Abstract
Gluten consumption can lead to severe health conditions in certain individuals, and following a strict gluten-free diet is often the only effective treatment option. Therefore, it is crucial to develop a gluten detection method that is accurate, sensitive, and specific to ensure the absence of gluten. An important aspect of developing effective gluten detection tests is the implementation of an efficient gluten extraction method. In this study, we conducted an evaluation of various buffer conditions for gliadin extraction from both heat-treated and non-heat-treated food samples. These buffer conditions included ethanol, 2-mercaptoethanol, guanidine hydrochloride, detergents, chelating agents, and deep eutectic solvents. Among the tested conditions, a combination of 2-mercaptoethanol and guanidine hydrochloride demonstrated significantly higher extraction efficacy compared to most other conditions. Furthermore, we explored the use of a less toxic extraction buffer, choline chloride, which exhibited a 1.4-fold higher extraction efficiency than the combination of 2-mercaptoethanol and guanidine hydrochloride (p < 0.05). Choline chloride showed great potential as a preferred buffer for commercial gliadin extraction kits, suitable for both heat-treated and non-heat-treated food samples. Overall, our findings highlight the importance of optimizing the gluten extraction process to improve the accuracy and reliability of gluten detection methods, ultimately contributing to the development of effective tools for individuals following a strict gluten-free diet.
Keywords: ELISA, Gluten, Gliadin extraction, Protein detection, Sample preparation
1. Introduction
Gluten, found in certain cereal grains, is a complex network of seed storage proteins. The majority of protein content in these grains is composed of gluten (approximately 85–90% in wheat kernels) [1,2]. Gluten primarily consists of gliadin and glutenin, which are categorized as prolamins—a class of water-insoluble proteins. Prolamins, including gliadin, hordein (in barley), and secalin (in rye), exhibit high levels of proline and glutamine amino acid residues and can be extracted using 60–70% ethanol. Notably, hordein and secalin, like gliadin, possess elevated levels of proline and amide nitrogen compared to the glutenin fraction of prolamins. Gliadins can be classified into alpha, beta, gamma, and omega (α, β, γ, and ω) structures based on their primary composition. The ω-gliadin contains significant cysteine levels. Cysteine is involved in the formation of disulphide bonds within or between proteins when oxidized. These disulphide crosslinks between gliadins are crucial for gluten polymerization and the baking process of bread [2,3]. Through covalent and non-covalent interactions, monomeric gliadins bind together. Due to these interactions, gluten is heat-stable and commonly used as a binding and extending agent in processed foods for improved texture, flavor, viscosity, and moisture retention [1].
Certain populations can develop specific health conditions as a result of gluten consumption. These conditions include gluten intolerance, autoimmune celiac disease (CD), wheat allergy, and non-celiac gluten sensitivity. CD is characterized by inflammation in the upper small intestine triggered by gluten intake in genetically susceptible individuals [1,4]. In the United States, CD is highly prevalent, affecting approximately 1% of the population [5]. Symptoms of CD include weight loss due to impaired nutrient absorption in the intestine, diarrhea, anemia resulting from inadequate iron absorption, and bone pain due to calcium malabsorption [5,6]. Gliadins, primarily due to their epitopes containing glutamine and proline residues that resist digestion by gastric, pancreatic, and intestinal proteolytic enzymes, are the major contributors to gluten-related disorders [1]. Unfortunately, currently there is no effective treatment for gluten intolerance-related disorders other than following strict gluten-free diet [7,8].
Food products are considered gluten-free according to the U.S. Food and Drug Administration (FDA) and the international standard, Codex Alimentarius, if they contain less than 20 parts per million (ppm) of gluten [9]. Foods such as bread, pasta, biscuits, cakes, breakfast cereals, soups, and bagels prepared using wheat, barley, and rye typically contain gluten [7]. Cross-contamination of gluten is a significant concern in other types of food due to the shared use of equipment with gluten-containing foods. Challenges in following a strict gluten-free diet arise from insufficient education, a lack of highly sensitive and specific gluten detection methods, and inadequate labeling [8].
To enhance the quality of life for individuals affected by gluten intolerance, there is an urgent need to implement precise, sensitive, and specific gluten detection tests [7,10]. Immune-based detection tests are commonly favored for gluten testing in food due to their simplicity, rapidity, sensitivity, and specificity [8]. Several commercially available enzyme-linked immunosorbent assays (ELISA) have been developed for gluten detection, utilizing different antibodies, such as polyclonal (pAb) or monoclonal (mAb), thereby exhibiting varying sensitivities and specificities [11]. However, a comparison of 14 commercially available ELISA kits demonstrated their inability to accurately quantify gluten across a wide range of food matrices, including the widely accepted R5 ELISA test endorsed by Codex Alimentarius [12]. This lack of accuracy in gluten testing and quantification is likely attributed to the inefficiency of gluten extraction during the sample preparation stage. Since gluten is insoluble in water, heat-stable, and capable of forming both covalent and non-covalent interactions within the food matrix, it is crucial to develop an effective method for gluten extraction.
