non-overlapping windows with an average sequence divergence greater than or equal to what is observed at the 72kb MUC19 region, and for homozygous individuals the P-value represents the proportion of windows with an average sequence divergence less than or equal to what is observed at the 72kb MUC19 region. After accounting for three multiple-comparisons using the Bonferroni correction, a P-value less than 0.0167 is considered significant.
Haplotype-Archaic Diplotype Divergence
We next wanted to assess if the 72kb MUC19 region is on average less divergent than expected between the two focal haplotype groups—i.e., Denisovan-like haplotypes (n = 287) and Human-like haplotypes (n = 4407)—and the four high-coverage archaic diplotypes. To do so, we calculated the average sequence divergence between each haplotype group and the archaic diplotypes for the 72kb MUC19 region. To assess significance, we compared it to a distribution of average sequence divergence from the genomic background of 72kb windows with comparable effective sequence length density—i.e., an effective sequence length of at least 40kb. To calculate P-values, we determined the proportion of windows from the genomic background with an average sequence divergence less than or equal to what we observed at the 72kb region. After correcting for four multiple comparisons per haplotype group using the Bonferroni correction, a P-value less than 0.0125 is considered significant.
Denisovan-Neanderthal Diplotype Divergence
To assess the extent of sequence divergence between the sequenced Denisovan and Neanderthal diplotypes, we calculated the sequence divergence for the 72kb MUC19 region between all high-coverage Neanderthal diplotypes and the sequenced Denisovan diplotype. To assess if the 72kb MUC19 region is significantly more diverged than expected among a pair of archaic individuals, we compared the observed sequence divergence to a distribution of sequence divergence from the genomic background of 72kb non-overlapping windows with comparable effective sequence length density—i.e., an effective sequence length of at least 40kb amongst the four archaic individuals. To calculate P-values, we determined the proportion of windows from the genomic background with a sequence divergence greater than or equal to what we observed at the 72kb MUC19 region. After correcting for three comparisons using the Bonferroni correction, a P-value less than 0.0167 is considered significant.
Site Patterns Tests of Introgression
To further corroborate claims of introgression based on our sequence divergence results we used the D+ statistic to formally test hypotheses about introgression [Lopez-Fang et al., 2022; Peede et al., 2022]. The D+ statistic utilizes observed site patterns from three populations and an outgroup—Newick format: (((P1, P2), P3), O); site pattern format: (P1’s allelic state, P2’s allelic state, P3’s allelic state, O’s allelic state)—as a proxy for gene tree frequencies, where P1 and P2 represent potential recipients of introgression from P3 and an outgroup is used to polarize the ancestral states. D+ specifically utilizes four site patterns: ABBA, BABA, BAAA, and ABAA (where an A denotes the ancestral allele and B denotes the derived allele) to test for asymmetries in site pattern frequencies. Under a scenario of no gene flow the D+ statistic is expected to be zero, a significant and positive D+ value indicates that P2 and P3 share more derived and ancestral alleles than expected, and a significant and negative D+ statistic indicates that P1 and P3 share more derived and ancestral alleles than expected. For all D+ tests we used the ancestral allele calls from the six primate alignment inferred from EPO pipeline to polarize ancestral states [Paten et al., 2008a; Paten et al., 2008b; Herrero et al., 2016]. Since the D+ statistic is normally distributed, to assess if observed D+ values significantly differed from zero for each test we constructed z-distributions of D+ values from 72kb windows of comparable effective sequence length and computed P-values using the scipy.stats.norm.sf function implemented in scipy v1.7.2 [Virtanen et al., 2020].
Gene Flow from Denisovans into non-African modern humans
We calculated D+ for the 72kb MUC19 region for all non-African TGP populations to test for introgression from the archaic humans. To test hypotheses about introgression on a population level we also calculated D+ for all combinations of P1 = {YRI}, P2 = {all non-African populations}, and P3 = {Denisovan, Altai Neanderthal, Chagyrskaya Neanderthal, Vindija Neanderthal}. To assess significance, we calculated D+ values from the genomic background of 72kb windows with comparable effective sequence length—i.e., an effective sequence length of at least 40kb amongst the combined dataset. After correcting for 19 multiple comparisons per P3 using the Bonferroni correction, a P-value less than 0.00263 is considered significant.
