Abstract
Purpose
Altered gene methylation precedes altered gene expression and the onset of disease. This study aimed to develop a potential model for predicting recurrence of early to mid-stage hepatocellular carcinoma (HCC) using methylation loci.
Methods
We used data from early to mid-stage HCC patients (TNM I-II) in the TCGA-LIHC dataset and lasso-cox regression model to identify an 18-DNA methylation site panel from which to calculate the riskScore of patients. The correlation of high/low riskScore with recurrence-free survival (RFS) and immune microenvironment in HCC patients was analyzed by bioinformatics. It was also validated in the GSE56588 dataset and the final dynamic nomogram was constructed.
Results
The results showed that riskScore was significantly correlated with RFS in HCC patients. The differential mutated genes between the two groups of HCC patients with high/low riskScore were mainly enriched in the TP53 signaling pathway. The immune microenvironment was better in HCC patients in the low-riskScore group compared to the high-riskScore group. This was validated in the GSE56588 dataset. Based on the subgroup stratification analysis of the relationship between high/low riskScore and RFS, as well as univariate and multivariate cox analyses, the riskScore was found to be independent of clinical indicators. We found that riskScore, vascular invasion and cirrhosis status could effectively differentiate RFS in HCC patients, and we also constructed prediction model based on these three factors. The model we constructed were validated in the TCGA-LIHC database and a web calculator was built for clinical use.
Conclusion
The methylation riskScore is a predictor of RFS independent of clinical factors and can be used as a marker to predict recurrence in HCC patients.
Keywords: Hepatocellular carcinoma, Methylation riskScore, Recurrence-free survival, Immune microenvironment
1. Introduction
Liver cancer is the sixth most commonly diagnosed cancer and the fourth leading cause of cancer death, with hepatocellular carcinoma (HCC) being the most common primary liver cancer [1]. HCC is an extremely heterogeneous tumor with a unique pattern of somatic genomic alterations [2]. In recent years, with the improvement of surveillance and imaging technology, more and more patients with hepatocellular carcinoma can be diagnosed in the early or mid-stage and undergo radical resection [3]. A study that analyzed a nationwide database in Japan showed that long-term recurrence-free survival (RFS) was achieved after HCC resection [4]. Furthermore, immune checkpoint inhibitor therapy increased the likelihood of achieving complete remissions [5]. However, the prognosis of HCC is poor, RFS is a key factor in developing individual treatment plans, and RFS varies among patients with HCC in the early to middle stages [6,7]. Therefore, it is crucial to stratify the risk of recurrence in patients who undergo surgery in early to mid-stage.
Emerging biomarkers such as PD-L1 expression, tumor mutational burden, microsatellite instability, gut microbiota, and DNA methylation have been advocated for their application in determining the immunotherapy response in HCC [8,9]. Aberrant DNA methylation is an early event in the development of cancer, and alterations in gene methylation precedes altered gene expression and disease onset [10]. Better predictions can be made by constructing DNA methylation models or finding relevant signatures. By constructing a diagnostic prediction model and a prognostic prediction model for survival, xu et al. demonstrated that ctDNA methylation markers have high diagnostic specificity and sensitivity in the diagnosis, monitoring, and prognosis of HCC [11]. In addition, it was shown that genome-wide DNA methylation profiles can accurately distinguish different histological stages of human hepatocarcinogenesis [12]. In a study by Yang and colleagues, DNA methylation of SOCS2-1-90 was found using next-generation sequencing to predict the recurrence of HCC after liver transplantation [13]. Similar methylation applications exist in other cancers, including cervical, breast and prostate cancers [[14], [15], [16]].
In this study, we used the TCGA database to construct a genome-wide model using methylation sites that can be used to diagnose and predict RFS in patients with early or mid-stage HCC. We also constructed a Nomogram based on methylation riskScore and clinical factors, which provides a new possible idea for clinical use in stratifying the risk of recurrence of HCC in the early and middle stages.
2. Method
2.1. Data collection and processing
Methylation data, DNA mutation data, expression data and related clinical information of patients with hepatocellular carcinoma were obtained from the TCGA database. Validation datasets GSE113019 (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc= GSE113019), GSE35069 (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc= GSE35069) and GSE56588 (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc= GSE56588) were downloaded from the GEO database. DNA methylation data were preprocessed and quality controlled using the ChAMP package. Methylation-based tumor purity calculations were performed using the InfiniumPurify package. Immune cell infiltration was calculated using the MethylCIBERSORT package. DNA mutation correlation analysis and visualization were done using the maftools package.
