Abstract
CDK4/6 inhibitors (CDK4/6i) plus endocrine therapy are now standard first-line therapy for advanced HR+/HER2− breast cancer, but developing resistance is just a matter of time in these patients. Here, we report that a cyclin E1–interacting lncRNA (EILA) is up-regulated in CDK4/6i-resistant breast cancer cells and contributes to CDK4/6i resistance by stabilizing cyclin E1 protein. EILA overexpression correlates with accelerated cell cycle progression and poor prognosis in breast cancer. Silencing EILA reduces cyclin E1 protein and restores CDK4/6i sensitivity both in vitro and in vivo. Mechanistically, hairpin A of EILA binds to the carboxyl terminus of cyclin E1 protein and hinders its binding to FBXW7, thereby blocking its ubiquitination and degradation. EILA is transcriptionally regulated by CTCF/CDK8/TFII-I complexes and can be inhibited by CDK8 inhibitors. This study unveils the role of EILA in regulating cyclin E1 stability and CDK4/6i resistance, which may serve as a biomarker to predict therapy response and a potential therapeutic target to overcome resistance.
EILA, a Cyclin E1-Interacting LncRNA, regulates Cyclin E1 stability and CDK4/6i resistance in breast cancer.
INTRODUCTION
Uncontrolled cell proliferation driven by aberrant cell cycle progression is one of the most important hallmarks of cancer, and thus, seeking agents to arrest cell cycle and halt cell proliferation is a rational way for cancer treatment (1). The cell cycle is under precise regulation of cyclin-dependent kinases (CDKs) and its partner cyclins (2). CDK4 and CDK6 are two major kinases that promote G1-S phase transition by phosphorylating and inactivating retinoblastoma (Rb), thereby enabling the S phase entry and DNA duplication. Targeting CDK4 and CDK6 with CDK4/6 inhibitors (CDK4/6i) arrests cell cycle at G1 phase and impedes cell proliferation (3, 4). Three CDK4/6 inhibitors (palbociclib, ribociclib, and abemaciclib) prolong progression-free survival (PFS) and overall survival (OS) of patients with advanced hormone receptor (HR)–positive and human epidermal growth factor receptor 2 (HER2)–negative breast cancer (5–7). As a result, CDK4/6 inhibitors in combination with endocrine therapy have become the standard treatment regimen for this type of breast cancer (8). Nevertheless, resistance to CDK4/6 inhibitors is inevitable and over 70% of patients undergo disease progression within 12 to 36 months, which presents a huge clinical challenge (9).
The major mechanisms of CDK4/6i resistance include aberrant activation of upstream oncogenic signaling and alterations in key cell cycle regulators (8). Cyclin E1, encoded by CCNE1 gene, activates CDK2 and promotes cell cycle progression. High expression of cyclin E1 not only predicts poor prognosis in breast cancer patient (10) but also promotes resistance to endocrine therapy and CDK4/6 inhibitors in preclinical models (11–13). Moreover, a large-scale gene expression analysis of tumor samples from the PALOMA-3 clinical trial showed that high cyclin E1 mRNA expression was associated with attenuated palbociclib efficacy, which was further validated in the preoperative-palbociclib (POP) trial (9, 14). Furthermore, the cytoplasmic low–molecular weight cyclin E1 also confers resistance to CDK4/6 inhibitors, indicating that the localization and proteostasis of cyclin E1 may also play a crucial role (15). On the contrary, biomarker analyses of MONALEESA-2 and PALOMA-2 trials showed that cyclin E1 mRNA expression was not predictive of palbociclib benefit (16, 17). These findings suggest that cyclin E1 protein, but not its mRNA, may play a key role in CDK4/6i resistance, and targeting cyclin E1 protein directly may be an effective way to overcome the resistance.
Long noncoding RNAs (lncRNAs) are transcripts (>200 nucleotides) without protein-coding potential, which are associated with various processes of malignant progressions in cancer, like proliferation, metastasis, and therapy resistance (18, 19). Our previous research has found that lncRNA DILA1 promotes tamoxifen resistance in breast cancer by stabilizing cyclin D1 protein, which reveals the role of lncRNAs in cell cycle regulation (20). However, the role of lncRNAs in CDK4/6i resistance remains largely unknown. Here, we investigate the potential role of lncRNAs in the maintenance of CDK4/6i resistance and elucidate the underlying mechanisms involved. First, we identify EILA as a cyclin E1–interacting lncRNA that facilitates CDK4/6i resistance by enhancing cyclin E1 stability. We also demonstrate that EILA is transcriptionally regulated by CTCF/CDK8/TFII-I complexes, which can be inhibited by CDK8 inhibitors. Furthermore, EILA is up-regulated in the CDK4/6i-resistant tumors and higher EILA expression is correlated with worse prognosis and less benefit from CDK4/6 inhibition in breast cancer. These findings indicate that EILA may serve as a biomarker to predict CDK4/6i response and as a therapeutic target to overcome CDK4/6i resistance.
RESULTS
Cyclin E1 dysregulation promotes CDK4/6 inhibitor resistance in breast cancer cells
CDK4/6i-resistant breast cancer cells (MCF7-palR and T47D-palR cells) were established as previously described (fig. S1, A and B) (21). To explore the role of cyclin E1 in the CDK4/6i-resistant cells, we first evaluated its mRNA and protein levels in the resistant cells and their parental cells. We found that the protein level of cyclin E1 was up-regulated in MCF7-palR and T47D-palR cells, while its mRNA level remained unchanged (Fig. 1A and fig. S1C). In addition, the cyclin E1 protein is more stable in the resistant cells than in their parental cells (Fig. 1B and fig. S1, D and E). These results suggest that cyclin E1 is dysregulated in posttranslational level. To determine whether the increased cyclin E1 expression contributes to CDK4/6i resistance, we silenced cyclin E1 using specific small interfering RNAs (siRNAs) in the resistant breast cancer cells and evaluated their palbociclib sensitivity with methylthiazolyldiphenyl-tetrazolium bromide (MTT) assays (fig. S1F). As expected, we found that the cyclin E1 knockdown restored palbociclib sensitivity in MCF7-palR and T47D-palR cells (Fig. 1C and fig. S1G). These results indicate that dysregulation of cyclin E1 protein is responsible for CDK4/6i resistance.
Fig. 1. CDK4/6i-resistant breast cancer cells express higher levels of cyclin E1 protein and cyclin E1–interacting lncRNA EILA.
(A) Western blot showing cyclin E1 protein levels in the parental (MCF7-pa and T47D-pa) and palbociclib-resistant (MCF7-palR and T47D-palR) breast cancer cells. (B) Western blot showing the protein degradation kinetics of cyclin E1 in MCF7-pa and MCF7-palR cells treated with cycloheximide (CHX; 20 μM) for the indicated time. (C) Cell viability assays of MCF7-palR cells treated with palbociclib when cyclin E1 was silenced. (D) Schematic overview for identifying candidate lncRNAs interacting with cyclin E1 and up-regulated in MCF7-palR cells. (E) qRT-PCR analysis of the five candidate lncRNAs in MCF7-pa and MCF7-palR cells. (F) qRT-PCR analysis of EILA expression in the cytoplasmic and nuclear fractions of resistant cells. (G) RNAscope ISH assays showing the subcellular localization of EILA in resistant cells. Scale bar, 10 μm. (H) RNA immunoprecipitation (RIP) assays followed by qRT-PCR assays showing the binding of EILA and cyclin E1. IgG was used as a negative control. (I and J) Western blot after RNA pull-down assay showing the binding of EILA and cyclin E1. Endogenous cyclin E1 protein in cell lysates of resistant cells (I) or recombinant human cyclin E1 protein (J) was used. S, sense; AS, antisense. (K) Representative images showing the colocalization of EILA and cyclin E1 in resistant cells. EILA and cyclin E1 were detected by fluorescence in situ hybridization (FISH) assay and immunofluorescence staining, respectively. Scale bars, 10 μm. In (G) and (K), representative images from three independent experiments are shown. In (B), (C), (E), (F), and (H), data are shown as means ± SD (n = 3). The P values were calculated by Student’s t test for two-group comparison and one-way ANOVAs for multiple-group comparison. ***P < 0.001, **P < 0.01, and N.S. for P > 0.05.
EILA is identified as a cyclin E1–interacting lncRNA
Recently, we have reported that lncRNA DILA1 promoted tamoxifen resistance by blocking cyclin D1 degradation, suggesting a critical role for lncRNAs in cell cycle regulation and drug resistance (20). To identify lncRNAs that may interact with cyclin E1 protein and promote CDK4/6i resistance, we carried out two high-throughput lncRNA sequencings. RNA immunoprecipitation (RIP) assays were performed with hemagglutinin (HA) antibody to pull down RNAs in MCF7-pa cells stably expressing HA-tagged cyclin E1 for sequencing. Fifty-two cyclin E1–interacting lncRNAs were identified (data S1), using immunoglobulin G (IgG) as a control. Additionally, we extracted RNAs from MCF7-pa and MCF7-palR cells for lncRNA sequencing. In total, 829 lncRNAs (data S2) up-regulated in MCF7-palR cells were identified, which potentially play a role in promoting CDK4/6i resistance. Among these lncRNAs, five lncRNAs (AC093297, LINC00664, PVT1, NRSN2-AS1, and SNHG17) were enriched in both sequencings, making them candidates for further study (Fig. 1D). To validate the sequence data, we performed real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay and found that MCF7-palR cells expressed significantly higher levels of AC093297, LINC00664, and SNHG17 than their parental cells (Fig. 1E). To determine whether these lncRNAs contribute to CDK4/6i resistance, we silenced them in MCF7-palR cells with specific siRNAs or antisense oligonucleotides (ASOs) and evaluated their palbociclib sensitivity. We found that only LINC00664 knockdown significantly restored CDK4/6i sensitivity (fig. S1, H to J). Therefore, LINC00664 is identified as a cyclin E1–interacting lncRNA (EILA) responsible for CDK4/6i resistance. According to the UCSC database and Ensemble database, EILA is a 3029–nucleotide (nt)–long ncRNA with 14 exons. The results of ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/) showed that EILA has no representative protein-coding open reading frame longer than 300 nt, indicating that its protein-coding ability is extremely low.