The conventional method for gluten extraction typically involves the use of 60% ethanol. Another commercially employed extraction method suggests the use of a cocktail solution, which contains 2 M guanidine hydrochloride as a disaggregating agent and 250 mM 2-mercaptoethanol as a reducing agent. These agents aid in breaking down gliadin homopolymers and heteropolymers, thereby exposing more epitopes for antibody binding. Additionally, this cocktail solution has been found to be more effective than 60% ethanol in recovering gliadin from various heat-treated or heat-untreated food samples [13]. Another recently introduced method, as described by Hnasko et al. [14], involves the extraction of gliadin prior to a lateral flow immunoassay (LFIA). This method utilizes a non-ionic detergent, 1% Triton X-100, diluted in phosphate buffer for rapid gliadin extraction from different food matrices specifically for LFIA application [14]. Furthermore, a sustainable approach was explored by Svigelj et al. [15], involving the use of a green extraction buffer composed of deep eutectic solvents (DESs) for gluten extraction. DESs are biodegradable and have low toxicity. In this application, DESs like ethaline and reline demonstrated the ability to extract gliadin protein from both heat-treated and untreated food samples as efficiently as the conventional 60% ethanol solution, with results compatible with a commercial ELISA kit [15].
In this study, we aimed to address the ongoing need for optimizing gluten extraction methods. Specifically, we compared the efficiency of gliadin extraction using the previously mentioned methods and explored alternative extraction approaches. Furthermore, we assessed the compatibility of these extraction methods with the subsequent sandwich ELISA analysis.
2. Materials & method
2.1. Determination of gluten concentration
The amount of gliadin in samples and the efficacy of the extraction methods are assessed using the in-house sandwich ELISA method. For the development of the sandwich ELISA, commercial antibodies, an anti-gliadin mouse monoclonal antibody (SAB42000864) and HRP-conjugated polyclonal antibody (Cat. No. A1052; Sigma-Aldrich, USA), are used. The sensitivity and specificity of these antibodies are initially assessed by direct and indirect ELISA by coating 96-well microtiter plates with gliadin (Cat. No. G3375; Sigma-Aldrich, USA). Since gliadin is an alcohol-soluble protein, gliadin stock solution is prepared in 70% ethanol and then diluted in a coating buffer. For coating buffer, we compared 70% ethanol, carbonate-bicarbonate buffer (0.1 M, pH 9.6), and a combination of both 70% ethanol and carbonate-bicarbonate buffer in direct ELISA. As the application of combination yielded superior results compared to the other two conditions (Fig. 1), this composition is used in subsequent ELISA experiments. For optimization of ELISA, two different compositions of blocking buffer (1% BSA (bovine serum albumin) (Sigma-Aldrich, USA) and SuperBlockTM (Cat. No. 37515; Thermo Scientific, USA) and three different starting gliadin concentrations (5, 10 and 20 μg/mL) in both direct and indirect ELISA are also tested.
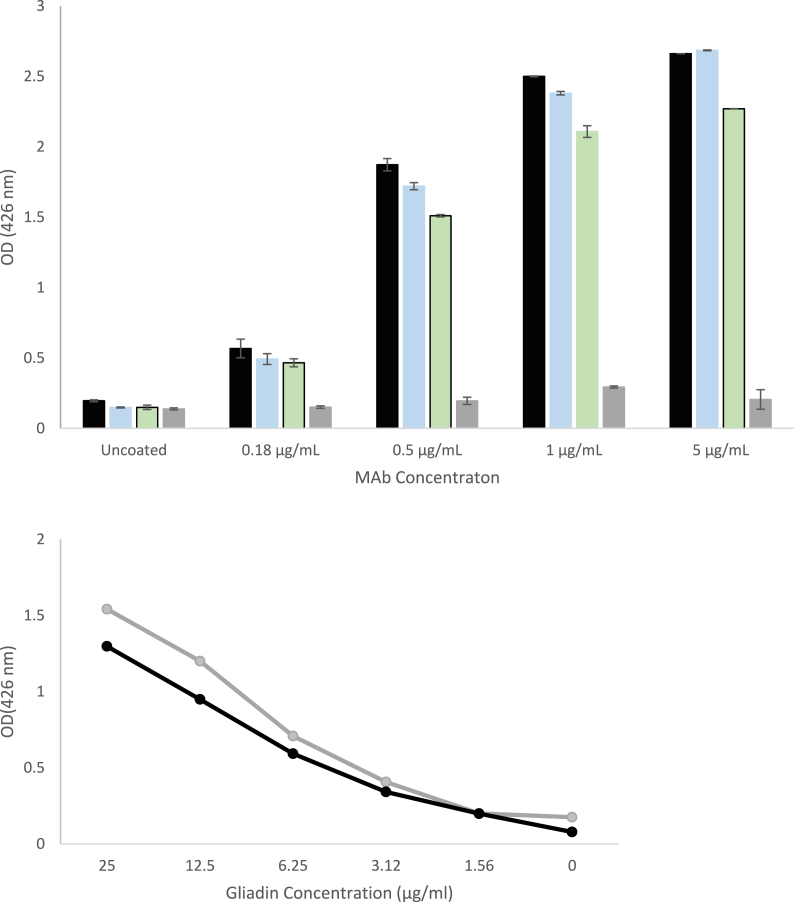
Fig. 1.
