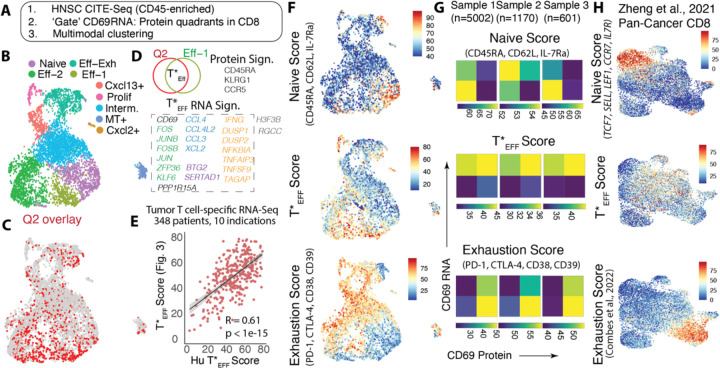
Fig. 5: De novo identification of potent effectors in human cancer by CITE-Seq.
(A) Schematic description of human HNSC tumor CITE-Seq analysis; (B) UMAP showing weighted nearest neighbor (WNN) determined clusters by multimodal RNA and Protein analysis; (C) overlay of Q2 cells (determined by gating on CD69 Protein and RNA – Fig.S13B) on the UMAP; (D) Differentially upregulated genes and proteins in the T*EFF (Q2 ᑎ Eff-1) cells vs. all others CD8 T cells, genes color-coded by functional category, box indicates genes used for the T*EFF gene signature; (E) Correlation between this human T*EFF gene signature and that from human orthologs of the gene signature in Fig. 3F (F) Naïve, T*EFF and Exhaustion scores overlayed on the WNN UMAP, and (G) Heatmap representation of median levels of the same scores in CD8 T cells from 3 different patient samples split into CD69 protein: CD69 mRNA quadrants; (n: # of CD8 T cells in each sample); (H) Naïve, T*EFF and Exhaustion scores overlaid onto the UMAP of combined CD8 T cells from a previously published pan-cancer atlas (30).