Abstract
Students often find neuroanatomy a daunting exercise of rote memorization in a dead language. This workshop was designed to enliven the teaching of neuroanatomy. We recast the topic by extending it to the cellular and sub-cellular levels, animating it by learning to build a brain, and infusing the topic with the lively arts. Due to COVID’s interference with the usual schedule of Society for Neuroscience (SfN) events, the 2021 Professional Development Workshop on Teaching was held as a webinar on April 12, 2022 with a follow-up question and answer session on June 7. In this workshop, not only were innovative teaching methods presented, but also the very definition of neuroanatomy was pushed to the limits—even reaching into the molecular and subcellular level. The presenters provided means of engaging students that were no cost, low cost, or well within the reach of most academic institutions. Judging by the attendance, this webinar was quite successful in its goals. Our speakers presented exciting and varied approaches to teaching neuroanatomy. Kaitlyn Casimo presented how the vast resources of the Allen Institute could be employed. Marc Nahmani described how open data resources could be utilized in creating a Course-Based Undergraduate Research Experience (CURE) on neural microanatomy. Erika Fanselow presented novel ways to overcome one of students’ big hurdles in grasping neuroanatomy: understanding 3-D relationships. Len White described a creative approach in teaching neuroanatomy by incorporating the humanities, particularly art and literature. This article presents synopses of the presentations, which are written by the four presenters. Additionally, prompted by questions from the viewers, we have constructed a table of our favorite resources.
A video of the original presentations as well as links to the subsequent Q & A sessions is available at https://neuronline.sfn.org/training/teaching-neuroscience-reviving-neuroanatomy/.
Keywords: Allen Brain Map, open education resources, open data, course-based research experiences, 3D modeling, art-making
Neuroanatomy oftentimes is seen to be as dead as its specimens. The stereotype of neuroanatomy courses is an arcane exercise taught by the likes of white-haired old men with indecipherable Latin words dribbling from their lips. Thus, undergraduates often see studying neuroanatomy as a necessary evil. Yet it is a subject that is central and even essential to the more complete understanding of the nervous system and behavior.
Indeed, neuroanatomy is not as dead as some would think, but rather it is an alive and vibrant aspect of neuroscience, particularly in the pedagogy of the topic. In this workshop, novel teaching methods were described, and the definition of neuroanatomy was expanded to extend to the microscopic, subcellular, and even molecular domains. Newer technologies and approaches provide for not only exciting means of teaching neuroanatomy but also the capability to do genuine research, allowing students to gain novel insights into this time-honored pursuit.
In the current article, Kaitlyn Casimo describes the many facets of Open Educational Resources using the Allen Brain Map. Many of these resources can be interwoven to teach neuroanatomy at many scales of understanding—including anatomical genetic expression and tract tracing as well as others. Further, these resources provide instructors and students a range of experiences from in-class curricula to course-based and individual research.
Marc Nahmani extended Kaitlyn Casimo’s overview into an examination of the subcellular domain using publicly available open databases, including data produced by the Allen Institute. More specifically, he provides his students with a CURE (Course-Based Research Experience; Corwin et al., 2015) based on open access websites that provide electron microscopy photomicrographs or other images at more macro scales. As he points out, these image libraries have removed the biggest barrier of undergraduate research, which is the tissue preparation and image acquisition. Secondly, barriers to research such as equipment and budgetary constraints are virtually removed. Thus genuine, inquiry-based research experiences are within easy grasp of both instructors and students. Further, he also describes where instructors may obtain the necessary expertise to use these resources.
Erika Fanselow moved the discussion to the more macro level and tackled one of the great challenges/barriers to students understanding neuroanatomy: creating a 3-D representation of the brain. Erika’s insight was to have students make a brain in order to learn a brain. Students also get the benefit of learning 3D modeling software, an experience somewhat rare in undergraduate curricula. Although the start-up costs are higher than other approaches described here, 3D printers are becoming common as colleges and universities create maker spaces. Regardless, the approach seems so much stronger than the 2D exploded diagrams that are offered by standard texts.
As we pick up STEAM in this century, Len White did a great job of incorporating the arts and other humanities into the study of neuroanatomy, thus enlivening the subject with both art and literature and providing a new perspective to this traditionally STEM topic. One of the sources of literature that Len recommends is Mary Shelly’s Frankenstein. Lore has it that Shelly was inspired to write a story of re-animation via electricity because she had read the work of Galvani on frog legs (Neuroscientifically Challenged, 2022). So, we come full circle; neuroscience inspired literature, which now inspires neuroscience.
TEACHING NEUROANATOMY WITH OPEN DATA FROM THE ALLEN BRAIN MAP
Presented by Kaitlyn Casimo
There are many potential sources for data, specimens, and other resources for teaching neuroscience. We summarize many of our favorite resources in Table 1. In this section, I (KC) will highlight the use of open data and associated tools from the Allen Institute for teaching neuroanatomy.
Table 1.
Resources we love.
