Abstract
We have previously developed a non-viral episomal vector based on matrix attachment region (MAR) that can facilitate plasmid replication episomally in mammal cells. In this study, we have focused on the development of an alternative tissue specific episomal vector by incorporating into cis-acting elements. We found that AAT promoter demonstrated the highest eGFP expression level in HepG2, Huh-7 and HL-7702 hepatic cells. Furthermore, hCMV enhancer when combined with AAT promoter significantly improved the eGFP expression level in the transfected HepG2 cells. The mean fluorescence intensity of eGFP in hCMV2 group was 1.33 fold, which was higher than that of the control (p < 0.01), followed by the hCMV1 group (1.21 fold). In addition, the percentages of eGFP-expressing cells in hCMV1 and hCMV2 groups were observed to be 49.3% and 57.2%, which were significantly higher than that of the enhancer-devoid control vector (44.3%) (p < 0.05). Moreover, the eGFP protein were up to 3.5 fold and 5.1 fold (p < 0.05), respectively. This observation could be related with the activities of some specific transcription factors (TFs) during the transcriptional process, such as SRF, REL and CREB1. The composite CMV/AAT promoter can be thus used for efficient transgene expression of MAR-based episomal vector in liver cells and as a potential gene transfer tools for the management of liver diseases.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-023-03774-x.
Keywords: Liver, Episomal vector, Promoter, Enhancer, Transgene expression
Introduction
Liver-directed gene therapy has been identified among the most promising candidate for several genetic and metabolic liver diseases (Nunes and Raper 1996; Ding et al. 2012; Maestro et al. 2021). However, an excellent gene therapy vector that can exhibit tissue-specificity, high transgenic expression efficiency and is relatively safe is required in clinical gene therapy. These are also considered as major parameters of vector performance. The non-viral episomal vectors have no viral components and could effectively replicate their genomes autonomously as extrachromosomal elements or as episomes so as to circumvent the potential risk of random integration of the vector into the host cells genome chromosomal DNA after the gene transfer, thus avoiding various deleterious or mutagenic effects (Mulia et al. 2021). Therefore, they display relatively lower toxicity and less immunogenicity for the host at both the cellular or somatic level. In addition, the episomal vectors are less expensive and can be conveniently produced in a large scale with consistent quality in comparison to packaging the viral vectors (Miao et al. 2001; Ehrhardt et al. 2008). Thus, it can at least theoretically ensure the long-desired increase in the safety profile to facilitate clinical gene therapy applications when compared to the viral ones.
The genomic AT-rich matrix attachment region (MAR) sequences are generally thought to facilitate the tethering of DNA to a subnuclear structure and reported to be critical for the organization of chromatin loops so that to define the boundaries of the various chromatin domains (Jackson et al. 2006; Bode et al. 2000). For instance, Argyros et al. (2008) confirmed that MAR was capable of preventing the transgene silencing as well as providing a sustained transgene expression and this plasmid can effectively replicate as an episomal entity. Similarly, we have previously reported an episomal system exploiting a 387 bp characteristic motif of 2200 bp MAR that could lead to markedly increased stable recombinant protein expression, mediate episomal replication and does not require any virally encoded trans-acting factors in the transfected Chinese hamster ovary, Chang liver and HEK-293 cells (Lin et al. 2015; Wang et al. 2017; Xu et al. 2019). Moreover, a MAR1 element that was incorporated into the downstream of expression cassette of the MAR-based episomal vector was found to significantly improve the transgenic ability in the hepatic cells (Wang et al. 2010, 2012; Zhao et al. 2017; Li et al. 2018; Zhang et al. 2019) (Supplementary Fig. 1). It was observed that the pEMM episomal vector that was employed as the backbone vector of this study as the high transgenic ability of vector was necessary for inhibiting transgene silencing of the foreign genes and stimulating innate immune response of the host cells in gene therapy.
In addition to the higher transgenic ability, liver-specific strategies for gene therapy could be potentially advantageous to minimize the possible adverse effects caused by the non-target gene expression for the development of safe and reproducible gene therapy approaches and are usually achieved by employing the expression cassettes including tissue-specific promoter. The α1-antitrypsin (AAT) is present in large amounts in the human serum and synthesized predominantly in the hepatocytes. The human AAT gene promoter could drive hepatoma-specific transcription from the heterologous SV40 promoter (Miao et al. 2001; De Simone and Cortese 1989). Additionally, it has been reported that the human tumor–specific alpha fetoprotein (AFP) promoter in combination with the hCMV enhancer element can act as a valuable tissue-specific promoter for targeting the hepatocellular carcinoma with a non-viral gene delivery system, thereby yielding significantly higher tissue-specificity with less undesired side effects (Haase et al. 2013). For example, it has been found that human apolipoprotein A-I (ApoAI) promoter can induce enhanced green fluorescent protein (eGFP) expression in cells originating from liver in vitro and in vivo, whereas the non-liver organ gene expression could be eliminated in the pApoAI-eGFP–treated mice (Hu et al. 2010).
However non-viral tissue-specific vectors displayed significantly lower delivery efficiency in comparison to the strong constitutive promoters that can substantially limit their application in gene therapy (Hu et al. 2010). Human cytomegalovirus major immediate-early gene (hCMV), Jaagsiekte sheep retrovirus (JSRV) or apolipoprotein E (ApoE) enhancers could be potentially incorporated to increase the expression level and persistence of protein of interest (Xu et al. 2001; De Geest et al. 2000; Lam et al. 2007; Yu et al. 2020; Suzuki et al. 2020). Moreover, the ability of these cis-acting elements could be affected by the plasmid backbone sequences which could be tightly related to the transcription activity of interesting gene (Giannakopoulos et al. 2009; Wang et al. 2016; Jia et al. 2019). Therefore, we have screened several enhancer elements to establish their suitability in our MAR-based episomal vector for further promoting the transgenic expression effect and safety.
Based on these findings, we have attempted to incorporate different liver–specific promoters (LSPs) and enhancer elements combination for improving the MAR-based episomal vector function. The adaptation and performance of these elements in our pEMM episomal vector contexts were evaluated in terms of the transgene expression magnitude in human hepatic cells and the underlying molecular mechanisms have been predicted by in silico analysis.