Gene Flow from Denisovans into Late Neanderthals
Lastly, we wanted to assess the possibility of an introgression event from the Denisovan population into the ancestral population of two late Neanderthals. To test this hypothesis we calculated D+ for our 72kb MUC19 region for the for two sets of configurations ((P1, P2), P3)): 1) ((Altai Neanderthal, Chagyrskaya Neanderthal), Denisovan)) and ((Altai Neanderthal, Vindija Neanderthal), Denisovan)), 2) ((Chagyrskaya Neanderthal, Vindija Neanderthal), Denisovan)), and ((Chagyrskaya Neanderthal, Vindija Neanderthal), Altai Neanderthal)). To assess significance, we calculated D+ values from the genomic background of 72kb windows with comparable effective sequence length—i.e., an effective sequence length of at least 40kb amongst the four archaic individuals. After correcting for two multiple comparison per set of configurations—i.e., set one where P1 is the Altai Neanderthal and P3 is the sequenced Denisovan and set two where P1 is the Chagyrskaya Neanderthal and P2 is the Vindija Neanderthal—using the Bonferroni correction, a P-value less than 0.025 is considered significant.
Heterozygosity in the Archaics and TGP
To understand if the 72kb MUC19 region is an outlier for heterozygosity in the four archaic genomes we counted the number of heterozygous sites for the 72kb MUC19 region and compared the observed number of heterozygous sites to a distribution of heterozygous site counts from the genomic background of 72kb windows with comparable effective sequence length density—i.e., an effective sequence length of at least 40kb amongst the four archaic individuals. To calculate P-values, we determined the proportion of windows from the genomic background where the number of heterozygous sites is greater than or equal to what we observed at the 72kb MUC19 region. After correcting for four multiple comparisons using the Bonferroni correction, a P-value less than 0.0125 is considered significant. Additionally, we were interested in understanding if African individuals and individuals carrying the Denisovan-like haplotype are also outliers for heterozygosity in our focal 72kb MUC19 region. To do so we computed the average number of heterozygous sites amongst all African individuals (n = 504), heterozygous individuals (n = 255) who carry exactly one copy of the Denisovan-like haplotype, and homozygous individuals (n = 16) who carry two copies of the Denisovan-like haplotype for the 72kb MUC19 region, and compared our observed values to distributions of average heterozygous site counts from the genomic background of 72kb windows with comparable effective sequence length density—i.e., an effective sequence length of at least 40kb amongst the combined dataset. To calculate P-values for the African and heterozygous individuals, we determine the proportion of windows from the genomic background with an average number of heterozygous sites greater than or equal to what is observed at the 72kb MUC19 region, and to calculate P-values for homozygous individuals we determine the proportion of windows from the genomic background with an average number of heterozygous sites less than or equal to what is observed at the 72kb MUC19 region. After accounting for three multiple-comparisons using the Bonferroni correction, a P-value less than 0.0167 is considered significant.
Psuedo-Ancestry Painting (PAP) in the late Neanderthals
As phasing is not currently possible for archaic genomes, we calculated pseudo-ancestry painting (PAP) scores in order to assign alleles from a target heterozygous individual to haplotypes with fixed differences from two source individuals. Specifically, for every heterozygous site in the Vindija and Chagyrskaya Neanderthals we asked if a pairing of source individuals can explain the observed heterozygous site—one source individual is fixed for the ancestral alleles and the other is fixed for the derived allele. The PAP score then corresponds to the number of heterozygous sites that can be explained by the two source individuals, normalized over the total number of heterozygous sites. We aimed to explain the excess of heterozygosity in the Chagyrskaya and Vindija Neanderthals by calculating PAP scores using a pairing of the Altai Neanderthal and Denisovan as sources, as well as a pairing of an MXL individual (i.e., NA19664) who is homozygous for the Denisovan-like haplotype and an YRI individual (i.e., NA19190) who is homozygous for the Human-like haplotype.
Copy number polymorphism of a 30bp tandem repeat motif between the Human-like and a Denisovan-like haplotypes
Repeat counts from short read data
We utilized previously employed methods to estimate repeat length from short-read data [Course et al., 2021]. The Samtools v1.9 view command was used to extract and count reads from TGP sample .cram files that map to hg38 Chr12:40482139–40491565. The process was repeated for two non-repetitive regions of the human genome (Chr7:5500000–5600000 and Chr12:6490000–6590000) to calculate average read density for each sample. The fraction of enrichment or depletion of reads was used to calculate the estimated repeat length compared to the reference human genome, which contains 287.5 copies of the 30bp repeat.
