Abstract
An approximately 60-kb transferable, vanB-carrying plasmid has been identified in a clinical Enterococcus faecium strain. A similar plasmid has been observed in an unrelated E. faecium strain, suggesting that plasmid transfer of vanB operons occurs in nature and plays a role in the dissemination of VanB-type resistance among strains of E. faecium.
The spread of glycopeptide-resistant enterococci through hospitals in the United States has been rapid and dramatic (3). To date, two types of transferable glycopeptide resistance phenotypes, VanA and VanB, have been described. The vanB operon is chromosomally located in most cases and in one case has been found within a 65-kb transposon (13). Three previous reports indicate that the vanB operon may be plasmid carried, but none have provided evidence suggesting that vanB-carrying plasmids have spread between clinical strains (2, 18, 19). We report herein transferable vanB plasmids from strains of Enterococcus faecium isolated in northeast Ohio.
The strains described in this report (E. faecium U37 and H11) were clinical isolates from two geographically distinct hospitals in northeast Ohio. Species identification was performed with the Vitek automated system. MICs of vancomycin, ampicillin, tetracycline, and minocycline were determined in duplicate by broth macrodilution with brain heart infusion (BHI) broth (12). Screening for associated resistances was performed by streaking clinical isolates (or transconjugants) onto BHI agar containing chloramphenicol (10 μg/ml), erythromycin (10 μg/ml), gentamicin (2,000 μg/ml) or streptomycin (2,000 μg/ml). Conjugation experiments were performed on nitrocellulose filters as described by Christie et al. (4). The fusidic acid- and rifampin-resistant recipients for conjugation experiments with clinical strains as donors were E. faecium GE-1 (7), E. faecium SF68 (9), Enterococcus faecalis JH2-2, and E. faecalis JH2-7 (10). Transconjugants were selected on BHI agar plates containing vancomycin (6 μg/ml) or tetracycline (10 μg/ml), fusidic acid (25 μg/ml), and rifampin (100 μg/ml). Plasmid stability was evaluated by growth of strains for 30 generations (starting from a single bacterium) and comparing colony counts on BHI agar plates or BHI agar plates with vancomycin (10 μg/ml).
Plasmid DNA was extracted as described by Ehrenfeld and Clewell (6). Genomic DNA was extracted as described by Storrs et al. (15). Digested DNA was transferred from agarose gels to nylon membranes by using a vacuum blotting apparatus (Pharmacia LKB, Uppsala, Sweden). Hybridization with digoxigenin-labeled probes was performed according to the specifications of the manufacturer (Boehringer-Mannheim Biochemicals, Indianapolis, Ind.). Probes included a 2.1-kb EcoRV fragment internal to the vanHBBXB operon (8), a 1.8-kb AccI fragment from plasmid pLRM19 (this study), and a restriction fragment from within transposon Tn5384 identical to the replication genes of broad-host-range plasmid pIP501 (1, 16). Amplification products derived from the joint of the circularized form of Tn916 (14) and from an internal region of the Tn916 tet(M) gene (11) were used as probes for Tn916-related sequences.
E. faecium U37 and H11 were confirmed as clonally distinct by IS6770 hybridization of genomic digests (data not shown) (17). MICs of vancomycin, teicoplanin, and ampicillin for U37 and H11 were 32 μg/liter, <0.5 μg/ml, and 256 μg/ml, respectively (Table 1). The two strains also expressed resistance to erythromycin, gentamicin, streptomycin, and tetracycline but were susceptible to chloramphenicol. Vancomycin resistance was transferable from both donors to GE-1 at rates of 10−7 to 2 × 10−7/recipient CFU. Transfer was also observed to E. faecium SF68. Attempts to transfer resistance to E. faecalis JH2-2 or JH2-7 were unsuccessful. E. faecium transconjugants from matings involving both donors expressed resistance to vancomycin and tetracycline-minocycline (Table 1) but were susceptible to ampicillin, erythromycin, gentamicin, and streptomycin. Vancomycin MICs for U37 and its transconjugant GE-1(pLRM19) correlated well. In contrast, the level of vancomycin resistance expressed by transconjugant GE-1(pLRM20) was less than observed in the donor strain H11 (Table 1).
TABLE 1.