2.2. Screening of candidate methylation sites and calculation of riskScore
Lasso-Cox analysis of all methylation sites was performed using the glmnet package to screen for methylation sites associated with RFS at p < 0.001. These candidate methylation sites were subjected to a multivariate Cox stepwise regression method to construct a model. The methylation values of individual patients were then used to calculate the risk score of each patient for subsequent analysis. By employing the following formula, we are able to calculate the risk score for each patient: , where Coefi represents the risk coefficient associated with the methylation site included in the final multivariate Cox stepwise regression model, and xi denotes the specific methylation value attributed to the incorporated methylation site. The median of all riskScores in the training cohort was used as the cutoff value to differentiate patients into high-risk and low-risk groups, and patients were grouped in the validation cohort using the same cutoff values.
Analysis of 28 cell subpopulations of immune cells and tumor immune-immune cycle based on mRNA calculation.
To quantify the proportions of immune cells in the tumor microenvironment, we applied single-sample gene set enrichment analysis (ssGSEA) [17], allowing the assessment of 28 immune cell types.
According to a previous study, the steps of the cancer-immunity cycle are described by eight axes of the immunogram score (IGS) as follows: IGS1, T cell immunity; IGS2, tumor antigenicity; IGS3, priming and activation; IGS4, trafficking and infiltration; IGS5, recognition of tumor cells; IGS6, inhibitor cells; IGS7, checkpoint expression; and IGS8, inhibitory molecules [18]. The gene sets IGS1, IGS2, IGS3, IGS4, IGS5, IGS6, IGS7, and IGS8 were used in a previous study [18].
2.3. Differential analysis and enrichment analysis of mRNA
The counts matrix of the expression data downloaded from the TCGA database for hepatocellular carcinoma was analyzed for differences using the DEseq2 package, and selected p value < 0.05 difference genes were analyzed for GO enrichment using the clusterProfiler package.
2.4. Evaluation of the construction of a nomogram and the construction of a web calculator
Univariate and multivariate Cox risk analysis of recurrence-free survival was performed by combining riskScore with all clinical information. Variables with p < 0.05 in the multivariate Cox analysis and cirrhosis status were included for multivariate Cox modeling. Nomogram were constructed using the model and the accuracy of the model was assessed using calibration curves, which were done using the rms package. To investigate the discriminatory efficacy of Nomogram in predicting recurrence at different times, we constructed time_ROC curves using the timeROC package. Dynamic Nomogram was generated using the DynNom package.
2.5. Statistical analysis
Statistical analyses were conducted using R software (version 3.4.2.). Data were expressed as the median and interquartile range (IQR). Two group values were analyzed for the differences by Wilcoxon rank sum test. The Kruskal-Wallis test was used in more than two group to test differences. A log-rank test and Kaplan-Meier curves were used for comparison Recurrence free survival (RFS) using. P < 0.05 was considered significant with two-sided.
3. Results
3.1. Selection of candidate methylation loci and building a predictive methylation riskScore model
The study was performed on 251 early-middle (TNM I and TNM II) HCC patients who were clinically and pathologically diagnosed with HCC. Of these patients, we randomly split these HCC patients with complete RFS information into training and validation data sets with an allocation of 1:1. Both cohorts have similar clinicopathological characteristics in many aspects except gender and histological grade (Table 1).
Table 1.
Clinicopathological characteristics of HCC patients from TCGA.
| Training dataset (n = 127) | Validation dataset (n = 124) | P-value | Total (n = 251) | |
|---|---|---|---|---|
| Age (years) | 0.944 | |||
| ≤ 60 | 62 (48.8%) | 59 (47.6%) | 121 (48.2%) | |
| >60 | 65 (51.2%) | 65 (52.4%) | 130 (51.8%) | |
| Gender | 0.0279 | |||
| female | 29 (22.8%) | 45 (36.3%) | 74 (29.5%) | |
| male | 98 (77.2%) | 79 (63.7%) | 177 (70.5%) | |