Materials and methods
Vector construction
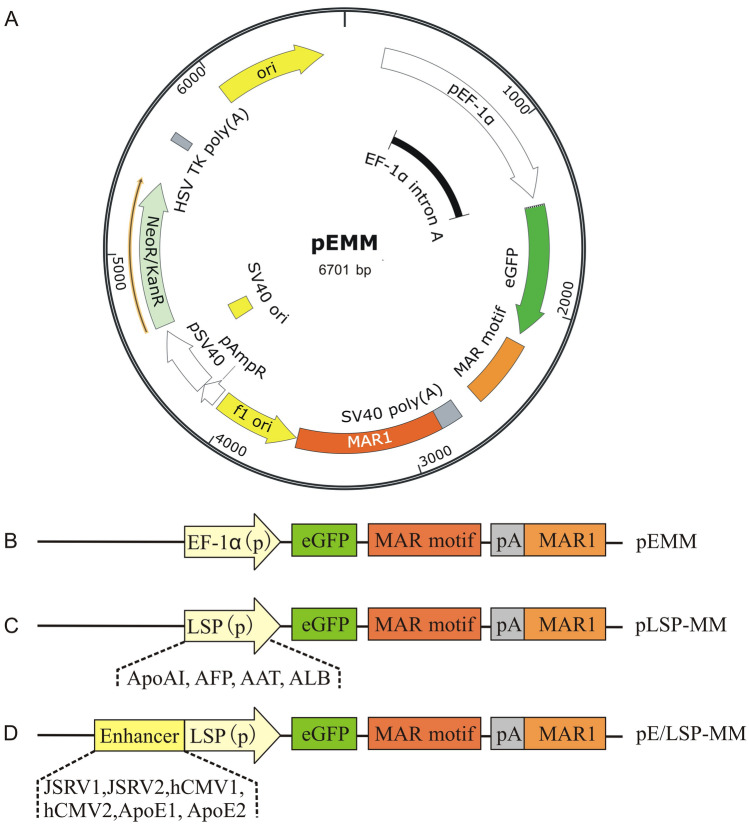
Based on a previously described pEM episomal vector (Wang et al. 2017) and a 791 bp MAR1 element (Genbank: AC117476.6 from 60,046 to 60,836) flanking at the 3′ end of the eGFP gene expression cassette was used to produce pEMM as a backbone vector (Fig. 1A and B) in this study. Four liver-specific promoter sequences, ApoAI, AFP, AAT and albumin (ALB), were first artificially synthesized by General Biosystems, Inc. (Chuzhou, China). They were digested with AgeI and then ligated into pEMM vector to replace the elongation factor 1α (EF-1α) promoter and thereby obtain the various vectors pApoAI-MM, pAFP-MM, pAAT-MM and pALB-MM respectively (Fig. 1C).
Fig. 1.
A schematic illustration of expression vectors employed in this study. The plasmids used in this study were based on pEMM (A and B), in which a MAR characteristic motif incorporating at the downstream of eGFP gene and a 770 bp MAR1 element flanking at the 3´end of the gene expression cassette driving by the constitutive EF-1α promoter. The liver tissue-specific vectors pLSP-MM was prepared (C) by replacing the EF-1α promoter with different LSPs, ApoAI, AFP, AAT and ALB, respectively. Incorporation of the different enhancer elements, JSRV1, JSRV2, CMV1, CMV2, ApoE1 and ApoE2, into the upstream of LSP to produce pE/LSP-MM vectors (D). p promoter, LSP liver tissue-specific promoter, eGFP enhanced green fluorescent protein, pA poly A
Six different enhancer sequences, JSRV1, JSRV2 (Yu et al. 2020), hCMV1 (GenBank: K03104.1, positions 214–620 bp), hCMV2 (positions 7–660 bp of pRL-CMV vector of Promega company), ApoE1 (GenBank: U32510.1) and ApoE2 (GenBank: U35114.1, positions 1 to 535), were artificially synthesized by General Biosystems, Inc. (Chuzhou, China). They were then cloned into the AAT promoter upstream of the selected pAAT-MM vector using Seamless Cloning and Assembly Kit (Tiandz, China) to produce enhancer-modified pAAT-MM vectors (Fig. 1D). The sequence alignment results of CMV1 and CMV2 enhancers are shown in Supplementary Fig. 2.
Cell culture
The human hepatocellular carcinoma cells HepG2, Huh-7, normal hepatic cells HL-7702 and the non-liver derived cells such as human colon cancer HCT116 and embryo kidney HEK-293E were provided by the Institute of Laboratory Animal Sciences (Beijing, China). They were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Proteineasy, Xinxiang, China) supplemented with 10% fetal bovine serum (Tianhang, Hangzhou, China) and 1% of a penicillin-streptomycin solution (Proteineasy, Xinxiang, China) and maintained in a humidified incubator at 37 °C with 5% CO2.
Transient and stable transfection
The exponentially growing cells were initially plated at a density of 1.25 × 105 cells/well in 24-well plates. After the cells have reached 70 ~ 80% confluence, duplicate transfections were performed for each vector using Lip2000 Transfection Reagent (Biosharp, Hefei, China). Non-treated and mock transfected cells (transfected without plasmid) were used as the controls. Transient eGFP expression was examined and validated under a Nikon Ti-s fluorescence microscope (Nikon, Tokyo, Japan) at 72 h after the transfection. Subsequently, the transient transfected cells were digested with 0.25% trypsin and collected for further analysis. For cell pools of stable HepG2-eGFP expression, cells were sub-cultured every three days till 24 days after the transfection. These cells were monitored under fluorescence microscope during the passaging and used for further analysis.
Flow cytometry analysis
The cells were collected after the pancreatic enzyme digestion, washed with PBS and then resuspended. The proportion of eGFP-positive cells and eGFP mean fluorescence intensity (MFI) of each sample was directly determined using a CytoFLEX Flow Cytometer (Beckman Coulter, Franklin Lakes, NJ, USA) at 72 h after the transfection. A total of 20,000 fluorescent events were acquired using a 530/15 bandpass filter for the green fluorescence protein signal obtained with a fluorescence emission wavelength at 530 nm. The results were finally analyzed using Flow Jo software (Tree Star, Ashland, OR, USA).
mRNA expression analysis
The total RNA was extracted from the transfected cells using TRIzol reagent (TaKaRa). The RNA (1 μg) was then reverse transcribed to cDNA with PrimeScript RT Master Mix (TaKaRa). Thereafter, cDNA (1 μL) was used as a template for PCR amplification with SYBR PCR Master Mix reagents (TaKaRa). The specific primers used were as following: eGFP, 5ʹ-CTACGTCCAGGAGCGCACCATCT-3ʹ and 5ʹ-GTTCTTCTGCTTGTCGGCCATGATAT-3ʹ; GAPDH, 5ʹ-CGACCCCTTCATTGACCTC-3ʹ and 5ʹ-CTCCACGACATACTCAGCACC-3ʹ. qPCR was carried out with ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) with an initial heating at 95 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s, respectively. The relative gene expression levels were calculated using the 2−ΔΔCT method with GAPDH as the internal reference gene. All the samples were used in duplicate, and the experiment was repeated twice.
Western blot analysis
The whole cell extracts were obtained by homogenization of the cells at 72 h after transient transfection with pE/AAT-MM plasmid (containing the different enhancer) in the cell lysate using Ultrasonic Cell Disruption System. The protein concentration was determined by Bicinchoninic Acid Assay (BCA) kit. Thereafter, equal amount of total protein was electrophoresed on 12% polyacrylamide gel (Beyotime, Shanghai, China) under denaturing conditions, and then transferred to a polyvinylidene fluoride membrane (Bio-Rad, USA). The transferred proteins were blocked in 5% defatted milk and then blotted with eGFP antibody (AbM59003-6E2-PU, BPI, China), or β-actin antibody (BM0627, Boster, China). The goat anti-mouse immunoglobulins (S0002, Affinity Biosciences, USA) were used as a secondary antibody. Finally, the membranes were treated with ECL luminescent reagent (Beyotime, China), and the bands were visualized in an ODYSSEY Fc (Li-COR) detector and quantified using Image J software (National Institutes of Health, Bethesda, MD, USA).