Broth dilution MICs for recipient and vancomycin-resistant transconjugants
| Strain | MIC (μg/ml)
|
||
|---|---|---|---|
| Ampicillin | Vancomycin | Minocycline | |
| U37 | 256 | 16–32 | 16 |
| GE-1(pLRM19) | ≤1 | 16–32 | 16 |
| H11 | 256 | 256 | 8 |
| GE-1(pLRM20) | ≤1 | 8–16 | 8 |
| GE-1 | ≤1 | ≤1 | ≤4 |
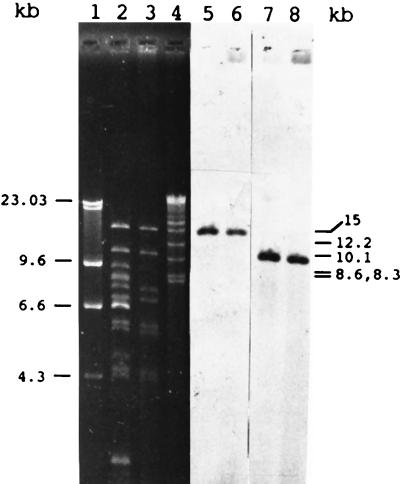
Plasmid preparations revealed a ca. 60-kb plasmid in transconjugants from matings involving U37 (plasmid designated pLRM19) and H11 (plasmid designated pLRM20). In addition to pLRM20, transconjugants from matings involving H11 possessed four additional plasmids (data not shown). A comparison of HincII digests of plasmid preparations from transconjugants from both U37 and H11 is shown in Fig. 1. Several identical bands are present in the two plasmids’ digests. vanB-hybridizing HincII fragments were identical in size (ca. 15 kb) in the two plasmids (Fig. 1), as were EcoRI fragments (data not shown). A 1.8-kb AccI fragment (external to the vanB operon) from pLRM19 hybridized to a similarly sized HincII band in both preparations, suggesting that pLRM19 and pLRM20 are related and possibly identical. Neither plasmid hybridized to the pIP501 replication region or to tet(M), and only very weak hybridization of the Tn916 joint to a single band (instead of the expected two) was observed. We therefore conclude that the tetracycline-minocycline resistance conferred by this plasmid is not due to the presence of a Tn916-like element.
FIG. 1.
Comparison of HincII digestions of plasmid preparations from GE-1(pLRM19) and GE-1(pLRM20). Lane 1, bacteriophage lambda digested with HindIII (size standard—sizes denoted on left side of figure); lane 2, GE-1(pLRM20); lane 3, GE-1(pLRM19); lane 4, High-molecular-weight size standards (Bethesda Research Laboratories, Gaithersburg, Md.) (relevant sizes denoted on right side of figure); lanes 5 and 6, Southern transfer of digests at left hybridized with 2.1-kb EcoRV fragment internal to the vanHBBXB operon; lanes 7 and 8, Southern transfer of digests at left hybridized with 1.8-kb AccI fragment of pLRM19 (see text).
The presence of highly similar vanB-carrying plasmids in clonally distinct E. faecium strains isolated at different hospitals suggests that plasmid-mediated transfer of vanB operons occurs in nature. Previous reports have confirmed or suggested the presence of the vanB operon on a plasmid in single E. faecalis or E. faecium isolates or in clonally related E. faecium strains (2, 19). No evidence that the plasmids had disseminated among different strains was presented in these reports. The data presented in this paper are consistent with the inter-enterococcal transfer of a vanB plasmid in nature.
The difference in expression of vancomycin resistance between clinical isolate H11 and its transconjugant GE-1(pLRM20) was not explainable by differences in plasmid stability between the two strains (data not shown). Hybridization studies indicated that H11 had an additional vanB operon located on the chromosome, which was not transferred with the plasmids. It is likely that this additional operon is the explanation for the increased expression of resistance in the donor compared to the transconjugant.
The rate of transfer of these plasmids is not consistent with rates reported for E. faecalis pheromone-responsive plasmids, which transfer to enterococcal recipients at rates of 10−1 to 10−2/recipient CFU (5). The failure to hybridize with an internal fragment from the repE gene of pIP501 and the failure of transfer to E. faecalis recipients argue against either plasmid belonging to the broad-host-range family. It is unclear why we were unable to transfer these plasmids to E. faecalis. It is possible that the host range of the plasmids is limited to E. faecium. A more extensive functional analysis of these plasmids is ongoing.
Acknowledgments
This study was supported in part by the medical research service of the Department of Veterans Affairs (L.B.R.).