| Alpha-fetoprotein (ng/ml) | 0.749 | |||
| ≤ 20 | 62 (48.8%) | 57 (46.0%) | 119 (47.4%) | |
| >20 | 45 (35.4%) | 47 (37.9%) | 92 (36.7%) | |
| Missing | 20 (15.7%) | 20 (16.1%) | 40 (15.9%) | |
| Liver cirrhosis | 0.804 | |||
| No | 52 (40.9%) | 47 (37.9%) | 99 (39.4%) | |
| Yes | 30 (23.6%) | 31 (25.0%) | 61 (24.3%) | |
| Missing | 45 (35.4%) | 46 (37.1%) | 91 (36.3%) | |
| Etiology | 0.999 | |||
| Alcohol consumption | 17 (13.4%) | 17 (13.7%) | 34 (13.5%) | |
| Hepatitis B | 42 (33.1%) | 42 (33.9%) | 84 (33.5%) | |
| Hepatitis C | 20 (15.7%) | 19 (15.3%) | 39 (15.5%) | |
| Other factors | 40 (31.5%) | 39 (31.5%) | 79 (31.5%) | |
| Missing | 8 (6.3%) | 7 (5.6%) | 15 (6.0%) | |
| Race | 0.223 | |||
| Asian | 55 (43.3%) | 60 (48.4%) | 115 (45.8%) | |
| Others | 4 (3.1%) | 9 (7.3%) | 13 (5.2%) | |
| White | 63 (49.6%) | 53 (42.7%) | 116 (46.2%) | |
| Missing | 5 (3.9%) | 2 (1.6%) | 7 (2.8%) | |
| Histological grade | 0.0482 | |||
| G1-G2 | 86 (67.7%) | 68 (54.8%) | 154 (61.4%) | |
| G3-G4 | 40 (31.5%) | 55 (44.4%) | 95 (37.8%) | |
| Missing | 1 (0.8%) | 1 (0.8%) | 2 (0.8%) | |
| TNM stage | 0.347 | |||
| I | 90 (70.9%) | 80 (64.5%) | 170 (67.7%) | |
| II | 37 (29.1%) | 44 (35.5%) | 81 (32.3%) | |
| Event | 0.425 | |||
| Yes | 46 (36.2%) | 52 (41.9%) | 98 (39.0%) | |
| No | 81 (63.8%) | 72 (58.1%) | 153 (61.0%) |
Then, methylation loci associated with prognosis were screened for modeling using Lasso-cox regression analysis (Fig. 1). The final inclusion of 18 methylation loci in the risk model was shown in Table S1. Correspondingly, we found that 13 of the 18 methylation loci were significantly different in the methylation high/low-riskScore group (Fig. S1). The risk score for each HCC patient was derived from the model.
Fig. 1.
Feature selection using least absolute shrinkage and selection operator (LASSO) COX regression. A. A coefficient profile plot was produced against the Log(lambda) sequence. B. Eighteen CpG sites with nonzero coefficients were selected by optimal lambda. By verifying the optimal parameter (lambda) in the LASSO model, the partial likelihood deviance curve was plotted versus log(lambda). At the optimal values log(lambda), where features are selected, dotted vertical lines are set using the minimum criteria and the one standard error of the minimum criteria.
3.2. Methylation riskScore can be used as a diagnostic biomarker for HCC patients
Firstly, we explored the potential role of methylation riskScore in diagnosing HCC patients. The results of paired tissue analysis of the included patients showed that methylation riskScore was higher in tumor tissues (Fig. 2A). Consistently, methylation riskScore was higher in tumor tissues among normal and tumor tissues in both groups of patients (Fig. 2B). Also, we obtained validation in the GSE113019 dataset, demonstrating a significant difference in riskScore in normal liver and hepatocellular carcinoma tissues (p = 0.00024, Fig. 2C). The ROC results showed promising methylation riskScore as a diagnostic biomarker for hepatocellular carcinoma (AUC = 0.742, Fig. 2D). Further analysis revealed that methylation riskScore also differed significantly in serum cfDNA of cirrhotic patients versus HCC patients with cirrhotic background (p = 0.00850, Fig. 2E). The ROC results showed that the optimal cut-off value of methylation riskScore in cfDNA was 23.927, and at the cut-off value riskScore had a good diagnostic ability for HCC patients (p < 0.001, Fig. 2F and G).
Fig. 2.
Analysis of RiskScore Differences in Hepatocellular Carcinoma A. Difference analysis of riskScore in TCGA-LIHC paired tissues; B. difference of riskScore in TCGA-LIHC normal tissues and tumor tissues; C. validation of riskScore differences in normal liver and hepatocellular carcinoma tissues in the GSE113019 dataset; D. ROC curves to analyze the ability of riskScore to differentiate TCGA-LIHC normal liver tissues from tumor tissues; E. analysis of riskScore differences in serum cfDNA in patients with cirrhosis versus hepatocellular carcinoma on a cirrhotic background in the GSE35069 dataset; F. Determination of the optimal cut-off value of riskScore in cfDNA using ROC curves; G. Diagnostic ability of riskScore in patients with hepatocellular carcinoma at the optimal cut-off value.
HCC patients with low methylation risk scores have better RFS.
