Abstract
Background
CAFs regulate the signaling of GC cells by promoting their migration, invasion, and proliferation and the function of immune cells as well as their location and migration in the TME by remodeling the extracellular matrix (ECM). This study explored the understanding of the heterogeneity of CAFs in TME and laid the groundwork for GC biomarker and precision treatment development.
Methods
The scRNA-seq and bulk RNA-seq datasets were obtained from GEO and TCGA. The prognostic significance of various CAFs subtypes was investigated using ssGSEA combined with Kaplan-Meier analysis. POSTN expression in GC tissues and CAFs was detected using immunohistochemistry, immunofluorescence, and Western blotting. Differential expression analysis identified the differentially expressed genes (DEGs) between normal and tumor samples in TCGA-STAD. Pearson correlation analysis identified DEGs associated with adverse prognosis CAF subtype, and univariate Cox regression analysis determined prognostic genes associated with CAFs. LASSO regression analysis and Multivariate Cox regression were used to build a prognosis model for CAFs.
Results
We identified five CAFs subtypes in GC, with the CAF_0 subtype associated with poor prognosis. The abundance of CAF_0 correlated with T stage, clinical stage, histological type, and immune cell infiltration levels. Periostin (POSTN) exhibited increased expression in both GC tissues and CAFs and was linked to poor prognosis in GC patients. Through LASSO and multivariate Cox regression analysis, three genes (CXCR4, MATN3, and KIF24) were selected to create the CAFs-score. We developed a nomogram to facilitate the clinical application of the CAFs-score. Notably, the CAFs signature showed significant correlations with immune cells, stromal components, and immunological scores, suggesting its pivotal role in the tumor microenvironment (TME). Furthermore, CAFs-score demonstrated prognostic value in assessing immunotherapy outcomes, highlighting its potential as a valuable biomarker to guide therapeutic decisions.
Conclusion
CAF_0 subtype in TME is the cause of poor prognosis in GC patients. Furthermore, CAFs-score constructed from the CAF_0 subtype can be used to determine the clinical prognosis, immune infiltration, clinicopathological characteristics, and assessment of personalized treatment of GC patients.
Keywords: Cancer associated fibroblasts, Single-cell RNA-Seq, Bulk RNA-Seq, CAFs-score, Immunotherapy, Gastric cancer
1. Introduction
Gastric cancer (GC) ranks as the fifth most common cancer globally [1]. Surgical resection, chemotherapy, radiation, targeted therapy, and immunotherapy remain the cornerstones of cancer treatment. Due to the metastases, highly intratumoral heterogeneity, and chemo-resistance characteristics of advanced GC, its treatment outcome is dismal, with only 25% overall survival (OS) for GC patients globally [2]. The Borrmann and Lauren classification based on the histology classification system and the subsequent TCGA classification based on genetic characteristics can better predict the prognosis of GC patients [[3], [4], [5]]. Due to its limited ability to predict response to drug therapy, TCGA molecular classification is not yet employed in clinical decision-making. Recent advances in omics technology have led to the development of multiple multi-gene signatures for predicting clinical outcomes and treatment effectiveness in GC patients [6,7].
The tumor microenvironment, consisting of fibroblasts, immune cells, and stroma, contributes to the heterogeneity observed among GC patients [8]. Cancer-associated fibroblasts (CAFs) derived from normal fibroblasts, bone marrow mesenchymal stem cells, adipocytes, endothelial cells, and epithelial cells are crucial components of the TME[9]. CAFs have been shown to specifically regulate GC cell signaling, promoting migration, invasion, and proliferation, as well as modulating their interactions with the stroma [10]. In addition, CAFs can regulate the function of immune cells as well as their location and migration in the TME by secreting cytokines and chemokines, expressing cell surface receptor proteins, and remodeling the extracellular matrix (ECM) [11]. Angiogenesis and hypoxia are also essential characteristics of the TME in GC. CAFs can stimulate either angiogenic or hypoxic conditions through diverse signaling mechanisms [12]. To enable accurate targeting of CAFs that contribute to carcinogenesis and progression, a deeper understanding of CAFs heterogeneity via the discovery of specific markers is necessary. Common CAFs markers such as α-smooth muscle actin (α-SMA), Fibroblast-activation protein (FAP), fibroblast-specific protein 1 (FSP-1/S100A4), and Platelet-derived growth factor receptor-β (PDGFR-β) can be used as a group of markers to characterize CAFs. Besides, the marker of CAFs includes matrix metalloproteinases (MMPs), Microfibril Associated Protein 5 (MFAP5), Podoplanin (PDPN), Tenascin-C (TN-C), Integrin (α11β1), Vimentin (VIM), Transgelin (TAGLN), and so on [13,14]. However, the expression of these cell surface markers is not homogeneous among CAF subtypes; not all CAFs exhibit these markers, and each marker may be expressed in CAFs from diverse origins. Analyzing scRNA-seq data from breast cancer, pancreatic cancer, colorectal cancer, and GC has led to the identification of distinct subtypes of CAFs, along with their corresponding functions and markers [[15], [16], [17], [18]]. In the TME of GC, subtypes of CAFs that co-expressed INHBA and FAP markers were found to be associated with a more advanced stage [18]. Four major subpopulations of fibroblasts were identified in GC tumor and adjacent mucosal samples by specific marker expression: myofibroblasts, pericytes, extracellular matrix CAFs (eCAFs), and inflammatory CAFs (iCAFs), with eCAFs being associated with a poor prognosis for GC[19]. Another study identified three CAFs subtypes based on gene expression profiles and pseudotemporal trajectory to trace CAFs differentiation: inflammatory (iCAFs), myofibroblastic (myCAFs), and intermediate CAFs (inCAFs) [20]. iCAFs have been shown to play a carcinogenic role in diffuse GC (DGC) and may be involved in the de novo carcinogenesis of DGC.
Conventional bulk RNA sequencing (bulk RNA-seq) is limited in its ability to fully grasp the heterogeneity among different cell subtypes in the TME, as it only provides average expression levels across all cells. The scRNA-seq could shed light on the heterogeneity of various cell subpopulations [21]. This study utilized GC scRNA-seq and bulk RNA-seq data from TCGA and GEO databases to identify and characterize CAFs subtypes associated with poor prognosis in GC patients. The CAFs-score based on these subtypes was established and its predictive efficacy was validated using internal and external cohorts. Comprehensive bioinformatics analyses were conducted to assess immune infiltration, chemotherapy sensitivity, and immunotherapy sensitivity between GC patients with high and low CAFs-scores. These findings enhance our understanding of CAFs heterogeneity in the TME and provide insights for the development of GC biomarkers and precision treatments.
2. Materials and methods
2.1. Data collection
The GEO database was used to download the GC scRNA-seq data. GSE183904 contained 26 primary gastric cancer tissues, 10 paired adjacent tissues, 3 metastatic peritoneal tissues, and 1 normal peritoneal tissue. The corresponding patient clinical information was obtained from the reference supplementary material [18].
The GC bulk RNA-seq transcriptome data and clinical data (including age, gender, survival time, survival status, TNM stage, and immunological subtype) were downloaded from The Cancer Genome Atlas (TCGA, https://portal.gc.cancer.gov/). We collected expression matrices and clinical information from ten GEO datasets (GSE26901, GSE26899, GSE26253, GSE13861, GSE66229, GSE15459, GSE34942, GSE38749, GSE29272, and GSE57303) and integrated them into a meta-GEO cohort. The “Combat” algorithm from the “sva” package was utilized to correct for non-biological technical biases. Additionally, we downloaded information from http://research-pub.gene.com/IMvigor210CoreBiologies containing the transcriptome and clinical data for the immune checkpoint blockades (ICBs) treatment cohort IMvigor210. The GSE176307 and GSE91061 cohorts contained transcriptome data from ICBs-treated melanoma and metastatic urothelial cancer, respectively. Using these three immunotherapy cohorts, the potential utility of CAFs-score in response to ICBs treatment was investigated.
2.2. The scRNA-seq data clustering dimension reduction and classification of CAFs subtypes
The raw data from GSE183904 includes 158,641 cells. We analyzed scRNA-seq data exclusively from 26 primary GC samples to develop a stringent CAFs system. Data quality control was conducted using the “Seurat” package in R version 4.1.1.1. After removing cells expressing more than 5000 genes or less than 200 genes, and log-normalizing the expressed genes, 117,102 cells remained. Batch effects among the 26 samples were eliminated using the “harmony” package. For non-linear dimension reduction, we applied the uniform manifold approximation and projection (UMAP) approach with 28 principal components and a resolution of 0.5. Subsequently, cells were clustered into distinct subpopulations using the FindNeighbors and FindClusters functions (dim = 1:30 and resolution = 0.5). t-Distributed Stochastic Neighbor Embedding (t-SNE) dimensional reduction was performed using the RunTSNE function. Fibroblasts were annotated using six marker genes (PDGFRA, PDGFRB, COL1A1, COL1A2, FAP, and DCN), resulting in a total of 10,741 fibroblasts. Further, re-clustering of fibroblasts and TSNE dimensionality reduction were performed to visualize fibroblast subtypes. Marker genes for each CAFs subtype were identified using the FindAllMarkers function, comparing markers between different subtypes of CAFs with logFC (fold change) = 0.585, minpct = 0.25, and adjusted p-value < 0.05. Additionally, the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was conducted for the DEGs of CAFs subtypes using the “Clusterprofiler” package.