Plasmid rescue experiments
For these experiments, the total cellular DNA (including extrachromosomal DNA) was extracted using a Universal Genomic DNA Kit (CWBIO, China) from the stable HepG2-eGFP cell pools at 24 d after the transfection. The DNA (1 μg) were then transformed into DH5α E. coli cells by heat shock. The transformed colonies were selected on agar plates containing 30 μg/ml kanamycin. DNA was isolated from the individual resistant clones and quantified using a NanoDrop spectrophotometer (LabTech), subjected to PCR amplification of eGFP (200 ng) and analysed by electrophoresis on 1% agarose gels.
In silico analysis of the transcription factor binding sites
The potential allele-specific transcription factor binding sites (TFBSs) in the hCMV2 sequences were identified using bio-informatics software JASPAR (http://jaspar.genereg.net/) (Fornes et al. 2020; Castro-Mondragon et al. 2022), Alibaba2.1 (http://gene-regulation.com/pub/programs/alibaba2/index.html) (Grabe 2002; Shyamala et al. 2022), and Match (http://gene-regulation.com/cgi-bin/pub/programs/match/bin/match.cgi) (Kel et al. 2003; Tossolini et al. 2022). JASPAR: using the vertebrata database of jasper core and setting “Homo sapiens” for searching the different profiles (transcription factors). Thereafter, all the profile(s) identified were added to Cart and hCMV2 sequence (GenBank: K03104.1 from 2 to 656 bp) was scanned with selected matrix models. Thereafter, the total putative sites were predicted by applying a minimum relative profile score threshold of 80% to minimize false positive matches and the symbol “ + ” was entered into the filter box to obtain the predicted sequence of profiles present only on the forward strand. Alibaba2.1, setting with a minimum matrix conservation value of 80%; and Match, using a vertebrate matrix and a cut-off value set to minimize false positive matches.
Statistical analysis
All the data have been presented as mean. The statistically significant difference was determined by LSD (L) Multiple Comparison Test of one-way ANOVA using SPSS 17.0 GraphPad Prism 5 (GraphPad Software Inc., California, USA). p < 0.05 was considered as statistically significant, whereas p < 0.01 indicated extremely significant difference for all the data.
Results
AAT promoter is more active compared to ApoAI, AFP and ALB in hepatic cells
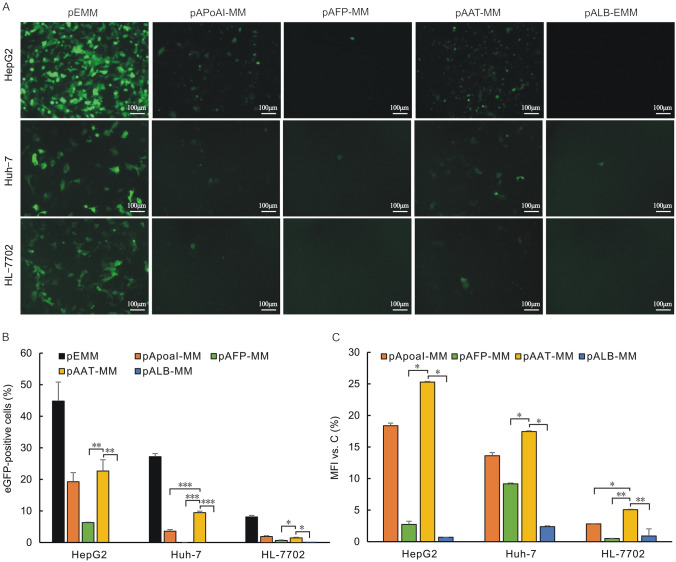
Four distinct LSPs, ApoAI, AFP, AAT and ALB, were evaluated for their effect as liver–specific episomal vectors. To explore the compatibility of pEMM eposimal vector with the different LSPs in hepatic cell lines, we first determined the eGFP expression levels by measuring the MFI and percentage of eGFP-positive cells of the transfected liver-derived cell lines. Flow cytometry analysis indicated that pAAT-MM displayed the highest eGFP expression levels in HepG2, Huh-7 and HL-7702 cells and approximately 25.27%, 17.43% and 5.06% of the control vector pEMM (LSP-devoid vector), followed by pApoAI (about 18.36%, 13.6%and 2.80%). However, the eGFP expression levels in AFP and ALB promoter groups were significantly weaker in comparison to of AAT promoter groups in HepG2 (p < 0.01), Huh-7 cells (p < 0.05) and HL-7702 cells respectively (p < 0.001) (Fig. 2A and C).
Fig. 2.
The eGFP expression levels of episomal vectors driven by different LSPs in the transfected liver cells. The vectors with LSPs, pApoAI-MM, pAFP-MM, pAAT-MM, pALB-MM, were transfected into HepG2, HL-7702 and Huh-7 human liver cells. The pEMM with EF-1α constitutive promoter (LSP-devoid) was also transfected as a potential control. The eGFP expression levels were detected under a fluorescence microscope with uniform exposure time 2 s at 72 h after transfection (A). The percentage of eGFP-positive cells and MFI were determined by flow cytometry (B and C). Relative MFI of eGFP expression were normalized to those which lacked LSP vector (pEMM). Significant differences are indicated by stars (*p < 0.05, **p < 0.01 and ***p < 0.001 relative to the pAAT-MM vector)
Synchronously, the percentage of eGFP-positive cells of the pAAT-MM vector was found to be at relatively high levels in liver cells and was approximately 22.6%, 9.4% and 1.5% in HepG2, Huh-7 as well as HL-7702 cells, respectively, and it was also increased significantly in comparison to pAFP and pALB groups in HepG2 (p < 0.05), Huh-7 (p < 0.05) and HL-7702 cells (p < 0.01) (Fig. 2A and B). Surprisingly, the eGFP transgene expressions were observed to be substantially attenuated in human normal hepatic cells HL-7702 in comparison to hepatocarcinoma cells HepG2 and Huh-7. These results suggested that this episomal vector may be more favorable for transgene therapy in hepatocellular carcinoma.
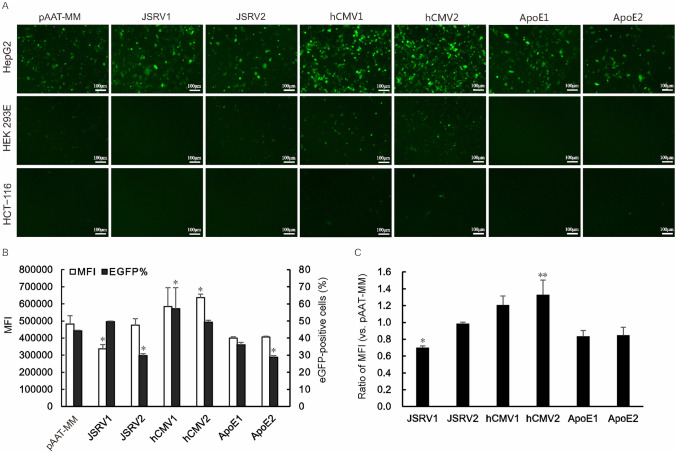
hCMV2/AAT promoter was found to be most active in hepatic cell lines, but also displayed weak expression levels in non-liver derived cells
To further enhance promoter activity, six different enhancer sequences were cloned in front of the AAT promoter sequence. The constructs featuring an hCMV enhancer, especially hCMV2, exhibited higher expression levels in comparison to others enhancer in HepG2 liver cells (Fig. 3A). The eGFP expression levels of the vectors containing hCMV2 enhancer were 1.33 fold and found to be significantly higher than that of the enhancer-devoid control vector (p < 0.01), followed by hCMV1 (1.21 fold). The percentages of eGFP-positive cells in hCMV1 groups were observed to be significantly higher than that of the control (57.2% vs. 44.3%, p < 0.05), followed by the hCMV2 (49.3%). Surprisingly, JSRV1 significantly decreased the eGFP expression level in HepG2 cells (p < 0.05). Except for these, other enhancers were comparable to MFI-eGFP or showed slightly lower activity in comparison to the devoid-enhancer vector (Fig. 3B and C). Similarly, the eGFP expression cells also displayed lower activity than the enhancer-devoid group (p < 0.05).