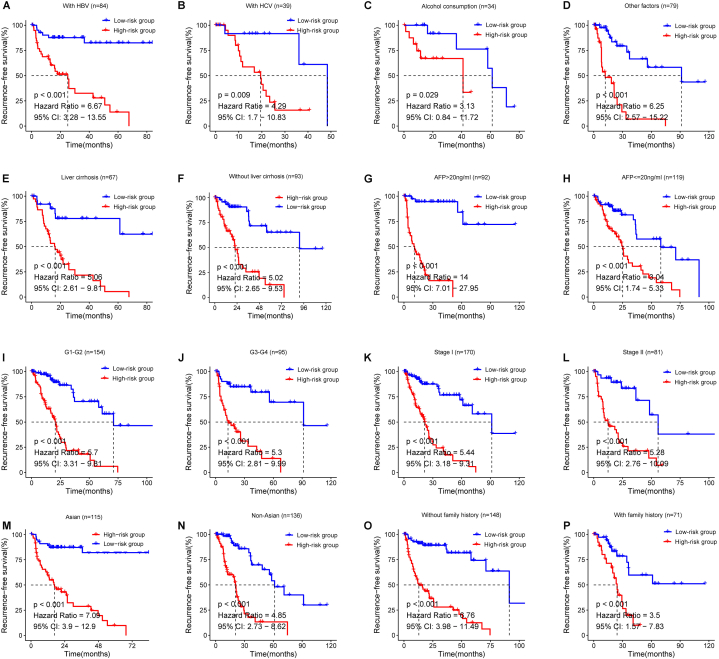
Then, we examined the effect of methylation risk score on RFS in HCC patients. The results showed that RFS in the low-riskScore group was significantly better than that in the high-riskScore group in the training set (p < 0.001), indicating that low-riskScore could be a good prognostic factor for HCC (Fig. 3A–C). Consistently, in the validation set, we further confirmed that RFS was significantly better in the low-risk score group than in the high-riskScore group (Fig. 3D–F). Excitingly, we examined the relationship between high/low-riskScore and RFS in each of the other subgroups. As shown in Fig. 4, patients in the low-risk group had significantly higher RFS than the high-risk group, regardless of other conditions such as concomitant HBV, HCV, alcohol consumption and so on. Moreover, the distribution of methylation riskScore among HCC patients was not related to any other clinical subgroups, such as AFP level, liver cirrhosis and race, except for differences in TNM classification (Fig. S2). Based on the hazard ratios in each subgroup, we concluded that the methylation riskScore could better differentiate patients with or without recurrence survival, especially in Asians with hepatitis B, cirrhosis, AFP>20 ng/ml and no family history in HCC patients.
Fig. 3.
Correlation of 18-DNA methylation panel in training set and validation set with RFS A. Distribution of riskScore in the training set; B. Relapse-free survival status of HCC patients with different riskScore in the training set; C. Relationship between high and low riskScore and RFS in the training set; D. Distribution of riskScore in the validation set; E. The status of relapse-free survival of HCC patients under different riskScore in the validation set; F. Relationship between high and low riskScore and RFS in the validation set.
Fig. 4.
Stratified analysis of the relationship between high/low riskScore and RFS. Kaplan-Meier analyses for RFS in subgroups stratified by different conditions as follows: A-D. different etiologies, E-F. presence or absence of liver cirrhosis, G-H. different levels of serum AFP, I-J. different grade, K-L. stage, M-N. race, and O–P. family history.
3.3. Altered biological processes and pathways in high- and low-risk subgroups
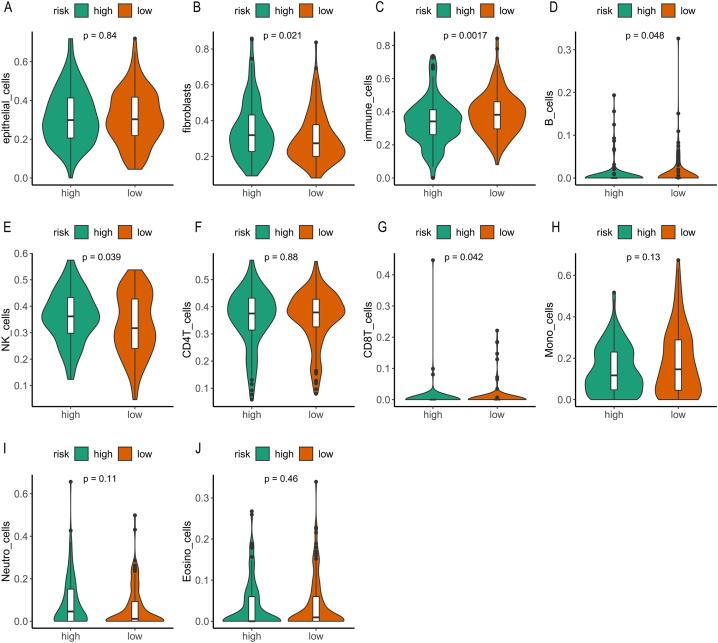
At the same time, we also explored the biological processes and possible mechanisms behind riskScore from the DNA and RNA levels. We used bioinformatics analysis to examine mutant genes in high/low - risk groups. The global landscape of differential mutated genes in the two groups is shown in Fig. 5A, and the main enrichment pathway of the significantly different driver genes was TP53 (Fig. 5B). In addition, the volcano map showed that differential gene expression was also present in the high/low-risk group (Fig. 5C), and then these differential genes were subjected to GO analysis. The differential genes mainly consisted of antigen binding, which was involved in the process of humoral immune response and had the function of immunoglobulin complex (Fig. 5D). Taken together, these results suggested that riskScore may be involved in and influence the regulation of the immune system. Therefore, further investigations should be conducted to explore the relationship between RiskScore and the immune system. Further analysis of the immune cycle between the two groups with high/low-riskScore revealed significant differences in IGS1, IGS3, IGS6, IGS7 and IGS8 between the two groups of patients (Fig. 6). In addition, there were significant differences in the immune microenvironment between the high/low-riskScore groups. As shown in Fig. 7, the contents of fibroblasts, immune cells, B cells, NK cells and CD8T cells were significantly different between the two groups, indicating that riskScore and the regulation of the immune microenvironment were related. Also, we validated the results in the GSE56588 dataset, which indicated that the contents of epithelial cells, fibroblasts, immune cells, B cells and CD8T cells were significantly different between the two groups of HCC patients with high/low -riskScore (Fig. S3).