| Resource name | URL | Created by | Description |
|---|---|---|---|
| 3D brain structure app | https://dnalc.cshl.edu/resources/products/3d-brain-app.html | Cold Spring Harbor Laboratory | 3D models of brain structures that can be displayed and rotated, with short descriptions (free) |
| Allen Institute education outreach | https://alleninstitute.org/learn | Allen Institute | Catalog of education resources from the Allen Institute, by institute staff and curated outside resources, which apply open data and tools as teaching resources (free) |
| Functional neuroanatomy online resource | https://www.neuroanatomy.ca/index.html | University of British Columbia | Comprehensive neuroanatomy online resource including learning modules, images of sliced brains, 3D models, videos and more (free) |
| MICrONS Explorer | https://www.microns-explorer.org/ | Princeton University (Seung Lab); Allen Institute for Brain Science; Baylor College of Medicine (Tolias Lab) | Large annotated open data image volumes from mouse visual cortex linked with correlated functional data (free) |
| Myscope microscopy training | https://myscope.training/index.html | Microscopy Australia | Interactive learning & training modules for numerous forms of microscopy (free) |
| NeuPrint | https://neuprint.janelia.org/ | Janelia Research Campus | Open-access imaging data from multiple imaging modalities (free) |
| Neurodata | https://neurodata.io/project/ocp/ | Open Connectome Project | Open-access repository for downloading & uploading 3D image volumes from various imaging modalities (free) |
| Neuronify | https://ovilab.net/neuronify/ | Dragly et al., 2017 | User friendly interactive plug-and-play software for simulating and modulating neuronal electrophysiology (free) |
| Neuroscience Information Framework | https://neuinfo.org/ | Neuroscience Information Framework | Large collection of open data neuroscience resources (free) |
| Neuroscience Online | https://nba.uth.tmc.edu/neuroscience/m/index.htm | Department of Neurobiology and Anatomy, McGovern Medical School at UTHealth | Open-access educational resource electronic textbook for the neurosciences (free) |
| Open Neuroscience Initiative textbook | https://www.austinlim.com/open-neuroscience-initiative | Austin Lim, Ph.D., DePaul University | Open-source comprehensive introductory neuroscience textbook (free) |
| Sylvius 4 Online | https://learninglink.oup.com/access/sylvius | S. Mark Williams and Leonard E. White, Duke University | An interactive atlases and visual glossary of human neuroanatomy (purchase through Oxford University Press) |
| Teaching with Open Neuroscience Datasets | https://sites.google.com/ucsd.edu/neuroedu/ | Ashley Juavinett, UCSD | Resources for teaching with Allen Institute datasets with a particular focus on coding (free) |
| The Human Brain | http://www.anatomie-amsterdam.nl/sub_sites/anatomie-zenuwwerking/123_neuro/start.htm | Floris Wouterlood, Department of Anatomy and Neurosciences, Amsterdam UMC | Detailed photographs of brain dissections, MRI images, histological sections (free) |
| Tinkercad® | https://www.tinkercad.com/ | Autodesk® | Cloud-based 3D modeling software (free) |
The Allen Institute is a biological sciences nonprofit with research focus areas in neuroscience, cell biology, and immunology. The institute practices open science, releasing data, analysis tools, and other resources publicly for others to use in their work, including for educational applications (Casimo, 2022). As of spring 2022, the institute has collected approximately 22 petabytes of data across dozens of discrete datasets. Many of these datasets are suitable for teaching neuroanatomy specifically, including some discussed in a previous workshop to the one summarized in this article, which focused on teaching neuroscience with big data (Grisham et al., 2017).
The Allen Institute education program, which I developed and lead, focuses on developing open educational resources (OERs) and supporting educators to develop their own resources, employing these open scientific resources in a classroom setting. Open data can be used to support a range of activities at various levels of intensity and engagement, from simple exploration, to a single lab module, to an extended course-based undergraduate research experience CURE, and beyond the classroom to independent research. The Allen Institute offers and curates some teaching resources that use the open datasets for teaching neuroanatomy and other topics.
Benefits of Teaching with Open Data
Because the students use open data that has been previously collected, fully virtual, hybrid, or fully in-person classroom formats are all feasible. Open data may be combined with student-collected data, students can transition from collecting a small amount of data themselves to honing their analysis skills on a much larger open dataset, or students can exclusively use the open data. In this last scenario, open data can also serve to expand access to high-impact opportunities at under-resourced institutions. Open datasets from the Allen Institute include techniques and tissues not accessible in undergraduate teaching settings even at the best-resourced institutions, such as human tissue samples, data from awake and behaving mice, and electron microscopy (use of EM data described by author MN below).
Importantly, the datasets have been extensively quality controlled before their release, but due to their big and comprehensive nature, have not been exhaustively analyzed. Consequently, students have the opportunity to generate novel findings while concentrating on developing their data analysis skills using high-quality data.
The use of open data in a classroom setting also creates opportunities for students to analyze and interpret data they would not otherwise have access to. Big datasets call for students to use more sophisticated statistics and informatics techniques. They may be best analyzed programmatically, which creates opportunities for students to learn how to code within a neuroscience context (Juavinett, 2020). Allen Institute open datasets come with an API (application programming interface; a programming toolkit to facilitate access and basic functions), and many have starter code examples that students can work through to learn how to analyze data programmatically without having to start from scratch.
Data for Teaching Neuroanatomy
Among the Allen Institute’s open datasets, the following resources are particularly useful for teaching neuroanatomy.
Allen Mouse Brain Atlas and other gene expression atlases
This atlas maps whole-brain, whole-genome expression across the adult mouse brain. Each gene’s expression is individually visualized using in situ hybridization (ISH), showing the relative position and amount of gene expression in its anatomical context. By toggling between the ISH image showing gene expression and neighboring Nissl-stained images using the online data viewer, students gain a visual intuition for the spatial relationships between structures and the interpretation of image-based data (Figure 1). The gene expression data is also accompanied by a detailed anatomical reference atlas, which includes annotated anatomy and an ontology of hundreds of individual regions and sub-regions (Figure 1). In the context of teaching neuroanatomy, the atlas can be used for simple exploration of anatomy, to examine specific genes’ expression across regions, or to underscore the link between structures, gene expression, and function.
Figure 1.
Allen Mouse Brain Atlas. Left: Serial ISH slices give anatomical context to gene expression; quantification of relative fluorescence is shown below in an interactive bar graph. Right: Detail viewer enables side-by-side exploration of annotated anatomy and ISH images.
Additional gene expression atlases cover the developing mouse brain, adult and developing human brain, and adult and developing nonhuman primate brain. The human atlases primarily use microarray rather than ISH, which has similar interpretation as relative gene expression level, but instead is visualized through heatmap plots or projected onto a cartoon anatomy. Because of the inaccessibility of human tissue in teaching labs, the adult Allen Human Brain Atlas provides a unique, comprehensive perspective that is otherwise not accessible to students.