To assess if Admixed American populations exhibit an elevated proportion of individuals whose repeat copies exceed the non-African average than all other non-African populations, we computed the proportion of non-African individuals who have repeat copies greater than or equal to the non-African average (~365 repeat copies) amongst all Admixed American populations and amongst all non-African populations excluding the Admixed American population. We then performed a Fisher’s exact test and computed the odds ratio and corresponding P-value using the scipy.stats.fisher_exact function implemented in scipy v1.7.2 [Virtanen et al., 2020]. Additionally, to assess if on average individuals who carry at least one Denisovan-like haplotype exhibit significantly elevated number repeat copies than individuals who carry no Denisovan-like haplotypes we performed a Welch’s T-test and calculated the corresponding P-value using the scipy.stats.ttest_ind function implemented in scipy v1.7.2 [Virtanen et al., 2020].
Repeat counts from long read data
Phased long-read genomes were obtained from the HPRC and HGSVC. A region corresponding to hg38 Chr12:40482543–40491249 was extracted from each of the .fasta files and used to report repeat length for each allele. The repeat length was divided by 30 and then averaged between the two haplotypes for each individual to calculate an estimated repeat length.
To ensure that the repeat copies inferred from long read data was comparable to our inferences from short read data we computed both Pearson’s and Spearman’s correlation coefficients and their corresponding P-values for the TGP individuals who had both type of sequencing data available, using the scipy.stats.pearsonr and scipy.stats.spearmanr functions implemented in scipy v1.7.2 [Virtanen et al., 2020].
Supplementary Material
Acknowledgements:
We would like to thank Alyssa Funk for contributing to the development of the PBS analysis, Ratchanon Pornmongkolsuk for early visualizations of global frequencies of MUC19, and Paolo Provero for his insightful comments and discussion. We would also like to thank the Crawford and Ramachandran and laboratories, especially Ria Vinod, Julian Stamp, Chibuikem Nwizu, Elizabeth Chevy, and Cole Williams for their invaluable feedback and support throughout the duration of this project. Part of this research was conducted using computational resources and services at the Center for Computation and Visualization, Brown University.
Funding:
This work was supported by The Leakey Foundation [to FAV], the National Institutes of Health [1R35GM128946-01 to EHS], and the Alfred P. Sloan Foundation [to EHS]. DP is also a trainee supported under the Brown University Predoctoral Training Program in Biological Data Science (NIH T32 GM128596). PM was funded by the National Institutes of Health (R35GM142978) and Burroughs Wellcome Fund (Career Award at the Scientific Interface). This work was supported, in part, by US National Institutes of Health (NIH) grant R01NS122766 (to PNV). EHS, FJ, and MAA are supported by the Human Frontier Science Program.
Funding Statement
This work was supported by The Leakey Foundation [to FAV], the National Institutes of Health [1R35GM128946-01 to EHS], and the Alfred P. Sloan Foundation [to EHS]. DP is also a trainee supported under the Brown University Predoctoral Training Program in Biological Data Science (NIH T32 GM128596). PM was funded by the National Institutes of Health (R35GM142978) and Burroughs Wellcome Fund (Career Award at the Scientific Interface). This work was supported, in part, by US National Institutes of Health (NIH) grant R01NS122766 (to PNV). EHS, FJ, and MAA are supported by the Human Frontier Science Program.
Data Availability:
The 1,000 Genomes Project Phase III, Simons Genome Diversity Project, high-coverage archaic genomes, Human Pangenome Reference Consortium, and Human Genome Structural Variant Consortium datasets are all publicly available. Ancient American genomes are available after signing data agreements from the original publications. All software used in this study is publicly available and all statistical tests are described in the methods. All the information needed to reproduce the results in this study is described in the methods and supplemental methods, additionally; the original code can be found: https://github.com/David-Peede/MUC19/tree/main.