Fig. 3.
The eGFP expression levels of episomal vectors driven by enhancer/AAT promoter. The vectors incorporating the different enhancers, JSRV1, JSRV2, hCMV1, hCMV2, ApoE1, ApoE2, were transfected into HepG2 liver cells, non-liver derived HEK-293E and HCT-116 cells. The pAAT-MM (enhancer-devoid) was transfected as a control. The eGFP expression levels were detected under a fluorescence microscope with uniform exposure time 2 s (A) at 72 h after transfection. The percentage of eGFP-positive cells and MFI were determined by flow cytometry (B and C). Relative MFI of eGFP expression were normalized to those of enhancer-devoid vector (pEMM). Significant differences are indicated by stars (*p < 0.05, and **p < 0.01 relative to the pAAT-MM vector)
For tissue-specific expression, we detected synchronically the eGFP expression in the non-liver derived cell lines. Both the quantity and fluorescence intensity of eGFP-expression cells in HEK-293E and HCT-116 cells were significantly weaker in comparison to the liver derived HepG2 cells (Fig. 3A), which demonstrated that the CMV enhancer did not markedly affect the liver–tissue-specific of the pATT-MM episomal vector.
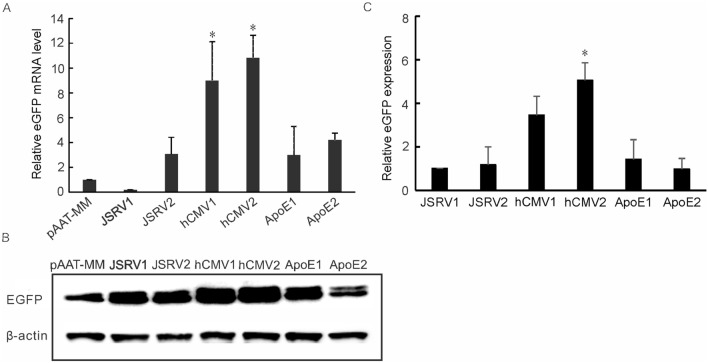
hCMV2/AAT promoter enhanced the eGFP expression levels of the MAR-based episomal vector
We further compared the eGFP expression levels in the transiently transfected cell pools of the episomal vectors incorporating into different enhancer elements. The results demonstrated that eGFP expression level of hCMV2 vector was substantially increased at the transcriptional level, and was approximately 10.8 fold compared to the control (AAT promoter without enhancer), followed by hCMV1 that was about 9.0 fold (p < 0.05) (Fig. 4A). Similarly, the eGFP protein in hCMV2 group was also exhibited high level and was about 5.1 fold which was found to be a significant difference in comparison to the control group (p < 0.05). This was followed by hCMV1 group (3.8 fold, p > 0.05) (Fig. 4B and C). However, other enhancer elements, JSRV1, JSRV2, ApoE1 and ApoE2, had little effect on the recombinant eGFP expression, with values approximately 1.0-, 1.2-, 1.4-and 1.0 fold compared to the control. These results suggested that hCMV2/AAT promoter in hepatic cells demonstrated positive effect on the efficiency of transgene expression.
Fig. 4.
EGFP expression in transiently transfected HepG2 cells with the episomal vectors incorporating into different enhancer elements. The vectors with the different enhancer elements, JSRV1, JSRV2, hCMV1, hCMV2, ApoE1 and ApoE2, were transfected into the HepG2 cells. The eGFP expression levels were detected by collecting the transfected cell pools at 72 h after the transfection. The pAAT-MM (enhancer-devoid) was used as a control vector. The relative eGFP mRNA expression level was measured by qRT-PCR using 2−∆∆Ct method, and GAPDH was as an internal reference gene (A). The eGFP protein expression levels were determined by Western blot analysis. The relative eGFP expression was normalized to those of non-containing enhancer pAAT-MM vector (pEMM) and β-actin was as an internal reference (B and C). Significant differences are indicated by stars (*p < 0.05 relative to the pAAT-MM control vector)
phCMV2/AAT-MM vector conferred persistent expression
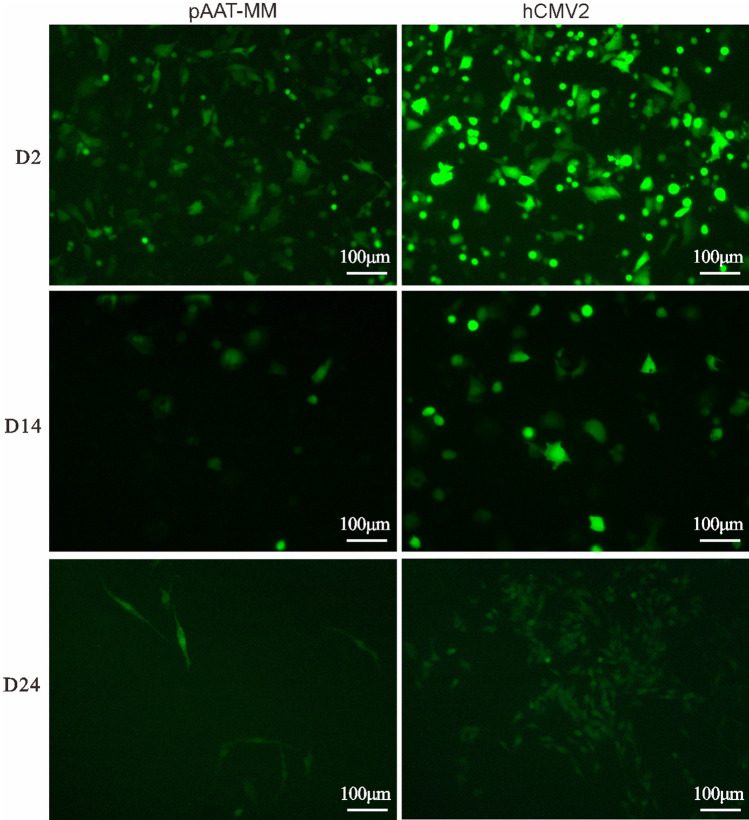
To further examine the effect of the hCMV2 enhancer element on the stability of transgene expression, the cells were cultured and passaged continuously until at the 24 days after transfection. A sustained eGFP expression as expected in phCMV2/AAT-MM-HepG2 was observed cell pools which was significantly greater markedly than that of the pAAT-MM control group (enhancer-devoid vector) at 14 and 24 days after the transfection. Although the hCMV2/AAT vectors retained transgene expression in the dividing cells, the fluorescence intensity of eGFP was found to be substantially attenuated (Fig. 5).