2.3. CAFs subtypes communication analysis with iTALK
The “iTALK” package (https://github.com/Coolgenome/iTALK) is a computational method used to describe and illustrate intercellular communication signals in the TME by capturing highly enriched ligand-receptor genes (or transcripts) from scRNA-seq data. With the “iTALK” package, we identified ligand-receptor communication between different CAFs subtypes and categorized these interactions into four groups: cytokines/chemokines, growth factors, immune checkpoints, and others.
2.4. Combination of bulk-Seq and scRNA-Seq data
The single-sample Gene Set Enrichment Analysis (ssGSEA) was a method used to estimate cell subpopulation proportions based on gene expression data [22]. We applied ssGSEA to assess CAF subtype abundance in TCGA-STAD and meta-GEO patients using the top five DEGs for each subtype. Patients were categorized into CAF high-abundance and CAF low-abundance groups based on the median subtype abundance. The prognostic value of CAF subtypes was evaluated in TCGA-STAD and meta-GEO using the Kaplan-Meier method. TCGA-STAD patients were divided into high and low groups for the adverse prognostic CAF subtype using the median ssGSEA value. CIBERSORT was used to compare infiltration levels of 22 immune cells between groups. Additionally, GISTIC 2.0 (https://cloud.genepattern.org/) was utilized to analyze CAF subtype-associated somatic copy number alterations (CNAs) and the threshold copy number of mutation peaks. The “maftools” package was also utilized to visualize somatic mutations in patients with varying CAF subtype abundance.
2.5. Identification of hub genes of CAFs and construction of CAFs-score for GC
Based on the R4.1.1.1 ″limma” package (FDR<0.05, |logFC>1|), we identified 3460 DEGs between tumor and normal tissues. Further analysis revealed 624 hub genes of CAFs by correlating DEGs with the CAF_0 subtype (P < 0.001 and |COR>0.4|). Subsequently, the “survminer” and “glmnet” packages were employed to identify 91 prognostic hub genes of CAFs using univariate Cox regression analysis. Lasso Cox regression analysis and stepwise multivariate Cox regression were performed to construct a CAFs signature. The final formula for the CAFs signature:
We calculated the CAFs-score for each GC patient using a formula and categorized them into two groups based on the median CAFs-score in TCGA-STAD. Similarly, CAFs-scores for each GC patient in the meta-GEO dataset were calculated. Firstly, we assessed the relationship between CAFs-score groups and survival status. A heatmap was used to visualize the differences in the expression of three prognostic hub genes of CAFs between high and low CAFs-score groups. Then, Kaplan-Meier analysis was used to compare the survival of GC patients with high and low CAFs-score. Finally, univariate and multivariate Cox proportional hazards regression (CPHR) analyses were performed to confirm CAFs-score as an independent risk factor for OS.
2.6. Nomogram
The “rms” package was used to construct a nomogram with CAFs-score and stage as independent prognostic factors, based on multivariate CPHR analysis. The nomogram's total score for each sample was calculated as the sum of scores for each factor [23]. We evaluated the predictive ability of the 1, 2, and 3-year models using the Kaplan-Meier method, calibration curves, and concordance analysis between predicted and actual OS rates. Model reliability was assessed using decision curve analysis (DCA).
2.7. Mutation landscape and signaling mechanisms of the CAFs-score model
Gene mutation analysis utilized the “Maftools” package to process somatic mutation data from TCGA-STAD, visualizing the top 20 high-frequency mutated genes in different CAFs-score groups. We assessed differences in tumor mutation burden (TMB), microsatellite stability (MSS), low-frequency microsatellite instability (MSI-L), and high-frequency microsatellite instability (MSI-H) between different CAFs-score groups. Additionally, the CSC index of each sample was determined using one-class logistic regression (OCLR) to investigate the correlation between the CSC index and the CAFs-score.
We used the “GSVA” and “GSEA” packages to conduct gene set variation analysis (GSVA) and Gene Set Enrichment Analysis (GSEA) enrichment analysis on gene sets downloaded from the Molecular Signatures Database (MSigDB) to compare the biological characteristics between different CAF-score subgroups. Additionally, GO enrichment analysis was performed using the “GSVA” package based on the 50 Hallmark Pathway signature gene sets from MSigDB.
2.8. Estimation of the immune landscape
The algorithms of CIBERSORT, EPIC, xCell, and MCPcounter employ a gene expression matrix to estimate the abundance of different subtypes of cells in a mixed cell population [[24], [25], [26], [27]]. We used CIBERSORT to compare infiltration levels of 22 immune cells between different CAFs-score groups. Additionally, we assessed the relationship between these immune cells and three prognostic hub genes of CAFs. Tumor-infiltrating immune cells (TIICs) were estimated using TIMER, QUNTISEQ, xCell, EPIC, CIBERSORT-ABS, and MCP-COUNTER based on TCGA-STAD. CAF_EPIC, CAF_MCPcounter, and CAF_xCell scores for all GC patients were calculated using “EPIC”, “xCell”, and “MCPcounter” packages in R 4.1.1.1. StromalScore scores for each sample were calculated using the “estimate” package. We visualized the correlations between the three prognostic hub genes of CAFs and the CAFs-score, as well as other CAF evaluation systems, using the “ggplot2” package. Additionally, common immune checkpoints between different CAFs-score groups were compared.
2.9. Immunotherapy evaluation based on CAFs-score
Previous studies analyzed 33 cancer types in TCGA, identifying six immune subtypes (C1–C6) - wound healing, IFN-γ dominant, inflammatory, lymphocyte depleted, immunologically quiet, and TGF-β dominant [28]. We assessed differences in immune subtypes between different CAFs-score groups. The Tumor Immune Dysfunction and Exclusion (TIDE) score estimates tumor immune evasion probability based on gene expression data [29]. TIDE score, merck18 (T-cell-inflamed signature) score, dysfunction score, and exclusion score were obtained from the TIDE website (http://tide.dfci.harvard.edu). Differences in TIDE score, MSI, dysfunction score, and exclusion score between different CAFs-score groups were compared using the “ggpubr” R package. CAFs-score rankings were performed in IMvigor210, GSE176307, and GSE91061 cohorts. OS curves and immunotherapy response levels were plotted using the “survival” and “ggplot2” packages.
2.10. Drug sensitivity analysis
We used the “oncopredict” software to predict drug sensitivity in GC patients based on the Genomics of Drug Sensitivity in Cancer (GDSC) website (https://www.cancerrxgene.org/) [30]. Drugs with median IC50 < 1 were screened and compared between different CAFs-score groups.
2.11. Collection of clinical samples and isolation of NFs/CAFs
Three pairs of fresh postoperative GC tissues and adjacent normal tissues were obtained from the Seventh Affiliated Hospital of Sun Yat-sen University. CAFs and NFs were isolated from tumors and adjacent normal tissues of GC patients by collagenase digestion. After three generations, all primary NFs and CAFs were identified with CAFs-specific markers (α-SMA and FAP) for the following experiment [31,32]. Additionally, ten postoperative specimens of GC and their paired paracancerous normal tissues were collected. After surgical resection, the fresh samples were stored in a refrigerator at −80 °C until use.
2.12. Immunohistochemistry (IHC) and immunofluorescence (IF)
Paraffin-embedded GC tumor tissue samples were sectioned into 4 μm-thick slices for IHC and IF staining. Deparaffinization, hydration, and EDTA treatment were performed, followed by quenching endogenous peroxidase activity with 3% H2O2 for 15 min at room temperature. The sections were probed overnight at 4 °C with rabbit monoclonal anti-POSTN (1:500, Abcam, ab215199). For IHC, secondary HRP-conjugated IgG and DAB color development were used. For IF, Alexa Fluor 488-conjugated secondary antibodies were used for 1 h at room temperature. DAPI counterstaining and imaging via confocal microscopy were performed. NFs and CAFs were cultured on Laser confocal cell culture dishes, fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.2% TritonX-100 at 37 °C for 15 min, and then treated with anti-α-SMA antibody (Proteintech, 14395-1-AP) and anti-FAP antibody (Proteintech, 15384-1-AP). All samples were stained with DAPI for 10 min in the dark, sealed with a fluorescent anti-quench agent, and observed under a fluorescence microscope.
2.13. Western blotting
NFs and CAFs were harvested and washed with PBS. Cells, GC tissues, and paracancerous normal tissues were lysed using RIPA buffer. Proteins were separated on SDS-PAGE gels, blocked with BSA, and probed with α-SMA, FAP, and POSTN antibodies. ECL luminescent solution was used for visualization.
2.14. Statistical analysis
Statistical analysis was conducted using R software (version 4.1.1), and specific R packages were utilized in each section. Student's t-test was applied for two-group comparisons, while one-way ANOVA was used for comparisons among more than two groups. The Pearson χ2 test was employed to assess the correlation between CAFs-score and clinicopathological parameters and other factors. Kaplan-Meier analysis was performed to evaluate survival, and statistical significance was set at p < 0.05.
3. Results
3.1. Identification of CAFs in scRNA-seq data
The flow chart of the study was depicted in Fig. S1. ScRNA-seq data from GSE183904 were obtained for 26 primary GC tissues. Fig. S2 shows the complete data preprocessing steps. After quality control based on cell signatures, mitochondrial and ribosomal gene expression, t-SNE and UMAP dimensionality reduction algorithms classified cells into eight major clusters and 28 more specific clusters (Figs. S3A and S3B). Specific gene markers were used to validate different cell types, including common CAFs gene markers (Figs. S3C and S3D). Fibroblasts displayed high expression of PDGFRA, PDGFRB, COL1A1, COL1A2, DCN, and FAP gene markers. Fig. S3E illustrates the cell types and proportions of several clusters, highlighting a large proportion of T-cells in each cell type.