References
- [1].Ahlquist KD, Banuelos Mayra M, Funk Alyssa, Lai Jiaying, Rong Stephen, Villanea Fernando A, and Witt Kelsey E. Our tangled family tree: new genomic methods offer insight into the legacy of archaic admixture. Genome biology and evolution, 13(7):evab115, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Browning Sharon R, Browning Brian L, Zhou Ying, Tucci Serena, and Akey Joshua M. Analysis of human sequence data reveals two pulses of archaic denisovan admixture. Cell, 173(1):53–61, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Villanea Fernando A and Schraiber Joshua G. Multiple episodes of interbreeding between neanderthal and modern humans. Nature ecology & evolution, 3(1):39, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yang Melinda A, Malaspinas Anna-Sapfo, Durand Eric Y, and Slatkin Montgomery. Ancient structure in Africa unlikely to explain neanderthal and non African genetic similarity. Molecular biology and evolution, 29(10):2987–2995, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lohse Konrad and Frantz Laurent AF. Neandertal admixture in eurasia confirmed by maximum-likelihood analysis of three genomes. Genetics, 196 (4):1241–1251, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Huerta-Sánchez Emilia, Jin Xin, Bianba Zhuoma, Peter Benjamin M, Vinckenbosch Nico las, Liang Yu, Yi Xin, He Mingze, Somel Mehmet, Ni Peixi ang, et al. Altitude adaptation in tibetans caused by introgression of denisovan-like DNA. Nature, 512(7513):194, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Racimo Fernando, Gokhman David, Fumagalli Matteo, Ko Amy, Hansen Tor ben, Moltke Ida, Albrechtsen Anders, Carmel Liran, Huerta-Sánchez Emilia, and Nielsen Rasmus. Archaic adaptive introgression in tbx15/wars2. Molecular biology and evolution, 34(3):509–524, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Racimo Fernando, Marnetto Davide, and Huerta-Sánchez Emilia. Signatures of archaic adaptive introgression in present-day human populations. Molecular biology and evolution, 34(2):296–317, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Witt Kelsey E, Funk Alyssa, Añorve-Garibay Valeria, Fang Lesly Lopez, and Huerta-Sánchez Emilia. The impact of modern admixture on archaic human ancestry in human populations. Genome Biology and Evolution, 15 (5):evad066, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Reynolds Austin W, Mata-Míguez Jaime, Miró-Herrans Aida, Briggs-Cloud Marcus, Sylestine Ana, Barajas-Olmos Francisco, Ortiz Humberto Garcia, Rzhetskaya Margarita, Orozco Lorena, Raff Jennifer A, et al. Comparing signals of natural selection between three indigenous North American populations. Proceedings of the National Academy of Sciences, page 201819467, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sankararaman Sriram, Mallick Swapan, Patterson Nick, and Reich David. The combined landscape of Denisovan and Neanderthal ancestry in present day humans. Current Biology, 26(9):1241–1247, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Petr Martin, Pääbo Svante, Kelso Janet, and Vernot Benjamin. Limits of long-term selection against Neandertal introgression. Proceedings of the National Academy of Sciences, 116(5):1639–1644, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Xinjun, Kim Bernard, Lohmueller Kirk E, and Sánchez Emilia Huerta. The impact of recessive deleterious variation on signals of adaptive introgression in human populations. Genetics, 215(3):799–812, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Racimo Fernando, Sankararaman Sriram, Nielsen Rasmus, and Huerta-Sánchez Emilia. Evidence for archaic adaptive introgression in humans. Nature Reviews Genetics, 16(6):359, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang Xinjun, Kim Bernard, Singh Armaan, Sankararaman Sriram, Durvasula Arun, and Lohmueller Kirk E. Maladapt reveals novel targets of adaptive introgression from neanderthals and denisovans in worldwide human populations. Molecular Biology and Evolution, 40(1):msad001, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mendez Fernando L, Watkins Joseph C, and Hammer Michael F. A haplotype at STAT2 introgressed from Neanderthals and serves as a candidate of positive selection in papua new guinea. The American Journal of Human Genetics, 91(2):265–274, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sankararaman Sriram, Mallick Swapan, Dannemann Michael, Prüfer Kay, Kelso Janet, Pääbo Svante, Patterson Nick, and Reich David. The genomic landscape of Neanderthal ancestry in present-day humans. Nature, 507 (7492):354–357, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Vernot Benjamin and Akey Joshua M. Resurrecting surviving Neandertal lineages from modern human genomes. Science, 343(6174):1017–1021, 2014. [DOI] [PubMed] [Google Scholar]