Optimization of direct ELISA. A. Three coating buffers (70% ethanol, carbonate-bicarbonate buffer, and 70% ethanol + carbonate-bicarbonate buffer) and two blocking buffer conditions (1% BSA and SuperBlock) are tested in combinations to find the optimum ELISA condition. The signal intensities were read at 426 nm. 1 (●): 70% ethanol + SuperBlock, 2 (▲): 70% ethanol + BSA, 3(■): carbonate-bicarbonate buffer + SuperBlock, 4 (▬): carbonate-bicarbonate buffer + BSA, 5 (♦): 70% ethanol + carbonate-bicarbonate buffer + SuperBlock, 6 (X): 70% ethanol + carbonate-bicarbonate buffer + BSA. B. Optimization of gliadin coating on 96-well plates: 20 μg/mL gliadin (black), 10 μg/mL gliadin (blue) and 5 μg/mL gliadin (green).
In the development of the sandwich ELISA, 96-well microtiter plates are coated with 50 μL of 1 μg/mL anti-gliadin mouse monoclonal antibody (SAB42000864) and incubated overnight at 4 °C. The plates are then washed three times with 250 μL of PBS-T (0.05% Tween20) and blocked with 250 μL of Superblock™, followed by an overnight incubation at 4 °C. For sample and standard addition, 50 μL of each sample and gliadin standards are added to their respective wells, followed by a 1-h incubation at room temperature. The reaction buffer in the sandwich ELISA is a 1:1 combination of PBS-T and Superblock™. Gliadin standards are diluted in the reaction buffer, while test samples undergo a food extraction procedure. After the sample incubation, the plates are washed three times with 250 μL of PBS-T. Next, 50 μL of a 1:1000 dilution of rabbit-anti-wheat HRP-conjugated polyclonal antibody (A1052) in reaction buffer is added to each well. The plates are incubated for 1 h at room temperature, followed by three washes with PBS-T. To initiate color development, 50 μL of the substrate, 3,3′,5,5′-Tetramethylbenzidine (TMB; Sigma-Aldrich, USA) is added to each well. The plates are incubated for approximately 15 min at room temperature. The reaction is stopped by adding 50 μL of a 1:10 dilution of sulfuric acid to each well. Finally, the plates are read at 426 nm using a microplate reader (SpectraMax iD3) to measure the absorbance and quantify the gliadin concentration in the samples. The in-house sandwich ELISA is compared with commercial R5 ELISA kit (R5 Codex; INgezim 30.GLH.K2) to assess its efficacy to detect gliadin.
2.2. Sample preparation and gluten extraction methods
Heat treatment of foods can cause gluten aggregation and reduce protein solubility [16]. Therefore, it is crucial to assess both heat-treated and heat-untreated samples to evaluate the effectiveness of gluten extraction protocols. In this study, we selected unbleached wheat flour as a representative heat-untreated sample and croutons as a representative heat-treated sample. To establish a reference method, we employed the gluten extraction protocol described by García et al. [13]. Additionally, we developed and tested derivatives of this protocol along with other extraction methods to assess their efficacy in gluten extraction. The efficiency of these extraction methods is evaluated using the sandwich ELISA method explained earlier.
2.2.1. Gliadin extraction method using 2-mercaptoethanol and guanidine hydrochloride
Briefly, 0.25 g of powdered food sample is weighed and added into a 15 mL polypropylene tube. Then, 2.5 mL extraction buffer is added and samples are vortexed for 5–10 s. Following 45 min of incubation at room temperature (Incubation-1), 7.5 mL of 80% ethanol is added and the sample is vortexed for up to 60 s. After 1-h incubation (Incubation-2), 600 μL of each sample is transferred to a vial and centrifuged at 2500 g for 5 min at room temperature [13]. Finally, the supernatants (∼400 μL) are transferred into clean vials and 1:10 dilution of each sample is tested in subsequent ELISA tests.
2.2.2. Impact of incubation time in gluten extraction
The length of exposure to extraction buffer and ethanol could impact the efficacy of the extraction of gluten. In the above-mentioned gluten extraction protocol, there are two incubation steps: Incubation-1 (with extraction buffer) and Incubation-2 (with ethanol). To evaluate the effect of incubation times with extraction buffer and ethanol, we evaluated 0, 15, and 30 min as alternatives for the 45 min (Incubation-1); and 5, 15, and 30 min as alternatives for the 1 h (Incubation-2) incubation steps.
2.2.3. Optimization of extraction buffer
The extraction buffer suggested by García et al. [13] contains; 250 mM 2-mercaptoethanol (Cat. No. 805740; Sigma-Aldrich, Germany) and 2 M guanidine hydrochloride (Cat. No. 59950; Sigma-Aldrich, Germany) (‘cocktail’), and this extraction buffer is recommended for the extraction of gliadin from heat-treated samples. In this study, the impact of the concentration of each component of this extraction buffer is also evaluated. Briefly, the gliadin extraction capacity of 2-mercaptaethanol (50 mM and 250 mM), guanidine hydrochloride (1 M and 2 M), and the combinations of the two chemicals with different concentrations are compared. We tested the efficacy of each of the above-mentioned buffer conditions for the extraction of gliadin in wheat flour and crouton.
2.2.4. Evaluation of detergents for extraction of gluten from food samples
As alternatives to the 50 mM 2-mercaptoethanol and 2 M guanidine hydrochloride, we also test a chelating agent, ethylenediaminetetraacetic acid (EDTA) (EDS, Germany), and three detergents (Tween® 20, Tween® 80 (P1754, France), and Triton™ X-100 (×100, USA)) for extraction of gliadin. These detergents and chelating agents are diluted in either PBS or 80% ethanol to a final concentration of 1%. These alternatives are tested using the above-mentioned extraction protocol, except that Incubation-1 is absent. The solutions and the samples are directly added into the corresponding tubes for 1-h incubation (Incubation-2).