To further investigate the biological feature of EILA, we first measured its RNA level in the resistant cells and their parental cells. The copy number of EILA was much higher in the resistant cells than in their parental cells (fig. S1K). In addition, Northern blot also confirmed that EILA was up-regulated in the resistant cells (fig. S1L). We then explored the subcellular localization of EILA by qRT-PCR analysis of cytoplasmic and nuclear fractions, and found that it was mostly located in the nucleus (over 70%), with minor localization (about 20%) in the cytoplasm (Fig. 1F). Consistent with these results, RNAscope in situ hybridization (ISH) also confirmed that EILA was predominantly localized in nucleus and was up-regulated in the resistant cells (Fig. 1G). RIP followed by qRT-PCR and RNA pull-down assays followed by Western blot showed that EILA can directly interact with cyclin E1 (Fig. 1, H and I). This finding was further verified using the tagged recombinant cyclin E1 protein (Fig. 1J). In addition, the RNA fluorescence in situ hybridization (FISH) assays of EILA followed by the immunofluorescence (IF) of cyclin E1 confirmed their colocalization in the nucleus of MCF7-palR and T47d-palR cells (Fig. 1K). In conclusion, these results indicate that EILA directly interacts with cyclin E1 in the nucleus of the resistant cells, which may play an important role in the resistance to CDK4/6 inhibitors.
EILA is clinically relevant in cell cycle regulation and associated with poor prognosis in breast cancer
To evaluate the clinical significance of EILA in breast cancer, we analyzed the RNA sequencing data of 109 paired normal and tumor tissues from The Cancer Genome Atlas Breast Invasive Carcinoma (TCGA-BRCA) database (Fig. 2A). The results showed that the expression of EILA in the tumor tissues was significantly higher than that in the normal tissues (P < 0.0001). Consistently, qRT-PCR analysis of 47 paired normal and tumor tissues from Sun Yat-Sen Memorial Hospital (SYSMH cohort 1, n = 47) also revealed that EILA was up-regulated in the tumor tissues (P = 0.0027) (Fig. 2B). These findings were further confirmed by RNA ISH, indicating aberrant expression of EILA in breast cancer (Fig. 2C). To assess the prognostic values of EILA, we analyzed the breast cancer transcriptomic data from bc-GenExMiner v4.7 database (http://bcgenex.ico.unicancer.fr/) (22). We found that higher expression of EILA was associated with shorter disease-free survival (DFS) in breast cancer patients [P = 0.030; hazard ratio (HR), 1.18] (Fig. 2D). In the subgroup analysis, higher expression of EILA predicted worse prognosis in estrogen receptor–positive (ER+) breast cancer patients (P < 0.011; HR, 1.26), but not in estrogen receptor–negative (ER−) breast cancer patients (P = 0.283; HR, 1.15), suggesting its potential as a biomarker in ER+ breast cancer (Fig. 2D). Consistent with the results of the online database, we confirmed that higher expression of EILA was associated with worse clinical outcome in ER+/HER2− breast cancer patient in an independent cohort (SYSMH cohort 2, n = 215) (Fig. 2E).
Fig. 2. EILA predicts poor prognosis in breast cancer patients.
(A) EILA expression in breast tumors compared to their paired normal tissues (TCGA-BRCA database, n = 109, Student’s t test). (B) qRT-PCR detection of EILA expression in paired breast tumors and normal tissues from SYSMH cohort 1 (n = 47, Student’s t test). ****P < 0.0001, **P < 0.01. (C) Representative images of EILA expression detected by in situ hybridization (ISH) staining paired tissues from SYSMH cohort 1. Scale bars, 200 μm for 100× images and 50 μm for 400× images. (D) Disease-free survival (DFS) curves of breast cancer patients based on EILA expression using Kaplan-Meier analysis in the bc-GenExMiner v4.7 database (for all BC patients, n = 2319; for ER+ patients, n = 1667; for ER− patients, n = 625; log-rank test, two-sided). (E) Kaplan-Meier survival curve of breast cancer patients with high (ISH staining score ≥ 4) and low (ISH staining score < 4) levels of EILA expression in SYSMH cohort 2 (n = 215, log-rank test, two-sided). (F) Representative images of ISH for EILA and IHC for cyclin E1 in breast cancer patients from SYSMH cohort 2. Scale bars, 200 μm for 100× images and 50 μm for 400× images. (G) Correlation analysis of EILA expression with cyclin E1 expression in breast cancer patients from SYSMH cohort 2. The r and P values were determined by Spearman analysis. (H and I) Distribution of breast cancer patients with high or low Ki67 (H) and pRb (I) expression in the low or high EILA expression groups from SYSMH cohort 2 (chi-square test, two-sided). ****P < 0.0001. (J) GSEA analysis showing different gene profiles between high (≥median, n = 554) or low (<median, n = 537) expression of EILA in TCGA-BRCA database. NES, normalized enrichment score. (K) Gene Ontology (GO) analysis of the top 500 genes correlated with EILA expression in TCGA-BRCA database.
Similarly, higher cyclin E1 expression also correlated with shorter DFS in ER+ breast cancer patients (fig. S2A). However, the RNA levels of EILA and cyclin E1 showed weak correlation (fig. S2B). To explore the correlation between EILA and cyclin E1 protein, we evaluated the protein level of cyclin E1 via immunohistochemical (IHC) staining. We found that EILA colocalized with cyclin E1 protein in breast cancer, and EILA expression is positively correlated with the protein level of cyclin E1 (r = 0.620, P < 0.0001) (Fig. 2, F and G). Moreover, higher expression of cyclin E1 protein also predicted worse prognosis (fig. S2C). Univariate and multivariate Cox proportional hazards regression analyses revealed that only the expressions of EILA and cyclin E1 were independent predictors of prognosis in ER+/HER2− breast cancer (table S1). In conclusion, these results suggest that EILA and cyclin E1 protein are both potential biomarkers to predict prognosis in breast cancer.
Rb is a tumor suppressor that is phosphorylated during the transition from G1 to S phase of the cell cycle, while Ki67 is a proliferation marker for cancer cells (23, 24). To determine whether EILA correlates with cell cycle progression and cell proliferation, we assessed the expressions of phosphorylated Rb (pRb) and Ki67 via IHC staining. We found that tumors in the EILA-high group expressed higher levels of pRb and Ki67 than those in the EILA-low group, suggesting that EILA overexpression is positively associated with cell cycle progression and cell proliferation (Fig. 2, H and I, and table S2). Similarly, cyclin E1–high group also expressed higher levels of pRb and Ki67 than did the cyclin E1–low group (fig. S2, D and E). In univariate and multivariate Cox proportional hazards regression model, higher EILA expression was significantly associated with Ki67 expression, but not with age, tumor size, lymph node metastasis, or clinical stage (table S3). However, none of the analyzed factors was found to have a significant association with cyclin E1 expression (table S4). To further explore the role of EILA in cell cycle regulation, we analyzed the transcriptomic data of breast cancer patients from TCGA-BRCA database. Patients were classified as EILA-high group (n = 554, >median) or EILA-low group (n = 537, ≤median) according to the expression level of EILA. The results of Gene Set Enrichment Analysis (GSEA) showed that the HALLMARK_E2F_TARGETS gene set and the GOBP_MEIOTIC_CELL_CYCLE_PROCESS gene set were significantly enriched in the EILA-high group than in the EILA-low group (Fig. 2J). We also analyzed the cyclin E1–high and cyclin E1–low groups and found similar results (fig. S2F). Additionally, Gene Ontology (GO) analysis on 500 genes most positively correlated with EILA expression showed high enrichment of positive regulation of cell cycle pathways (Fig. 2K). Collectively, these findings suggest that EILA is a prognostic biomarker for survival and is correlated with positive regulation of cell cycle progression in breast cancer.
EILA promotes cell proliferation and CDK4/6 inhibitor resistance in vitro
EILA is identified as a cyclin E1–interacting lncRNA that plays an important role in CDK4/6i resistance, and is tightly associated with cell cycle progression and unfavorable prognosis. To further explore the mechanism of EILA in CDK4/6i resistance, we silenced EILA with specific ASOs in the CDK4/6i-resistant cells (fig. S3A). EILA knockdown reduced cell proliferation and restored palbociclib sensitivity in both MCF7-palR and T47D-palR cells (Fig. 3, A and B, and fig. S4, A and B). In addition, 5-ethynyl-2'-deoxyuridine (EdU) incorporation assay and cell cycle analysis by flow cytometry demonstrated that EILA knockdown reduced DNA synthesis and caused G1-S phase arrest, and the reduction was in a greater degree with palbociclib treatment (Fig. 3, C and D, and fig. S4, C and D). Senescence-associated β-galactosidas (SA-β-gal) activity assays showed that EILA knockdown induced senescence in the resistant cells, which was further enhanced by palbociclib (Fig. 3E and fig. S4E). Together, these results indicate that EILA is necessary for drug resistance in the CDK4/6i-resistant cells.
Fig. 3. EILA is required for CDK4/6 inhibitor resistance.
(A to E) MCF7-palR cells were transfected with negative control antisense oligonucleotides (NC) or specific antisense oligonucleotides for EILA (ASO#1 and ASO#2). (A and B) Cell proliferation assays (A) and colony formation (B) assays revealing the growth of transfected cells under palbociclib treatment. (C) EdU incorporation assays displaying the portion of DNA-replicating cells. The incorporated EdU was labeled with Azide Alexa Fluor 555 (red color), and the cell nucleus was stained with DAPI (blue color). Scale bar, 100 μm. (D) Cell cycle distribution of the transfected cells under palbociclib treatment. The cells were stained with propidium iodide (PI) and analyzed by flow cytometry. (E) Representative images of SA-β-gal staining of the cells. Scale bar, 100 μm. (F to J) MCF7-pa cells were transfected with empty vector (EV) or EILA overexpression vector (EILA) under palbociclib treatment. Cell proliferation assays (F), colony formation assays (G), EdU incorporation assays (H), cell cycle distribution (I), and SA-β-gal staining assays (J) were performed. For (A) to (J), data from three independent experiments were expressed as means ± SD and the P values were calculated by Student’s t test for two-group comparison and one-way ANOVAs for multiple-group comparison. ****P < 0.0001, ***P < 0.001, **P < 0.01, and N.S. for P > 0.05.