- [19].Gittelman Rachel M, Schraiber Joshua G, Vernot Benjamin, Mikacenic Carmen, Wurfel Mark M, and Akey Joshua M. Archaic hominin admixture facilitated adaptation to out-of-africa environments. Current Biology, 26(24):3375–3382, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sams Aaron J, Dumaine Anne, Nédélec Yohann, Yotova Vania, Alfieri Carolina, Tanner Jerome E, Messer Philipp W, and Barreiro Luis B. Adaptively introgressed Neandertal haplotype at the OAS locus functionally impacts innate immune responses in humans. Genome biology, 17(1):1–15, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dannemann Michael and Kelso Janet. The contribution of Neanderthals to phenotypic variation in modern humans. The American journal of human genetics, 101(4):578–589, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Marnetto Davide and Huerta-Sánchez Emilia. Haplostrips: revealing population structure through haplotype visualization. Methods in Ecology and Evolution, 8(10):1389–1392, 2017. [Google Scholar]
- [23].Zhang Xinjun, Witt Kelsey E, Bañuelos Mayra M, Ko Amy, Yuan Kai, Xu Shuhua, Nielsen Rasmus, and Huerta-Sánchez Emilia. The history and evolution of the denisovan-epas1 haplotype in tibetans. Proceedings of the National Academy of Sciences, 118(22):e2020803118, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fan Shaohua, Hansen Matthew EB, Lo Yancy, and Tishkoff Sarah A. Going global by adapting local: A review of recent human adaptation. Science, 354(6308):54–59, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tamm Erika, Kivisild Toomas, Reidla Maere, Metspalu Mait, Glenn Smith David, Mulligan Connie J, Bravi Claudio M, Rickards Olga, Martinez-Labarga Cristina, Khusnutdinova Elsa K, et al. Beringian standstill and spread of Native American founders. PloS one, 2(9):e829, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Beck Hylke E, Zimmermann Niklaus E, McVicar Tim R, Vergopolan Noemi, Berg Alexis, and Wood Eric F. Present and future köppen-geiger climate classification maps at 1-km resolution. Scientific data, 5(1):1–12, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yi Xin, Liang Yu, Huerta-Sánchez Emilia, Jin Xin, Cuo Zha Xi Ping, Pool John E, Xu Xun, Jiang Hui, Vinckenbosch Nicolas, Korneliussen Thorfinn Sand, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. science, 329(5987):75–78, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Skov Laurits, Hui Ruoyun, Shchur Vladimir, Hobolth Asger, Scally Aylwyn, Schierup Mikkel Heide, and Durbin Richard. Detecting archaic introgression using an unadmixed outgroup. PLoS Genetics, 14(9):e1007641, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Martin Alicia R, Gignoux Christopher R, Walters Raymond K, Wojcik Genevieve L, Neale Benjamin M, Gravel Simon, Daly Mark J, Bustamante Carlos D, and Kenny Eimear E. Human demographic history impacts genetic risk prediction across diverse populations. The American Journal of Human Genetics, 100(4):635–649, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fang Lesly Lopez, Vecchyo Diego Ortega-Del, McTavish Emily Jane, and Huerta-Sánchez Emilia. Leveraging shared ancestral variation to detect local introgression. bioRxiv, 2022. doi: 10.1101/2022.03.21.485082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Peede David, Vecchyo Diego Ortega-Del, and Huerta-Sánchez Emilia. The utility of ancestral and derived allele sharing for genome-wide inferences of introgression. bioRxiv, 2022. doi: 10.1101/2022.12.02.518851. [DOI] [Google Scholar]
- [32].Mafessoni Fabrizio, Grote Steffi, de Filippo Cesare, Slon Viviane, Kolobova Kseniya A., Viola Bence, Markin Sergey V., Chintalapati Man jusha, Peyrégne Stephane, Skov Laurits, Skoglund Pontus, Krivoshapkin Andrey I., Derevianko Anatoly P., Meyer Matthias, Kelso Janet, Peter Benjamin, Prüfer Kay, and Pääbo Svante. A high-coverage Neandertal genome from Chagyrskaya cave. Proceedings of the National Academy of Sciences, 117(26):15132–15136, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Slon Viviane, Mafessoni Fabrizio, Vernot Benjamin, De Filippo Cesare, Grote Steffi, Viola Bence, Hajdinjak Mateja, Peyrégne Stéphane, Nagel Sarah, Brown Samantha, et al. The genome of the offspring of a Neanderthal mother and a Denisovan father. Nature, 561(7721):113–116, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Peter Benjamin M. 100,000 years of gene flow between Neandertals and denisovans in the Altai mountains. bioRxiv, 2020. doi: 10.1101/2020.03.13.990523. [DOI] [Google Scholar]