2.2.5. Rapid gliadin extraction method using Triton X-100
In this extraction method, 0.1 g of sample is added into 4 mL of 1% Triton-X 100 dissolved in 0.2 M phosphate buffer. After the solution is mixed, the sample is incubated for 1 min [14] and the sandwich ELISA described above is performed to determine the gliadin concentration.
2.2.6. Gliadin extraction method with pure choline chloride-based deep eutectic solvents (ChCl-DESs)
For the ChCl-DESs-based method, we follow the protocol described by Svigelj et al. [15]. Briefly, 3.5 mL of the ChCI-DES solution is added into a tube and mixed with 0.35 g of the milled sample of wheat flour and crouton. Then, the tube is vortexed for 2 min followed by 45 min of incubation in a water bath at 55 °C. After one additional vortexing for 2 min, the sample is transferred into a vial and centrifuged at 2500 g for 10 min. The supernatant (∼400 μL) is transferred into a new vial and diluted in PBS at 1:10 ratio for subsequent ELISA analysis. In addition, each component of ChCl-DESs (choline chloride, ethylene glycol, and urea) is tested individually.
2.3. Statistical analysis
The statistical analyses are conducted utilizing SPSS Statistics 23 software. To ensure robustness, each treatment condition is replicated at least twice. The effects of the treatments are then compared using a one-way ANOVA test. A significance level of p < 0.05 is used to determine statistical significance.
3. Results and discussion
3.1. Characterization of antibodies, optimization of assay conditions, and development of ELISA
The performance of commercial anti-gliadin antibodies used in this study is tested using direct and indirect ELISA procedures. As a first step, the ELISA procedure is optimized by testing different coating buffers and blocking buffer conditions (Fig. 1). Gliadin is a water-insoluble protein and the coating buffer used in coating ELISA plates needs to be optimized for properly dissolving the gliadin. Three coating buffer conditions tested include; 70% ethanol, carbonate-bicarbonate buffer, and a mixture of both. In addition, two blocking buffer conditions, 1% BSA and SuperBlock™ are evaluated. We saw higher signals with the Superblock™ solution as a blocking buffer compared to 1% BSA (p < 0.05), and no significant difference between different gliadin concentrations (p > 0.05). Therefore, Superblock™ was used as a blocking buffer and 10 μg/mL as starting gliadin concentration in subsequent ELISA tests (Fig. 1).
The combination of 70% ethanol and carbonate-bicarbonate as coating buffer gave the highest signal with Superblock™ as blocking buffer (p < 0.05). This construct allows more efficient capture of anti-gliadin polyclonal antibody by the coated gliadin (10 μg/mL) on the plate compared to the other test conditions (Fig. 1A). For example, the signal of this combination was 2.5-fold higher than that of BSA and bicarbonate-carbonate combination at 1:4000 dilution of the polyclonal antibody. Furthermore, the highest signal for detection of 10 μg/mL gliadin on the microtiter plate was obtained following the 70% ethanol plus carbonate-bicarbonate buffer with Superblock™ , which had a 1.97-fold higher signal than that of BSA and bicarbonate-carbonate combination (Fig. 1A). The reason behind this effect could be the need for 70% ethanol for properly dissolving the gliadin [[17], [18], [19]], yielding a homogenous solution, and effect of carbonate-bicarbonate buffer on adherence of gliadin onto the polystyrene plate surface by passive adsorption [20].
We further compared the blocking buffer conditions in indirect ELISA, where mouse monoclonal anti-wheat antibody was used as the capture antibody and the goat anti-mouse polyclonal antibody as the detector. The result of the assay showed that at three different starting concentrations of the coated gliadin (5, 10, and 20 μg/mL), using Superblock™ as the blocking buffer led to higher signals in all of the concentrations (Fig. 1B). Therefore, we optimized the coating and blocking conditions as 70% ethanol and carbonate-bicarbonate buffer and Superblock™, respectively in subsequent ELISA procedures.
In sandwich ELISA, higher concentrations of monoclonal antibody (Mab) are required to cover the surface of each well of the 96-well plate for coating, compared to the above-mentioned procedure. Therefore, we tested four different coating concentrations of Mab (0.18, 0.5, 1, and 5 μg/mL), starting from the recommended concentration for the indirect ELISA by the manufacturer (0.18 μg/mL) (Fig. 2A). Coating of the wells with 1 μg/mL Mab yield significantly higher signals compared to 0.18 and 0.5 μg/mL coating for detection of 0.3 μg/mL gliadin (p < 0.05). The signal intensity for coating the plates with 5 μg/mL was statistically comparable to coating the plates with 1 μg/mL (p > 0.05). Therefore, coating the plates with 1 μg/mL Mab was selected as the coating concentration for the subsequent sandwich ELISA tests.
Fig. 2.
Optimization of Mab coating concentration for sandwich ELISA. A. Four different Mab coating concentrations tested on 96-well plate: 0.18, 0.5, 1, and 5 μg/mL. Absorbance was read at 426 nm. Columns for each Mab concentration represents the dilution rates of gliadin (5 μg/mL (black), 1.25 μg/mL (blue), 0.31 μg/mL (green), and blank (grey), respectively). B. Comparison of in-house sandwich ELISA (grey) and commercial R5 ELISA detection kit (black).