3.2. Clustering of CAFs based on scRNA-seq data in GC
GSE183904 raw data consists of 158,641 cells and 26,571 genes. We utilized scRNA-seq data from 26 primary GC samples to establish a stringent CAFs framework, identifying CAF populations using six marker genes: PDGFRA, PDGFRB, COL1A1, COL1A2, DCN, and FAP (Fig. S4). The lack of expression of epithelial cell-specific genes confirmed accurate CAFs identification (Fig. S5).
The t-SNE plot of 26 samples, revealed five distinct CAFs subtypes (Fig. 1A and B). The CAF_0 subtype was prevalent in most GC patients (Fig. 1C). Intercluster comparison identified 1665 DEGs: 291 in CAF_0, 443 in CAF_1, 590 in CAF_2, 196 in CAF_3, and 145 in CAF_4. Fig. 1D displays the top five DEGs, serving as marker genes for each CAFs subtype. Additionally, Fig. 1E–I presented the percentage distribution of gender, age, stage, TCGA subtypes, and pathology in the five CAFs subtypes. The CAF_0 subtype showed higher proportions in females, the elderly, and mixed intestinal and diffuse pathological types. These DEGs showed significant enrichment in pathways such as focal adhesion, complement, coagulation cascades, and ECM-receptor interaction, as indicated by KEGG analysis (Fig. 1J).
Fig. 1.
Identification of CAFs subtypes in the TME of gastric cancer. (A) t-Stochastic Neighbor Embedding (t-SNE) plot for the 10,741 CAFs in 26 primary gastric cancer tissues. (B) The t-SNE plot of the distribution of 5 subtypes of CAFs after clustering. (C) Cell number and proportions of subtypes of CAFs in 26 primary gastric cancer tissues. (D) Bubble plot showing the expression of the top 5 marker genes in the CAFs subtypes. The proportion of CAFs subgroups with various clinical characteristics, including gender (E), age (F), stage (G), TCGA classification (H), and Lauren classification (I). (J) KEGG enrichment analysis of 5 subtypes of CAFs.
3.3. Cell communication analysis of CAFs in GC
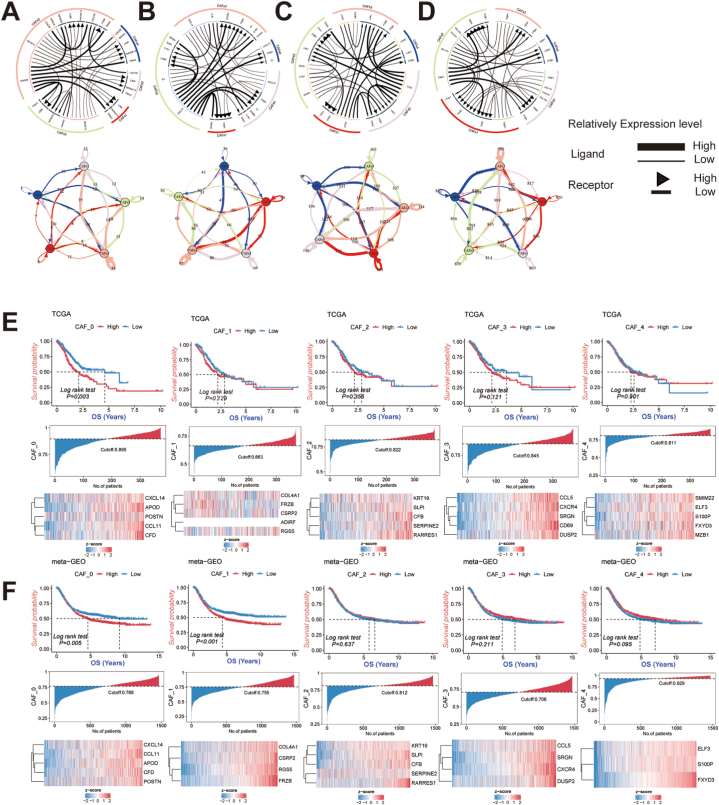
We utilized iTALK, a tool for analyzing cell communication signals, to explore the intercellular communication between CAFs subtypes. Specifically, we investigated checkpoints, cytokines, and growth factors to better understand the interactions among CAFs subtypes (Fig. 2A–D). For immune checkpoint-related receptor ligands, TNFSF14-LTBR and TNFSF9-TRAF2 gene pairs showed high expression between CAFs subtypes. Cytokine-related receptor ligands IL-6 and CXCL12 were highly expressed, as were growth factor-related receptor ligands CTGF and ITGB1, across all CAFs subtypes. The gene pair TIPM1-CD63 is substantially expressed for other similar receptor ligands.
Fig. 2.
Exploring cellular communication between 5 subtypes of CAFs and the relationship with gastric cancer prognosis. The circos plots show the top 20 most highly expressed ligand-receptors for CAFs subtypes interactions; the network plots depict the number of ligand-receptor interactions between subtypes of CAFs. Ligand-receptors-related gene pairs include immune checkpoint (A), cytokines/chemokines (B), growth factors (C), and others (D). The ssGSEA combined with Kaplan-Meier analysis was used to analyze the prognostic significance of different CAFs subtypes, including the CAF_0 subtype, CAF_1 subtype, CAF_2 subtype, CAF_3 subtype and CAF_4 subtype in TCGA-STAD (E) and meta-GEO (F) cohorts.
3.4. Identification of adverse prognosis CAFs subtype
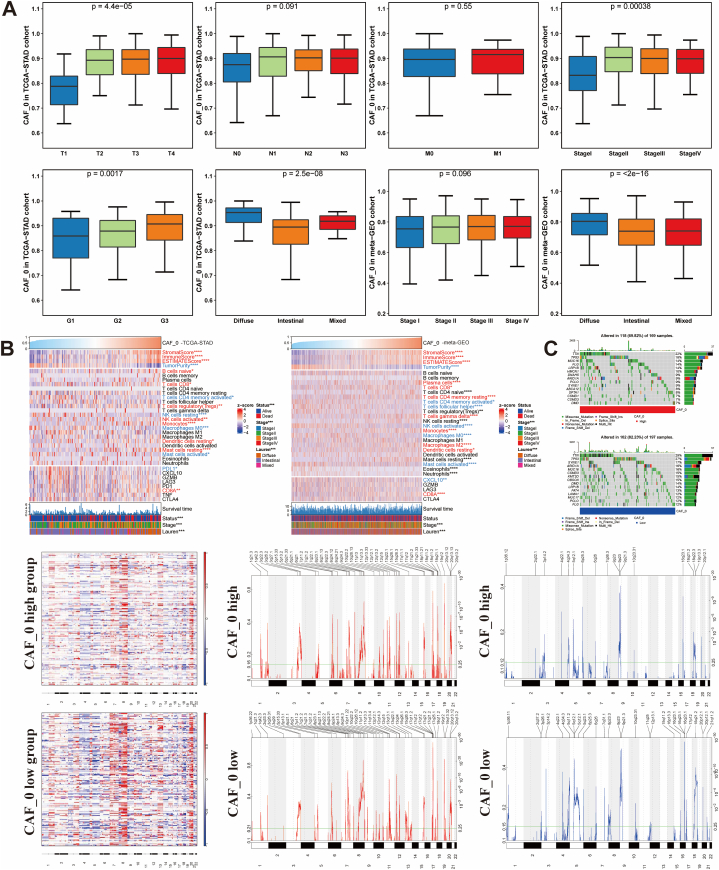
We conducted Kaplan-Meier survival analyses to investigate the prognostic value of different subtypes of CAFs in both TCGA-STAD and the meta-GEO cohort. Our study revealed that the high-abundance CAF_0 subtype was linked to a poor prognosis in GC (Fig. 2E and F). Further analysis of clinical characteristics demonstrated that GC patients with a high abundance of CAF_0 were more likely to have a higher T stage, advanced stage, and poorer tumor grade. Additionally, the CAF_0 subtype was found to be more prevalent in diffuse-type GC, while being less common in intestinal and mixed-type GC, suggesting its detrimental impact on GC (Fig. 3A). Moreover, immune infiltration analysis showed a significant positive correlation between the infiltration level of the CAF_0 subtype and immune scores as well as stromal scores, while exhibiting a negative correlation with tumor purity (Fig. 3B). Specifically, we observed that the infiltration level of the CAF_0 subtype was positively correlated with monocytes and CD8+ T cells, but negatively correlated with activated mast cells and memory CD4+ T cells, indicating its complex role within the tumor microenvironment (TME). Fig. S6 shows the differences in immune infiltration among other CAFs subtypes. Furthermore, in-depth mutation spectrum analysis revealed significant differences between the CAF_0 high-infiltration and CAF_0 low-infiltration groups (Fig. 3C). The high-infiltration group showed elevated somatic mutation frequencies in TTN (22%), TP53 (18%), and MUC16 (14%) genes, while the low-infiltration group exhibited higher rates of mutations in TTN (29%), TP53 (29%), and SYNE1 (17%) genes. GISTIC analysis compared CNV profiles between the two groups, highlighting the most pronounced amplification peak at 19q12 and a notable deletion at 9p21.3 in the high-infiltration group. Conversely, the low-infiltration group displayed the highest amplification peak at 17q21.2 and frequent deletions in the chromosomal region 16q23.1.