Fig. 5.
EGFP expression in cell pools of the stably transfected HepG2 cells with the episomal vectors mediated by hCMV2 enhancer element. For pools of stable HepG2 cells expressing eGFP, transient transfected cells were sub-cultured every there days till 24 days after the transfection. These transfected cell pools were photographed under a fluorescence microscope at 2, 14 and 24 days after the transfection
phCMV2/AAT-MM vector was retained extrachromosomally in stably transfected HepG2 cell pools
The cellular DNA extraction was performed to recover extrachromosomal DNA from HepG2 cell pools transfected with the phCMV2/AAT-MM vector. The plasmid rescue assay was conducted as described previously (Argyros et al. 2011) and the rescued plasmids were identified by PCR amplification of eGFP gene which was expected to yield 205 bp fragments (Fig. 6). These results demonstrated that the plasmid existed as circular episomes in liver cells and was not directly integrated into the genomic DNA of the host cell.
Fig. 6.
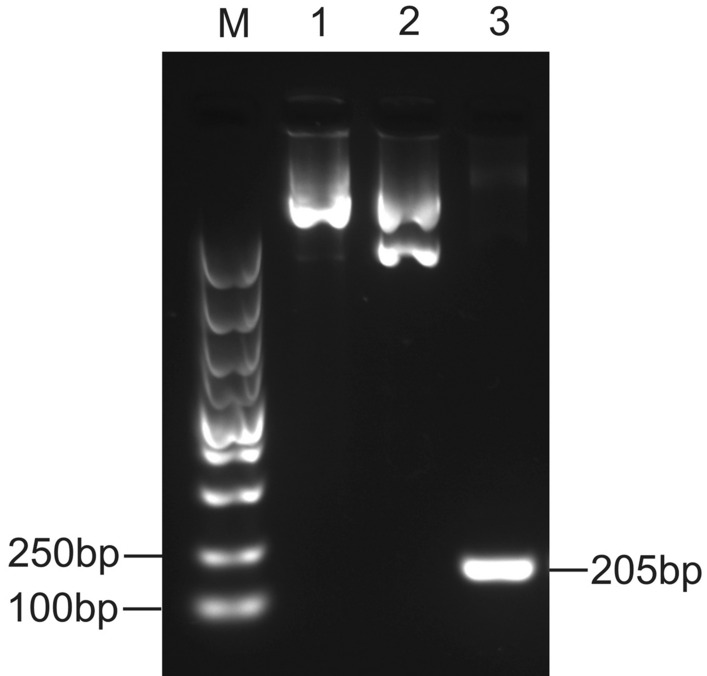
The determination of the episomal status of the pCMV2/AAT-MM plasmid in the transfected HepG2 cells. DNA was extracted from HepG2 cells transfected with pCMV2/AAT-MM. The extracted DNA was then used to transform DH5α E. coli cells by heat shock treatment. The transformants were selected and the plasmid DNA was prepared and subjected to PCR amplification. M: DL5000 marker; Lane 1, pCMV2/AAT-MM plasmid; Lane 2, DNA isolated from a single bacterial clone; Lane 3, PCR results obtained with eGFP primers from isolated extrachromosomal DNA which showed a 205-bp PCR product for eGFP
Transcription factor binding motifs of hCMV2 enhancer via in silico analysis
It has been established that enhancers can act as cis-regulatory elements to control transcriptional regulation by recruiting the DNA-binding transcription factors (TFs) in a tissue-specific manner. In the present study, the potential TFBSs of hCMV2 enhancer were determined using a bioinformatics approach. The results revealed that a total of 2467 predicted sequences of TFBSs were presumably present in the hCMV sequence. Based on the value of relative score item (> 0.99), we selected 28 predicted binding site sequence which have been referred as 19 TFs. Eleven TFs among all these regulatory elements had the highest score (1.00). These TFs included SRF, REL, CREB1, FOS::JUN, HOXA4, HOXB4, HOXC4, HOXD4, NR2C1, NFIC and GATA2 (Table 1). In addition, the scores of these TFBSs were > 10.00 except for NFIC and GATA2. Moreover, CREB1, NFIC and NFKB1 were identified several times (4, 5 and 3 times, respectively) at the different sites with identical predicted sequences. The findings indicate that these TFREs may have a synergistic effect on the contribution of enhance the transcriptional activity of exogenous genes.
Table 1.
Putative transcription factor binding sites of hCMV2 sequence
| TFs | Score | Relative score | Start | End | Predicted sequence |
|---|---|---|---|---|---|
| SRF | 20.00 | 1.00 | 195 | 206 | GCCCATATATGG |
| REL | 12.96 | 1.00 | 570 | 579 | GGGGATTTCC |
| CREB1 | 11.57 | 1.00 | 273 | 280 | TGACGTCA |
| 326 | 333 | TGACGTCA | |||
| 409 | 416 | TGACGTCA | |||
| 595 | 602 | TGACGTCA | |||
| FOS::JUN | 10.66 | 1.00 | 562 | 568 | TGACTCA |
| HOXB4 | 12.07 | 1.00 | 181 | 188 | GTCATTAG |
| HOXD4 | 11.31 | 1.00 | 181 | 188 | GTCATTAG |
| HOXA4 | 11.20 | 1.00 | 181 | 188 | GTCATTAG |
| HOXC4 | 11.15 | 1.00 | 181 | 188 | GTCATTAG |
| NR2C1 | 11.68 | 1.00 | 177 | 185 | CGGGGTCAT |
| NFIC | 9.70 | 1.00 | 136 | 141 | TTGGCA |
| 365 | 370 | TTGGCA | |||
| 478 | 483 | TTGGCA | |||
| 529 | 534 | TTGGCA | |||
| 616 | 621 | TTGGCA | |||
| GATA2 | 6.65 | 1.00 | 551 | 555 | GGATA |
| PAX3 | 16.93 | 0.99 | 167 | 176 | TAATCAATTA |
| NFKB1 | 14.55 | 0.99 | 314 | 323 | GGGACTTTCC |
| 465 | 474 | GGGACTTTCC | |||
| 633 | 642 | GGGACTTTCC | |||
| NFIC | 14.03 | 0.99 | 475 | 485 | TACTTGGCAGT |
| NFIA | 12.61 | 0.99 | 388 | 397 | TATGCCAAGT |
| GSX2 | 11.11 | 0.99 | 181 | 188 | GTCATTAG |
| NR2C2 | 9.94 | 0.99 | 178 | 185 | GGGGTCAT |
| ZNF354C | 8.72 | 0.99 | 584 | 589 | CTCCAC |
| ETS1 | 7.63 | 0.99 | 470 | 475 | TTTCCT |
TFBSs were predicted using JASPAR, Alibaba2.1 and Match database online software
Discussion
Liver-targeted episomal vectors for gene therapy possess broad application prospects for alternative protein replacement therapy of rare monogenic diseases and liver cancer. In a previous study, Miao et al. (2001) demonstrated that episomal maintenance of plasmids in the liver cells can lead to persistent gene expression without eliciting substantial long-term immune response or toxicity. We have previously designed a pEM episomal vector using MAR characteristic motif that exhibited high transgene efficiency and transgene stability in CHO-K1 cells (Wang et al. 2017), as well as in human Chang-liver cells (Xu et al. 2019; Wang et al. 2016). This could be attributed to the truncated MAR motif thus allowing reduced plasmid size which can further increase gene transfer efficiency.