In addition, we performed the gain-of-function assays by overexpressing EILA in MCF-7 and T47D parental (MCF7-pa and T47D-pa) cells (fig. S3B). As the results showed, EILA overexpression rendered resistance to palbociclib (Fig. 3, F and G, and fig. S4, F and G). Moreover, the enforced expression of EILA in both MCF7-pa and T47D-pa cells increased DNA synthesis and promoted G1-S transition in the presence of palbociclib treatment (Fig. 3, H and I, and fig. S4, H and I). The ectopically expressed EILA also reduced palbociclib-induced cell senescence (Fig. 3J and fig. S4J). In conclusion, these results suggest that EILA is able to promote CDK4/6i resistance in breast cancer cells.
Hairpin A of EILA stabilizes cyclin E1 protein by interacting with its C-terminal domain
To explore how EILA interacts with and regulates cyclin E1, we silenced EILA in MCF7-palR and T47D-palR cells and examined cyclin E1 expression with qRT-PCR and Western blot assays, respectively. We found that EILA knockdown reduced the protein level of cyclin E1 but had little influence on its mRNA level (Fig. 4, A and B). In addition, silencing EILA significantly shortened the half-life of cyclin E1 protein (Fig. 4C and fig. S5, A and B), indicating that EILA knockdown facilitates the degradation of cyclin E1. On the other hand, enforced expression of EILA up-regulated the protein level of cyclin E1 and prolonged its half-life but did not affect its mRNA level (Fig. 4, D to F, and fig. S5, C and D). These results reveal that EILA upregulates cyclin E1 expression by protecting it from degradation.
Fig. 4. Hairpin A of EILA stabilizes cyclin E1 protein by interacting with its C-terminal domain.
(A and B) qRT-PCR detection (A) and Western blot (B) of cyclin E1 in MCF7-palR and T47D-palR cells after EILA knockdown. (C) Western blot showing the degradation kinetics of cyclin E1 protein in MCF7-palR cells transfected with NC or EILA ASOs followed by cycloheximide treatment for indicated time. The degradation rate of cyclin E1 protein was quantified by band intensity. (D and E) qRT-PCR detection (D) and Western blot (E) of the cyclin E1 expression in MCF7-pa and T47D-pa cells transfected with empty vector (EV) or EILA overexpression vector (EILA). (F) Western blot showing the degradation kinetics of cyclin E1 protein in MCF7-pa cells expressing EV or EILA. (G and H) RNA pull-down assays followed by Western blot showing the interaction between sequential truncated EILA fragments and endogenous cyclin E1 in MCF7-palR cell lysates. (I) Western blot of cyclin E1 protein in MCF7-pa cells transfected with empty EV or hairpin A overexpression vector (hairpin A). (J) Overview of cyclin E1 truncated mutants and its domain. NLS, nuclear localization signal. (K) RNA pull-down followed by Western blot showing the interaction of cyclin E1 truncated mutants and EILA. (L) RIP followed by qRT-PCR assays revealing the binding of EILA and cyclin E1 truncated mutants. For (A), (C), (D), (F), and (L), data from three independent experiments were expressed as means ± SD and the P values were calculated by Student’s t test for two-group comparison and one-way ANOVAs for multiple-group comparison. ****P < 0.0001, ***P < 0.001, and N.S. for P > 0.05.
Recent studies have shown that specific lncRNAs may have well-defined secondary structures, which could play a crucial role in their interactions with proteins (25, 26). To determine the nucleotide sequence of EILA that interacts with cyclin E1, we performed RNA pull-down assays using a series of EILA deletion mutation, followed by Western blot of the products. As shown in the results, EILA mutants that contained nucleotides 1 to 180 bound to cyclin E1 protein as efficiently as the full-length EILA, while mutants with the nucleotide 1 to 180 deletion lose the ability of binding to cyclin E1 protein (Fig. 4, G and H). According to the RNAfold software, there is a stable hairpin structure within nucleotides 1 to 180, termed hairpin A (fig. S5E). We found that enforced expression of hairpin A increased cyclin E1 protein expression and prolonged its half-life without influencing its mRNA level (Fig. 4I and fig. S5, F to H). To investigate the importance of EILA binding for increasing cyclin E1 stability, we created a mutant form of EILA with impaired interaction by mutating hairpin A (EILAmA). We then evaluated the effects of EILAmA on cyclin E1 protein expression and found that EILAmA was unable to bind to cyclin E1 or increase its expression in MCF7-palR cells (fig. S5, I and J). Furthermore, EILAmA failed to prolong cyclin E1’s half-life compared to full-length EILA (fig. S5K). These results indicated that hairpin A of EILA is necessary and responsible for the interaction with cyclin E1 protein to regulate its stability.
To determine the precise interacting domain of cyclin E1 with EILA, we constructed HA-tagged full-length and truncated cyclin E1 overexpression vectors. As the results of RNA pull-down assays demonstrated, only the F2 truncated mutant could be pulled down by biotin-labeled EILA, which indicates that the C terminus is the interacting region of cyclin E1 with EILA (Fig. 4, J and K). As expected, the RIP-qPCR assays also revealed that EILA was enriched by F2 truncated mutant (Fig. 4L). These results indicate that EILA interacts with cyclin E1 protein at its C terminus.
EILA hinders the binding of cyclin E1 and FBXW7
It has been well established that the protein stability of cyclin E1 is under precise control of ubiquitination-mediated proteasomal degradation, and the phosphorylation sites of cyclin E1 (like T62, T380, and S384) regulate its ubiquitination (27). Since EILA interacts with the C terminus of cyclin E1 and stabilizes the protein, we then hypothesize that EILA may influence its C-terminal phosphorylation sites and ubiquitination-mediated degradation. Therefore, we examined the C-terminal phosphorylation sites (T380 and S384) of cyclin E1 in the resistant and parental cells. We found that the resistant cells expressed lower levels of S384-phosphorylated cyclin E1 than did the parental cells (Fig. 5A). The phosphorylation sites of cyclin E1 are also regulated by CDK2 and glycogen synthase kinase 3β (GSK3β) (27). To exclude the effects of CDK2 and GSK3β on cyclin E1 phosphorylation, we detected their expression in the resistant and parental cells and found that both cells expressed similar levels of CDK2 and GSK3β (Fig. 5A). Moreover, we performed the coimmunoprecipitation and ubiquitination assay with specific cyclin E1 antibody to detect the levels of ubiquitinated cyclin E1 in MCF7-pa and MCF7-palR cells. In line with our hypothesis, the ubiquitination-mediated degradation level of cyclin E1 is reduced in the resistant cells rather than in the parental cells (Fig. 5B). These findings suggest that the reduced S384-phosphorylated cyclin E1 in the resistant cells may inhibit its ubiquitination-mediated degradation. To determine the role of EILA in regulating S384-phosphorylated cyclin E1 and the ubiquitination-mediated degradation of cyclin E1, we silenced EILA with specific ASOs and found that EILA knockdown markedly increased the level of S384-phosphorylated cyclin E1 and ubiquitinated cyclin E1 (Fig. 5, C and D). On the other hand, EILA overexpression decreased phosphorylated cyclin E1 and the ubiquitination-mediated degradation of cyclin E1 (Fig. 5, E and F). These results indicate that EILA regulates the phosphorylation of cyclin E1 on S384 and its ubiquitination, and therefore increases its stability.
Fig. 5. EILA hinders the ubiquitination-mediated proteasomal degradation of cyclin E1.
(A) Western blot analysis of phospho-cyclin E1 (S384), phospho-cyclin E1 (T380), cyclin E1, CDK2, and GSK3β expression levels in the parental cells (MCF7-pa and T47D-pa) and palbociclib-resistant cells (MCF7-palR and T47D-palR). (B) Coimmunoprecipitation (Co-IP) analysis of ubiquitinated cyclin E1 protein in MCF7-pa and MCF7-palR cells. (C) Western blot analysis of phospho-cyclin E1 (S384), cyclin E1, CDK2, and GSK3β expression levels in MCF7-palR cells after EILA knockdown. (D) Co-IP analysis of ubiquitinated cyclin E1 protein in MCF7-palR cells after EILA knockdown. (E) Western blot showing the expression levels of phospho-cyclin E1 (S384), cyclin E1, CDK2, and GSK3β in MCF7-pa cells transfected with empty vector (EV) or EILA overexpression vector (EILA). (F) Co-IP analysis of ubiquitinated cyclin E1 protein in MCF7-pa cells transfected with EV or EILA. (G) Co-IP and Western blot analysis showing the association of FBXW7 and cyclin E1 in MCF7-pa and MCF7-palR cells. (H) Co-IP and Western blot analysis revealing the association of FBXW7 and cyclin E1 in MCF7-palR cells with EILA knockdown. (I) Western blot analysis of cyclin E1 and Fxbw7 in MCF7-palR cells transfected with NC or FBXW7 siRNAs for 24 hours and with/without EILA knockdown. (J) Western blot analysis of cyclin E1 expression in MCF7-palR and T47D-palR cells with EILA knockdown and with DMSO or ubiquitination inhibitor MG-132 treatment for 24 hours.
The phosphorylated cyclin E1 is recognized and ubiquitinated by FBXW7, an E3 ubiquitin ligase, followed by ubiquitination-mediated proteasomal degradation (27, 28). To examine whether FBXW7 regulates cyclin E1 stability in the CDK4/6i-resistant cells, we detected the binding between cyclin E1 and FBXW7, and found that less cyclin E1 was bound to FBXW7 in MCF7-palR cells than in MCF7-pa cells (Fig. 5G). To determine whether FBXW7 plays a role in the regulation of cyclin E1 stability mediated by EILA, we silenced EILA in MCF7-palR cells and detected the binding between cyclin E1 and FBXW7 by coimmunoprecipitation assay. We found that EILA knockdown rendered more cyclin E1 bound to FBXW7 (Fig. 5H), which indicates that EILA hinders the binding between cyclin E1 and FBXW7. Moreover, silencing EILA expression was unable to reduce cyclin E1 in MCF7-palR cells with FBXW7 knockdown, suggesting that FBXW7 participates in the regulation of cyclin E1 stability mediated by EILA (Fig. 5I). We further explored the role of ubiquitination-mediated proteasomal degradation in the regulation of cyclin E1 stability mediated by EILA, and found that EILA knockdown was unable to reduce cyclin E1 in MCF7-palR cells treated with the proteasome inhibitor MG-132 (Fig. 5J). Together, these results reveal that EILA stabilizes cyclin E1 by impairing its binding with FBXW7 and inhibiting its ubiquitination-mediated proteasomal degradation.