Fig. 3.
Correlation analysis of CAF_0 with other clinical characteristics, and exploration of CAF_0 immune infiltration and mutation spectrum in gastric cancer (GC). Kruskal-Wallis test revealed that increasing CAF_0 infiltration correlated with higher T stages, poor pathological stages, progressive tumor grades, and Lauren subtype (A). The difference of immune infiltration in GC patients with different CAF_0 abundance (B). Differences in mutation spectrum among GC patients with varying levels of CAF_0 abundance (C).
3.5. Construction and validation of the CAFs-score model
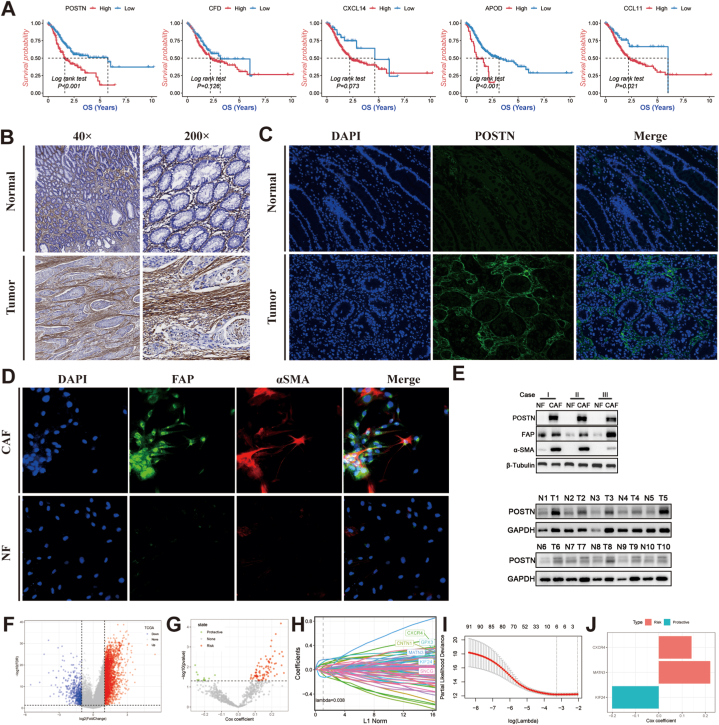
The marker genes of the CAF_0 subtype, including POSTN, CXCL14, APOD, CFD, and CCL11, on the prognosis of GC, were also evaluated (Fig. 4A). High expression of these marker genes was associated with a poor prognosis, especially the marker gene of POSTN. In contrast to the normal stomach tissue, IHC and IF revealed that POSTN is primarily expressed in the stromal cells (Fig. 4B and C). Hence, we concentrated on the POSTN expression pattern in the stromal compartment. In addition, the high expression of α-SMA and FAP were detected in CAFs under a fluorescence microscope (Fig. 4D). Through Western blot analysis, we observed distinct expression levels of POSTN between GC tumor tissues and normal tissues. Additionally, CAFs derived from gastric cancer showed significantly higher levels of POSTN expression. (Fig. 4E). By comparing the TCGA-STAD tumor tissue to the control, 3460 DEGs were identified (Fig. 4F). The discovery of 624 CAFs hub genes was the outcome of further investigation into relationships between DEGs and adverse prognostic CAF_0 subtype. Univariate Cox regression analysis identified 91 prognostic hub genes of the CAF_0 subtype (Fig. 4G). After LASSO regression analysis, 6 genes remained, as determined by the least partial likelihood of deviance (Fig. 4H and I). Three genes (CXCR4, MATN3, and KIF24) were ultimately retrieved after multivariate Cox regression analysis to create the risk score, known as the “CAFs-score” (Fig. 4J).
Fig. 4.
Identification of CAFs hub genes for the building of CAFs-score. (A) The Kaplan-Meier analysis of marker genes for subtypes of CAFs with a poor prognosis, including POSTN, CCL11, APOD, CXCL14, and CFD. (B) Expression of POSTN proteins in human gastric normal and cancer tissues by Immunohistochemical analysis. (C) Expression of POSTN proteins in human gastric normal and cancer tissues by immunofluorescence staining. (D) The expressions of α-SMA and FAP in NF and CAF were detected by immunofluorescence staining. (E) Western blot analysis was conducted to assess the expression of POSTN in gastric cancer tissue, adjacent non-cancerous tissue, as well as in normal fibroblasts and cancer-associated fibroblasts. (F) Volcano plot of differentially expressed genes of normal and cancer tissues in TCGA-STAD. (G) Volcano plot of CAFs prognostic hub genes identified by univariate Cox regression analysis. (H) The lambda trajectory of each independent variable. (I) Plots illustrating the calculated coefficient distributions for the logarithmic (lambda) series used for parameter selection (lambda). (J) Coefficients of multivariate Cox for each gene in the CAFs-score.
Finally, we re-distinguished different CAFs-score groups in the TCGA-STAD and meta-GEO (Fig. 5A and B). Patients with lower CAFs-scores exhibited significantly better OS than those with higher CAFs-scores (p < 0.05). Additionally, our AUC analyses on the CAFs-score model showed that OS had high accuracy at 1–5 years. Furthermore, GSVA enrichment analysis was conducted to investigate the biological differences between the two CAFs-score subgroups (Fig. S7). The high CAFs-score subgroup exhibited enrichment in pathways associated with calcium, cell adhesion molecules, focal adhesion, vascular smooth muscle contraction, neuroactive ligand-receptor interaction signaling, and more. Additionally, we demonstrated a strong correlation between the CAFs-score and the top 10 classic tumor pathways, including epithelial-mesenchymal transition, angiogenesis, coagulation, hedgehog signaling, and others.
Fig. 5.
Validation of the prognostic CAFs-related signature based on CAFs-score (KM survival analysis based on the median CAFs-score). The upper left parts are distribution plots for the relationship between CAFs-score and survival status; the lower left parts are heatmaps for the three prognostic CAFs-related genes in the cohorts; the upper right parts are ROC curve for the CAFs-score in the different cohorts; the lower right parts are survival curves between high- and low-CAFs-score groups. Internal cohorts: (A) TCGA-STAD; External cohorts: (B)meta-GEO.
3.6. Clinicopathologic characteristics and establishment nomogram based on CAFs-score
In both TCGA-STAD and meta-GEO cohorts, we conducted Univariate Cox regression and multivariate Cox regression analyses, incorporating CAFs-score, age, gender, stage, and Lauren classification. The results indicated that CAFs-score and stage were the only independent prognostic factors for gastric cancer patients (Fig. 6A). To enhance the clinical utility of CAFs-score in predicting OS, we developed a nomogram that included CAFs-score and stage to predict 1-, 2-, and 3-year OS rates in GC patients (Fig. 6B). With increasing CAFs-score, patients experience a progressively worse prognosis. The calibration curve demonstrates a good fit for predicting OS. Furthermore, DCA reveals that the nomogram outperforms stage-based assessment in distinguishing high-risk patients and their stages. TimeROC analysis shows higher AUC values for CAFs-score and nomogram compared to the stage. The results highlight the clinical relevance of nomograms combining CAFs-score and stage (Fig. 6C and D).
Fig. 6.
The establishment of a nomogram for predicting the prognosis of gastric cancer. (A) Univariate and multivariate Cox analyses of CAFs-score and clinicopathological characteristics in TCGA-STAD and meta-GEO. (B) The nomogram model incorporating CAFs-score and stage was established. (C) The time-dependent ROC curves, calibration plots, decision curves, and Comparison curves of the predictive capacity of clinicopathological features of the nomogram for 1-, 2-, and 3-years OS in TCGA-STAD. (D) The time-dependent ROC curves, calibration plots, decision curves, and Comparison curves of the predictive capacity of clinicopathological features of the nomogram for 1-, 2-, and 3-years OS in meta-GEO.
3.7. Immune infiltration in CAFs-score
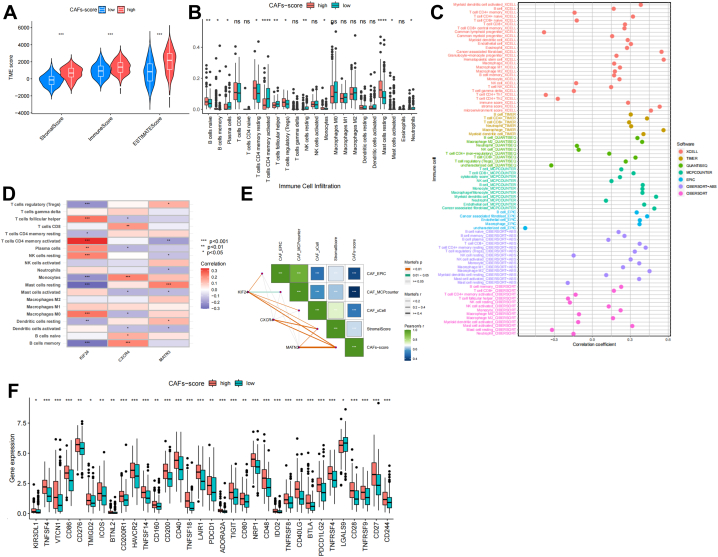
High immunological, stromal, and estimate Scores were linked to high CAFs-scores (Fig. 7A). The fractions of 22 immune cells were estimated using the CIBERSORT online tool. Then, we analyzed the tumor-infiltrating immune cells (TIICs) composition among the different CAFs-score groups of TCGA-STAD. The results showed that while plasma cells, T cells CD4 memory activated, NK cells resting, neutrophils, Mast cells activated, and T cells follicular helper were more prevalent in the low CAFs-score subgroup, the patients in the high CAFs-score subgroup had significantly higher proportions of B cells naive, B cells memory, T cells CD4 memory resting, T cells regulatory (Tregs), and mast cells resting (P < 0.05) (Fig. 7B and S8A). Additionally, we assessed the abundance of TIICs using various models: TIMER, QUNTISEQ, XCELL, EPIC, CIBERSORT-ABS, and MCP-COUNTER (Fig. 7C). The bulk of immune cells was also demonstrated to be strongly associated with the three genes (Fig. 7D). Strong relationships exist between the CAFs-score and the EPIC, MCP-COUNTER, XCELL, and Stromal Score (Fig. 7E and Fig. S8B). The results showed that, except for LGALS9, several immunological checkpoints were expressed more strongly in the high CAFs-score group (Fig. 7F).