Fig. 5.
A. Lanscape of differentially significant mutant gene mutations in high/low riskScore grouping; B. Enrichment analysis of mutations in significantly different driver gene. C. Differential analysis of gene expression in high/low riskScore grouping; D. GO enrichment analysis of differential genes.
Fig. 6.
Differences in immune circulation between the two groups. The violin diagram showing the differences in every step of immune circulation between the two groups with high/low riskScore.
Fig. 7.
Methylation data assess differences in the immune microenvironment between the two groups. The violin diagram showing the differences in different cell subpopulations between the two groups with high/low riskScore.
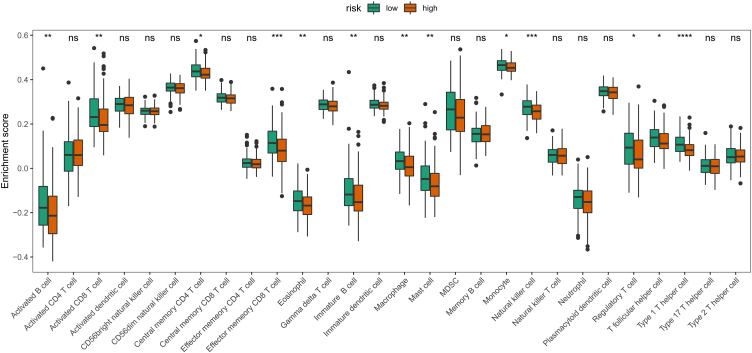
To characterize in more detail the changes in immune subpopulations in different risk subgroups, we assessed the contents of 28 immune cell subsets in the high/low-riskScore groups using ssGSEA, and the results showed that the expression of 13 immune cell subsets, including activated B cells, activated CD8T cells, and effector memory CD8T cells, were significantly different between the two groups (Fig. 8).
Fig. 8.
ssGSEA assesses differences in 28 immune cell subpopulations between the two groups. The boxplot diagram showing the differences in different immune cell subpopulations between the two groups with high/low riskScore.
3.4. Establishment of nomogram based on the riskScore
Finally, the results of univariate and multivariate cox analysis showed that the methylation risk score and vascular invasion were significantly associated with RFS in HCC patients (Table 2). Cirrhosis is a recognized recurrence-related factor in HCC. Although cirrhosis was not statistically different in the two groups, we included it in our analysis. Then, we used the TCGA-LIHC dataset to establish a model for predicting HCC recurrence based on riskScore, vascular invasion and cirrhosis status. After that, we produced the corresponding nomogram and calibrated the actual 1–3 years RFS, showing that the model had high accuracy (Fig. 9A–D). Survival curve results showed that RFS was significantly higher in the low-risk group of HCC patients than in the high-risk and intermediate-risk groups (Fig. 9E). In addition, the model had less time-dependence and higher accuracy in predicting the recurrence of recurrence in HCC patients at 1, 2, and 3 years (Fig. 9F). We also constructed a dynamic nomogram for predicting recurrence to facilitate subsequent use (Fig. S4) (https://zhaolihua2008.shinyapps.io/DynNomapp1/). The likelihood of recurrence in HCC patients can be calculated by entering the corresponding content correct values in the dynamic nomogram.
Table 2.