Allen Mouse Brain Connectivity Database
This dataset uses a series of thousands of individual injections of anterograde tracers to visualize projections from sites across the brain and retina. Each individual experiment began with the injection of a fluorescent tracer into a particular brain region, showing the projection patterns across the whole brain from that region. Students who already have a grounding in the various regions of the brain can use this dataset to explore long-range connectivity patterns and the anatomical relationships between regions. This dataset, as well as the gene expression Allen Mouse Brain Atlas, is featured in the Allen Institute’s lesson Exploring Pathways in the Brain.
Allen Cell Types Database
This multimodal collection of datasets aims to define and characterize the cell types of the brain, focusing on human and mouse. In the context of teaching neuroanatomy, the single-cell morphology data contained in this collection demonstrate cutting-edge techniques in cellular anatomy. The datasets include stained images of single neurons, 3D digital reconstructions, and quantification of the morphology. These are linked to traditional morphology-based cell types, as well as updated multimodal cell type definitions that also incorporate transcriptomic and electrophysiological data. The morphology data and its interpretation are featured in the Allen Institute’s lesson Neurons: Beyond the Textbook.
MICrONS Electron Microscopy Datasets
These datasets include serial electron microscopy images of the mouse brain (a segment of layer 2/3 of visual cortex and a full cubic millimeter of cortex). In addition to the raw data, the resource also includes 3D digital reconstructions, interactive analysis and visualization tools, segmentation and labeling of individual cells, and functional data. In the next section, Marc Nahmani describes his use of this data resource for a course-based research experience.
In conclusion, open datasets from the Allen Brain Map can be used to support teaching neuroanatomy and provide valuable access to data types, tissues, and data quantities not otherwise accessible in undergraduate neuroscience labs. These highlighted data resources are those most useful for teaching neuroanatomy in particular; however, the Allen Brain Map includes many other datasets and analysis tools. Additional datasets are also added frequently.
USING FREELY AVAILABLE IMAGE VOLUME DATA FOR NEUROANATOMY COURSE-BASED RESEARCH EXPERIENCES
Presented by Marc Nahmani
Advances in microscopy and computing power have dramatically increased the complexity of current imaging datasets (Lichtman et al., 2014; Jorgenson et al., 2015). Consequently, modern microscope image volumes contain myriad structures and/or comparisons that are ripe for exploration. Increasingly, these image volumes are made freely available in data repositories, representing a virtual gold mine of open data resources for undergraduate-driven research projects. Indeed, freely available open data image volumes can be used to bypass the time-consuming data acquisition step within neuroanatomy-based CUREs, affording students time to focus on understanding the rationale for their experiments and how they fit into larger contexts (Nahmani, 2019). Here, I briefly relay some key points in the design and implementation of neuroanatomy-based CUREs using freely available open data image volumes.
Design and Benefits
Designing a semester or quarter-length CURE in which students gain data acquisition, data analysis, and critical thinking skills is challenging. Instructors must weigh demands for resources, space, cost, and time when attempting to link a CURE’s learning objectives with a set of desired learning outcomes. Yet, if we forgo the time-consuming data acquisition steps, CUREs can be primarily focused on strengthening our students’ analytical and critical thinking skills. That is, these CUREs are aimed at helping students build the essential cognitive tools they will need to tackle scientific questions on their own.
Using open data to combine a CURE’s acquisition and analysis steps has numerous benefits, including: (1) all the data are already digitized and analyses can be done remotely, (2) the only necessary resources are a basic laptop and freely downloadable software and image volume(s), (3) it focuses the learning objectives and outcomes on strengthening critical analysis skills, and (4) it affords undergraduates the opportunity to transform their analyses into novel projects and publications (Figure 2). Furthermore, these CUREs are inherently flexible, course size and length can be scaled up or down based on the academic unit constraints and the depth of the research question(s). At UW Tacoma, we typically run our open data CUREs as 7–15 student 3-credit courses. Our courses have one instructor who meets with students once or twice a week, for a total of six contact hours per week. We have successfully run these courses in-person in a basic computer lab, and in a fully remote context with students using their own computers. The main constraint on the size of open data CUREs is the necessity to iteratively train students to locate and analyze the target structure(s) within the image volume. Thus, a single instructor may want to cap their open data CURE at ~15 students, while academic units with teaching assistants may be able to scale up to larger class sizes.
Figure 2.
Typical steps in a student-driven open data CURE. (A) Python command line scripts are used to download portions of large image volumes. (B) Students learn how to recognize, analyze, and quantify neuronal features within their image volume. This single image from an open data focused ion beam scanning electron microscopy image volume of CA1 hippocampus (https://www.epfl.ch/labs/cvlab/data/dataem/) shows the traces made by a student around portions of two dendrites and their spines (yellow and cyan outlines). (C) Student 3D reconstructions of the dendrites shown in B, as well as an axon and a presynaptic bouton (both purple) that form synapses with the postsynaptic spines of these dendrites (gray). Students in this open data CURE analyzed this image volume used Fiji (Schindelin et al., 2012) and Reconstruct (Fiala, 2005) open-source software to determine the prevalence and sizes of synaptic spinules (green) within presynaptic boutons in CA1 hippocampus. Synaptic spinules are protrusions from one neuron into the pre or postsynaptic compartment of another neuron. Note the green synaptic spinules seen protruding from postsynaptic spines into the presynaptic bouton of the axon above (top red arrow), and into the bouton below (bottom red arrow). Postsynaptic densities delineating the size of synaptic connections are shown in red. Scale bar = 1 μm for B; Scale cube = 0.5 μm per side for C.