Fig. 7.
The TIME of gastric cancer based on the CAFs-score. (A) Evaluation of TIME in different CAFs-score groups based on ESTIMATE algorithm. (B) The relative level of immune infiltration of 22 immune cells between groups with different CAFs-score. (C) The correlation between CAFs-score and different immune cells and stromal cells was evaluated based on multiple algorithms. (D) Correlation of immune cells with the three genes used to calculate the CAFs-score. (E) CAFs-score correlation with other CAFs or stromal scoring systems. (F) Expression of selected immune checkpoint genes in different CAFs-score groups. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
3.8. Characteristics in TMB, MSI, and CSC index
We analyzed the gene variants to elucidate the molecular biology characteristics of the different CAFs-score subgroups. The top 20 genes with the highest mutational frequencies in the different CAFs-score groups are shown in Fig. 8A–B, respectively. The findings revealed missense mutation to be the most common kind of mutation. Both TTN and TP53 exhibited mutation rates of over 35% and were the most frequently mutated genes. Additionally, we investigated the correlation between TMB and the CAFs-score. TMB was significantly higher in the low CAFs-score group compared to the high CAFs-score group (Fig. 8C). Fig. 8D also demonstrated a significant association between the CAFs-score and TMB (R = −0.44, p < 0.05). In addition, we discovered that a high CAFs-score was strongly associated with MSS status while a low CAFs-score was strongly associated with MSI-H status (Fig. 8E and F). Fig. 8G demonstrates that there was a strong negative association between the CAFs-score and the CSC index (R = −0.64, p < 0.05).
Fig. 8.
The mutation landscape in different CAFs-score groups. (A–B) Significantly mutated genes in the TCGA-STAD samples of the subgroups with low and high CAFs-score, respectively. Mutated genes (rows, top 20) are ranked by mutation rate; samples (columns) are grouped to emphasize mutual exclusivity among mutations. The percentage of mutations is presented on the right, while the total number of mutations is shown at the top. The color coding indicates the sort of mutation. (C–D) Distribution of tumor mutational burden (TMB) among different CAFs-score groups and correlation with CAFs-score. (E–F) Distribution of microsatellite instability (MSI) in different CAFs-score groups and CAFs-score in MSS/MSI-L/MSI-H groups. (G) The relationships between the CSC index and CAFs-score.
3.9. The predictive responsiveness of CAFs-score in immunotherapy
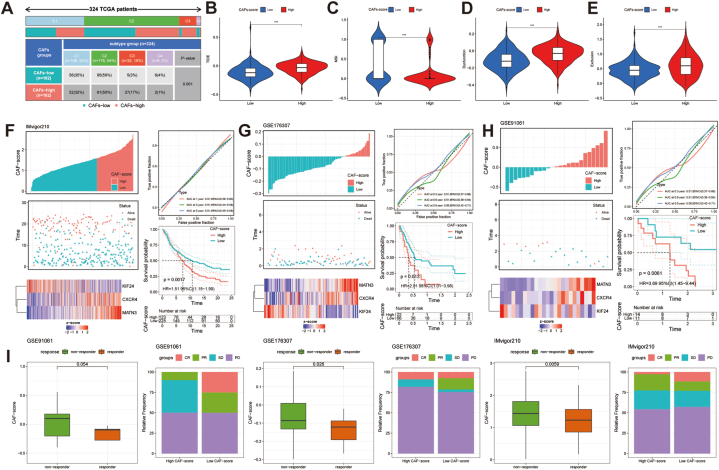
Considering the differences in immune infiltration, TMB, and MSI among different CAFs-score subgroups, we wanted to investigate the CAFs-score in evaluating the efficacy of immunotherapy. First, we discovered that there was a significant relationship between the immunological subtypes and the CAFs-score in the two groupings (Fig. 9A, p < 0.05) to evaluate the potential effectiveness of immunotherapy in various CAFs-score groupings under clinical conditions. The findings indicated that patients with higher TIDE prediction scores were less likely to benefit from immunotherapy, as it was associated with a higher probability of immune evasion. Conversely, patients with low CAFs-scores were more likely to benefit from immune checkpoint inhibitor (ICI) treatment, as the low-risk subgroup exhibited lower TIDE scores compared to the high CAFs-score subgroup (Fig. 9B). Additionally, we observed a statistically significant difference between the two risk subgroups in MSI score (Fig. 9C), T cell dysfunction (Fig. 9D), and T cell exclusion score (Fig. 9E).
Fig. 9.
The responsiveness of CAFs-score to immunotherapy in different cohorts. (A) Distribution of immune subtypes within different CAFs-score groups in TCGA-STAD. (B–E) TIDE, MSI, T cell dysfunction, and T cell exclusion score in different CAFs-score subgroups within TCGA-STAD, respectively. (F–H) The CAFs-score was used to evaluate the differences in prognosis across the different CAFs-score subgroups in the IMvigor210, GSE176307, and GSE91061 cohorts. (I) Evaluation of differences in immunotherapy response between CAFs-score subgroups within the IMvigor210, GSE176307, and GSE91061 cohorts. ***P < 0.001.
We further validated the CAFs-score model using the immunotherapy cohort (IMvigor210, GSE176307, GSE91061) and observed that patients with a low CAFs-score had a longer survival time and a more favorable prognosis compared to those with a high CAFs-score (Fig. 9F–H). The low CAFs-score group exhibited a higher proportion of patients with complete response (CR) and partial response (PR), while the high CAFs-score group had a higher proportion of patients with stable disease (SD) and progressive disease (PD) (Fig. 9I). In TCGA-STAD and the immunotherapy cohorts, we consistently observed that the low CAFs-score group had a higher response to immunotherapy.
3.10. Assessment of the CAFs-score in chemotherapy
Potential anticancer drugs with IC50 < 1, indicating significant inhibitory effects on GC, were identified (Fig. 10A). There were statistically significant differences in medication response between different risk categories (Fig. 10B–K). Notably, chemotherapeutic drugs such as Paclitaxel, Docetaxel, and Vinblastine exhibited high sensitivity in the high CAFs-score group, except for AZD8055. These results suggest that the CAFs-score may serve as a promising predictor of chemosensitivity.
Fig. 10.
Drug sensitivity analysis based on CAFs-score. (A) IC50 testing results for drugs with IC50 < 1 in different CAFs-score subgroups. (B–K) Potential drugs with significant treatment differences in the different CAFs-score subgroups.
4. Discussion
The clinical challenges of recurrence, metastasis, chemotherapy resistance, and immunotherapy insensitivity in advanced GC have led to more research focusing on TME. Numerous studies have shown that tumor-stromal cell interactions in the TME play a crucial role in GC development and progression [33]. As the most abundant and essential stromal cells, CAFs can secrete a wide range of extracellular proteins to build ECM and numerous growth factors, cytokines, and chemokines to promote tumor development and the construction of an immunosuppressive microenvironment [11]. However, CAFs have been reported to be heterogeneous and complex cell populations. The plasticity of CAF subtypes within the TME is well-documented in various cancer types, including colorectal, pancreatic, and breast cancers. However, conventional bulk RNA-Seq lacks the ability to distinguish between CAF subtypes at the single-cell level. We integrated scRNA-seq data and bulk RNA-seq data to identify five distinct CAF subtypes from 26 primary GC tissues. None of these CAFs subtypes express epithelial cell biomarker (EPCAM), and the combined KEGG results demonstrated that this dimension reduction result could more appropriately represent the heterogeneity of CAFs in the TME of these 26 GC samples. Notably, the ECM-receptor interaction pathway was enriched in four CAFs subtypes. We further applied iTALK to analyze cell-to-cell communication between CAFs subtypes. TNFSF14-LTBR and TNFSF9-TRAF2 gene pairs were widely expressed between CAFs subtypes for immune checkpoint-related receptor ligands. Intratumoral TNFSF14 expression has essential effects on anti-tumor immune response and TME remodeling. TNFSF14 can normalize blood vessels in the TME as well as shift a cold TME to an immunologically hot TME that may be susceptible to appropriate therapy interventions [34]. TNFSF9 has involved in the regulation of T cell anti-tumor immune response. Several therapeutic adjuvants that target TNFSF9 have been made to boost the proliferation of CD8+ T cells and kill cancer cells [35]. In addition, the high expression of TNFSF9 in tumor cells can stimulate tumor proliferation and metastasis via specific signaling pathways, and TNFSF9 inhibition can reduce tumor proliferation and metastasis [36]. However, the effect of TNFSF9 expression in CAFs on TME requires further investigation. For cytokine-related receptor ligands, IL-6 and CXCL12 were expressed in high abundance. IL-6 promotes tumorigenesis by regulating multiple signaling pathways [37]. Through the IL-6/JAK2/STAT3 signaling pathway, CAFs have been demonstrated to encourage the progression of GC[38]. A complementary strategy may play the role of stromal fibroblasts in IL-6-targeted therapy against GC. Likewise, CAFs, but not normal fibroblasts, promote tumor progression by secreting CXCL12([39]; [40]). CXCL12 also plays a vital role in the proliferation, migration, peritoneal dissemination, and treatment resistance of GC [41]. CTGF and ITGB1 were expressed in all CAFs subtypes for growth factor-related receptor ligands. Recent research has shown that CTGF expression is higher in CAFs and that CTGF level is negatively associated with disease-free survival (DFS) [42]. CAFs subtypes co-expressing DDR2 and POSTN in ovarian cancer dramatically boosted tumor cell proliferation and migration, and DDR2 regulated POSTN expression mechanistically through ITGB1[43]. Similar to this study, POSTN was proved that highly expressed in CAF_0 subtypes, indicating that ITGB1 may enhance GC progression in TME via a related mechanism. For other associated receptor-ligands, gene pair TIPM1-CD63 is highly expressed. Numerous studies have demonstrated that elevated TIMP-1 expression is associated with a poor prognosis in various cancer types, including GC([44]; [45]). TIPM1 regulates the phenotype of tumor cells via interacting with CD63 and β1-integrin. The formation of the TIMP1/CD63/β1-Integrin complex has become a spotlight of cancer research. Exosomes released by CD63+ CAFs rendered breast cancer cells resistant to tamoxifen, indicating a role for this CAFs subtype in supporting tumor progression in breast cancer [46].