CTCF/CDK8/TFII-I complexes regulate EILA expression
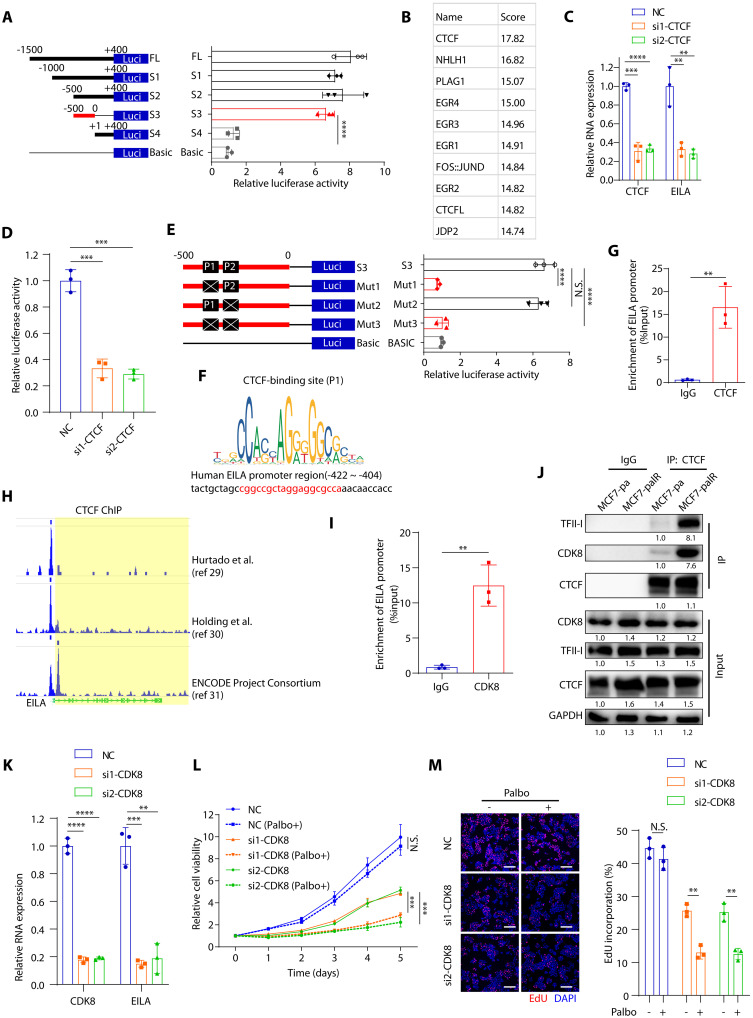
To determine the transcription factor that modulates EILA expression, we performed luciferase reporter assays with various truncated mutants of its promoter region [1.5 kb upstream and 0.4 kb downstream of the transcription start site (TSS)]. We found that the −500- to 0-bp (base pair) region is essential for the luciferase activity, which indicates that the transcription factor of EILA may bind to this region (Fig. 6A). Next, we used the JASPAR database to predict the previously unknown transcription factor of EILA, and found that CTCF ranked first in the binding score (Fig. 6B). Moreover, CTCF knockdown significantly reduced EILA expression and its luciferase promoter activity (Fig. 6, C and D). There are two predicted binding sites in the −500- to 0-bp region for CTCF transcription factor, named P1 (−422 to −404) and P2 (−389 to −371) site. Mutation of P1, but not P2, site significantly reduced the luciferase promoter activity of EILA, indicating that the P1 site is the major binding site of CTCF (Fig. 6, E and F). In addition, the chromatin immunoprecipitation (ChIP)–PCR assay also confirmed that the EILA promoter region contained CTCF-binding site (Fig. 6G). These findings were further confirmed by the ChIP-sequencing (ChIP-seq) data on Cistrome Data Browser (http://cistrome.org/db/#/) (29–31), which showed that the EILA promoter contains CTCF-binding sites (Fig. 6H).
Fig. 6. EILA expression is regulated by CTCF/CDK8/TFII-I complexes.
(A) Luciferase reporter assays showing the transcriptional activity in MCF7-palR cells transfected with truncated fragments of the EILA promoter region. (B) Predicted transcriptional factors of EILA in the JASPAR online software based on the −500- to 0-bp sequence. (C) qRT-PCR analysis of CTCF and EILA expression in MCF7-palR cells after CTCF knockdown. (D) Luciferase reporter assays showing the EILA transcriptional activity in MCF7-palR cells after CTCF knockdown. (E) Luciferase reporter assays performed in MCF7-palR cells transfected with mutated fragments of the EILA promoter region (−500 to 0 bp). (F) Predicted CTCF-binding site in the EILA promoter region. (G) ChIP-qPCR analysis revealing the localization of CTCF at EILA promoter in MCF7-palR cells. (H) Analysis of ChIP-seq binding peaks in the Cistrome Database and UCSC Genome Browser. (I) ChIP-qPCR analysis showing the localization of CDK8 at EILA promoter in MCF7-palR cells. (J) Co-IP assays and Western blot analysis showing the interaction between CTCF and CDK8/TFII-I in MCF7-palR cells. In (K to M), CDK8 was silenced in MCF7-palR cells. (K) qRT-PCR detection of CTCF and EILA in MCF7-palR cells after CTCF knockdown. (L and M) Cell proliferation assays (L) and EdU incorporation (M) assays showing the growth of transfected cells under palbociclib treatment. For (A), (C) to (E), (G), (I), and (K) to (M), data from three independent experiments were expressed as means ± SD and the P values were calculated by Student’s t test for two-group comparison and one-way ANOVAs for multiple-group comparison. ****P < 0.0001, ***P < 0.001, **P < 0.01, and N.S. for P > 0.05.
CTCF is a multifunctional regulator in chromatin structure and gene expression, including transcriptional activation and repression (32). It recruits and binds to protein partners to regulate gene expression, like TFII-I and CDK8 (33). According to the Cistrome Data Browser (34), the general transcriptional factor TFII-I can also bind to the promoter region of EILA (fig. S6A). In addition, ChIP-PCR assay also showed that the EILA promoter region contained CDK8-binding site (Fig. 6I). We also observed more robust interaction of TFII-I, CDK8, and CTCF in the resistant cells than in their parental cells (Fig. 6J), which indicates that CTCF may cooperate with TFII-I and CDK8 to promote EILA expression. To explore the roles of CDK8 and TFII-I in the regulation of EILA expression, we silenced CDK8 and TFII-I in the resistant cells and found that knockdown of CDK8 and TFII-I significantly reduced EILA expression (Fig. 6K and fig. S6B). To determine whether CDK8 and TFII-I play roles in CDK4/6i resistance, we evaluated the palbociclib sensitivity after CDK8 and TFII-1 knockdown, and found that both CDK8 and TFII-I knockdown restored CDK4/6i sensitivity in the resistant cells (Fig. 6, L and M, and fig. S6, C to E). These results suggest that CTCF/CDK8/TFII-I complexes regulate EILA expression by binding to its promoter region and maintain resistance to CDK4/6 inhibitors.
CDK8 plays an oncogenic role in various kinds of cancers (35). It regulates gene expression by associating with mediator complex or by phosphorylating transcription factors (36). CDK8 inhibitors have shown promising efficacy in acute melanoma leukemia (AML), breast cancer, and colon cancer, which indicates that CDK8 is a potential therapeutic target for cancer treatment (30). To determine whether CDK8 inhibition reduces EILA expression and restores CDK4/6 inhibitor sensitivity, we treated the resistant cells with two CDK8 inhibitors [SEL120 and Senexin A (SENA)] and then measured the EILA expression and their sensitivity to palbociclib. We found that both CDK8 inhibitors significantly reduced EILA expression in the resistant cells, suggesting that CDK8 is responsible for EILA expression (fig. S6F). In addition, the two CDK8 inhibitors also markedly resensitized the resistant cells to palbociclib treatment (fig. S6, G to I). The parental cells showed less sensitivity to the CDK8 inhibitors, possibly due to their lower expression of EILA (fig. S6, J and K). Together, these results reveal that targeting EILA expression by CDK8 inhibitors may be a promising strategy in post-CDK4/6 inhibitor setting.
EILA promotes CDK4/6 inhibitor resistance in vivo
To further determine whether EILA promotes CDK4/6i resistance of breast cancer cells in vivo, we inoculated MCF7-palR cells into the mammary fat pads of nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice. After tumors were palpable, they were randomly assigned into six groups and treated with control solution, palbociclib, EILA-specific ASOs, control ASOs, and their combinations. Consistent with the previous findings in vitro, EILA-specific ASOs significantly reduced tumor growth and the combinations of palbociclib and EILA-specific ASOs further shrank the tumors (Fig. 7, A and B), indicating that EILA-specific ASOs resensitizes the resistant tumors to CDK4/6 inhibitors. Moreover, the combinations of palbociclib and EILA-specific ASOs had little influence on mouse weight (Fig. 7C), suggesting that these combinations are tolerable.
Fig. 7. EILA promotes CDK4/6 inhibitor resistance in vivo.
The xenografts (n = 6 per group) of MCF7-palR cells were intratumorally injected with NC or EILA-ASOs and then treated with control or palbociclib. (A to C) The harvested tumors (A) and the tumor volume (B) and mouse weight (C) were measured. (D) Representative images showing the expression of Ki67, pRb, and cyclin E1 analyzed by immunohistochemistry (IHC) staining and the EILA expression analyzed by ISH staining. (E) IHC score for Ki67, pRb, and cyclin E1 and ISH score for EILA. For (B), (C), and (E), data were shown as means ± SD and the P values were calculated by Student’s t test for two-group comparison and one-way ANOVAs for multiple-group comparison. ****P < 0.0001, ***P < 0.001, **P < 0.01, and N.S. for P > 0.05. Scale bar, 50 μm.