Sample Scientific Questions
While the large-scale features within most open data volumes have been analyzed and annotated, a host of subcellular structures and cell/synapse-specific comparisons often remain unexplored and unpublished. For example, within large electron microscope (EM) image volumes, subcellular features that have lower contrast, such as smooth endoplasmic reticula, small subcellular protrusions, synaptic vesicles, and subcellular membrane-bound compartments (e.g., lysosomes, endosomes), are often left unanalyzed by automated reconstruction algorithms. As such, students can form hypotheses related to innumerable questions – such as the number of docked vesicles within subsets of synapses, or the spatial relationship of endoplasmic reticula within inhibitory versus excitatory boutons relative to their synaptic vesicle pool size. Moreover, given the scope and scale of many of these image volumes, some of the comparisons of the annotated neuronal features (e.g., spatial relationship of inhibitory to excitatory boutons) may remain unanalyzed and/or unpublished and thus be amenable for a student project. Importantly, given the direct relationship between the size of boutons, dendritic spines, and synapses with their physiological strength (Murthy et al., 2001; Ostroff et al., 2002; Branco et al., 2010; Holderith et al., 2012; Araya et al., 2014; Meyer et al., 2014; Quinn et al., 2019; Holler et al., 2021), students can often make interesting interpretations on neuronal function based on their in-depth structural analyses.
Acquiring Open Data Image Volumes
There is a wealth of open data image repositories with freely available volumes spanning numerous imaging modalities (e.g., bright field, confocal, EM, cryo tomography, fMRI, etc.). We have provided links to some of these repositories in Table 1. Many of these repositories use the neuroglancer online software tool ( https://github.com/google/neuroglancer) to visualize the image volume. Using the neuroglancer tool, the instructor can peruse the image volume to locate a suitable region for analysis, noting the coordinates of the area and the number of images required. Larger image volumes are often housed on cloud servers and use a Python-based command line interface to download entire, or portions of, open data image volumes. To perform these downloads: (1) download the latest version of Python: www.python.org (some operating systems have Python 3 preinstalled), (2) create a virtual environment: https://docs.python.org/3/library/venv.html, (3) install the Python client called cloudvolume, to read and write volumes from neuroglancer ( https://github.com/seung-lab/cloud-volume), and (4) follow the instructions on the repository’s page for downloading from their cloud servers, (e.g., https://neurodata.io/help/download for the open connectome project repository). Once you have created a virtual environment, direct downloads of portions of large image volumes are as simple as a cut and paste operation. If you are new to coding, contact a knowledgeable colleague. A few minutes of assistance is likely all you will need.
What if I don’t know how to analyze EM images?
A concern instructors may have in developing a CURE using open data image volumes is the lack of experience analyzing EM images and/or images from other microscopy modalities. There are numerous image volume modalities available for free download (Table 1), and while the types of questions one can ask will differ based on the imaging modality, the same benefits discussed above apply. Thus, each instructor can choose the imaging modality that matches their expertise. There are, however, also numerous training resources for instructor and student alike to understand how EM images were acquired (e.g., https://myscope.training/index.html), and to learn how to recognize and analyze distinct neurites and subcellular structures within EM images (e.g., McCoy and Nahmani, 2021; https://synapseweb.clm.utexas.edu/atlas).
CREATING 3D DIGITAL MODELS OF NERVOUS SYSTEM STRUCTURES: A HANDS-ON UNDERGRADUATE NEUROANATOMY COURSE PROJECT
Presented by Erika E. Fanselow
One of the most challenging skills to master when learning neuroanatomy is to extrapolate from 2D images used in many instructional settings, as well as many brain imaging modalities, to the 3D shapes of nervous system structures and their spatial relationships. For example, what shapes are the ventricles and which structures surround them? Where do axons of sensory and motor neuron pathways travel and decussate? What do the dural reflections and venous sinuses look like and where are they located?
Viewing real brains from humans, sheep, and other species, as well as commercially available 3D models, can be useful when learning the shapes of structures and their spatial relationships. Brains, however, can be expensive and/or difficult to obtain, and commercially available models are also expensive, cannot be modified, and often are not sufficiently detailed for in-depth neuroanatomy courses. To enhance learning the 3D shapes of nervous system structures, I developed a project for an undergraduate neuroanatomy course in which students construct digital 3D models of nervous system components. This project has been implemented for over 5 years in a small-enrollment honors neuroanatomy practicum course in the Department of Neuroscience at the University of Pittsburgh.
Implementation of Digital 3D Modeling Techniques in the neuroanatomy classroom
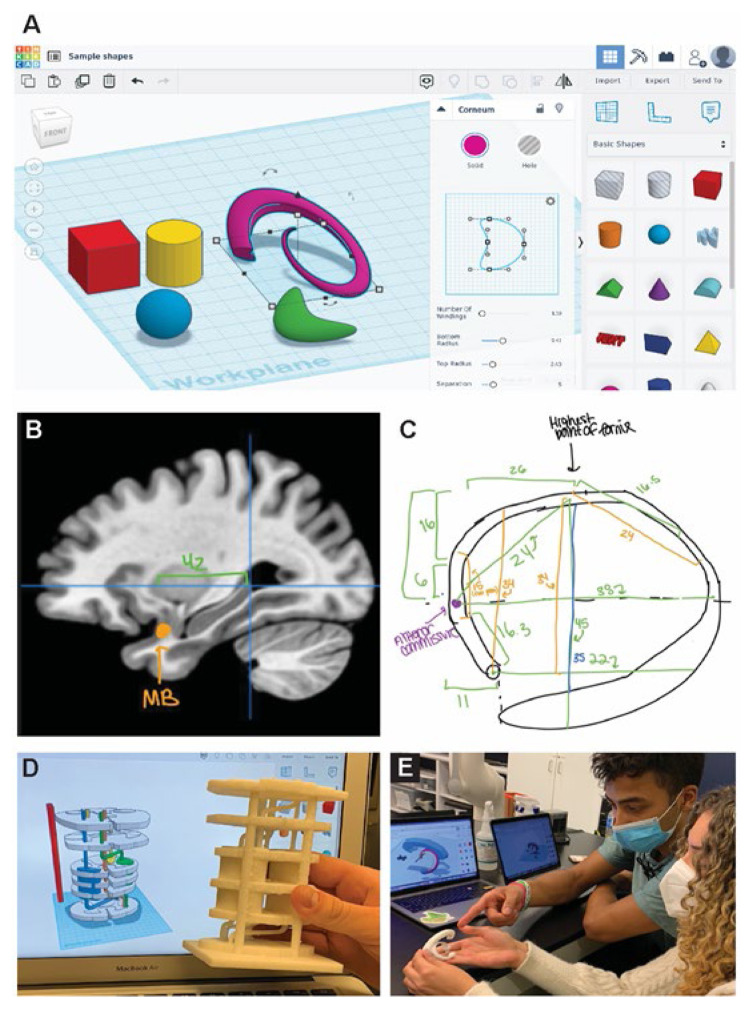
This project spans most of a semester, interleaved with other instruction and projects. The first step is to help students become familiar with using 3D modeling software (e.g., Tinkercad® software shown in Figure 3A), which is done using in-class demonstrations and short out-of-class tutorials. Next, groups of 3–4 students make measurements and sketches (Figure 3B) of the structure or structures they chose to model, based on 2D resources such as brain atlases, magnetic resonance images (Figure 3C), diffusion tensor images, and other information from published scientific resources. Students then manipulate and combine shapes using 3D modeling software to create a prototype model, which is 3D printed and assessed to further refine the model (Figure 3D, E). At the end of the term, the student groups present their model to the class and describe how they would utilize it in a lecture-based course as an improvement over learning neuroanatomy based mainly on 2D images.