To further explore the prognostic value of these CAFs subtypes, combined with the TCGA-STAD cohort, the adverse prognostic impact of CAF_0 subtypes was found. CAF_0 marker genes POSTN, CXCL14, APOD, and CCL11 have been confirmed in various CAFs. The marker gene of POSTN was highly expression in the CAFs compared with the NF according to our experiment results. It is proved by our results that POSTN is primarily expressed in stromal cells. We believe that the overexpression of POSTN in CAFs of GC is correlated with metastasis and tumor progression. CAF-derived POSTN activates the PI3K/Akt pathway, leading to the promotion of ovarian cancer cell migration and invasion through the induction of epithelial-mesenchymal transition (EMT) [47]. Analysis of GC scRNA-seq data revealed that POSTN-expressing eCAFs were related to M2 macrophages and poor OS(19). A subtype of CXCL14-overexpressing CAFs promotes tumor cell proliferation, migration, and invasion via different signaling pathways in breast and prostate cancers [48,49]. Previous studies have demonstrated increased expression of APOD in various fibroblast subtypes [50,51]. Through scRNA-seq analysis in pancreatic cancer, it was found that certain CAFs expressing COL11A1 were associated with cancer invasiveness and derived from adipose stromal cells expressing APOD. This reveals the heterogeneity in the origin and function of CAFs subtypes and suggests potential targets for optimizing anti-cancer therapies [52]. In head and neck cancer, CCL11 was overexpressed in CAFs by isolating and cultivating primary CAFs and normal fibroblasts (NFs) [53]. CCL11 boosted cancer cell migration and invasion by improving cancer stem cell-like properties and inducing EMT. Previous studies have found that CFD plays a detrimental impact on the regulation of ECM-related genes in NFs [54]. Still, the regulatory role of CFD on CAFs and its influence on TME has not been reported. Kaplan-Meier analysis showed that five CAF_0 subtype markers, including POSTN, CCL11, and APOD, were significantly associated with poor prognosis in GC patients. Further investigations are needed to verify and characterize the specific mechanisms of these markers in CAFs. In light of the prognostic significance of CAF_0 subtypes, we developed a CAFs-score based on three genes, consisting of one protective gene (KIF24) and two risk genes (CXCR4 and MATN3). In Pancreatic ductal adenocarcinoma (PDAC), KIF24 knockdown induced hyperproliferation and enhanced mitotic progression [55]. CAFs-secreted SDF-1 promotes malignant tumor progression via SDF-1/CXCR4/SATB-1 and SDF-1/CXCR4/PPARδ in pancreatic and lung adenocarcinomas, respectively [56,57]. In peritoneal metastatic GC, targeting the CXCR4/mTOR signaling pathway exhibits anti-metastatic properties and induces autophagic cell death in CER cells [58]. CXCR4 exhibits elevated expression in GC and is inversely correlated with GC prognosis [59]. Reducing CXCR4 significantly inhibits in vitro GC cell proliferation, migration, and invasion [60]. To offer a theoretical foundation for targeting CXCR4, more research is needed to elucidate how CAFs influence GC development through CXCR4 in TME. Several studies have indicated that MATN3, an ECM protein family member, serves as an independent predictor of poor prognosis in GC([61]; [62]). Given that CAFs are a significant contributor to the ECM protein, further bench studies are still needed to demonstrate how CAF influences the TME of GC via MATN3.
We initially validated the CAFs-score in the internal (TCGA-STAD) and external (mate-GEO) datasets. There were significant differences in OS between the groups with high and low CAFs-scores in both the internal and external cohorts. It can serve as an independent prognostic factor for GC patients. Additionally, the nomogram, integrating CAFs-score and stage, showed improved accuracy in predicting the prognosis of GC patients.
The extensive analysis of immune correlations with CAFs-score demonstrated higher stromal score and immune score in the high CAFs-score group, and various algorithms confirmed a positive correlation between CAFs-score and CAFs infiltration level. Moreover, CAFs-score exhibited significant associations with the infiltration levels of CD4 T cells, Tregs, and mast cells. It has been reported that there are more Treg and mast cells in the TME of GC, which is associated with cancer progression and suggests poor OS([63]; [64]). These findings imply that GC patients with a high CAFs-score have a more suppressed TME. MSI-H and higher TMB levels are both immunotherapy sensitivity indicators [65,66]. We found that patients with a low CAFs-score had higher TMB and MSI-H/MSI-L levels. Even though many GC patients exhibited innate or acquired immunotherapy resistance [67]. Our data demonstrated the robustness of the CAFs-score in distinguishing GC patients with diverse immunophenotypes, with high CAFs-score patients showing elevated TIDE scores. External validation further confirmed its predictive potential for immunotherapy response. These findings provide novel insights into the involvement of CAFs in reshaping the cancer niche and influencing the immune response.
Chemotherapy and targeted medication therapy can prolong the lifespan of advanced GC patients. Besides 5-fluorouracil (5-FU) and platinum, paclitaxel is also utilized in GC management. Notably, both docetaxel and paclitaxel have demonstrated survival advantages in the first- and second-line treatment of metastatic GC, respectively [68]. To broaden the clinical application of CAFs-score, our drug sensitivity data show that docetaxel and paclitaxel may be good choices for treating GC patients with a low CAFs-score and may have better therapeutic outcomes.
In summary, we have comprehensively characterized five distinct CAFs subtypes in the GC TME. Notably, we discovered that the CAF_0 subtype, identified by the marker genes POSTN, CDF, CXCL14, APOD, and CCL11, was strongly associated with poor prognosis. Furthermore, the CAFs-score based on this subtype demonstrated accurate prognostic and treatment response prediction for GC patients. In the future, widespread utilization of single-cell technologies will be crucial in characterizing diverse cell subtypes within the TME and providing novel insights for further bench research.
Nevertheless, it is essential to acknowledge several limitations in our study. Since our study is based on bioinformatics analysis of comprehensive omics data, only POSTN has been experimentally validated as a marker of CAFs in GC. Further investigations are necessary to characterize and validate other GC CAFs markers newly identified in our study. Furthermore, to strengthen the robustness of our findings, it is crucial to validate the identification of CAFs subtypes and the CAFs-scores based on the poor prognostic CAF_0 subtype using prospective data from multiple GC cohorts, in addition to the retrospective data obtained from public databases.
Author contribution statement
Zidan Zhao: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper. Tsz Kin Mak: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper. Yuntao Shi: Performed the experiments; Analyzed and interpreted the data. Kuan Li: Performed the experiments. Mingyu Huo; Changhua Zhang: Contributed reagents, materials, analysis tools, or data.
Data availability statement
Data associated with this study has been deposited at The GEO database was used to download the GC scRNA-seq data. GSE183904 contained 26 primary gastric cancer tissues, 10 paired adjacent tissues, 3 metastatic peritoneal tissues, and 1 normal peritoneal tissue. The corresponding patient clinical information was obtained from the reference supplementary materials.
The GC bulk RNA-seq transcriptome data and clinical data (including age, gender, survival time, survival status, TNM stage, and immunological subtype) were downloaded from The Cancer Genome Atlas (TCGA, https://portal.gc.cancer.gov/). We collected the expression matrices and clinical information of GSE26901, GSE26899, GSE26253, GSE13861, GSE66229, GSE15459, GSE34942, GSE38749, GSE29272, and GSE57303 from the GEO database and integrated them into a meta-GEO cohort. The " Combat” algorithm from the “sva” package was employed to correct non-biological technical biases. Additionally, we downloaded information from http://research-pub.gene.com/IMvigor210CoreBiologies containing the transcriptome and clinical data for the immune checkpoint blockades (ICBs) treatment cohort IMvigor210. The GSE176307 and GSE91061 cohorts contained transcriptome data from ICBs-treated melanoma and metastatic urothelial cancer, respectively. Using these three immunotherapy cohorts, the potential utility of CAFs-score in response to ICBs treatment was investigated.