EILA-specific ASOs markedly reduced EILA expression and the protein level of cyclin E1 (Fig. 7, D and E). In addition, EILA knockdown also significantly lowered the expressions of Ki67 and pRb, and the combinations of palbociclib and EILA-specific ASOs reduced their expressions in a greater degree (Fig. 7, D and E). Collectively, these results suggest that targeting EILA expression with specific ASOs overcomes CDK4/6i resistance in vivo.
EILA predicts CDK4/6 inhibitor response and is up-regulated after progression on CDK4/6 inhibitors
To further demonstrate the clinical significance of EILA in CDK4/6i resistance, we detected the EILA expression in 37 ER+ advanced breast cancer patients treated with CDK4/6 inhibitors (SYSMH cohort 3). These patients were divided into two groups (EILA-high and EILA-low) according to the ISH score of EILA. Analyses of Kaplan-Meier plot revealed that high EILA expression predicted shortened PFS in these patients (Fig. 8A). Similarly, high cyclin E1 protein expression was also associated with poor clinical outcome in these patients (Fig. 8B). Moreover, seven paired biopsies were taken from these patients before treatment and after progression on CDK4/6 inhibitors. Analysis of these paired biopsies showed that the resistant tumors expressed higher levels of EILA than the naïve tumors, with up-regulation of cyclin E1 protein (Fig. 8, C and D). These results indicate that EILA expression is associated with CDK4/6 inhibitor response and is up-regulated after progression, which may serve as a biomarker for CDK4/6i resistance and as a therapeutic target to overcome the resistance.
Fig. 8. EILA predicts poor response to CDK4/6i treatment and is up-regulated in CDK4/6i-resistant breast cancers.
(A) Kaplan-Meier survival curve of breast cancer patients with high (ISH staining score ≥ 4) and low (ISH staining score < 4) expression of EILA in SYSMH cohort 3 (n = 37, log-rank test, two-sided). (B) Kaplan-Meier survival curve of breast cancer patients with high (IHC staining score ≥ 4) and low (IHC staining score < 4) expression of cyclin E1 in SYSMH cohort 3 (n = 37, log-rank test, two-sided). (C and D) ISH for EILA and IHC for cyclin E1 in breast cancer patients before CDK4/6i treatment and after progression (n = 7). Representative images were shown. Scale bars, 50 μm. Paired Wilcoxon signed-rank test determines the P values. *P < 0.05.
DISCUSSION
Here, we prove that the dysregulation of cyclin E1 protein contributes to CDK4/6i resistance in breast cancer cells. We identify EILA as a cyclin E1–interacting lncRNA that stabilizes cyclin E1 and induces CDK4/6i resistance in breast cancer cells. Bioinformatics analysis reveals that high expression of EILA is associated with cell cycle progression and predicts poor prognosis in breast cancer, which is further verified by IHC and ISH analysis. In addition, targeting EILA expression with specific ASOs restores CDK4/6i sensitivity both in vitro and in vivo. We also investigate the regulation of EILA expression and find that the CTCF/CDK8/TFII-I complexes bind to the promoter region of EILA and facilitate its expression. CDK8 inhibitors reduce EILA expression and overcome CDK4/6i resistance. In patients receiving CDK4/6 inhibitor treatment, higher expression of EILA is associated with shorter PFS. Furthermore, the expression of EILA is increased after progression on CDK4/6 inhibitors.
CDK4/6 inhibitors plus endocrine therapy are now the standard treatment for advanced HR+/HER2 breast cancer (8). However, resistance to CDK4/6 inhibitors is inevitable and therapeutic strategies to override the resistance attract great attention. CCNE1 amplification and cyclin E1 overexpression are commonly seen in many malignancies, including ovarian, endometrial, esophagogastric, and breast cancer (37). Mechanistically, cyclin E1 cooperates with CDK2 to promote Rb phosphorylation and facilitates G1-S transition of the cell cycle (27). In breast cancer, cyclin E1 overexpression not only predicts poor prognosis (10) but also correlates with resistance to endocrine therapy, anti-HER2 therapy, and CDK4/6 inhibitors (13, 15, 38). Apart from binding to CDK2 to form cyclin E1–CDK2 complexes, cyclin E1 also promotes cell cycle progression in a CDK2-independent manner (39). These findings indicate that cyclin E1 may serve as a potential target to reduce cell proliferation and overcome drug resistance. However, by now, there are no drugs targeting cyclin E1 directly. Although CDK2 inhibitors suppress the function of cyclin E1–CDK2 complexes and arrest cell cycle progression, most of them are not specific to CDK2. Meanwhile, they inhibit CDK1 or CDK9 and bring unexpected off-target effects, which restrict their application (40).
Here, we report that targeting EILA with specific ASOs facilitates the ubiquitination-mediated degradation of cyclin E1 protein, which provides a possible solution to reduce cyclin E1. The performance of ASOs represents a potential technology for therapeutic application, with high specificity and manageable adverse events (41). So far, multiple clinical trials have validated the safety and activity of ASOs in a broad range of diseases, including amyotrophic lateral sclerosis, and chronic hepatitis B and atherosclerosis (42–44). Besides, ASOs also showed potential antitumor activity in many malignancies (45). Here, we prove that targeting EILA with ASOs overcomes CDK4/6i resistance in vitro and in vivo, which shows the antitumor activity of EILA-ASOs in preclinical models. However, as a systematic disease, the intratumoral drug administration of EILA-ASOs limits its application in cancer treatment. We are going to optimize the delivery of EILA-ASOs and its structure in the future.
Up to day, there are few biomarkers to predict CDK4/6i response beside ER status (16, 46). Biomarker analyses of 302 patients from the PALOMA-3 trial showed that higher cyclin E1 mRNA expression was associated with resistance to CDK4/6 inhibitors in the PALOMA-3 trial, which was further confirmed by the POP trial (9, 14). However, biomarker analyses of the MONALEESA-2 and PALOMA-2 trials did not show such association between cyclin E1 mRNA level and PFS from CDK4/6 inhibition (16, 17). These contradictory findings suggest that the role of cyclin E1 in CDK4/6i resistance remains to be elucidated. In addition to cyclin E1 mRNA, mRNAs of other cell cycle regulators, like cyclin D1, CDK4, CKD6, and Rb, failed to predict CDK4/6i treatment response in these clinical trials (16, 17). Therefore, it is of great clinical significance and translational value to seek biomarkers to distinguish those who can benefit from CDK4/6 inhibition. No previous research has focused on the predictive value of lncRNA in CDK4/6i response. We report that overexpression of EILA predicts poor prognosis in breast cancer patients with CDK4/6i treatment. Meanwhile, we found that the EILA expression is positively correlated with the expression of cyclin E1 protein. Moreover, the overexpression of cyclin E1 protein is associated with shortened DFS, which indicates the necessity to detect the expression of cyclin E1 protein with IHC in patients receiving CDK4/6i treatment.
CDK8 is reported as an oncogene in many malignancies, which regulates gene expression by binding to mediator complexes or phosphorylating transcription factors (36). CDK8 inhibitors show promising anticancer activity in acute myeloid leukemia (AML), breast cancer, and colon cancer, indicating that CDK8 is a potential therapeutic target for cancer treatment (35). We demonstrate that CDK8 cooperates with CTCF to promote EILA expression, which can be blocked by CDK8 inhibitors. Both CDK8 inhibitors, SEL120 and SENA, reduce EILA expression and resensitize the resistant cells to CDK4/6 inhibition, suggesting the application of CDK8 inhibitors in post-CDK4/6i settings.
In conclusion, our study reveals that the cyclin E1–interacting lncRNA EILA plays an important role in regulating cyclin E1 stability and maintaining the resistance to CDK4/6 inhibitors. We report that EILA is a predictive biomarker for CDK4/6i treatment, and it is also a potential therapeutic target for post-CDK4/6i progression.
MATERIALS AND METHODS
Cell culture, reagents, transfection, and transduction
The parental cell lines (MCF7-pa and T47D-pa) were obtained from the American Type Culture Collection (ATCC; Rockville, MD, USA), and the palbociclib-resistant cell lines (MCF7-palR and T47D-palR) were established as previously described (21). MCF7-pa, MCF7-palR, and human embryonic kidney (HEK) 293T cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA) with 10% fetal bovine serum (FBS) (HyClone, USA). T47D-pa and T47D-palR cell lines were cultured in RPMI 1640 medium (Gibco, USA). All the cell lines were identified with short tandem repeat profiling and were free of mycoplasma. The cell lines were maintained at 37°C and in 5% CO2.
Palbociclib was provided by Pfizer Inc., and SEL120 and SENA were purchased from Selleck (Houston, TX). These reagents were dissolved with dimethyl sulfoxide (DMSO) for in vitro experiments.
For siRNA or ASO transfection, cells (1 × 105) were seeded in the six-well plate and then transfected with specific siRNAs or ASOs using RNAiMax (13778150, Invitrogen). The sequence of siRNA and ASO was listed in table S5.
For lentivirus production, the full-length sequence of EILA, HA-tagged cyclin E1, and its truncated mutations were cloned into the pCDH-puro plasmids and then were cotransfected with pMD2.G and pSPAX2 plasmids into the HEK293T cells to generate lentivirus. The lentivirus-containing supernatant was collected and filtered with a 0.25-μm filter (SLGP033RB, Millipore). We then transduced the cells with the indicated lentivirus and polybrene (5 μg/ml). After 72 hours, we selected the transduced cells with puromycin (5 μg/ml).
RNA immunoprecipitation
The RIP assays were conducted following the manufacturer’s instructions of the Magna RIP kit (17-700, Millipore). In brief, the cultured cells were harvested and lysed with RIP lysis buffer. After centrifugation, the supernatant of the lysates was incubated with antibody-conjugated magnetic beads at 4°C overnight. The beads were washed, and the coprecipitated RNAs were extracted for qRT-PCR or sequencing. Negative control IgG and human anti-HA antibody [1:100, Cell Signaling Technology (CST), 3724] were used in the research.
RIP sequencing and lncRNA sequencing
For RIP sequencing, MCF7-pa cells stably expressing HA-tagged cyclin E1 were harvested to perform RIP assay with IgG or HA antibody. After the coprecipitated RNAs were extracted, ribosomal RNAs (rRNAs) were removed. The retained mRNA and ncRNAs were fragmented into short fragments with fragmentation buffer and then reverse-transcribed into cDNA library for sequencing. The RIP sequencing was performed using Illumina HiSeq6000 platform in Gene Denovo Biotechnology Co. (Guangzhou, China).