Figure 3.
(A) Screenshot showing Tinkercad® online 3D modeling software used in this project (Autodesk screenshot reprinted courtesy of Autodesk, Inc.). (B, C) Students made sketches of the structures they will model, using 2D resources such as MRI (e.g., the MNI space structural MR images from neurovault.org shown in C). (B, C courtesy of Mackenzie Bultman.) (D, E) After manipulating and joining shapes in Tinkercad to form digital models based on their measurements, students’ model prototypes are 3D printed and used to refine the models.
Resources and Instructor Skills Involved in Teaching this Project
To implement this project in an undergraduate course, the instructor should have basic knowledge of 3D printing technologies, including an understanding of the capabilities and limitations of 3D printing techniques and materials. Though 3D printing technology continues to innovate rapidly, it’s not (yet?) possible or practical to simply design and print anything one can imagine. Thus, it is helpful to have a working understanding of how various types of 3D printers function and the properties of available printing materials. This allows the instructor to discuss practical constraints on their students’ models and help them design printable structures.
Required resources include 3D digital modeling software (see below), 3D printer(s), and printing supplies (e.g., filament, resin). At the time of writing, the costs of adequate printers and materials are substantially less prohibitive than when this project was initially developed, making it reasonable to have a dedicated printer for the course/project. Additionally, it is increasingly common for universities to have 3D printing resources available on campus, such as in machine shops and maker spaces.
Digital 3D Modeling Software Used to Date
There are multiple types of 3D modeling software available, but there are some trade-offs to be made when choosing which of these to use in an undergraduate neuroanatomy course. The main trade-off is between the complexity of modeling functions vs. the time and effort to learn the software; many programs have highly sophisticated molding functions, but at the cost of being difficult to learn to use. Another practical concern is software availability/cost. To date, this project has mainly utilized the free, cloud-based, 3D modeling software Tinkercad® (Figure 3A) by Autodesk® (see link in Table 1), in part because it is intuitive to use and includes pre-made, modifiable “organic” shapes, in addition to standard CAD shapes. Most students who have participated in this project had no prior digital 3D modeling experience but nonetheless learned how to use the software fairly rapidly. A downside of Tinkercad for use in this project is that its functionality is less complex than some other available programs. Occasionally, students have used other modeling programs to smooth shapes or form them in ways that cannot be accomplished in Tinkercad, but these programs have not been explicitly taught as part of the project to date.
Impacts of this Project on Student Learning
The main benefit of this project is in teaching students to relate images of the brain, such as MRI and tissue viewed in brain atlases, to the actual 3D structures of nervous system components. In addition, each student group learns about a nervous system structure or set of structures in substantial detail. Another benefit is the high degree of student engagement in and out of class. Most students who have participated were excited to work on this project and took great pride in their final product. Finally, students also acquire a rudimentary understanding of 3D digital modeling software and printing capabilities.
Adaptations of this Project that Don’t Require 3D Printing
Once a digital model has been made (Figure 4A), it can be utilized in multiple ways. Though holding a detailed, printed model in one’s hands (Figure 4B) is very helpful for prototyping, refining the model, and/or learning about the structures involved, it is not strictly necessary to print the 3D structures students design. For example, digital models can be visualized using augmented reality (Figure 4C) and virtual reality settings, modalities that students could use to interact with the models during classroom and/or online instruction. In addition, the essence of this project is the translation from 2D images into 3D structures, which also could be accomplished using household materials such as clay, pipe cleaners, drinking straws, and similar. A potential drawback to this approach is a reduction in the accuracy of the shapes and relative sizes of structures, but the overall shapes and relationships between structures could nonetheless be learned.
Figure 4.
(A) Completed digital 3D model viewed in a Tinkercad workspace. (B) The same model printed using stereolithography (“resin”) 3D printing technology and then painted. (C) Two versions of the digital model, with and without the brain slab, projected onto a desk using augmented reality software on a smartphone.
Use of the Constructed 3D Models in Large Lecture-Based Courses
Since the models generated in this project were designed with classroom instruction in mind, some will be printed and tested as learning tools in a large, lecture-based neuroanatomy course during upcoming semesters. In particular, some models were created for use in active learning activities and their utility in enhancing instruction will be assessed. Additionally, some of the models, with accompanying printing instructions and suggested lesson plans, will be made available as open education resources, licensed for free use and modification under a Creative Commons license.