Funding
This study was supported by the Guangdong Provincial Key Laboratory of Digestive Cancer Research (No. 2021B1212040006), Sanming Project of Medicine in Shenzhen (No. SZSM201911010), and Shenzhen Key Medical Discipline Construction Fund (No. SZXK016).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e19217.
Contributor Information
Mingyu Huo, Email: mingyu9318@163.com.
Changhua Zhang, Email: zhchangh@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Smyth E.C., Nilsson M., Grabsch H.I., van Grieken N.C., Lordick F. Gastric cancer. Lancet. 2020;396(10251):635–648. doi: 10.1016/s0140-6736(20)31288-5. eng. [DOI] [PubMed] [Google Scholar]
- 2.Sexton R.E., Al Hallak M.N., Diab M., Azmi A.S. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020;39(4):1179–1203. doi: 10.1007/s10555-020-09925-3. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y.C., Fang W.L., Wang R.F., Liu C.A., Yang M.H., Lo S.S., et al. Clinicopathological variation of lauren classification in gastric cancer. Pathol. Oncol. Res. 2016;22(1):197–202. doi: 10.1007/s12253-015-9996-6. eng. [DOI] [PubMed] [Google Scholar]
- 4.Luo Y., Gao P., Song Y., Sun J., Huang X., Zhao J., et al. Clinicopathologic characteristics and prognosis of Borrmann type IV gastric cancer: a meta-analysis. World J. Surg. Oncol. 2016;14(1):49. doi: 10.1186/s12957-016-0805-9. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Y., Xie J., Huang W., Chen H., Xi S., Han Z., et al. Tumor immune microenvironment and chemosensitivity signature for predicting response to chemotherapy in gastric cancer. Cancer Immunol. Res. 2019;7(12):2065–2073. doi: 10.1158/2326-6066.Cir-19-0311. eng. [DOI] [PubMed] [Google Scholar]
- 7.Zeng D., Li M., Zhou R., Zhang J., Sun H., Shi M., et al. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol. Res. 2019;7(5):737–750. doi: 10.1158/2326-6066.Cir-18-0436. eng. [DOI] [PubMed] [Google Scholar]
- 8.Oya Y., Hayakawa Y., Koike K. Tumor microenvironment in gastric cancers. Cancer Sci. 2020;111(8):2696–2707. doi: 10.1111/cas.14521. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biffi G., Tuveson D.A. Diversity and biology of cancer-associated fibroblasts. Physiol. Rev. 2021;101(1):147–176. doi: 10.1152/physrev.00048.2019. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun H., Wang X., Wang X., Xu M., Sheng W. The role of cancer-associated fibroblasts in tumorigenesis of gastric cancer. Cell Death Dis. 2022;13(10):874. doi: 10.1038/s41419-022-05320-8. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamauchi M., Gibbons D.L., Zong C., Fradette J.J., Bota-Rabassedas N., Kurie J.M. Fibroblast heterogeneity and its impact on extracellular matrix and immune landscape remodeling in cancer. Matrix Biol. 2020:91–92. doi: 10.1016/j.matbio.2020.05.001. 8-18. eng. [DOI] [PubMed] [Google Scholar]
- 12.Mao X., Xu J., Wang W., Liang C., Hua J., Liu J., et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol. Cancer. 2021;20(1):131. doi: 10.1186/s12943-021-01428-1. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Öhlund D., Elyada E., Tuveson D. Fibroblast heterogeneity in the cancer wound. J. Exp. Med. 2014;211(8):1503–1523. doi: 10.1084/jem.20140692. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Togo S., Polanska U.M., Horimoto Y., Orimo A. Carcinoma-associated fibroblasts are a promising therapeutic target. Cancers. 2013;5(1):149–169. doi: 10.3390/cancers5010149. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartoschek M., Oskolkov N., Bocci M., Lövrot J., Larsson C., Sommarin M., et al. Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat. Commun. 2018;9(1):5150. doi: 10.1038/s41467-018-07582-3. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elyada E., Bolisetty M., Laise P., Flynn W.F., Courtois E.T., Burkhart R.A., et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9(8):1102–1123. doi: 10.1158/2159-8290.Cd-19-0094. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H.O., Hong Y., Etlioglu H.E., Cho Y.B., Pomella V., Van den Bosch B., et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 2020;52(6):594–603. doi: 10.1038/s41588-020-0636-z. eng. [DOI] [PubMed] [Google Scholar]
- 18.Kumar V., Ramnarayanan K., Sundar R., Padmanabhan N., Srivastava S., Koiwa M., et al. Single-cell Atlas of lineage states, tumor microenvironment, and subtype-specific expression programs in gastric cancer. Cancer Discov. 2022;12(3):670–691. doi: 10.1158/2159-8290.Cd-21-0683. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X., Sun Z., Peng G., Xiao Y., Guo J., Wu B., et al. Single-cell RNA sequencing reveals a pro-invasive cancer-associated fibroblast subgroup associated with poor clinical outcomes in patients with gastric cancer. Theranostics. 2022;12(2):620–638. doi: 10.7150/thno.60540. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J., Park C., Kim K.H., Kim E.H., Kim H., Woo J.K., et al. Single-cell analysis of gastric pre-cancerous and cancer lesions reveals cell lineage diversity and intratumoral heterogeneity. npj Precis. Oncol. 2022;6(1):9. doi: 10.1038/s41698-022-00251-1. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papalexi E., Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018;18(1):35–45. doi: 10.1038/nri.2017.76. eng. [DOI] [PubMed] [Google Scholar]
- 22.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iasonos A., Schrag D., Raj G.V., Panageas K.S. How to build and interpret a nomogram for cancer prognosis. J. Clin. Oncol. 2008;26(8):1364–1370. doi: 10.1200/jco.2007.12.9791. eng. [DOI] [PubMed] [Google Scholar]
- 24.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Racle J., de Jonge K., Baumgaertner P., Speiser D.E., Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife. 2017:6. doi: 10.7554/eLife.26476. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becht E., Giraldo N.A., Lacroix L., Buttard B., Elarouci N., Petitprez F., et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. doi: 10.1186/s13059-016-1070-5. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830.e14. doi: 10.1016/j.immuni.2018.03.023. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–D961. doi: 10.1093/nar/gks1111. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H., Deng T., Liu R., Ning T., Yang H., Liu D., et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer. 2020;19(1):43. doi: 10.1186/s12943-020-01168-8. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nurmik M., Ullmann P., Rodriguez F., Haan S., Letellier E. In search of definitions: cancer-associated fibroblasts and their markers. Int. J. Cancer. 2020;146(4):895–905. doi: 10.1002/ijc.32193. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rojas A., Araya P., Gonzalez I., Morales E. Gastric tumor microenvironment. Adv. Exp. Med. Biol. 2020;1226:23–35. doi: 10.1007/978-3-030-36214-0_2. eng. [DOI] [PubMed] [Google Scholar]
- 34.Skeate J.G., Otsmaa M.E., Prins R., Fernandez D.J., Da Silva D.M., Kast W.M. TNFSF14: LIGHTing the way for effective cancer immunotherapy. Front. Immunol. 2020;11:922. doi: 10.3389/fimmu.2020.00922. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J., Wang Y. Role of TNFSF9 bidirectional signal transduction in antitumor immunotherapy. Eur. J. Pharmacol. 2022;928 doi: 10.1016/j.ejphar.2022.175097. eng. [DOI] [PubMed] [Google Scholar]
- 36.Wu J., Wang Y., Yang Y., Liu F., Jiang Z., Jiang Z. TNFSF9 promotes metastasis of pancreatic cancer by regulating M2 polarization of macrophages through Src/FAK/p-Akt/IL-1β signaling. Int. Immunopharm. 2022;102 doi: 10.1016/j.intimp.2021.108429. eng. [DOI] [PubMed] [Google Scholar]
- 37.Hirano T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021;33(3):127–148. doi: 10.1093/intimm/dxaa078. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X., Tao P., Zhou Q., Li J., Yu Z., Wang X., et al. IL-6 secreted by cancer-associated fibroblasts promotes epithelial-mesenchymal transition and metastasis of gastric cancer via JAK2/STAT3 signaling pathway. Oncotarget. 2017;8(13):20741–20750. doi: 10.18632/oncotarget.15119. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J., Liu X., Wu J., Lou C., Zhang Q., Chen H., et al. CXCL12 derived from CD248-expressing cancer-associated fibroblasts mediates M2-polarized macrophages to promote nonsmall cell lung cancer progression. Biochim. Biophys. Acta, Mol. Basis Dis. 2022;1868(11) doi: 10.1016/j.bbadis.2022.166521. eng. [DOI] [PubMed] [Google Scholar]
- 40.Izumi D., Ishimoto T., Miyake K., Sugihara H., Eto K., Sawayama H., et al. CXCL12/CXCR4 activation by cancer-associated fibroblasts promotes integrin β1 clustering and invasiveness in gastric cancer. Int. J. Cancer. 2016;138(5):1207–1219. doi: 10.1002/ijc.29864. eng. [DOI] [PubMed] [Google Scholar]
- 41.Lee H.J., Jo D.Y. The role of the CXCR4/CXCL12 axis and its clinical implications in gastric cancer. Histol. Histopathol. 2012;27(9):1155–1161. doi: 10.14670/hh-27.1155. eng. [DOI] [PubMed] [Google Scholar]