For lncRNA sequencing, the total RNAs from MCF7-pa and MCF7-palR cells were extracted with TRIzol reagent (15596026, Invitrogen) according to the manufacturer’s instructions. The RNA quality was checked by the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and by the ribonuclease (RNase)–free agarose gel electrophoresis. After removal of rRNAs, the enriched mRNAs and ncRNAs were fragmented into short fragments and then reverse-transcribed into cDNA library for sequencing. The lncRNA sequencing was performed using Illumina HiSeq6000 platform in Gene Denovo Biotechnology Co. (Guangzhou, China).
RNA ISH and IHC
For RNA ISH, the digoxin-conjugated oligonucleotide probe (/5DiGN/AAGAAATCGGCCAGAGCCTCA/3DiG_N/) for EILA detection was designed and synthesized by Exiqon (306584156, QIAGEN). EILA expression was measured in the sections from paraffin-embedded tissue samples. Briefly, the sections were digested with proteinase K (20 μg/ml) at 37°C for 10 min and hybridized with the probe (200 nM) at 54°C overnight. After hybridization, these sections were washed using 2× SSC added with 25% deionized formamide at 54°C for 10 min and 2× SSC at 54°C for 10 min. After washing, these sections were incubated with peroxidase (POD)–conjugated anti-digoxin monoclonal antibody (200-032-156, Jackson ImmunoResearch, 1:400) at 4°C overnight and subsequently dyed with diaminobenzidine (DAB) (GK500710, Dako) and hematoxylin according to the manufacturer’s instructions.
For IHC, the sections were dewaxed, rehydrated, and boiled in sodium citrate buffer (P0083, Beyotime) for antigens retrieval. After incubation in H2O2 (3%) and blocking with bovine serum albumin (1%), the sections were incubated with primary antibodies against pRb (8516, CST, 1:200), Ki67 (ZM0166, ZSGB-BIO, ready to use), and cyclin E1 (A301-566A, Bethyl Laboratories, 1:100) at 4°C overnight and then stained with DAB (GK500710, Dako) and hematoxylin according to the manufacturer’s instructions.
The ISH and IHC score were evaluated by two independent observers based on both the proportion and intensity of positively stained tumor cells in 10 random fields under a 40× objective. The proportion of positively stained tumor cells were divided into four levels: 0 (no positive cells), 1 (<10%), 2 (10% to 50%), and 3 (>50%). The staining intensity of positive signal was graded as follows: 0 (no staining), 1 (light brown), 2 (brown), and 3 (dark brown). The staining score was calculated as follows: staining score = proportion of positively stained tumor cells × staining intensity. Using this method, the expression of indicated markers was scored as 0, 1, 2, 3, 4, 6, 8, 9, or 12, with their median values as the cutoff points to divide patients into high or low expression group.
RNA FISH and IF
Cells (5 × 104 per dish) seeded on the confocal dishes were washed with phosphate-buffered saline (PBS) and fixed with 4% formaldehyde at room temperature for 15 min. Then, the cells were digested with 0.4% trypsin at room temperature for 5 min and permeabilized with PBS containing 0.5% Triton X-100 on ice for 5 min. The hybridization was performed by incubating the cells with EILA probes at 54°C overnight. After washing off the probes, the cells were coincubated with the fluorescein-conjugated antibody against digoxin (Roche) and the primary antibody against cyclin E1 (A301-566A, Bethyl Laboratories, 1:100) at 4°C overnight. Then, the cells were incubated with Alexa Fluor 647 secondary antibodies (A32733, Invitrogen, 1:200) for 1 hour and stained with DAPI (4′,6-diamidino-2-phenylindole) at room temperature for 10 min. The images were taken with a confocal microscope (LSM800, Zeiss) to analyze the colocalization of EILA and cyclin E1.
RNAscope
The seven ZZ probes (Hs-EILA-O1) targeting 2 to 842 nt of EILA were designed and synthesized by Advanced Cell Diagnostics (ACD; CA, USA). The RNAscope assay was performed to detect the expression and location of EILA in the parental and resistant cells using an RNAscope 2.5 High Definition (HD)—RED Assay kit (322350, ACD) according to the manufacturer’s instruction. Briefly, the cells seeded on the confocal dishes were rinsed with PBS and fixed with 10% neutral formalin at room temperature for 30 min. After fixation, cells were dehydrated, rehydrated, incubated with H2O2 at room temperature for 10 min, and digested with proteinase III at room temperature for 10 min. Hybridization was performed with the probes at 40°C for 2 hours, and the probes were furthered labeled with TSA Plus Cy3 fluorescence and blocked with the horseradish peroxidase (HRP) blocking solution. The cells were dyed with DAPI and analyzed with a confocal microscope.
Western blot
The cells were harvested and lysed in radioimmunoprecipitation assay lysis buffer (P0013B, Beyotime) containing 1% protease and phosphatase inhibitors (78442, Thermo Fisher Scientific). Sample proteins were separated by SDS–polyacrylamide gel electrophoresis and transferred onto the polyvinylidene difluoride membranes (Merck Millipore). The membranes were blocked with 5% nonfat milk and incubated with primary antibodies against cyclin E1 (sc-247, Santa Cruz Biotechnology, 1:1000), HA (3724, CST, 1:1000), p-E1(S384) (PA5-106061, Thermo Fisher Scientific, 1:1000), p-E1(T380) (PA5-36636, Thermo Fisher Scientific, 1:1000), CDK2 (sc-6248, Santa Cruz Biotechnology, 1:1000), GSK3β (12456, CST, 1:1000), ubiquitin (3936, CST, 1:1000), FBXW7 (MA5-26563, Thermo Fisher Scientific, 1:1000), TFII-I (610942, BD Biosciences, 1:1000), CDK8 (sc-1521, Santa Cruz Biotechnology, 1:1000), and CTCF (3418, CST, 1:1000). After washing, the membranes were further incubated with HRP-conjugated secondary antibodies at room temperature for 1 hour. The blots were then detected with enhanced chemiluminescence (34095, Pierce). All the results of Western blot were quantified by ImageJ (version 1.54d), and the band intensity was labeled below every blot, with the first lane as a control.
Coimmunoprecipitation and ubiquitination assay
The cells were harvested and lysed in IP lysis buffer (87788, Thermo Fisher Scientific) containing 1% protease and phosphatase inhibitors (78442, Thermo Fisher Scientific). The lysates were incubated with antibodies against cyclin E1 (A301-566A, Bethyl Laboratories, 1:50), CTCF (3418, CST, 1:50), or rabbit IgG (negative control) at 4°C overnight. Then, the Dynabeads protein G (10004D, Invitrogen) was added into the lysates and incubated at room temperature for 1 hour. After washing, the precipitated proteins were boiled in lithium dodecyl sulfate (LDS) buffer (NP0007, Invitrogen) and detected by Western blot. For ubiquitination assay, the cells were treated with 20 μM MG-132 (C2211, Sigma-Aldrich) for 12 hours before harvesting. The treated cells were then lysed and subjected to coimmunoprecipitation assays, and the precipitated proteins were detected with Western blot.
Quantitative real-time PCR
RNAs were reverse-transcribed into cDNA using the PrimeScript RT Master Mix (RR036A, Takara) according to the manufacturer’s instruction. qRT-PCR assays were performed by using TB Green Premix ExTaq II (RR820A, Takara) according to the manufacturer’s recommendations. All the primers used in this article were listed in table S6.
Chromatin immunoprecipitation
ChIP assays were performed using the EZ-Magna ChIP A/G Chromatin Immunoprecipitation Kit (17-10086, Merck Millipore) according to the manufacturer’s recommendations. In brief, the harvested MCF7-palR cells were cross-linked with 1% formaldehyde at 37°C for 20 min, lysed, and sonicated to generate ~200 to 1000 bp DNA fragments. Antibodies against CTCF (3418, CST, 1:50), CDK8 (sc-1521, Santa Cruz Biotechnology, 1:50), and rabbit/mouse IgG were used for immunoprecipitation, and the precipitated DNA fragments were subjected to qPCR amplification. The primers used were listed in table S6.
Subcellular fractionation of RNAs
The RNAs were isolated from nuclear and cytoplasmic extracts by a protein and RNA isolation system kit (AM1921, Invitrogen) according to the manufacturer’s instructions. The expression of EILA in the nuclear and cytoplasmic fractionations was detected by qRT-PCR.
Northern blot
The Northern blot assay was performed using a NorthernMax kit (AM1940, Invitrogen) according to the manufacturer’s recommendations. In brief, the RNAs mixed with RNA loading buffer were heated at 65°C for 10 min and cooled on ice. The RNAs were then separated on agarose gels, transferred to the positively charged nylon membrane, and crosslinked with ultraviolet. The membrane was incubated with the digoxin-conjugated oligonucleotide probe at 54°C overnight for detection of EILA and ACTB. After washing, the membrane was further incubated with alkaline phosphatase (AP)–conjugated antibody (11093274910, Roche, 1:5000) against digoxin for 30 min at room temperature and detected with the chemiluminescent substrate for alkaline phosphatase detection (CSPD) substrate.
RNA pull-down
The biotin-labeled RNAs were transcribed using a TranscriptAid T7 High Yield Transcription kit (K0441, Thermo Fisher Scientific) and biotinylated using a Magnetic RNA-Protein Pull-Down kit (20164, Thermo Fisher Scientific) according to the manufacturer’s instructions. After purifying using a GeneJET RNA Purification kit (K0731, Thermo Fisher Scientific), the biotin-labeled RNAs were heated at 95°C for 2 min, cooled on ice for 3 min, and then kept at room temperature for 30 min to form secondary structure. Then, the folded biotin-labeled RNAs were captured with streptavidin magnetic beads at room temperature for 1 hour and incubated with cell lysates or recombinant cyclin E1 protein (ab119719, Abcam) at room temperature for 2 hours. After washing, the eluted proteins were subjected to Western blot.