The performance of in-house developed sandwich ELISA is compared with commercial R5 ELISA kit and it yielded similar detection efficacy for pure gliadin as the commercial gliadin detection kit (p > 0.05; Fig. 2B).
3.2. Gliadin extraction from food samples by different methods
3.2.1. Combination of 2-mercaptoethanol and guanidine hydrochloride
The protocol reported by García et al. [13] using 2-mercaptoethanol and guanidine hydrochloride is evaluated for extraction of gliadin from wheat flour. In addition, the combination of the components with different concentrations is tested to evaluate the extraction efficiency of each. One of the main components of gluten, gliadin, which is used as a target analyte in the majority of gluten detection assays contain a number of intramolecular disulfide bonds within its structures. These disulfide bonds should be cleaved for effective extraction of gliadin from a sample and as a reductant 2-mercaptoethanol could serve this purpose [21]. 2-mercaptoethanol has formerly and extensively been used as a component of gliadin extraction buffers within a variety of gluten detection assays [[22], [23], [24]]. Guanidine hydrochloride can denature proteins and it is used for different applications, including DNA extraction. Especially, in heat-processed foods, extraction of the gliadins could not be achieved as effectively as in non-heated processed foods. This is mainly because of the denaturation of proteins (particularly most of the α/β- and γ-gliadins) due to exposure to high heat and the formation of insoluble aggregates. A few of the gliadin proteins are heat stable, including ω-gliadins that do not contain disulfide bridges. A simple ethanol-based extraction buffer is not sufficient for disintegrating the aggregation of denatured gliadin proteins (α/β- and γ-gliadins), therefore, additional reducing and disaggregating agents are needed in the extraction buffer [13,25,26]. The gliadin extraction method reported by García et al. [13] is used as the gold standard and other developed methods were compared to it.
First, different concentrations and combinations of 2-mercaptoethanol and guanidine hydrochloride are tested. Specifically, 50 mM 2-mercaptoethanol, 250 mM 2-mercaptoethanol, 1 M guanidine hydrochloride, 2 M guanidine hydrochloride, and combinations of them are tested for the extraction of gliadin from wheat and crouton (Fig. 3). In the extraction of gliadin from a wheat sample, the inclusion of guanidine hydrochloride as a denaturing agent into extraction buffer has a significant effect on the extraction of gliadin compared to other tested conditions (p < 0.05; Fig. 3A). On the other hand, the inclusion of 2-mercaptoethanol doesn't impact the gliadin extraction efficacy of the extraction buffer (p > 0.05; Fig. 3A). These findings are expected as there is no heat denaturation of the gluten molecules within the wheat flour samples that would negatively impact gliadin extraction from this sample. The inclusion of guanidine hydrochloride only improves the extraction by denaturing the gliadin molecules that wouldn't otherwise dissolve in the extraction buffer.
Fig. 3.
Evaluation of different gliadin extraction conditions using 2-mercaptoethanol and guanidine hydrochloride from wheat flour (A) and crouton (B). Gliadin extraction from wheat flour (A) and crouton (B) [1]: gliadin standard (starting concentration: 5 μg/mL) [2], combination of PBS and 60% ethanol [3], 1 M guanidine hydrochloride [4], 2 M guanidine hydrochloride [5], 50 mM 2-mercaptoethanol [6], combination of 50 mM 2-mercaptoethanol and 1 M guanidine hydrochloride [7], combination of 50 mM 2-mercaptoethanol and 2 M guanidine hydrochloride [8], 250 mM 2-mercaptoethanol [9], combination of 250 mM 2-mercaptoethanol and 1 M guanidine hydrochloride [10], combination of 250 mM 2-mercaptoethanol and 2 M guanidine hydrochloride. Columns for each condition represents the dilution rates of gliadin. Dilution rates are 1:4 (black) and 1:16 (blue) for gliadin standard and 1:10 (black) and 1:100 (blue) for other conditions.
In a heat-processed food sample (crouton), the inclusion of 2-mercaptoethanol and guanidine hydrochloride significantly improve the extraction of gliadin from the sample (p < 0.05; Fig. 3B). Compared to ethanol only extraction method, the inclusion of only guanidine hydrochloride to the extraction buffer doesn't significantly improve the gliadin extraction efficacy (p > 0.05; Fig. 3B). On the other hand, the inclusion of the reducing agent 2-mercaptoethanol significantly improves gliadin extraction from the heat-processed sample (p < 0.05; Fig. 3B). Moreover, when the combination of guanidine hydrochloride and 2-mercaptoethanol is applied in the extraction buffer the gliadin extraction is significantly enhanced compared to other tested conditions (p < 0.05; Fig. 3B). This improvement on the extraction efficacy is observed even in the lowest tested concentrations of guanidine hydrochloride and 2-mercaptoethanol. These observations on the impact of these two reagents on the extraction of gliadin are parallel to the previous studies [13,25,26]. Based on these results, we select the PBS solutions with 1 M guanidine hydrochloride, 50 mM 2-mercaptoethanol and the PBS solution with 2 M guanidine hydrochloride and 50 mM 2-mercaptoethanol as the optimum conditions for improved extraction for both heat-processed and non-heat-processed food.