For liver-targeted gene therapy, the expression of therapeutic genes in the non-liver organs is not expected and could be potentially eliminated by optimal use of LSPs. Interestingly, DNA vectors driven by AAT promoter in combination with the entire genomic loci or viral elements, have demonstrated that a longer duration of transgene expression could be possible in the liver (Aliño et al. 2003; Ehrhardt et al. 2003). For instance, Cabrera-Pérez et al. confirmed in a previous report that adeno-associated vector (AAV) transcriptionally targeted to liver under control of the AAT promoter was relatively more effective than that of liver-specific promoters TBG, HLP and constitutive PGK promoter (Cabrera-Pérez et al. 2019). In the current study, pAAT-EMM transfected HepG2 cells exhibited optimal eGFP expression level in our pEMM episomal vector system in comparison to others LSPs, such as ApoAI, AFP and ALB. In addition, liver-specificity was further confirmed which was consistent with the previous reports.
Tissue-specific promoters generally possess low transfection efficiency which is often less than 20% in comparison to the viral vector. Similarly, transient transfection efficiency of our pEMM episomal vector system driven by AAT promoter was approximately 22.6% and 9.4% in two human hepatocarcinoma cell lines, respectively. However, an ideal gene therapy primarily depends on the high transfection efficiency that can be achieved by selecting an optimal promoter/enhancer combination or modified by different chromatin modification elements such MARs. Interestingly, previous studies have demonstrated the transfection efficiency of GFP positive cells reached around 10.73% after transfection with MAR-based episomal vector driving by human AAT promoter and enhancer in Huh-7 human hepatic cells, whereas the vector lack of MAR displayed the values around 5.31% (Quiviger et al. 2018). In our MAR-based episomal vector, the percentage of eGFP positive cells displayed about 50 ~ 60% activity which was predominantly driven by the liver–tissue-specific CMV/AAT combination in HepG2 cells. Synchronously, the MFI of pCMV/AAT-MM-treated cells also increased significantly than that of the control group (lacking of CMV enhancer). This could be potentially attributed to the well-compatible hCMV/AAT promoter in our episomal vector system, especially hCMV enhancer incorporation to markedly increase the gene expression level and persistence. Surprisingly, Quiviger et al. (2018) also reported that MAR element that was introduced downstream of SV40 polyadenylation signal (the expression cassette) showed the lowest transfection efficiency (< 4%). This was inconsistent with the results of the previous studies (Jia et al. 2019) which suggested that the transgene expression level was significantly enhanced when incorporated a MAR1 sequences into the downstream of expression cassette of pEM episomal vector. Moreover, episomal vector system exhibited stronger transgene effect in human hepatocarcinoma cells, but not in the normal hepatic cells. These results may be more conducive to the transgenic treatment of liver cancer but have to be validated under in vivo settings. The actual reason still remains ambiguous and probably related to the configuration of particular DNA sequences or elements on the vector backbone.
It has been found that AAT promoter when combined with CMV enhancer can lead to higher eGFP expression levels than that of AAT promoter without this enhancer or other enhancers. The TFs in hepatic cells could essentially bind to the CMV/AAT promoter with high probabilities. Episomal plasmid DNAs can form nucleosomes (Reeves et al. 1985; Hebbar and Archer 2008) and DNA sequences that can regulate histone positioning to alter both the strength and longevity of target transgene expression obtained from plasmid DNAs (Nishikawa et al. 2003; Sumida et al. 2006; Kamiya et al. 2007, 2009; Fukunaga et al. 2012). Consequently, sustainable access of TFs to CMV/AAT promoter might maintain a histone positioning/modification status that could be vital for the continued access. Although, the nucleosome structures in plasmid DNAs exclusively containing AAT promoter and in genome are slightly different which might affect the histone status at/near the AAT promoter of our episomal vector, thereby resulting in the incompetent access of TFs and declined transgene express. Thus, through online bioinformatics analysis of the TFBSs between hCMV enhancer and TFs (Inukai et al. 2017), several TFs were identified that could be related to the regulation of transgenic expression such as SRF, REL and CREB1 etc., which need to be further verified by experimental analysis.
Overall, it was observed that hCMV/AAT promoter achieved better transgene expression rates, expression levels, as well as the stability in hepatocarcinoma cell lines. We have confirmed for the first time the usefulness of this promoter as a cis-regulatory element of liver targeting episomal vector. Nevertheless, only in-vitro studies were performed for the transgene expression of the episomal vectors and the effect should be evaluated under in vivo settings in our subsequent work. In addition, sustained transgenic expression could also be an important research focus of shared concern at both endogenously equivalent and clinically relevant levels for optimal application of clinical gene therapy.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Fig. 1: The eGFP expression levels of MAR-based episomal vectors driven by EF-1α constitutive promoter in the transfected HepG2 cells. The pEGFP-C1 vector exploited a 387 bp characteristic motif of 2200 bp MAR is to produce an episomal vector pEM driven by the constitutive promoter EF-1α. pEMM employed a 791 bp MAR1 element incorporating into pEM at the 3´end of eGFP expression cassette. A The plasmids of the two vectors were transfected into HepG2 cells respectively and the eGFP expression levels were detected under a fluorescence microscope with uniform exposure time 2S at 48 h after the transfection. B MFI were determined by flow cytometry and relative eGFP MFI levels were normalized in comparison to that of control vector (pEM). Significant differences are indicated by stars (*p < 0.05) (JPG 3071 KB)
Supplementary Fig. 2: The sequence alignment results of CMV1 and CMV2 enhancers. The size of CMV1 and CMV2 enhancers were derived from the enhancer of human cytomegalovirus major immediate-early gene (GenBank: K03104.1) and pRL-CMV vector of Promega company, which were 407 and 654 bp in size, respectively. The CMV2 contains the CMV1 sequence, but with a three-base mutation as shown in the red font (JPG 1973 KB)
Acknowledgements
This work was funded by Natural Science Foundation of Henan Province of China (212300410384, 232300421115), and Scientific and Technological Research Project of Henan Province of China (212102310318, 212102310651 and 222102110118)
Author contributions
