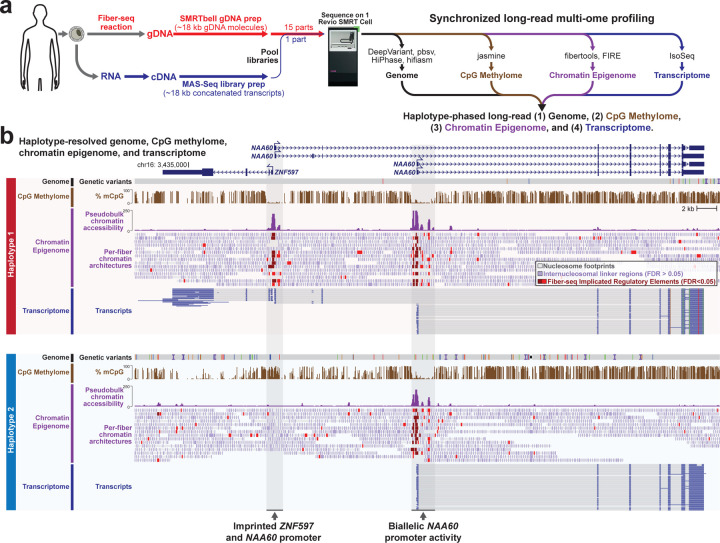
Figure 1 |. Synchronized long-read genome, methylome, epigenome and transcriptome sequencing.
a, Schematic describing the experimental and computational workflow for synchronized multi-ome profiling. Specifically, cells are subjected to a Fiber-seq reaction followed by genomic DNA extraction and SMRTbell library preparation, and in parallel cells are subjected to an RNA extraction followed by complementary DNA (cDNA) synthesis and MAS-Seq library preparation. The two libraries are then mixed together and sequenced simultaneously using a single sequencing run, enabling the simultaneous detection of the genome, CpG methylome, chromatin epigenome, and transcriptome from the sample. b, Example genomic region showing the haplotype-resolved genome, CpG methylome, chromatin epigenome, and transcriptome from GM12878 cells at a known imprinted locus.