3.2.2. Evaluation of detergents and chelating agent
Based on the above-mentioned tests, 50 mM 2-mercaptoethanol with 2 M guanidine hydrochloride was selected as the control extraction buffer composition to compare the subsequent extraction methods. In this section, a chelating agent (EDTA) and three non-ionic detergents (Tween-20, Tween-80, and TritonX-100) were utilized. Chelating agents mainly interact with metal ions and enable a stable, water-soluble complex. EDTA particularly binds to divalent metal ions (including calcium and magnesium). Detergents are amphiphilic by their nature and could interact with hydrophobic and hydrophilic compounds and improve their dispersion [27]. Therefore, both chelating agents, EDTA, and detergents are readily used in a variety of different extraction buffers to improve extraction efficacy. To see if the similar efficiency of gliadin extraction from the heat-processed food sample (crouton) could be achieved with chelating agents or detergents, we compare four different solution compositions (EDTA, Tween-20, Tween-80, and Triton X-100). These chelating agents and detergents are diluted in either PBS or 80% ethanol. All of the tested solutions yielded very low signals compared to the controls (p < 0.05; Table-1). Therefore, the application of EDTA or these detergents didn't provide any improvement in the extraction of gliadin from heat-processed foods. Moreover, we could not obtain an efficient extraction of gliadin from a heat-processed food sample by using the extraction method recently reported by Hnasko et al. [14], although it is reported to be well compatible with the developed lateral flow assay (Fig. 3A). All of these extraction conditions yield low signals in gluten-free food sample (Fig. 4).
Table 1.
Comparison of gliadin extraction conditions: Gliadin extraction ability of different buffer conditions prepared with detergents assessed in wheat flour are measured by OD at 426 nm. Different letters indicate significant differences between treatments. Treatment conditions include [1]: PBS [2], combination of PBS and ethanol [3], combination of 50 mM 2-mercaptoethanol and 2 M guanidine hydrochloride [4], 0.1% EDTA in PBS [5], 0.1% EDTA in ethanol [6], 0.1% Tween 20 in PBS [7], 0.1% Tween 20 in ethanol [8], 0.1% Tween 80 in PBS [9], 0.1% Tween 80 in ethanol [10], 0.1% Triton X-100 in PBS [11], 0.1% Triton X-100 in ethanol. Columns for each condition represents the dilution rates (1:10 and 1:100 for other conditions).
| Sample Dilution Extraction Buffer |
1:10 | 1:100 |
|---|---|---|
| 1- only PBS | 1.36 ± 0.28 b | 0.77 ± 0.26 b, |
| 2- PBS and EtOH | 1.21 ± 0.25 b, c | 0.69 ± 0.13 b, c |
| 3–50 mM 2-mercaptoethanol and 2 M guanidine hydrochloride in EtOH | 2.54 ± 0.08 a | 1.59 ± 0.09 a |
| 4–0.1% EDTA in PBS | 0.75 ± 0.13 d, e | 0.44 ± 0.05 b, c |
| 5–0.1% EDTA in EtOH | 0.92 ± 0.13 c, d | 0.78 ± 0.12 b |
| 6–0.1% Tween-20 in PBS | 0.58 ± 0.06 d, e | 0.37 ± 0.10 c |
| 7–0.1% Tween-20 in EtOH | 0.57 ± 0.02 d, e | 0.53 ± 0.01 b, c |
| 8–0.1% Tween-80 in PBS | 0.43 ± 0.09 d, e | 0.36 ± 0.10 c |
| 9–0.1% Tween-80 in EtOH | 0.70 ± 0.11 d, e | 0.60 ± 0.23 b, c |
| 10–0.1% Triton X-100 in PBS | 0.37 ± 0.19 e | 0.51 ± 0.13 b, c |
| 11–0.1% Triton X-100 in EtOH | 0.47 ± 0.04 d, e | 0.42 ± 0.02 b, c |
Fig. 4.
Gliadin extraction from gluten free food sample. (1) gliadin standard (starting concentration: 10 μg/mL) [2], only PBS [3], combination of PBS and ethanol [4], combination of 50 mM 2-mercaptoethanol, 2 M guanidine hydrochloride and ethanol [5], 0.1% EDTA in PBS [6], 0.1% EDTA in ethanol [7], 0.1% Tween 20 in PBS [8], 0.1% Tween 20 in ethanol [9], 0.1% Tween 80 in PBS [10], 0.1% Tween 80 in ethanol [11], 0.1% Triton X-100 in PBS, and [12] 0.1% Triton X-100 in ethanol. Dilution rates are 1:1.