INFUSING HUMANITIES INTO THE TEACHING AND LEARNING OF HUMAN NEUROANATOMY
Presented by Leonard E. White
Students studying neuroscience increasingly learn at an accelerated pace that emphasizes knowledge acquisition over reflection and introspection. This is too often the case at most of our institutions when it comes to taking seriously the anatomy of the human brain or the brain of any other species–even the so-called simpler brains of invertebrates. Indeed, the wealth of anatomical detail discovered and described since the first systematic studies of human neuroanatomy began in the 16th century is simply staggering. With the constant introduction of powerful new neuroanatomical techniques, the advent of anatomical and functional connectomics, and the U.S. federal BRAIN initiative (Bargmann et al., 2014), details are arriving at an increasing rate—and there seems to be no slowing to the expansion of knowledge of the mammalian brain. Obviously, instructors must carefully focus the neuroanatomical content of their courses on the learning outcomes they aim to achieve for their students. There are, however, other approaches to helping students consume and digest the relevant neuroanatomical knowledge that would support their learning. One such productive approach to tempering the “fast learning” (firehose or fast-food metaphors) that seems to attend most courses or modules on neuroanatomy is to enrich the experience by infusion of the “slow learning” (or slow-cooking metaphor) inherent in the humanities. I (LEW) have been attempting to encourage such slow learning with reflection and introspection through two means: (1) integrations of selected passages and readings from classic and contemporary literature into lessons on neuroanatomy; and (2) creation of co-curricular opportunity for art-making while examining human brain specimens at the gross and microanatomical levels.
Two great works of literature that have numerous passages worthy of interpreting through neuroanatomical and neurobiological lenses are Mary Shelley’s 1818 novel, Frankenstein; or, The Modern Prometheus; and the other is Herman Melville’s 1851 novel, Moby-Dick; or, The Whale (both works are now in the public domain). For example, one of the very best descriptions of the challenges of binocular vision that this educator (LEW) has encountered (in neuroscientific literature or otherwise) is found in Melville’s novel, Chapter 74. The Sperm Whale’s Head—Contrasted View. Consider this excerpt and how it might be situated in a neuroanatomical lesson on visual fields, visual neuroanatomy or visual psychophysics:
So long as a man’s eyes are open in the light, the act of seeing is involuntary; that is, he cannot then help mechanically seeing whatever objects are before him. Nevertheless, any one’s experience will teach him, that though he can take in an undiscriminating sweep of things at one glance, it is quite impossible for him, attentively, and completely, to examine any two things—however large or however small—at one and the same instant of time; never mind if they lie side by side and touch each other. But if you now come to separate these two objects, and surround each by a circle of profound darkness; then, in order to see one of them, in such a manner as to bring your mind to bear on it, the other will be utterly excluded from your contemporary consciousness. How is it, then, with the whale? True, both his eyes, in themselves, must simultaneously act but is his brain so much more comprehensive, combining, and subtle than man’s, that he can at the same moment of time attentively examine two distinct prospects, one on one side of him, and the other in an exactly opposite direction? If he can, then is it as marvellous a thing in him, as if a man were able simultaneously to go through the demonstrations of two distinct problems in Euclid. Nor, strictly investigated, is there any incongruity in this comparison.
Such a passage has led to rich conversations infused with curiosity and wonder in and out of classroom setting that illuminated both the neuroanatomical and neurophysiological consequences (e.g., stereopsis; binocular rivalry) associated with the placement of the eyes in the cranium, whether the consideration is specific to humans, sperm whales, or any other vertebrate.
When approaching topics pertaining to neurodiversity and the unique architecture of cerebral networks in people (or the variability and heritability of neural architecture in non-human animal models; see Wang et al., 2020), students have been enriched by reading contemporary creative works by non-speaking autists, including those who that directly address the exterosensory, proprioceptive, and attentive processes underlying such authors’ experience of their bodies, the spaces and places around them, and their introspections and reflections on their own consciousness (see, for example, Tito Rajarshi Mukhopadhyay’s The Mind Tree: A Miraculous Child Breaks the Silence of Autism or Naoki Higashida’s The Reason I Jump: The Inner Voice of a Thirteen-Year-Old Boy with Autism). Introducing such original writing into discussions of neurodiversity and autism spectrum disorders honors the adage, “nothing about us without us”, while shedding considerable light on how students of neuroanatomy should approach difference when examining individual brain specimens at the gross and microscopic levels.
Another productive means for slow learning with reflection and introspection is art-making. I have approached doing so through both curricular and co-curricular initiatives. Within courses, as students transition from the readiness assurance to the application phase in a team-based learning framework, students are challenged to work collaboratively to visualize their collective knowledge through impromptu whiteboard sketches of brain structure and neuroanatomical pathways. Outside of class, students are challenged to visualize their knowledge with certain parameters set to encourage reflection and creativity. At times, this is as simple as directing students to craft representations of the human cerebral hemispheres from produce or other found objects; or capturing photographs of nature that bear visible and/or aesthetic resemblance to cerebral or neuronal structure. During the past two winter breaks (conceived originally as a means for building community among Neuroscience majors during our first full Covid-19-impacted year) a student-organized neuroscience-inspired art contest was initiated. Many remarkable works were created, annotated, and submitted; several representative submissions are displayed in Figure 5.
Figure 5.
Examples of original, neuroscience-inspired artwork solicited in winter break art contests for undergraduate students at Duke University. (A) “Anxiety”, by Hanna Varga (ink on paper). (B) “Fragmented Black Memory”, by Lenique Huggins (pencil, alcohol marker on paper). (C) “Inseparable”, by Marie Cheng (digital). All works reproduced here with the permission of the artists.