Univariate and multivariate analyses of clinicopathological characteristics and Methylation risk score with RFS in the entire TCGA-LIHC dataset.
| Variables | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Methylation risk score (High/Low) | 3.47(2.49–4.83) | 1.824e-13* | 3.70 (2.57–5.34) | <0.001* |
| Vascular invasion (Yes/No) | 1.41(1.03–1.93) | 0.035* | 1.52(1.01–2.28) | 0.046 |
| Adjacent tissue inflammation (Yes/No) | 1.32(0.95–1.83) | 0.010 | ||
| Age (>60/≤60 years) | 0.97(0.73–1.28) | 0.813 | ||
| Albumin (≥4.0/<4.0 g/dl) | 0.79(0.59–1.06) | 0.112 | ||
| Creatinine (≥1.1/<1.1 mg/dl) | 0.83(0.61–1.13) | 0.239 | ||
| AFP (>20/≤20 ng/ml) | 0.98(0.72–1.33) | 0.881 | ||
| Liver cirrhosis (Yes/No) | 1.38(1.00–1.93) | 0.054 | ||
| Histological grade (G3-4/G1-2) | 1.06(0.80–1.41) | 0.689 | ||
| Platelet (≥200/<200 × 109/L) | 1.03(0.77–1.38) | 0.834 | ||
| Family history (Yes/No) | 0.93(0.67–1.29) | 0.667 | ||
| Gender (male/female) | 1.09(0.8–1.485) | 0.584 | ||
| Race (Asian/not Asian) | 1.04(0.79–1.39) | 0.765 | ||
| TNM stage (II/I) | 1.40(1.05–1.87) | 0.023* | 0.96(0.64–1.43) | 0.836 |
| BMI (≥25/<25 kg/m2) | 0.95(0.71–1.27) | 0.712 | ||
| HBV (Yes/No) | 0.88(0.65–1.20) | 0.416 | ||
| Alcohol consumption (Yes/No) | 0.83 (0.53–1.30) | 0.421 | ||
Abbreviations: RFS, recurrence-free survival; HR, hazard ratio; 95% CI, 95% confidence interval.
*Statistically significant; AFP, Alpha-fetoprotein; BMI, body mass index.
Fig. 9.
RiskScore-based nomogram created and performance validated using the TCGA-LIHC dataset. A. Nomogram constructed using riskScore, vascular invasion, and liver cirrhosis for patients in the TCGA-LIHC dataset; B. Calibration curves for 1- year, 2- year, and 3-year RFS; C. RFS curve of high/medium/low risk HCC patients evaluated by column line plot; D. Validation of time-dependent ROC curve.
4. Discussion
Scientific and medical developments have led to major advances in the clinical management of HCC, with five-year survival rates of 40%–60% after radical surgery. However, the high recurrence rate after surgery has become a major obstacle to further improving the prognosis, with a 5-year recurrence rate of 60% [19]. Several models have been developed to effectively predict survival and prognosis after radical resection of early HCC, including a metastasis prediction model based on 153 hepatocellular carcinomas and a metastasis prediction model based on 17 validated immune factors [20,21]. In addition, it was shown that DNA hypermethylation is characteristic in the early development of HCC [22]. Long et al. successfully developed a diagnostic, prognostic and recurrence model for HCC through a comprehensive analysis of gene expression and DNA methylation data [23]. Unusually, the model we developed was based on the methylation riskScore, which is a predictor of RFS independent of clinical factors. This model is associated with the immune microenvironment as an independent risk predictor, and at the same time this provides a new possible idea for the stratification of the risk of recurrence of HCC in the early and middle stages.
The generation of methylation gene risk scores is an evolving field. Recent studies showed that adding methylation risk scores to a metabolic syndrome risk assessment model further improved model fitting in studies used for score validation [24,25]. Our results showed that HCC patients with a high methylation riskScore had significantly shorter recurrence-free survival than the low-risk group, suggesting that it was a predictor of recurrence-free survival. Moreover, the distribution of methylation riskScore among HCC patients was not related to any other clinical subgroups except for differences in TNM classification. To explore the generalizability and applicability of the model to patients with HCC, different subgroups of patients with early or mid-stage in our TCGA-LIHC dataset were analyzed. The model was effective in distinguishing their RFS in a population of patients with different etiologies, with or without cirrhotic background, different serum AFP levels, different degrees of differentiation, pathological grading, race and with or without family history. However, we can see from the results that the differentiation is better in Asian patients with hepatitis B, cirrhotic background, AFP >20 ng/ml and no family history.