3.2.3. Evaluation of choline chloride-based deep eutectic solvents (ChCl-DESs)
We further tested an alternative method developed by Svigelj et al. [15] that utilizes choline chloride-based deep eutectic solvents (ChCl-DESs) to make a green and effective gluten extraction. This approach includes the use of ethaline and reline due to their optimum viscosity and dipolar and hydrogen bond donor properties, which result in high gliadin recovery from unprocessed and heat-treated food samples without affecting the subsequent immunoassay-based detection [15]. DESs are safe green solvents composed of a hydrogen bond acceptor and a hydrogen bond donor that end up with a lower melting point when mixed. While ethaline is the mixture of choline chloride as the acceptor and ethylene glycol as the donor in a 1:2 M ratio, reline is composed of choline chloride as the acceptor and urea as the donor in a 1:2 M ratio [15]. We tested combinations of ChCI-DESs, which are reline (1:2 choline chloride: urea), ethaline (1:2 choline chloride: ethylene glycol), 2 M ethylene glycol, 1 M choline chloride, and 2 M urea. Based on our results, all the combinations of ChCI-DESs gave superior signals compared to the controls that include an efficient extraction buffer composed of 50 mM 2-mercaptoethanol and 2 M guanidine hydrochloride (Fig. 5). Using 1 M choline chloride; 1.4-fold more signal than the control extraction method using 50 mM 2-mercaptoethanol, 2 M guanidine hydrochloride and ethanol, and 3-fold more signal than the controls using PBS-only and PBS plus ethanol is achieved. This is a promising finding in an effort to make less toxic and efficient gluten extraction from heat-treated food samples for detection and this method could be incorporated in commercial ELISA kits or rapid gluten detection tests upon further optimizations.
Fig. 5.
Testing different combinations of ChCl-DESs for extraction of gliadin from crouton. [1] blank [2], only PBS [3], combination of PBS + ethanol [4], combination of 50 mM 2-mercaptoethanol, 2 M guanidine hydrochloride and ethanol [5], 2 M ethylene glycol [6], combination of 2 M ethylene glycol and 1 M choline chloride [7], 1 M choline chloride [8], 2 M urea [9], combination of 2 M urea and 1 M choline chloride. Dilution rates are 1:1000 except 1 (blank).
4. Conclusion
In conclusion, this study aimed to optimize the ELISA procedure and develop effective gliadin extraction methods for gluten detection. The results demonstrate the following key findings: Firstly, the ELISA procedure was successfully optimized by selecting the combination of 70% ethanol and carbonate-bicarbonate buffer as the coating buffer and Superblock™ as the blocking buffer. These conditions provided higher signals compared to other tested conditions, ensuring reliable and reproducible measurements. Secondly, the combination of 2-mercaptoethanol and guanidine hydrochloride proved to be an efficient extraction method for gliadin from both heat-treated and non-heat-treated food samples. The inclusion of guanidine hydrochloride as a denaturing agent and 2-mercaptoethanol as a reducing agent significantly improved gliadin extraction efficacy, particularly in heat-treated samples where protein denaturation and aggregation occur.
Furthermore, the evaluation of detergents and chelating agents did not show significant improvements in gliadin extraction efficiency compared to the control extraction buffer. Excitingly, the use of choline chloride-based deep eutectic solvents (ChCl-DESs) showed promising results, with significantly higher signals obtained compared to the control extraction method. ChCl-DESs, such as reline and ethaline, proved to be effective in extracting gliadin from both heat-treated and non-heat-treated food samples, offering a greener and less toxic alternative for gluten extraction.
Overall, the optimized ELISA procedure and the identified effective gliadin extraction methods hold great potential for improving gluten detection accuracy and efficiency. These findings contribute to the development of reliable gluten detection techniques, benefiting individuals with gluten-related health conditions who rely on strict gluten-free diets. Future studies should further optimize these extraction methods and explore their integration into commercial gluten detection kits or rapid tests.
Author contribution statement
Ece Güven, Reha Onur Azizoglu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
Dr. Reha Azizoglu was supported by Türkiye Bilimsel ve Teknolojik Araştırma Kurumu {TUBITAK, 118C377}. The financial support received from TUBITAK does not mean that the content of the publication is approved in a scientific sense by TUBITAK.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Biesiekierski J.R. What is gluten? J. Gastroenterol. Hepatol. 2017;32:78–81. doi: 10.1111/jgh.13703. [DOI] [PubMed] [Google Scholar]
- 2.Shewry P. What is gluten—why is it special? Front. Nutr. 2019:101. doi: 10.3389/fnut.2019.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wieser H. Chemistry of gluten proteins. Food Microbiol. 2007;24(2):115–119. doi: 10.1016/j.fm.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Balakireva A.V., Zamyatnin A.A., Jr. Properties of gluten intolerance: gluten structure, evolution, pathogenicity and detoxification capabilities. Nutrients. 2016;8(10):644. doi: 10.3390/nu8100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fasano A., Berti I., Gerarduzzi T., Not T., Colletti R.B., Drago S.…Horvath K. Prevalence of celiac disease in at-risk and not-at-risk groups in the United States: a large multicenter study. Arch. Intern. Med. 2003;163(3):286–292. doi: 10.1001/archinte.163.3.286. [DOI] [PubMed] [Google Scholar]