Another art-making initiative takes the form of optional class sessions when students have an opportunity to examine a variety of human (and non-human) brain specimens at the gross and microanatomical levels and make visual representation of what they observe. This approach stemmed from an existing co-curricular activity in the Duke University School of Medicine, called the “Anatomy Drawing Program”, which accompanied the learning of human cadaveric and functional anatomy. My initial goal was to extend this program by creating additional sessions when students were learning medical neuroscience and clinical neuroanatomy. These sessions were designed to encourage engagement with human brain specimens as objects worthy of close reading—like passages of prose or poetry—and re-presentation artistically in visual media. My primary goal was to invite learners to see the brain through their metacognitive lenses of perception, attention, reflection and empathy (Eichbaum, 2014). Secondarily, I aspired to bring learners to a space where imagination and subjectivity engage objectivity in the creation of visual art in various two dimensional media. Art-making materials were provided, including charcoal, pastel chalks, acrylic and watercolor paints, and a variety of paper stocks. Along the way, students were guided by a community-based visual artist, advanced medical and graduate students, and other neuroscience faculty. Typically, two such brain drawing sessions are offered during the semester. In the first session, students are given opportunity to inspect visually and haptically whole human brain specimens, human brains that have been bisected in the midsagittal plane, human brains that have been sectioned into coronal, axial or sagittal slabs, and a variety of non-human mammalian and non-mammalian vertebrate brains. In addition, light boxes, compound microscopes and dissecting scopes are available for visual inspection at variable level of magnification neurohistological slides of brain regions stained to reveal Nissl substance, myelin, or specific histochemical or cytoarchitectonic markers. Students are encouraged to create art inspired by their observations of these specimens in whatever media and through whatever perspective they wish to explore (Figure 6).
Figure 6.
Brain drawing sessions at Duke University and examples of original art-work created by students. (A)–(C) Students engaged in art-marking while closely examining human CNS specimens and meningeal samples. (D) Painting created by student shown in C. (E) Students examined human brain specimens with visible pathology and invited to make art reflecting their observation and their emotional, humanistic response. Here, as student is sketching a hemisphere with marked cortical atrophy consistent with advanced Alzheimer’s disease. (F) Coronal slab from a brain with severe cortical atrophy and secondary ventricular enlargement. (G) Student’s painting of the slab shown in F done in black to reflect the imagined “dark shadows” the disease may have cast over the individual and their experience living and dying with advanced neurodegenerative disease.
In a second brain drawing session, students are challenged to engage with the brain in more humanistic terms. Specifically, specimens from individuals who died with grossly visible pathologic alterations in the brain are made available for examination (see Figure 6E and F). Students are encouraged to reflect deeply and imaginatively on the human impact of brain injury and neurological disability (see Figure 6G). Thus, in these sessions when pathology is visible, the brains are not merely objects of “awe-full” fascination (Eichbaum et al., 2019). They may be that for most students; but they are also a means to engage students in the depth and meaning of human experience contextualized by the pathological transformation of brain structure and function.
CONCLUSIONS AND FUTURE DIRECTIONS
Historically, it is fair to say that neuroanatomy was the first academic study of the nervous system, and it was primarily a descriptive endeavor. Since neuroanatomy is still largely descriptive, it has not always enjoyed the esteem afforded to other aspects of neuroscience. Nonetheless, descriptive studies have an important role. Without a rich descriptive framework, other studies make no sense.
In this workshop, educators were provided with modern methods of inquiry using big data, methods using newer technology, and methods enriching the study of neural structure that intertwine the humanities with the study of structure. The next generation of neuroscientists and clinicians absolutely must have a firm foundation in neuroanatomy in order to effectively delve deeper and understand phenomena that they encounter. We believe the approaches described here will engage students more, result in superior understanding of the material, and provide a bedrock for their future careers. Indeed, we have already reported positive outcomes for students utilizing open data from the Allen Brain Map (Casimo, 2022) and other repositories (Nahmani, 2019; McCoy and Nahmani, 2021). Future educational research designed and conducted by us and those who would adopt these and other adjacent practices should assess their proximal impact on student learning of neuroanatomy and the broader development of neuroscientific efficacy.
We sincerely hope that this workshop rejuvenated pedagogical practices in neuroanatomy as it is delivered both online and in person. A video of the workshop has been generously provided by SfN’s Neuronline at https://neuronline.sfn.org/training/teaching-neuroscience-reviving-neuroanatomy/.
ACKNOWLEDGEMENTS
KC: The Allen Institute wishes to thank our founder, Paul G. Allen, for his vision, encouragement, and support.
EF: I would like to thank the students who participated in my NROSCI 1112 course and helped to improve this project over the last several years. I would also like to acknowledge the indispensable institutional support of the University of Pittsburgh’s Open Lab and their personnel, including Will Hinson and Sera Thornton, who have given technological and pedagogical support for this project. Funding for ongoing development of digital 3D models as open education resources was provided by a University of Pittsburgh Provost’s Open Education Resource funding award. Tinkercad is a registered trademark of Autodesk, Inc., and/or its subsidiaries and/or affiliates in the USA and other countries.
MN: I would like to thank all the students who participated in my TBIOMD 495 capstone research courses and did such fantastic work. In addition, I appreciate the support of the UW Bridge fund and the UW Tacoma SIAS Scholarship & Teaching awards that aided the development of these courses.
LEW: I would like to thank students at Duke University and in the global learning community gathered around the Medical Neuroscience course (Coursera) for sharing their artistic vision of human neuroanatomy and the impact of neurological injury and disease. The work described was partially supported by a Chancellor’s Fellowship award to the student-led Anatomy Drawing Program in the Duke University School of Medicine.
WG: We acknowledge the help of Natalie A. Pham-Schottler for her help in editing and formatting this manuscript.
We further thank the Society for Neuroscience (SfN) staff, particularly Professional Development Associate Vlera Kojcini, Senior Manager of Professional Development Ava Onalaja, and SfN Digital Learning team members, Bianca Williams and Eli Kane, for making these webinars possible.