- 42.Wells J.E., Howlett M., Cole C.H., Kees U.R. Deregulated expression of connective tissue growth factor (CTGF/CCN2) is linked to poor outcome in human cancer. Int. J. Cancer. 2015;137(3):504–511. doi: 10.1002/ijc.28972. eng. [DOI] [PubMed] [Google Scholar]
- 43.Akinjiyan F.A., Dave R.M., Alpert E., Longmore G.D., Fuh K.C. DDR2 expression in cancer-associated fibroblasts promotes ovarian cancer tumor invasion and metastasis through periostin-ITGB1. Cancers. 2022;14(14) doi: 10.3390/cancers14143482. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Justo B.L., Jasiulionis M.G. Characteristics of TIMP1, CD63, and β1-integrin and the functional impact of their interaction in cancer. Int. J. Mol. Sci. 2021;22(17) doi: 10.3390/ijms22179319. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H., Xiang Y., Zong Q.B., Zhang X.Y., Wang Z.W., Fang S.Q., et al. miR-6745-TIMP1 axis inhibits cell growth and metastasis in gastric cancer. Aging (Albany NY) 2021;13(21):24402–24416. doi: 10.18632/aging.203688. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao Y., Li X., Zeng C., Liu C., Hao Q., Li W., et al. CD63(+) cancer-associated fibroblasts confer tamoxifen resistance to breast cancer cells through exosomal miR-22. Adv. Sci. 2020;7(21) doi: 10.1002/advs.202002518. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yue H., Li W., Chen R., Wang J., Lu X., Li J. Stromal POSTN induced by TGF-β1 facilitates the migration and invasion of ovarian cancer. Gynecol. Oncol. 2021;160(2):530–538. doi: 10.1016/j.ygyno.2020.11.026. eng. [DOI] [PubMed] [Google Scholar]
- 48.Sjöberg E., Meyrath M., Milde L., Herrera M., Lövrot J., Hägerstrand D., et al. A novel ACKR2-dependent role of fibroblast-derived CXCL14 in epithelial-to-mesenchymal transition and metastasis of breast cancer. Clin. Cancer Res. 2019;25(12):3702–3717. doi: 10.1158/1078-0432.Ccr-18-1294. eng. [DOI] [PubMed] [Google Scholar]
- 49.Augsten M., Hägglöf C., Olsson E., Stolz C., Tsagozis P., Levchenko T., et al. CXCL14 is an autocrine growth factor for fibroblasts and acts as a multi-modal stimulator of prostate tumor growth. Proc. Natl. Acad. Sci. U. S. A. 2009;106(9):3414–3419. doi: 10.1073/pnas.0813144106. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takaya K., Asou T., Kishi K. Identification of Apolipoprotein D as a dermal fibroblast marker of human aging for development of skin rejuvenation therapy. Rejuvenation Res. 2022 doi: 10.1089/rej.2022.0056. eng. [DOI] [PubMed] [Google Scholar]
- 51.Joseph D.B., Henry G.H., Malewska A., Reese J.C., Mauck R.J., Gahan J.C., et al. Single-cell analysis of mouse and human prostate reveals novel fibroblasts with specialized distribution and microenvironment interactions. J. Pathol. 2021;255(2):141–154. doi: 10.1002/path.5751. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu K., Cai L., Cui C., de Los Toyos J.R., Anastassiou D. Single-cell analysis reveals the pan-cancer invasiveness-associated transition of adipose-derived stromal cells into COL11A1-expressing cancer-associated fibroblasts. PLoS Comput. Biol. 2021;17(7) doi: 10.1371/journal.pcbi.1009228. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang W.Y., Lin Y.S., Lin Y.C., Nieh S., Chang Y.M., Lee T.Y., et al. Cancer-associated fibroblasts promote tumor aggressiveness in head and neck cancer through chemokine ligand 11 and C-C motif chemokine receptor 3 signaling circuit. Cancers. 2022;14(13) doi: 10.3390/cancers14133141. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ezure T., Sugahara M., Amano S. Senescent dermal fibroblasts negatively influence fibroblast extracellular matrix-related gene expression partly via secretion of complement factor D. Biofactors. 2019;45(4):556–562. doi: 10.1002/biof.1512. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mashima Y., Nohira H., Sugihara H., Dynlacht B.D., Kobayashi T., Itoh H. Life Sci Alliance; 2022. KIF24 Depletion Induces Clustering of Supernumerary Centrosomes in PDAC Cells; p. 5. 11. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei L., Ye H., Li G., Lu Y., Zhou Q., Zheng S., et al. Cancer-associated fibroblasts promote progression and gemcitabine resistance via the SDF-1/SATB-1 pathway in pancreatic cancer. Cell Death Dis. 2018;9(11):1065. doi: 10.1038/s41419-018-1104-x. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Lan W., Xu M., Song J., Mao J., Li C., et al. Cancer-associated fibroblast-derived SDF-1 induces epithelial-mesenchymal transition of lung adenocarcinoma via CXCR4/β-catenin/PPARδ signalling. Cell Death Dis. 2021;12(2):214. doi: 10.1038/s41419-021-03509-x. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hashimoto I., Koizumi K., Tatematsu M., Minami T., Cho S., Takeno N., et al. Blocking on the CXCR4/mTOR signalling pathway induces the anti-metastatic properties and autophagic cell death in peritoneal disseminated gastric cancer cells. Eur. J. Cancer. 2008;44(7):1022–1029. doi: 10.1016/j.ejca.2008.02.043. eng. [DOI] [PubMed] [Google Scholar]
- 59.Wen F., Lu X., Huang W., Chen X., Ruan S., Gu S., et al. Characteristics of immunophenotypes and immunological in tumor microenvironment and analysis of immune implication of CXCR4 in gastric cancer. Sci. Rep. 2022;12(1):5720. doi: 10.1038/s41598-022-08622-1. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xue S., Ma M., Bei S., Li F., Wu C., Li H., et al. Identification and validation of the immune regulator CXCR4 as a novel promising target for gastric cancer. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.702615. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu P.L., He Y.F., Yao H.H., Hu B. Martrilin-3 (MATN3) overexpression in gastric adenocarcinoma and its prognostic significance. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2018;24:348–355. doi: 10.12659/msm.908447. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang P., Xiao W.S., Li Y.H., Wu X.P., Zhu H.B., Tan Y.R. Identification of MATN3 as a novel prognostic biomarker for gastric cancer through comprehensive TCGA and GEO data mining. Dis. Markers. 2021;2021 doi: 10.1155/2021/1769635. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lv Y., Zhao Y., Wang X., Chen N., Mao F., Teng Y., et al. Increased intratumoral mast cells foster immune suppression and gastric cancer progression through TNF-α-PD-L1 pathway. J Immunother Cancer. 2019;7(1):54. doi: 10.1186/s40425-019-0530-3. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qu Y., Wang X., Bai S., Niu L., Zhao G., Yao Y., et al. The effects of TNF-α/TNFR2 in regulatory T cells on the microenvironment and progression of gastric cancer. Int. J. Cancer. 2022;150(8):1373–1391. doi: 10.1002/ijc.33873. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ganesh K., Stadler Z.K., Cercek A., Mendelsohn R.B., Shia J., Segal N.H., et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019;16(6):361–375. doi: 10.1038/s41575-019-0126-x. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jardim D.L., Goodman A., de Melo Gagliato D., Kurzrock R. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell. 2021;39(2):154–173. doi: 10.1016/j.ccell.2020.10.001. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kawazoe A., Shitara K., Boku N., Yoshikawa T., Terashima M. Current status of immunotherapy for advanced gastric cancer. Jpn. J. Clin. Oncol. 2021;51(1):20–27. doi: 10.1093/jjco/hyaa202. eng. [DOI] [PubMed] [Google Scholar]
- 68.Kanagavel D., Fedyanin M., Tryakin A., Tjulandin S. Second-line treatment of metastatic gastric cancer: current options and future directions. World J. Gastroenterol. 2015;21(41):11621–11635. doi: 10.3748/wjg.v21.i41.11621. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at The GEO database was used to download the GC scRNA-seq data. GSE183904 contained 26 primary gastric cancer tissues, 10 paired adjacent tissues, 3 metastatic peritoneal tissues, and 1 normal peritoneal tissue. The corresponding patient clinical information was obtained from the reference supplementary materials.
The GC bulk RNA-seq transcriptome data and clinical data (including age, gender, survival time, survival status, TNM stage, and immunological subtype) were downloaded from The Cancer Genome Atlas (TCGA, https://portal.gc.cancer.gov/). We collected the expression matrices and clinical information of GSE26901, GSE26899, GSE26253, GSE13861, GSE66229, GSE15459, GSE34942, GSE38749, GSE29272, and GSE57303 from the GEO database and integrated them into a meta-GEO cohort. The " Combat” algorithm from the “sva” package was employed to correct non-biological technical biases. Additionally, we downloaded information from http://research-pub.gene.com/IMvigor210CoreBiologies containing the transcriptome and clinical data for the immune checkpoint blockades (ICBs) treatment cohort IMvigor210. The GSE176307 and GSE91061 cohorts contained transcriptome data from ICBs-treated melanoma and metastatic urothelial cancer, respectively. Using these three immunotherapy cohorts, the potential utility of CAFs-score in response to ICBs treatment was investigated.