We also explored the biological processes and possible mechanisms behind the riskScore at the DNA and RNA levels. We found that the mutation genes that were significantly different in the two groups of patients with high/low riskScore were mainly enriched in TP53, Hippo, wnt and Notch signaling pathways, suggesting that there are differences in immune aspects between the two groups of patients with high/low riskScore. Song et al. showed that high TP53 mutation frequency was associated with TSC2 mutation, which was independently associated with recurrence and poorer RFS within 1 year after hepatectomy [26]. The lncRNAs in a lncRNAs-based risk scoring system are mainly involved in Wnt signaling and can effectively predict RFS in cirrhotic HCC patients [27]. It was shown that Notch pathway are closely associated with finding prognostic biomarkers to predict RFS in HCC patients [28]. Meanwhile, this inference was supported by RNA differential analysis, enrichment analysis and tumor immune cycle analysis. We analyzed the differences in the immune microenvironment between the two groups, and found that patients in the low-riskScore group had a better immune microenvironment and were more inclined to "hot" tumors. In addition, we validated the effect of riskScore on the immune microenvironment in the dataset GSE56588. HCC exhibits an extremely heterogeneous immune microenvironment, leading to inconsistent outcomes in immunotherapy [29]. This study confirms the involvement of methylation risk scoring in influencing the immune microenvironment, thereby potentially aiding in maximizing the benefits of immune therapy for patients. Based on this, we built a dynamic nomogram based on riskScore, vascular invasion and cirrhosis status to facilitate its application in the clinical setting. This model focuses on early-stage HCC patients, providing them with more targeted diagnosis, treatment, and care, particularly for those at a higher risk of recurrence. Additionally, the model helps to avoid unnecessary medical interventions for low-risk HCC patients, ensuring a balanced approach to their clinical management. Although we have developed and validated a risk score for distinguishing HCC patients with recurrence-free survival and demonstrated its relationship with the immune microenvironment, there are still some limitations that should be acknowledged. Firstly, the methylation panel used in this study should be further optimized and validated in prospective cohorts. Secondly, the detailed mechanisms by which these methylation sites affect the immune microenvironment need to be explored further.
In conclusion, we used the TCGA database to construct a model that can be used to diagnose and predict recurrence-free survival of HCC, which is related to the immune microenvironment. We also construct a nomogram based on riskScore and clinical factors, which gives a new possible idea for clinical use to stratify the risk of recurrence of HCC in the early and middle stages. Of course, the generalization and use of the model needs to be validated by a large number of subsequent clinical trials. Such validation studies will provide crucial insights into the model's reliability and effectiveness in clinical settings.
Author contribution statement
Ledu Zhou, Guo Long, Xingyu Mi: Conceived and designed the experiments.
Lihua Zhao, Liang Xiao: Conceived and designed the experiments; Wrote the paper.>
Biao Tang: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.>
Wenxin Su: Conceived and designed the experiments; Performed the experiments; Analyzed and interpr>
Funding statement
Ledu Zhou was supported by Hunan Provincial Natural Science Foundation of China {2021JJ31132].
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e19434.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Schulze K., Nault J.C., Villanueva A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J. Hepatol. 2016;65(5):1031–1042. doi: 10.1016/j.jhep.2016.05.035. [DOI] [PubMed] [Google Scholar]
- 3.Pinero F., Dirchwolf M., Pessoa M.G. Biomarkers in hepatocellular carcinoma: diagnosis, prognosis and treatment response assessment. Cells. 2020;9(6) doi: 10.3390/cells9061370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eguchi S., Kanematsu T., Arii S., Omata M., Kudo M., Sakamoto M., et al. Recurrence-free survival more than 10 years after liver resection for hepatocellular carcinoma. Br. J. Surg. 2011;98(4):552–557. doi: 10.1002/bjs.7393. [DOI] [PubMed] [Google Scholar]
- 5.Santoni M., Rizzo A., Kucharz J., Mollica V., Rosellini M., Marchetti A., et al. Complete remissions following immunotherapy or immuno-oncology combinations in cancer patients: the MOUSEION-03 meta-analysis. Cancer Immunol. Immunother. 2023;72(6):1365–1379. doi: 10.1007/s00262-022-03349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu J.X., Zhang X., Miao R.C., Xiang X.H., Fu Y.N., Zhang J.Y., et al. Six-long non-coding RNA signature predicts recurrence-free survival in hepatocellular carcinoma. World J. Gastroenterol. 2019;25(2):220–232. doi: 10.3748/wjg.v25.i2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizzo A., Ricci A.D., Brandi G. Systemic adjuvant treatment in hepatocellular carcinoma: tempted to do something rather than nothing. Future Oncol. 2020;16(32):2587–2589. doi: 10.2217/fon-2020-0669. [DOI] [PubMed] [Google Scholar]