Luciferase reporter assay
A series of truncated DNA fragments of the EILA promoter region were cloned into the pGL3 reporter plasmid. The resistant cells were transfected with these pGL3 reporter plasmids together with pRL-TK plasmids at 50:1. Forty-eight hours after transfection, these cells were harvested and the Firefly and Renilla luciferase activity was measured using a Dual-Luciferase Reporter Gene Assay kit (RG027, Beyotime). The pGL3-basic plasmid was used as a control, and the Firefly luciferase activity was normalized to Renilla activity luciferase activity.
Tumor xenograft formation in mice
The animal experiments were approved by the Sun Yat-Sen University laboratory animal care and use committee. Female NOD/SCID mice aged 4 weeks were housed under standard condition (25°C, 50% humidity) at the specific pathogen–free (SPF) animal facility. The mice were subcutaneously implanted with 17β-estrogen pellets (0.72 mg, 60-day release, Innovative Research of America), and 5 days later, MCF7-palR cells (1 × 107/0.1 ml of PBS) were injected into the mammary fat pad of the mice. When the tumors were palpable, the mice were randomly assigned into indicated groups (n = 6 per group). For palbociclib and EILA ASOs combinational treatment assay, the xenograft mice were randomized to six groups and treated with the following: (i) negative control ASOs (intratumoral injection, 5 nmol per injection, every 2 days, RiboBio, China) + control lactate buffer, (ii) negative control ASOs + palbociclib (oral gavage, 100 mg/kg, every day, 50 mM, lactate buffer at pH 4.0), (iii) EILA-ASO#1 + lactate buffer, (iv) EILA-ASO#1 + palbociclib, (v) EILA-ASO#2 + lactate buffer, (vi) EILA-ASO#2 + palbociclib. The mouse weights and xenograft sizes were monitored every 7 days. Tumor volumes (mm3) were calculated by the following formula: volume (mm3) = length × width2 × 0.5. At the endpoints, the mice were euthanized and the tumors were harvested for IHC staining and RNA ISH assay.
Determination of EILA copy numbers
The total RNAs were extracted from parental and resistant cells and detected by qRT-PCR. To establish the standard curve, in vitro transcribed EILA RNAs were diluted into a series of dilution and detected by qRT-PCR. According to the threshold cycle (CT) value and copy number of the standard curve, EILA copy numbers per cell were calculated.
Colony formation unit assay and EdU incorporation assay
For colony formation unit assay, cells were seeded in six-well plates (1000 cells per well) followed by the indicated treatment. The medium was replenished every 3 days. After 15 days, the cells were fixed with 4% paraformaldehyde and incubated with crystal violet for staining. For EdU incorporation assay, cells were seeded in the 96-well plates (4000 cells per well) and treated with indicated agents. The cells were further incubated with EdU (10 mM, C0075L, Beyotime, China) for 2 hours before harvest. These cells were fixed with 4% paraformaldehyde and dyed with Azide Alexa Fluor 555 (C0075L, Beyotime, China) and DAPI following the manufacturer’s protocol.
Flow cytometry for cell cycle
For cell cycle analysis, cells were seeded in six-well plates (3 × 105 per well) and treated with indicated agents. After harvest, the cells were fixed with 70% cold ethanol and dyed with propidium iodide (PI; Sigma-Aldrich, Germany). The samples were analyzed with a flow cytometer (Becton Dickinson, USA), and the data were processed with DNA modeling software.
Patients and tumor specimens
Fresh paired breast cancer and normal breast tissue were obtained from 47 early-stage breast cancer patients (SYSMH cohort 1) diagnosed between 1 January 2020 and 30 December 2020. In SYSMH cohort 2, paraffin-embedded primary tumor samples were obtained from 215 ER+/HER2− positive female breast cancer patients (age 32 to 92 years, median 58 years) at the Breast Tumor Center of Sun Yat-Sen Memorial Hospital between 1 January 2012 and 30 December 2018. These patients were confirmed to have ER+ breast cancer by postoperative pathological diagnosis and received adjuvant endocrine treatment and regular follow-up (median DFS, 53.8 months). Detailed clinicopathological information is provided in tables S4 to S6. In SYSMH cohort 3, paraffin-embedded recurrent tumors were obtained from 37 advanced ER+/HER2− positive female breast cancer patients (age 36 to 72 years, median 49 years) who received CDK4/6 inhibitors and regular follow-up (median PFS, 7.5 months). All samples were collected with signed informed consent and ethics approval from Sun Yat-Sen Memorial Hospital.
Statistics and reproducibility
All in vitro experiments and statistical analyses were performed using GraphPad Prism version 8.0 or Excel software. Unless otherwise noted, results were expressed as means ± SD [****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05 and N.S. (not significant) for P > 0.05], and the P values were calculated by Student’s t test for two-group comparison and one-way analyses of variance (ANOVAs) for multiple-group comparison.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (82061148016, 82230057, 82272859, 82203087, 82071859, and 82102729), Guangdong Natural Science Foundation (2020B1515120007 and 2023A1515010127), Science and Technology Foundation of the Guangdong Province (2021B1515230001), Guangdong Innovation and Entrepreneurship Team Projects (2019BT02Y198), Science and Technology Program of Guangzhou (202201020486), and Beijing Medical Award Foundation (YXJL-2020-0941-0760).
Author contributions: Conceptualization: Z.C., Q.S., Y.L., and Q.L. Methodology: Z.C., L.J., S.L., L.L.W., J.W., X.J., and M.Z. Visualization: Z.C., Q.S., J.L., Q.W., and W.Y. Supervision: Y.L., J.Z., C.G., H.Y., Y.Y., and Q.L. Writing—original draft: Z.C., Q.S., and Y.L. Writing—review and editing: Y.Y. and Q.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 to S6
Legends for data S1 and S2
Other Supplementary Material for this manuscript includes the following:
Data S1 and S2
REFERENCES AND NOTES
- 1.D. Hanahan, R. A. Weinberg, Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 2.M. Malumbres, M. Barbacid, Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 9, 153–166 (2009). [DOI] [PubMed] [Google Scholar]
- 3.B. O'Leary, R. S. Finn, N. C. Turner, Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 13, 417–430 (2016). [DOI] [PubMed] [Google Scholar]
- 4.C. J. Sherr, D. Beach, G. I. Shapiro, Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov. 6, 353–367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.G. N. Hortobagyi, S. M. Stemmer, H. A. Burris, Y. S. Yap, G. S. Sonke, L. Hart, M. Campone, K. Petrakova, E. P. Winer, W. Janni, P. Conte, D. A. Cameron, F. Andre, C. L. Arteaga, J. P. Zarate, A. Chakravartty, T. Taran, F. Le Gac, P. Serra, J. O'Shaughnessy, Overall survival with ribociclib plus letrozole in advanced breast cancer. N. Engl. J. Med. 386, 942–950 (2022). [DOI] [PubMed] [Google Scholar]
- 6.M. P. Goetz, M. Toi, M. Campone, J. Sohn, S. Paluch-Shimon, J. Huober, I. H. Park, O. Tredan, S. C. Chen, L. Manso, O. C. Freedman, G. G. Jaliffe, T. Forrester, M. Frenzel, S. Barriga, I. C. Smith, N. Bourayou, A. Di Leo, MONARCH 3: Abemaciclib as initial therapy for advanced breast cancer. J. Clin. Oncol. 35, 3638–3646 (2017). [DOI] [PubMed] [Google Scholar]
- 7.R. S. Finn, M. Martin, H. S. Rugo, S. Jones, S. A. Im, K. Gelmon, N. Harbeck, O. N. Lipatov, J. M. Walshe, S. Moulder, E. Gauthier, D. R. Lu, S. Randolph, V. Dieras, D. J. Slamon, Palbociclib and letrozole in advanced breast cancer. N. Engl. J. Med. 375, 1925–1936 (2016). [DOI] [PubMed] [Google Scholar]
- 8.L. M. Spring, S. A. Wander, F. Andre, B. Moy, N. C. Turner, A. Bardia, Cyclin-dependent kinase 4 and 6 inhibitors for hormone receptor-positive breast cancer: Past, present, and future. Lancet 395, 817–827 (2020). [DOI] [PubMed] [Google Scholar]
- 9.N. C. Turner, Y. Liu, Z. Zhu, S. Loi, M. Colleoni, S. Loibl, A. DeMichele, N. Harbeck, F. Andre, M. A. Bayar, S. Michiels, Z. Zhang, C. Giorgetti, M. Arnedos, C. Huang Bartlett, M. Cristofanilli, Cyclin E1 expression and palbociclib efficacy in previously treated hormone receptor-positive metastatic breast cancer. J. Clin. Oncol. 37, 1169–1178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.K. Keyomarsi, S. L. Tucker, T. A. Buchholz, M. Callister, Y. Ding, G. N. Hortobagyi, I. Bedrosian, C. Knickerbocker, W. Toyofuku, M. Lowe, T. W. Herliczek, S. S. Bacus, Cyclin E and survival in patients with breast cancer. N. Engl. J. Med. 347, 1566–1575 (2002). [DOI] [PubMed] [Google Scholar]