REFERENCES
- 1.Bonafede M E, Carias L L, Rice L B. Enterococcal transposon Tn5384: evolution of a composite transposon through cointegration of enterococcal and staphylococcal plasmids. Antimicrob Agents Chemother. 1997;41:1854–1858. doi: 10.1128/aac.41.9.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyce J M, Opal S M, Chow J W, Zervos M J, Potter-Bynoe G, Sherman C B, Romulo R L, Fortna S, Medeiros A A. Outbreak of multidrug-resistant Enterococcus faecium with transferable vanB class vancomycin resistance. J Clin Microbiol. 1994;32:1148–1153. doi: 10.1128/jcm.32.5.1148-1153.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin—United States, 1989–1993. Morbid Mortal Weekly Rep. 1993;42:597–599. [PubMed] [Google Scholar]
- 4.Christie P J, Korman R Z, Zahler S A, Adsit J C, Dunny G M. Two conjugation systems associated with plasmid pCF10: identification of a conjugative transposon that transfers between Streptococcus faecalis and Bacillus subtilis. J Bacteriol. 1987;169:2529–2536. doi: 10.1128/jb.169.6.2529-2536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clewell D B. Plasmids, drug resistance and gene transfer in genus Streptococcus. Microbiol Rev. 1981;45:409–436. doi: 10.1128/mr.45.3.409-436.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrenfeld E E, Clewell D B. Transfer functions of the Streptococcus faecalis plasmid pAD1: organization of plasmid DNA encoding response to sex pheromone. J Bacteriol. 1987;169:3473–3481. doi: 10.1128/jb.169.8.3473-3481.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eliopoulos G M, Wennersten C, Zighelboim-Daum S, Reiszner E, Goldmann D, Moellering R C., Jr High-level resistance to gentamicin in clinical isolates of Streptococcus (Enterococcus) faecium. Antimicrob Agents Chemother. 1988;32:1528–1532. doi: 10.1128/aac.32.10.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evers S, Courvalin P. Regulation of VanB-type vancomycin resistance gene expression by the VanSB-VanRB two-component regulatory system in Enterococcus faecalis V583. J Bacteriol. 1996;178:1302–1309. doi: 10.1128/jb.178.5.1302-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heaton M P, Discotto L F, Pucci M J, Handwerger S. Mobilization of vancomycin resistance by transposon-mediated fusion of a VanA plasmid with an Enterococcus faecium sex pheromone-response plasmid. Gene. 1996;171:9–17. doi: 10.1016/0378-1119(96)00022-4. [DOI] [PubMed] [Google Scholar]
- 10.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lacroix J M, Walker C B. Detection and incidence of the tetracycline resistance determinant tet(M) in the microflora associated with adult perodontitis. J Periodontol. 1995;66:102–108. doi: 10.1902/jop.1995.66.2.102. [DOI] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. M7-A3. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 13.Quintiliani R, Jr, Courvalin P. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecium BM4281. Gene. 1996;172:1–8. doi: 10.1016/0378-1119(96)00110-2. [DOI] [PubMed] [Google Scholar]
- 14.Rice L B, Marshall S H, Carias L L. Tn5381, a conjugative transposon identifiable as a circular form in Enterococcus faecalis. J Bacteriol. 1992;174:7308–7315. doi: 10.1128/jb.174.22.7308-7315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Storrs M J, Poyart-Salmeron C, Trieu-Cuot P, Courvalin P. Conjugative transposition of Tn916 requires the excisive and integrative activities of the transposon-encoded integrase. J Bacteriol. 1991;173:4347–4352. doi: 10.1128/jb.173.14.4347-4352.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swinfield T-J, Oultram J D, Thompson D E, Brehm J K, Minton N P. Physical characterization of the replication region of Streptococcus faecalis plasmid pAMβ1. Gene. 1990;87:79–90. [PubMed] [Google Scholar]
- 17.Thorisdottir A S, Carias L L, Marshall S H, Green M, Zervos M J, Giorgio C, Mermel L A, Boyce J M, Medeiros A A, Fraimow H, Rice L B. IS6770, an enterococcal insertion-like element useful for determining the clonal relationship of clinical enterococcal isolates. J Infect Dis. 1994;170:1539–1548. doi: 10.1093/infdis/170.6.1539. [DOI] [PubMed] [Google Scholar]
- 18.Woodford N, Chadwick P R, Morrison D, Cookson B D. Strains of glycopeptide-resistant Enterococcus faecium can alter their van genotypes during an outbreak. J Clin Microbiol. 1997;35:2966–2968. doi: 10.1128/jcm.35.11.2966-2968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodford N, Jones B L, Baccus Z, Ludlam H A, Brown D F. Linkage of vancomycin and high-level gentamicin resistance genes on the same plasmid in a clinical isolate of Enterococcus faecalis. J Antimicrob Chemother. 1995;35:179–184. doi: 10.1093/jac/35.1.179. [DOI] [PubMed] [Google Scholar]