- [35].Kuhlwilm Martin, Gronau Ilan, Hubisz Melissa J, De Filippo Cesare, Prado-Martinez Javier, Kircher Martin, Fu Qiaomei, Burbano Hernán A, Lalueza-Fox Carles, de La Rasilla Marco, et al. Ancient gene flow from early modern humans into eastern neanderthals. Nature, 530(7591):429–433, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Prüfer Kay, Racimo Fernando, Patterson Nick, Jay Flora, Sankararaman Sriram, Sawyer Susanna, Heinze Anja, Renaud Gabriel, Sudmant Peter H, De Filippo Cesare, et al. The complete genome sequence of a neanderthal from the altai mountains. Nature, 505(7481):43, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hubisz Melissa J, Williams Amy L, and Siepel Adam. Mapping gene flow between ancient hominins through demography-aware inference of the ancestral recombination graph. PLoS genetics, 16(8):e1008895, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Grantham Richard. Amino acid difference formula to help explain protein evolution. science, 185(4154):862–864, 1974. [DOI] [PubMed] [Google Scholar]
- [39].Li Wen-Hsiung, Wu Chung-I, and Luo Chi-Cheng. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Molecular biology and evolution, 2(2):150–174, 1985. [DOI] [PubMed] [Google Scholar]
- [40].Pollard Katherine S, Hubisz Melissa J, Rosenbloom Kate R, and Siepel Adam. Detection of nonneutral substitution rates on mammalian phylogenies. Genome research, 20(1):110–121, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Javitt Gabriel, Khmelnitsky Lev, Albert Lis, Lavi Shlomo Bigman Nadav Elad, Morgenstern David, Ilani Tal, Levy Yaakov, Diskin Ron, and Fass Deborah. Assembly mechanism of mucin and von willebrand factor polymers. Cell, 183(3):717–729, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Xu Duo, Pavlidis Pavlos, Thamadilok Supaporn, Redwood Emilie, Fox Sara, Blekhman Ran, Ruhl Stefan, and Gokcumen Omer. Recent evolution of the salivary mucin MUC7. Scientific reports, 6(1):31791, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Xu Duo, Pavlidis Pavlos, Taskent Recep Ozgur, Alachiotis Nikolaos, Flanagan Colin, DeGiorgio Michael, Blekhman Ran, Ruhl Stefan, and Gokcumen Omer. Archaic hominin introgression in Africa contributes to functional salivary MUC7 genetic variation. Molecular Biology and Evolution, 34(10):2704–2715, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Viscardi Lucas Henriques, Paixão-Cˆortes Vanessa Rodrigues, Comas David, Salzano Francisco Mauro, Rovaris Diego, Bau Claiton Dotto, Amorim Carlos Eduardo G, and Bortolini Maria C´atira. Searching for ancient balanced polymorphisms shared between neanderthals and modern humans. Genetics and Molecular Biology, 41:67–81, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Posth Cosimo, Nakatsuka Nathan, Lazaridis Iosif, Skoglund Pontus, Mallick Swapan, Lamnidis Thiseas C, Rohland Nadin, Nägele Kathrin, Adamski Nicole, Bertolini Emilie, et al. Reconstructing the deep population history of central and south america. Cell, 175(5):1185–1197, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Peyrégne Stéphane, Kelso Janet, Peter Benjamin M, and Pääbo Svante. The evolutionary history of human spindle genes includes back-and-forth gene flow with Neandertals. Elife, 11:e75464, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pajic Petar, Shen Shichen, Qu Jun, May Alison J, Knox Sarah, Ruhl Stefan, and Gokcumen Omer. A mechanism of gene evolution generating mucin function. Science advances, 8(34):eabm8757, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Villanea Fernando Aand Witt Kelsey E. Underrepresented populations at the archaic introgression frontier. Frontiers in Genetics, 13:821170, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature, 526(7571):68, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wong Karen HY, Ma Walfred, Wei Chun-Yu, Yeh Erh-Chan, Lin Wan-Jia, Wang Elin HF, Su Jen-Ping, Hsieh Feng-Jen, Kao Hsiao-Jung, Chen Hsiao Huei, et al. Towards a reference genome that captures global genetic diversity. Nature communications, 11(1):5482, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Paten Benedict, Herrero Javier, Beal Kathryn, Fitzgerald Stephen, and Birney Ewan. Enredo and pecan: genome-wide mammalian consistency based multiple alignment with paralogs. Genome research, 18(11):1814–1828, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Paten Benedict, Herrero Javier, Fitzgerald Stephen, Beal Kathryn, Flicek Paul, Holmes Ian, and Birney Ewan. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome research, 18(11):1829–1843, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Herrero Javier, Muffato Matthieu, Beal Kathryn, Fitzgerald Stephen, Gordon Leo, Pignatelli Miguel, Albert J Vilella Stephen MJ Searle, Amode Ridwan, Brent Simon, et al. Ensembl comparative genomics resources. Database, 2016:bav096, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cingolani Pablo, Platts Adrian, Wang Le Lily, Coon Melissa, Nguyen Tung, Wang Luan, Land Susan J, Lu Xiangyi, and Ruden Douglas M. A program for annotating and predicting the effects of single nucleotide polymorphisms, snpeff: Snps in the genome of drosophila melanogaster strain w1118; iso-2; iso-3. fly, 6(2):80–92, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Scheib CL, Li Hongjie, Desai Tariq, Link Vivian, Kendall Christopher, Dewar Genevieve, Griffith Peter William, Mörseburg Alexander, Johnson John R, Potter Amiee, et al. Ancient human parallel lineages within North America contributed to a coastal expansion. Science, 360(6392):1024–1027, 2018. [DOI] [PubMed] [Google Scholar]
- [56].Lindo John, Haas Randall, Hofman Courtney, Apata Mario, Moraga Mauricio, Verdugo Ricardo A, Watson James T, Llave Carlos Viviano, Witonsky David, Beall Cynthia, et al. The genetic prehistory of the andean highlands 7000 years bp though european contact. Science advances, 4(11): eaau4921, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Fuente Constanza De la, Ávila-Arcos María C, Galimany Jacqueline, Carpenter Meredith L, Homburger Julian R, Blanco Alejandro, Contreras Paloma, Dávalos Diana Cruz, Reyes Omar, Roman Manuel San, et al. Genomic insights into the origin and diversification of late maritime hunter gatherers from the chilean patagonia. Proceedings of the National Academy of Sciences, 115(17):E4006–E4012, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Moreno-Mayar J Víctor, Potter Ben A, Vinner Lasse, Steinrücken Matthias, Rasmussen Simon, Terhorst Jonathan, Kamm John A, Albrechtsen Anders, Malaspinas Anna-Sapfo, Sikora Martin, et al. Terminal pleistocene alaskan genome reveals first founding population of native americans. Nature, 553 (7687):203, 2018. [DOI] [PubMed] [Google Scholar]
- [59].Rasmussen Morten, Anzick Sarah L, Waters Michael R, Skoglund Pontus, DeGiorgio Michael, Stafford Thomas W Jr, Rasmussen Simon, Moltke Ida, Albrechtsen Anders, Doyle Shane M, et al. The genome of a late pleistocene human from a clovis burial site in western montana. Nature, 506(7487): 225, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Villa-Islas Viridiana, Izarraras-Gomez Alan, Larena Maximilian, Campos Elizabeth Mejía Perez, Sandoval-Velasco Marcela, Rodríguez Juan Esteban Rodríguez, Bravo-Lopez Miriam, Moguel Barbara, Fregel Rosa, Garfias-Morales Ernesto, et al. Demographic history and genetic structure in pre hispanic central mexico. Science, 380(6645):eadd6142, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Li Heng and Durbin Richard. Fast and accurate short read alignment with burrows–wheeler transform. bioinformatics, 25(14):1754–1760, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Li Heng, Handsaker Bob, Wysoker Alec, Fennell Tim, Ruan Jue, Homer Nils, Marth Gabor, Abecasis Goncalo, and Durbin Richard. The sequence alignment/map format and samtools. Bioinformatics, 25(16):2078–2079, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Bhatia Gaurav, Patterson Nick, Sankararaman Sriram, and Price Alkes L. Estimating and interpreting fst: the impact of rare variants. Genome research, 23(9):1514–1521, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Baumdicker Franz, Bisschop Gertjan, Goldstein Daniel, Gower Graham, Aaron P Ragsdale Georgia Tsambos, Zhu Sha, Eldon Bjarki, Ellerman E Castedo, Galloway Jared G, et al. Efficient ancestry and mutation simulation with msprime 1.0. Genetics, 220(3):iyab229, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Virtanen Pauli, Gommers Ralf, Oliphant Travis E, Haberland Matt, Reddy Tyler, Cournapeau David, Burovski Evgeni, Peterson Pearu, Weckesser Warren, Bright Jonathan, et al. Scipy 1.0: fundamental algorithms for scientific computing in python. Nature methods, 17(3):261–272, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Course Meredith M, Sulovari Arvis, Gudsnuk Kathryn, Eichler Evan E, and Valdmanis Paul N. Characterizing nucleotide variation and expansion dynamics in human-specific variable number tandem repeats. Genome Research, 31(8):1313–1324, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chen Yin, Zhao Yu Hua, Kalaslavadi Tejas Baba, Hamati Edward, Nehrke Keith, Le Anh Dao, Ann David K, and Wu Reen. Genome-wide search and identification of a novel gel-forming mucin muc19/muc19 in glandular tissues. American journal of respiratory cell and molecular biology, 30(2): 155–165, 2004. [DOI] [PubMed] [Google Scholar]