- 6.Taylor S.L., Nordlee J.A., Jayasena S., Baumert J.L. Evaluation of a handheld gluten detection device. J. Food Protect. 2018;81(10):1723–1728. doi: 10.4315/0362-028X.JFP-18-184. [DOI] [PubMed] [Google Scholar]
- 7.Jnawali P., Kumar V., Tanwar B. Celiac disease: overview and considerations for development of gluten-free foods. Food Sci. Hum. Wellness. 2016;5(4):169–176. [Google Scholar]
- 8.Zhang J., Portela S.B., Horrell J.B., Leung A., Weitmann D.R., Artiuch J.B.…Yates S.N. An integrated, accurate, rapid, and economical handheld consumer gluten detector. Food Chem. 2019;275:446–456. doi: 10.1016/j.foodchem.2018.08.117. [DOI] [PubMed] [Google Scholar]
- 9.Food and Drug Administration, HHS. Food labeling: gluten-free labeling of foods. Final rule. Fed. Regist. 2013;78(150):47154–47179. [PubMed] [Google Scholar]
- 10.Rubio-Tapia A., Hill I.D., Kelly C.P., Calderwood A.H., Murray J.A. ACG clinical guidelines: diagnosis and management of celiac disease. Off. J. Am. Coll. Gastroenterol.| ACG. 2013;108(5):656–676. doi: 10.1038/ajg.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lexhaller B., Tompos C., Scherf K.A. Fundamental study on reactivities of gluten protein types from wheat, rye and barley with five sandwich ELISA test kits. Food Chem. 2017;237:320–330. doi: 10.1016/j.foodchem.2017.05.121. [DOI] [PubMed] [Google Scholar]
- 12.Bruins Slot I.D., Bremer M.G., van der Fels‐Klerx I., Hamer R.J. Evaluating the performance of gluten ELISA test kits: the numbers do not tell the tale. Cereal Chem. 2015;92(5):513–521. [Google Scholar]
- 13.García E., Llorente M., Hernando A., Kieffer R., Wieser H., Méndez E. Development of a general procedure for complete extraction of gliadins for heat processed and unheated foods. Eur. J. Gastroenterol. Hepatol. 2005;17(5):529–539. doi: 10.1097/00042737-200505000-00010. [DOI] [PubMed] [Google Scholar]
- 14.Hnasko R.M., Jackson E.S., Lin A.V., Haff R.P., McGarvey J.A. A rapid and sensitive lateral flow immunoassay (LFIA) for the detection of gluten in foods. Food Chem. 2021;355 doi: 10.1016/j.foodchem.2021.129514. [DOI] [PubMed] [Google Scholar]
- 15.Svigelj R., Bortolomeazzi R., Dossi N., Giacomino A., Bontempelli G., Toniolo R. An effective gluten extraction method exploiting pure choline chloride-based deep eutectic solvents (ChCl-DESs) Food Anal. Methods. 2017;10:4079–4085. [Google Scholar]
- 16.Marston K., Khouryieh H., Aramouni F. Effect of heat treatment of sorghum flour on the functional properties of gluten-free bread and cake. LWT--Food Sci. Technol. 2016;65:637–644. [Google Scholar]
- 17.Bietz J.A., Wall J.S. 1973. Isolation and Characterization of Gliadin-like Subunits from Glutenin. [Google Scholar]
- 18.Byers M., Miflin B.J., Smith S.J. A quantitative comparison of the extraction of protein fractions from wheat grain by different solvents, and of the polypeptide and amino acid composition of the alcohol‐soluble proteins. J. Sci. Food Agric. 1983;34(5):447–462. doi: 10.1002/jsfa.2740340506. [DOI] [PubMed] [Google Scholar]
- 19.Urade R., Sato N., Sugiyama M. Gliadins from wheat grain: an overview, from primary structure to nanostructures of aggregates. Biophys. Rev. 2018;10:435–443. doi: 10.1007/s12551-017-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konstantinou G.N. Enzyme-linked immunosorbent assay (ELISA) Food Allergens: Methods Protocols. 2017:79–94. doi: 10.1007/978-1-4939-6925-8_7. [DOI] [PubMed] [Google Scholar]
- 21.Kwak H.W., Yun N.K., Lee D.J., Lee K.H. Effect of mercaptoethanol on the wet spinning of wheat gliadin fiber. Textile Sci. Eng. 2012;49(3):189–193. [Google Scholar]
- 22.Shewry P.R., Miflin B.J., Lew E.J., Kasarda D.D. The preparation and characterization of an aggregated gliadin fraction from wheat. J. Exp. Bot. 1983;34(11):1403–1410. [Google Scholar]
- 23.Doña V.V., Fossati C.A., Chirdo F.G. Interference of denaturing and reducing agents on the antigen/antibody interaction. Impact on the performance of quantitative immunoassays in gliadin analysis. Eur. Food Res. Technol. 2008;226:591–602. [Google Scholar]
- 24.Lacorn M., Dubois T., Weiss T., Zimmermann L., Schinabeck T.M., Loos-Theisen S., Scherf K. Determination of gliadin as a measure of gluten in food by R5 sandwich ELISA RIDASCREEN® gliadin matrix extension: collaborative study 2012.01. J. AOAC Int. 2022;105(2):442–455. doi: 10.1093/jaoacint/qsab148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wieser H. Investigations on the extractability of gluten proteins from wheat bread in comparison with flour. Z. Lebensm. Unters. Forsch. A. 1998;207:128–132. [Google Scholar]
- 26.Mothes T., Stern M. How gluten-free is gluten-free, and what does this mean to coeliac patients? Eur. J. Gastroenterol. Hepatol. 2003;15(5):461–463. doi: 10.1097/01.meg.0000059124.68845.8c. [DOI] [PubMed] [Google Scholar]
- 27.Stevenson S.G., Preston K.R. Effects of surfactants on wheat protein fractionation by flow field-flow fractionation. J. Liq. Chromatogr. Relat. Technol. 1997;20(16–17):2835–2842. [Google Scholar]