- 8.Yu M.C., Lee C.W., Lin C.H., Wu C.H., Lee Y.S., Tsai C.L., et al. Differential hypermethylation of the VTRNA2-1 promoter in hepatocellular carcinoma as a prognostic factor: tumor marker prognostic study. Int. J. Surg. 2020;79:282–289. doi: 10.1016/j.ijsu.2020.05.016. [DOI] [PubMed] [Google Scholar]
- 9.Rizzo A., Cusmai A., Gadaleta-Caldarola G., Palmiotti G. Which role for predictors of response to immune checkpoint inhibitors in hepatocellular carcinoma? Expert Rev Gastroenterol Hepatol. 2022;16(4):333–339. doi: 10.1080/17474124.2022.2064273. [DOI] [PubMed] [Google Scholar]
- 10.Moore L.D., Le T., Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu R.H., Wei W., Krawczyk M., Wang W., Luo H., Flagg K., et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017;16(11):1155–1161. doi: 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez-Meza G., von Felden J., Gonzalez-Kozlova E.E., Garcia-Lezana T., Peix J., Portela A., et al. DNA methylation profiling of human hepatocarcinogenesis. Hepatology. 2021;74(1):183–199. doi: 10.1002/hep.31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z., Zhu H., Zhang L., Wei Q., Zhou L., Xu X., et al. DNA methylation of SOCS1/2/3 predicts hepatocellular carcinoma recurrence after liver transplantation. Mol. Biol. Rep. 2020;47(3):1773–1782. doi: 10.1007/s11033-020-05271-3. [DOI] [PubMed] [Google Scholar]
- 14.Ma J.H., Huang Y., Liu L.Y., Feng Z. An 8-gene DNA methylation signature predicts the recurrence risk of cervical cancer. J. Int. Med. Res. 2021;49(5) doi: 10.1177/03000605211018443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macias-Garcia L., Martinez-Ballesteros M., Luna-Romera J.M., Garcia-Heredia J.M., Garcia-Gutierrez J., Riquelme-Santos J.C. Autoencoded DNA methylation data to predict breast cancer recurrence: Machine learning models and gene-weight significance. Artif. Intell. Med. 2020;110 doi: 10.1016/j.artmed.2020.101976. [DOI] [PubMed] [Google Scholar]
- 16.Jeyapala R., Kamdar S., Olkhov-Mitsel E., Savio A.J., Zhao F., Cuizon C., et al. An integrative DNA methylation model for improved prognostication of postsurgery recurrence and therapy in prostate cancer patients. Urol. Oncol. 2020;38(2):e1–e9. doi: 10.1016/j.urolonc.2019.08.017. 39. [DOI] [PubMed] [Google Scholar]
- 17.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Karasaki T., Nagayama K., Kuwano H., Nitadori J.I., Sato M., Anraku M., et al. An immunogram for the cancer-immunity cycle: towards personalized immunotherapy of lung cancer. J. Thorac. Oncol. : official publication of the International Association for the Study of Lung Cancer. 2017;12(5):791–803. doi: 10.1016/j.jtho.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Villanueva A. Hepatocellular carcinoma. N. Engl. J. Med. 2019;380(15):1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 20.Roessler S., Jia H.L., Budhu A., Forgues M., Ye Q.H., Lee J.S., et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budhu A., Forgues M., Ye Q.H., Jia H.L., He P., Zanetti K.A., et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10(2):99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 22.Kondo Y., Kanai Y., Sakamoto M., Mizokami M., Ueda R., Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis--A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32(5):970–979. doi: 10.1053/jhep.2000.19797. [DOI] [PubMed] [Google Scholar]
- 23.Long J., Chen P., Lin J., Bai Y., Yang X., Bian J., et al. DNA methylation-driven genes for constructing diagnostic, prognostic, and recurrence models for hepatocellular carcinoma. Theranostics. 2019;9(24):7251–7267. doi: 10.7150/thno.31155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hidalgo B.A., Minniefield B., Patki A., Tanner R., Bagheri M., Tiwari H.K., et al. A 6-CpG validated methylation risk score model for metabolic syndrome: the HyperGEN and GOLDN studies. PLoS One. 2021;16(11) doi: 10.1371/journal.pone.0259836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S., Luo L., He Y., Li R., Xiang Y., Xing Z., et al. Dental pulp stem cell-derived exosomes alleviate cerebral ischaemia-reperfusion injury through suppressing inflammatory response. Cell Prolif. 2021;54(8) doi: 10.1111/cpr.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song K., He F., Xin Y., Guan G., Huo J., Zhu Q., et al. TSC2 mutations were associated with the early recurrence of patients with HCC underwent hepatectomy. Pharmgenomics Pers Med. 2021;14:269–278. doi: 10.2147/PGPM.S294307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye J., Li H., Wei J., Luo Y., Liu H., Zhang J., et al. Risk scoring system based on lncRNA expression for predicting survival in hepatocellular carcinoma with cirrhosis. Asian Pac J Cancer Prev. 2020;21(6):1787–1795. doi: 10.31557/APJCP.2020.21.6.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu T., Han C., Zhu G., Liao X., Qin W., Yang C., et al. Prognostic value of Notch receptors in postsurgical patients with hepatitis B virus-related hepatocellular carcinoma. Cancer Med. 2017;6(7):1587–1600. doi: 10.1002/cam4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Federico A., Rizzo A., Carloni R., De Giglio A., Bruno R., Ricci D., et al. Atezolizumab-bevacizumab plus Y-90 TARE for the treatment of hepatocellular carcinoma: preclinical rationale and ongoing clinical trials. Expert Opin Investig Drugs. 2022;31(4):361–369. doi: 10.1080/13543784.2022.2009455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.