- 11.U. S. Asghar, A. R. Barr, R. Cutts, M. Beaney, I. Babina, D. Sampath, J. Giltnane, J. A. Lacap, L. Crocker, A. Young, A. Pearson, M. T. Herrera-Abreu, C. Bakal, N. C. Turner, Single-cell dynamics determines response to CDK4/6 inhibition in triple-negative breast cancer. Clin. Cancer Res. 23, 5561–5572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. T. Herrera-Abreu, M. Palafox, U. Asghar, M. A. Rivas, R. J. Cutts, I. Garcia-Murillas, A. Pearson, M. Guzman, O. Rodriguez, J. Grueso, M. Bellet, J. Cortes, R. Elliott, S. Pancholi, J. Baselga, M. Dowsett, L. A. Martin, N. C. Turner, V. Serra, Early adaptation and acquired resistance to CDK4/6 inhibition in estrogen receptor-positive breast cancer. Cancer Res. 76, 2301–2313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.C. E. Caldon, C. M. Sergio, J. Kang, A. Muthukaruppan, M. N. Boersma, A. Stone, J. Barraclough, C. S. Lee, M. A. Black, L. D. Miller, J. M. Gee, R. I. Nicholson, R. L. Sutherland, C. G. Print, E. A. Musgrove, Cyclin E2 overexpression is associated with endocrine resistance but not insensitivity to CDK2 inhibition in human breast cancer cells. Mol. Cancer Ther. 11, 1488–1499 (2012). [DOI] [PubMed] [Google Scholar]
- 14.M. Arnedos, M. A. Bayar, B. Cheaib, V. Scott, I. Bouakka, A. Valent, J. Adam, V. Leroux-Kozal, V. Marty, A. Rapinat, C. Mazouni, B. Sarfati, I. Bieche, C. Balleyguier, D. Gentien, S. Delaloge, M. Lacroix-Triki, S. Michiels, F. Andre, Modulation of Rb phosphorylation and antiproliferative response to palbociclib: The preoperative-palbociclib (POP) randomized clinical trial. Ann. Oncol. 29, 1755–1762 (2018). [DOI] [PubMed] [Google Scholar]
- 15.S. Vijayaraghavan, C. Karakas, I. Doostan, X. Chen, T. Bui, M. Yi, A. S. Raghavendra, Y. Zhao, S. I. Bashour, N. K. Ibrahim, M. Karuturi, J. Wang, J. D. Winkler, R. K. Amaravadi, K. K. Hunt, D. Tripathy, K. Keyomarsi, CDK4/6 and autophagy inhibitors synergistically induce senescence in Rb positive cytoplasmic cyclin E negative cancers. Nat. Commun. 8, 15916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R. S. Finn, Y. Liu, Z. Zhu, M. Martin, H. S. Rugo, V. Dieras, S. A. Im, K. A. Gelmon, N. Harbeck, D. R. Lu, E. Gauthier, C. Huang Bartlett, D. J. Slamon, Biomarker analyses of response to cyclin-dependent kinase 4/6 inhibition and endocrine therapy in women with treatment-naïve metastatic breast cancer. Clin. Cancer Res. 26, 110–121 (2020). [DOI] [PubMed] [Google Scholar]
- 17.G. N. Hortobagyi, S. M. Stemmer, H. A. Burris, Y. S. Yap, G. S. Sonke, S. Paluch-Shimon, M. Campone, K. Petrakova, K. L. Blackwell, E. P. Winer, W. Janni, S. Verma, P. Conte, C. L. Arteaga, D. A. Cameron, S. Mondal, F. Su, M. Miller, M. Elmeliegy, C. Germa, J. O'Shaughnessy, Updated results from MONALEESA-2, a phase III trial of first-line ribociclib plus letrozole versus placebo plus letrozole in hormone receptor-positive, HER2-negative advanced breast cancer. Ann. Oncol. 29, 1541–1547 (2018). [DOI] [PubMed] [Google Scholar]
- 18.L. Statello, C. J. Guo, L. L. Chen, M. Huarte, Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. M. Schmitt, H. Y. Chang, Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Q. Shi, Y. Li, S. Li, L. Jin, H. Lai, Y. Wu, Z. Cai, M. Zhu, Q. Li, Y. Li, J. Wang, Y. Liu, Z. Wu, E. Song, Q. Liu, LncRNA DILA1 inhibits Cyclin D1 degradation and contributes to tamoxifen resistance in breast cancer. Nat. Commun. 11, 5513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Z. Cai, J. Wang, Y. Li, Q. Shi, L. Jin, S. Li, M. Zhu, Q. Wang, L. L. Wong, W. Yang, H. Lai, C. Gong, Y. Yao, Y. Liu, J. Zhang, H. Yao, Q. Liu, Overexpressed cyclin D1 and CDK4 proteins are responsible for the resistance to CDK4/6 inhibitor in breast cancer that can be reversed by PI3K/mTOR inhibitors. Sci. China Life Sci. 66, 94–109 (2023). [DOI] [PubMed] [Google Scholar]
- 22.P. Jezequel, M. Campone, W. Gouraud, C. Guerin-Charbonnel, C. Leux, G. Ricolleau, L. Campion, bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res. Treat. 131, 765–775 (2012). [DOI] [PubMed] [Google Scholar]
- 23.E. S. Knudsen, K. E. Knudsen, Tailoring to RB: Tumour suppressor status and therapeutic response. Nat. Rev. Cancer 8, 714–724 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.J. Gerdes, Ki-67 and other proliferation markers useful for immunohistological diagnostic and prognostic evaluations in human malignancies. Semin. Cancer Biol. 1, 199–206 (1990). [PubMed] [Google Scholar]
- 25.J. K. Kulski, Long noncoding RNA HCP5, a hybrid HLA class I endogenous retroviral gene: Structure, expression, and disease associations. Cell 8, 480 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.C. Lin, L. Yang, Long noncoding RNA in cancer: Wiring signaling circuitry. Trends Cell Biol. 28, 287–301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.H. C. Hwang, B. E. Clurman, Cyclin E in normal and neoplastic cell cycles. Oncogene 24, 2776–2786 (2005). [DOI] [PubMed] [Google Scholar]
- 28.D. M. Koepp, L. K. Schaefer, X. Ye, K. Keyomarsi, C. Chu, J. W. Harper, S. J. Elledge, Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294, 173–177 (2001). [DOI] [PubMed] [Google Scholar]
- 29.A. Hurtado, K. A. Holmes, C. S. Ross-Innes, D. Schmidt, J. S. Carroll, FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43, 27–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.A. N. Holding, A. E. Cullen, F. Markowetz, Genome-wide estrogen receptor-α activation is sustained, not cyclical. eLife 7, e40854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The ENCODE Project Consortium , An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. E. Phillips, V. G. Corces, CTCF: Master weaver of the genome. Cell 137, 1194–1211 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R. Pena-Hernandez, M. Marques, K. Hilmi, T. Zhao, A. Saad, M. A. Alaoui-Jamali, S. V. del Rincon, T. Ashworth, A. L. Roy, B. M. Emerson, M. Witcher, Genome-wide targeting of the epigenetic regulatory protein CTCF to gene promoters by the transcription factor TFII-I. Proc. Natl. Acad. Sci. U.S.A. 112, E677–E686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A. Adamo, S. Atashpaz, P. L. Germain, M. Zanella, G. D'Agostino, V. Albertin, J. Chenoweth, L. Micale, C. Fusco, C. Unger, B. Augello, O. Palumbo, B. Hamilton, M. Carella, E. Donti, G. Pruneri, A. Selicorni, E. Biamino, P. Prontera, R. McKay, G. Merla, G. Testa, 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat. Genet. 47, 132–141 (2015). [DOI] [PubMed] [Google Scholar]
- 35.J. Chou, D. A. Quigley, T. M. Robinson, F. Y. Feng, A. Ashworth, Transcription-associated cyclin-dependent kinases as targets and biomarkers for cancer therapy. Cancer Discov. 10, 351–370 (2020). [DOI] [PubMed] [Google Scholar]
- 36.J. Soutourina, Transcription regulation by the mediator complex. Nat. Rev. Mol. Cell Biol. 19, 262–274 (2018). [DOI] [PubMed] [Google Scholar]
- 37.C. Chu, Y. Geng, Y. Zhou, P. Sicinski, Cyclin E in normal physiology and disease states. Trends Cell Biol. 31, 732–746 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M. Scaltriti, P. J. Eichhorn, J. Cortes, L. Prudkin, C. Aura, J. Jimenez, S. Chandarlapaty, V. Serra, A. Prat, Y. H. Ibrahim, M. Guzman, M. Gili, O. Rodriguez, S. Rodriguez, J. Perez, S. R. Green, S. Mai, N. Rosen, C. Hudis, J. Baselga, Cyclin E amplification/overexpression is a mechanism of trastuzumab resistance in HER2+ breast cancer patients. Proc. Natl. Acad. Sci. U.S.A. 108, 3761–3766 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Y. Matsumoto, J. L. Maller, A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science 306, 885–888 (2004). [DOI] [PubMed] [Google Scholar]
- 40.U. Asghar, A. K. Witkiewicz, N. C. Turner, E. S. Knudsen, The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 14, 130–146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.S. T. Crooke, B. F. Baker, R. M. Crooke, X. H. Liang, Antisense technology: An overview and prospectus. Nat. Rev. Drug Discov. 20, 427–453 (2021). [DOI] [PubMed] [Google Scholar]
- 42.M. F. Yuen, J. Heo, J. W. Jang, J. H. Yoon, Y. O. Kweon, S. J. Park, Y. Tami, S. You, P. Yates, Y. Tao, J. Cremer, F. Campbell, R. Elston, D. Theodore, M. Paff, C. F. Bennett, T. J. Kwoh, Safety, tolerability and antiviral activity of the antisense oligonucleotide bepirovirsen in patients with chronic hepatitis B: A phase 2 randomized controlled trial. Nat. Med. 27, 1725–1734 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.T. Miller, M. Cudkowicz, P. J. Shaw, P. M. Andersen, N. Atassi, R. C. Bucelli, A. Genge, J. Glass, S. Ladha, A. L. Ludolph, N. J. Maragakis, C. J. McDermott, A. Pestronk, J. Ravits, F. Salachas, R. Trudell, P. Van Damme, L. Zinman, C. F. Bennett, R. Lane, A. Sandrock, H. Runz, D. Graham, H. Houshyar, A. McCampbell, I. Nestorov, I. Chang, M. McNeill, L. Fanning, S. Fradette, T. A. Ferguson, Phase 1-2 trial of antisense oligonucleotide tofersen for SOD1 ALS. N. Engl. J. Med. 383, 109–119 (2020). [DOI] [PubMed] [Google Scholar]
- 44.M. J. Graham, R. G. Lee, T. A. Brandt, L. J. Tai, W. Fu, R. Peralta, R. Yu, E. Hurh, E. Paz, B. W. McEvoy, B. F. Baker, N. C. Pham, A. Digenio, S. G. Hughes, R. S. Geary, J. L. Witztum, R. M. Crooke, S. Tsimikas, Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. N. Engl. J. Med. 377, 222–232 (2017). [DOI] [PubMed] [Google Scholar]
- 45.I. Tamm, B. Dorken, G. Hartmann, Antisense therapy in oncology: New hope for an old idea? Lancet 358, 489–497 (2001). [DOI] [PubMed] [Google Scholar]
- 46.M. Anurag, S. Haricharan, M. J. Ellis, CDK4/6 inhibitor biomarker research: Are we barking up the wrong tree? Clin. Cancer Res. 26, 3–5 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S6
Legends for data S1 and S2
Data S1 and S2