JZ designed, analyzed the experiments and wrote the manuscript. TW contributed to revising the manuscript, and supervising the work. JZ, XW, CZ, CM, SG and YT helped revise this article. All authors read and approved the final manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author, [Tianyun Wang], upon reasonable request.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Aliño SF, Crespo A, Dasí F. Long-term therapeutic levels of human alpha-1 antitrypsin in plasma after hydrodynamic injection of nonviral DNA. Gene Ther. 2003;10(19):1672–1679. doi: 10.1038/sj.gt.3302065. [DOI] [PubMed] [Google Scholar]
- Argyros O, Wong SP, Niceta M, Waddington SN, Howe SJ, Coutelle C, Miller AD, Harbottle RP. Persistent episomal transgene expression in liver following delivery of a scaffold/matrix attachment region containing non-viral vector. Gene Ther. 2008;15(24):1593–1605. doi: 10.1038/gt.2008.113. [DOI] [PubMed] [Google Scholar]
- Argyros O, Wong SP, Fedonidis C, Tolmachov O, Waddington SN, Howe SJ, Niceta M, Coutelle C, Harbottle RP. Development of S/MAR minicircles for enhanced and persistent transgene expression in the mouse liver. J Mol Med. 2011;89(5):515–529. doi: 10.1007/s00109-010-0713-3. [DOI] [PubMed] [Google Scholar]
- Bode J, Benham C, Knopp A, Mielke C. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements) Crit Rev Eukaryot Gene Expr. 2000;10:73–90. doi: 10.1615/CritRevEukarGeneExpr.v10.i1.90. [DOI] [PubMed] [Google Scholar]
- Cabrera-Pérez R, Vila-Julià F, Hirano M, Mingozzi F, Torres-Torronteras J, Martí R. Alpha-1-antitrypsin promoter improves the efficacy of an adeno-associated virus vector for the treatment of mitochondrial neurogastrointestinal encephalomyopathy. Hum Gene Ther. 2019;30(8):985–998. doi: 10.1089/hum.2018.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Berhanu Lemma R, Turchi L, Blanc-Mathieu R, Lucas J, Boddie P, Khan A, Manosalva Pérez N, Fornes O, Leung TY, Aguirre A, Hammal F, Schmelter D, Baranasic D, Ballester B, Sandelin A, Lenhard B, Vandepoele K, Wasserman WW, Parcy F, Mathelier A. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50(D1):D165–D173. doi: 10.1093/nar/gkab1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone V, Cortese R. A negative regulatory element in the promoter of the human alpha 1-antitrypsin gene. Nucleic Acids Res. 1989;17(22):9407–9415. doi: 10.1093/nar/17.22.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Geest B, Van Linthout S, Lox M, Collen D, Holvoet P. Sustained expression of human apolipoprotein A-I after adenoviral gene transfer in C57BL/6 mice: role of apolipoprotein A-I promoter, apolipoprotein A-I introns, and human apolipoprotein E enhancer. Hum Gene Ther. 2000;11:101–112. doi: 10.1089/10430340050016193. [DOI] [PubMed] [Google Scholar]
- Ding B, Li T, Zhang J, Zhao L, Zhai G. Advances in liver-directed gene therapy for hepatocellular carcinoma by non-viral delivery systems. Curr Gene Ther. 2012;12(2):92–102. doi: 10.2174/156652312800099625. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A, Peng PD, Xu H, Meuse L, Kay MA. Optimization of cis-acting elements for gene expression from nonviral vectors in vivo. Hum Gene Ther. 2003;14:215–225. doi: 10.1089/10430340360535779. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A, Haase R, Schepers A, Deutsch MJ, Lipps HJ, Baiker A. Episomal vectors for gene therapy. Curr Gene Ther. 2008;8(3):147–161. doi: 10.2174/156652308784746440. [DOI] [PubMed] [Google Scholar]
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, Santana-Garcia W, Tan G, Chèneby J, Ballester B, Parcy F, Sandelin A, Lenhard B, Wasserman WW, Mathelier A. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48(D1):D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga S, Kanda G, Tanase J, Harashima H, Ohyama T, Kamiya H. A designed curved DNA sequence remarkably enhances transgene expression from plasmid DNA in mouse liver. Gene Ther. 2012;19:828–835. doi: 10.1038/gt.2011.127. [DOI] [PubMed] [Google Scholar]
- Giannakopoulos A, Stavrou EF, Zarkadis I, Zoumbos N, Thrasher AJ, Athanassiadou A. The functional role of S/MARs in episomal vectors as defined by the stress-induced destabilization profile of the vector sequences. J Mol Biol. 2009;387(5):1239–1249. doi: 10.1016/j.jmb.2009.02.043. [DOI] [PubMed] [Google Scholar]
- Grabe N. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. 2002;2(1):S1–S15. [PubMed] [Google Scholar]
- Haase R, Magnusson T, Su B, Kopp F, Wagner E, Lipps H, Baiker A, Ogris M. Generation of a tumor- and tissue-specific episomal non-viral vector system. BMC Biotechnol. 2013;13:49. doi: 10.1186/1472-6750-13-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbar PB, Archer TK. Altered histone H1 stoichiometry and an absence of nucleosome positioning on transfected DNA. J Biol Chem. 2008;283:4595–4601. doi: 10.1074/jbc.M709121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YR, Ren XL, Wang H, Ma Y, Wang L, Shen YY, Oka K, Zhang ZZ, Zhang Y. Liver-specific expression of an exogenous gene controlled by human apolipoprotein A-I promoter. Int J Pharmacol. 2010;398(1–2):161–164. doi: 10.1016/j.ijpharm.2010.07.023. [DOI] [PubMed] [Google Scholar]
- Inukai S, Kock KH, Bulyk ML. Transcription factor-DNA binding: beyond binding site motifs. Curr Opin Genetics Dev. 2017;43:110–119. doi: 10.1016/j.gde.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Juranek S, Lipps HJ. Designing nonviral vectors for efficient gene transfer and long-term gene expression. Mol Ther. 2006;14:613–626. doi: 10.1016/j.ymthe.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Jia YL, Guo X, Ni TJ, Lu JT, Wang XY, Wang TY. Novel short synthetic matrix attachment region for enhancing transgenic expression in recombinant Chinese hamster ovary cells. J Cell Biochem. 2019;120:18478–18486. doi: 10.1002/jcb.29165. [DOI] [PubMed] [Google Scholar]
- Kamiya H, Fukunaga S, Ohyama T, Harashima H. The location of the left-handedly curved DNA sequence affects exogenous DNA expression in vivo. Arch Biochem Biophys. 2007;461:7–12. doi: 10.1016/j.abb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Kamiya H, Goto H, Harashima H. Effects of non-B DNA sequences on transgene expression. J Biosci Bioeng. 2009;108:20–23. doi: 10.1016/j.jbiosc.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Kel AE, Gössling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31(13):3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PY, Sia KC, Khong JH, Geest B, Lim KS, Ho IA, Wang GY, Miao L, Huynh H, Hui KM. An efficient and safe herpes simplex virus type 1 amplicon vector for transcriptionally targeted therapy of human hepatocellular carcinomas. Mol Ther. 2007;15(6):1129–1136. doi: 10.1038/sj.mt.6300165. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang XY, Zhao CP, Tian ZW, Xu DH, Wang TY, Zhang JH. Effects of different promoters and MAR combinations on transgene expression of recombinant CHO cells. Sichuan Da Xue Xue Bao Yi Xue Ban. 2018;49(1):18–23. [PubMed] [Google Scholar]