REFERENCES
- Araya R, Vogels TP, Yuste R. Activity-dependent dendritic spine neck changes are correlated with synaptic strength. Proc Natl Acad Sci U S A. 2014;111(28):E2895–2904. doi: 10.1073/pnas.1321869111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C, Newsome W, Anderson D, Brown E, Deisseroth K, Donoghue J, MacLeish P, Marder E, Normann R, Sanes J, Schnitzer M, Sejnowski T, Tank D, Tsien R, Ugurbil K, Hudson K, Ling G, Wingfield J, Peña C, Jorgenson L. The BRAIN Initiative. Bethesda, MD: National Institutes of Health, U.S. Department of Health and Human Services; 2014. Jun 5, Brain Research Through Advancing Innovative Neurotechnologies (BRAIN) Working Group Report to the Advisory Committee to the Director, NIH. [Google Scholar]
- Branco T, Marra V, Staras K. Examining size-strength relationships at hippocampal synapses using an ultrastructural measurement of synaptic release probability. J Struct Biol. 2010;172(2):203–210. doi: 10.1016/j.jsb.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimo K. Enhancing Student Research Experiences with Open Data from the Allen Brain Map. J Undergrad Neurosci Educ. 2022;20(2):A177–A182. doi: 10.59390/NWXO4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin LA, Graham MJ, Dolan EL. Modeling course-based undergraduate research experiences: an agenda for future research and evaluation. CBE Life Sci Educ. 2015;14(1):es1. doi: 10.1187/cbe.14-10-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragly SA, Hobbi Mobarhan M, Våvang Solbrå A, Tennøe S, Hafreager A, Malthe-Sørenssen A, Fyhn M, Hafting T, Einevoll GT. Neuronify: An Educational Simulator for Neural Circuits. eNeuro. 2017;2017;4(2):ENEURO.0022–17. doi: 10.1523/ENEURO.0022-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichbaum QG. Thinking about thinking and emotion: the metacognitive approach to the medical humanities that integrates the basic and clinical sciences. Perm J. 2014;18(4):64–75. doi: 10.7812/TPP/14-027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichbaum QG, Pena G, White LE, Masukume G. In: The ‘awe-full’ fascination of pathology. Bleakley A, editor. Routledge: Handbook of the Medical Humanities; London, England and New York, NY: Routledge; 2019. pp. 321–331. [DOI] [Google Scholar]
- Fiala JC. Reconstruct: a free editor for serial section microscopy. Journal of Microscopy. 2005;218:52–61. doi: 10.1111/j.1365-2818.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- Grisham W, Brumberg JC, Gilbert T, Lanyon L, Williams RW, Olivo R. Teaching with Big Data: Report from the 2016 Society for Neuroscience Teaching Workshop. J Undergrad Neurosci Educ. 2017;16(1):A68–A76. [PMC free article] [PubMed] [Google Scholar]
- Holderith N, Lorincz A, Katona G, Rozsa B, Kulik A, Watanabe M, Nusser Z. Release probability of hippocampal glutamatergic terminals scales with the size of the active zone. Nat Neurosci. 2012;15(7):988–997. doi: 10.1038/nn.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler S, Köstinger G, Martin KAC, Schuhknecht GFP, Stratford KJ. Structure and function of a neocortical synapse. Nature. 2021;591(7848):111–116. doi: 10.1038/s41586-020-03134-2. [DOI] [PubMed] [Google Scholar]
- Jorgenson LA, Newsome WT, Anderson DJ, Bargmann CI, Brown EN, Deisseroth K, Donoghue JP, Hudson KL, Ling GSF, MacLeish PR, Marder E, Normann RA, Sanes JR, Schnitzer MJ, Sejnowski TJ, Tank DW, Tsien RY, Ugurbil K, Wingfield JC. The BRAIN Initiative: developing technology to catalyse neuroscience discovery. Philos Trans R Soc Lond B Biol Sci. 2015;370(1668):20140164. doi: 10.1098/rstb.2014.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juavinett A. Learning How to Code While Analyzing an Open Access Electrophysiology Dataset. J Undergrad Neurosci Educ. 2020;19(1):A94–A104. [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Pfister H, Shavit N. The big data challenges of connectomics. Nat Neurosci. 2014;17(11):1448–1454. doi: 10.1038/nn.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy J, Nahmani M. 3D Reconstruction and Analysis of Thin Subcellular Neuronal Structures using Focused-Ion Beam Scanning Electron Microscopy Data. J Vis Exp. 2021;175:e63030. doi: 10.3791/63030. [DOI] [PubMed] [Google Scholar]
- Meyer D, Bonhoeffer T, Scheuss V. Balance and stability of synaptic structures during synaptic plasticity. Neuron. 2014;82(2):430–443. doi: 10.1016/j.neuron.2014.02.031. [DOI] [PubMed] [Google Scholar]
- Murthy VN, Schikorski T, Stevens CF, Zhu Y. Inactivity produces increases in neurotransmitter release and synapse size. Neuron. 2001;32(4):673–682. doi: 10.1016/s0896-6273(01)00500-1. [DOI] [PubMed] [Google Scholar]
- Nahmani M. Versatile Undergraduate Neurobiology Course-Based Research Experiences Using Open Access 3D Electron Microscopy Image Volumes. J Undergrad Neurosci Educ. 2019;18(1):A65–A74. [PMC free article] [PubMed] [Google Scholar]
- Neuroscientifically Challenged. History of Neuroscience: Luigi Galvani. Neuroscientifically Challenged. 2022. Available at https://neuroscientificallychallenged.com/posts/history-of-neuroscience-luigi-galvani.
- Ostroff LE, Fiala JC, Allwardt B, Harris KM. Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron. 2002;35(3):535–545. doi: 10.1016/s0896-6273(02)00785-7. [DOI] [PubMed] [Google Scholar]
- Quinn DP, Kolar A, Harris SA, Wigerius M, Fawcett JP, Krueger SR. The Stability of Glutamatergic Synapses Is Independent of Activity Level, but Predicted by Synapse Size. Front Cell Neurosci. 2019;13:291. doi: 10.3389/fncel.2019.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Anderson RJ, Ashbrook DG, Gopalakrishnan V, Park Y, Priebe CE, Qi Y, Laoprasert R, Vogelstein JT, Williams RW, Johnson GA. Variability and heritability of mouse brain structure: Microscopic MRI atlases and connectomes for diverse strains. Neuroimage. 2020;222:117274. doi: 10.1016/j.neuroimage.2020.117274. [DOI] [PMC free article] [PubMed] [Google Scholar]