- [68].Kerschner Joseph Edward. Mucin gene expression in human middle ear epithelium. The Laryngoscope, 117(9):1666–1676, 2007. [DOI] [PubMed] [Google Scholar]
- [69].Yu DF, Chen Y, Han JM, Zhang H, Chen XP, Zou WJ, Liang LY, Xu CC, and Liu ZG. Muc19 expression in human ocular surface and lacrimal gland and its alteration in Sjögren syndrome patients. Experimental eye research, 86(2):403–411, 2008. [DOI] [PubMed] [Google Scholar]
- [70].Haller Benjamin Cand Messer Philipp W. Slim 3: Forward genetic simulations beyond the wright–fisher model. Molecular biology and evolution, 36(3):632–637, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Harrow Jennifer, Frankish Adam, Gonzalez Jose M, Tapanari Electra, Diekhans Mark, Kokocinski Felix, Aken Bronwen L, Barrell Daniel, Zadissa Amonida, Searle Stephen, et al. Gencode: the reference human genome an notation for the encode project. Genome research, 22(9):1760–1774, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Gutenkunst Ryan N, Hernandez Ryan D, Williamson Scott H, and Bustamante Car los D. Inferring the joint demographic history of multiple populations from multidimensional snp frequency data. PLoS genetics, 5 (10):e1000695, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Reich David, Green Richard E, Kircher Martin, Krause Johannes, Patterson Nick, Durand Eric Y, Viola Bence, Briggs Adrian W, Stenzel Udo, Johnson Philip LF, et al. Genetic history of an archaic hominin group from denisova cave in siberia. Nature, 468(7327):1053–1060, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kim Bernard Y, Huber Christian D, and Lohmueller Kirk E. Inference of the distribution of selection coefficients for new nonsynonymous mutations using large samples. Genetics, 206(1):345–361, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hinch Anjali G, Tandon Arti, Patterson Nick, Song Yunli, Rohland Nadin, Palmer Cameron D, Chen Gary K, Wang Kai, Buxbaum Sarah G, Akylbekova Ermeg L, et al. The landscape of recombination in african americans. Nature, 476(7359):170–175, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Medina-Munoz Santiago G, Vecchyo Diego Ortega-Del, Cruz Hervert Luis Pablo, Ferreyra-Reyes Leticia, Garcia-Garcia Lourdes, Estrada Andres Moreno, and Ragsdale Aaron. Demographic modeling of admixed latin american populations from whole genomes. bioRxiv, pages 2023–03, 2023. [DOI] [PMC free article] [PubMed]
- [77].Miles Alistair and Harding NJ. scikit-allel: A python package for exploring and analysing genetic variation data, 2016.
- [78].Jacobs Guy S, Hudjashov Georgi, Saag Lauri, Kusuma Pradiptajati, Darusallam Chelzie C, Lawson Daniel J, Mondal Mayukh, Pagani Luca, Ricaut Fran¸cois-Xavier, Stoneking Mark, et al. Multiple deeply divergent denisovan ancestries in papuans. Cell, 2019. [DOI] [PubMed] [Google Scholar]
- [79].Malaspinas Anna-Sapfo, Westaway Michael C, Muller Craig, Sousa Vitor C, Lao Oscar, Alves Isabel, Bergström Anders, Athanasiadis Georgios, Cheng Jade Y, Crawford Jacob E, et al. A genomic history of aboriginal australia. Nature, 538(7624):207–214, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Gravel Simon, Henn Brenna M, Gutenkunst Ryan N, Indap Amit R, Marth Ga bor T, Clark Andrew G, Yu Fuli, Gibbs Richard A, 1000 Genomes Project, Bustamante Carlos D, et al. Demographic history and rare allele sharing among human populations. Proceedings of the National Academy of Sciences, 108(29):11983–11988, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 1,000 Genomes Project Phase III, Simons Genome Diversity Project, high-coverage archaic genomes, Human Pangenome Reference Consortium, and Human Genome Structural Variant Consortium datasets are all publicly available. Ancient American genomes are available after signing data agreements from the original publications. All software used in this study is publicly available and all statistical tests are described in the methods. All the information needed to reproduce the results in this study is described in the methods and supplemental methods, additionally; the original code can be found: https://github.com/David-Peede/MUC19/tree/main.