- Lin Y, Li Z, Wang T, Wang X, Wang L, Dong W, Jing C, Yang X. MAR characteristic motifs mediate episomal vector in CHO cells. Gene. 2015;559(2):137–143. doi: 10.1016/j.gene.2015.01.032. [DOI] [PubMed] [Google Scholar]
- Maestro S, Weber ND, Zabaleta N, Aldabe R, Gonzalez-Aseguinolaza G. Novel vectors and approaches for gene therapy in liver diseases. JHEP Reports. 2021;3(4):100300. doi: 10.1016/j.jhepr.2021.100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao CH, Thompson AR, Loeb K, Ye X. Long-term and therapeutic-level hepatic gene expression of human factor IX after naked plasmid transfer in vivo. Mol Ther. 2001;3(6):947–957. doi: 10.1006/mthe.2001.0333. [DOI] [PubMed] [Google Scholar]
- Mulia GE, Picanço-Castro V, Stavrou EF, Athanassiadou A, Figueiredo ML. Advances in the development and the applications of nonviral, episomal vectors for gene therapy. Hum Gene Ther. 2021;32(19–20):1076–1095. doi: 10.1089/hum.2020.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa J, Amano M, Fukue Y, Tanaka S, Kishi H, Hirota Y, Yoda K, Ohyama T. Left-handedly curved DNA regulates accessibility to cis-DNA elements in chromatin. Nucleic Acids Res. 2003;31:6651–6662. doi: 10.1093/nar/gkg854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes FA, Raper SE. Liver-directed gene therapy. Med Clin North Am. 1996;80(5):1201–1213. doi: 10.1016/S0025-7125(05)70486-7. [DOI] [PubMed] [Google Scholar]
- Quiviger M, Giannakopoulos A, Verhenne S, Marie C, Stavrou EF, Vanhoorelbeke K, Izsvák Z, De Meyer SF, Athanassiadou A, Scherman D. Improved molecular platform for the gene therapy of rare diseases by liver protein secretion. Eur J Med Genet. 2018;61(11):723–728. doi: 10.1016/j.ejmg.2018.04.010. [DOI] [PubMed] [Google Scholar]
- Reeves R, Gorman CM, Howard B. Minichromosome assembly of non-integrated plasmid DNA transfected into mammalian cells. Nucleic Acids Res. 1985;1:3599–3615. doi: 10.1093/nar/13.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamala N, Kongettira CL, Puranam K, Kupsal K, Kummari R, Padala C, Hanumanth SR. In silico identification of single nucleotide variations at CpG sites regulating CpG island existence and size. Sci Rep. 2022;12(1):3574. doi: 10.1038/s41598-022-05198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumida N, Nishikawa J, Kishi H, Amano M, Furuya T, Sonobe H, Ohyama T. A designed curved DNA segment that is a remarkable activator of eukaryotic transcription. FEBS J. 2006;273:5691–5702. doi: 10.1111/j.1742-4658.2006.05557.x. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Wakao Y, Goda T, Kamiya H. Conventional plasmid DNAs with a CpG-containing backbone achieve durable transgene expression in mouse liver. J Gene Med. 2020;22(1):e3138. doi: 10.1002/jgm.3138. [DOI] [PubMed] [Google Scholar]
- Tossolini I, Gugliotta A, López Díaz F, Kratje R, Prieto C. Screening of CHO-K1 endogenous promoters for expressing recombinant proteins in mammalian cell cultures. Plasmid. 2022;119–120:102620. doi: 10.1016/j.plasmid.2022.102620. [DOI] [PubMed] [Google Scholar]
- Wang TY, Zhang JH, Jing CQ, Yang XJ, Lin JT. Positional effects of the matrix attachment region on transgene expression in stably transfected CHO cells. Cell Biol Int. 2010;34(2):141–145. doi: 10.1042/CBI20090017. [DOI] [PubMed] [Google Scholar]
- Wang F, Wang TY, Tang YY, Zhang JH, Yang XJ. Different matrix attachment regions flanking a transgene effectively enhance gene expression in stably transfected Chinese hamster ovary cells. Gene. 2012;500(1):59–62. doi: 10.1016/j.gene.2012.03.049. [DOI] [PubMed] [Google Scholar]
- Wang TY, Wang L, Yang YX, Zhao CP, Jia YL, Li Q, Zhang JH, Peng YY, Wang M, Xu HY, Wang XY. Cell compatibility of an eposimal vector mediated by the characteristic motifs of matrix attachment regions. Curr Gene Ther. 2016;16(4):271–277. doi: 10.2174/1566523216666161202160936. [DOI] [PubMed] [Google Scholar]
- Wang X, Xu Z, Tian Z, Zhang X, Xu D, Li Q, Zhang J, Wang T. The EF-1alpha promoter maintains high-level transgene expression from episomal vectors in transfected CHO-K1 cells. J Cell Mol Med. 2017;21(11):3044–3054. doi: 10.1111/jcmm.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZL, Mizuguchi H, Ishii-Watabe A, Uchida E, Mayumi T, Hayakawa T. Optimization of transcriptional regulatory elements for constructing plasmid vectors. Gene. 2001;272:149–156. doi: 10.1016/S0378-1119(01)00550-9. [DOI] [PubMed] [Google Scholar]
- Xu ZJ, Jia YL, Wang M, Yi DD, Zhang WL, Wang XY, Zhang JH. Effect of promoter, promoter mutation and enhancer on transgene expression mediated by episomal vectors in transfected HEK293, change liver and primary cells. Bioengineered. 2019;10(1):548–560. doi: 10.1080/21655979.2019.1684863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DL, Chow N, Wootton SK. JSRV intragenic enhancer element increases expression from a heterologous promoter and promotes high level AAV-mediated transgene expression in the lung and liver of mice. Viruses. 2020;12(11):1266. doi: 10.3390/v12111266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang J, Cheng S, Yang W, Li S. Enhanced transgene expression using two β-globin MARs flanking expression cassettes in stably transfected CHO-K1 cells. 3 Biotech. 2019;9(11):435. doi: 10.1007/s13205-019-1971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CP, Guo X, Chen SJ, Li CZ, Yang Y, Zhang JH, Chen SN, Jia YL, Wang TY. Matrix attachment region combinations increase transgene expression in transfected Chinese hamster ovary cells. Sci Rep. 2017;7:42805. doi: 10.1038/srep42805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1: The eGFP expression levels of MAR-based episomal vectors driven by EF-1α constitutive promoter in the transfected HepG2 cells. The pEGFP-C1 vector exploited a 387 bp characteristic motif of 2200 bp MAR is to produce an episomal vector pEM driven by the constitutive promoter EF-1α. pEMM employed a 791 bp MAR1 element incorporating into pEM at the 3´end of eGFP expression cassette. A The plasmids of the two vectors were transfected into HepG2 cells respectively and the eGFP expression levels were detected under a fluorescence microscope with uniform exposure time 2S at 48 h after the transfection. B MFI were determined by flow cytometry and relative eGFP MFI levels were normalized in comparison to that of control vector (pEM). Significant differences are indicated by stars (*p < 0.05) (JPG 3071 KB)
Supplementary Fig. 2: The sequence alignment results of CMV1 and CMV2 enhancers. The size of CMV1 and CMV2 enhancers were derived from the enhancer of human cytomegalovirus major immediate-early gene (GenBank: K03104.1) and pRL-CMV vector of Promega company, which were 407 and 654 bp in size, respectively. The CMV2 contains the CMV1 sequence, but with a three-base mutation as shown in the red font (JPG 1973 KB)
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, [Tianyun Wang], upon reasonable request.