Abstract
IBD is considered a relapsing disease with relapsing phases. Probiotics are beneficial microorganisms that modulate inflammatory signaling pathways. Our aim was to identify the precise molecular effects of probiotics on inflammatory signaling pathways during the presence of inflammation. Evaluation of the expression of JAK/STAT and inflammatory genes after treatment of the HT -29 cell line with the sonicated pathogens and probiotics, simultaneously was performed by quantitative real-time polymerase chain reaction (qPCR) assay. The production of IL-6 and IL-1β after administration of probiotics was conducted by means of cytokine assay. The probiotic cocktail resulted in the downregulation of TIRAP, IRAK4, NEMO, and RIP genes in the NF-кB pathway compared with Sonicat-treated cells. The expression of JAK/STAT genes was various after probiotic treatment. The application of probiotics has been observed to result in a notable decrease in the production of IL-6 and IL-1β. The investigated probiotic cocktail, especially Bifidobacterium spp. showed anti-inflammatory effects on HT -29 cells via modulation of JAK/STAT and NF-кB signaling pathways. The use of probiotics with the least side effects could be considered a suitable treatment for patients with inflammatory bowel disease, even at the beginning of inflammation.
Keywords: Probiotic, JAK/STAT, NF-kB, Inflammation
1. Introduction
Probiotics are live microorganisms that have beneficial effects with favorable effects on health [1]. Several studies reported the therapeutic properties of Lactobacillus spp. and Bifidobacterium spp. in inflammatory diseases, colorectal cancers, microbial infections, and acute diarrhea [2]. Besides, one of the most important properties of probiotics is their ability to modulate innate and adaptive immune responses [3]. Abnormal activities are usually observed in inflammatory bowel disease (IBD). In IBD, immune cells produce a large number of cytokines, especially inflammatory cytokines, including TNF- α, IL-6, IL-1β, and IFN- γ [4]. In this situation, probiotics can play an important role in reducing the inflammatory status. Several studies have been conducted to demonstrate the anti-inflammatory effects of probiotics. Animal models and human studies show that the use of probiotics, such as Bifidobacterium longum, can increase the production of IL-10 in serum [5]. In addition, patients with ulcerative colitis, a subtype of inflammatory bowel disease, have been shown to have a decrease in C-reactive protein when taking Bifidobacterium longum [5]. Different studies also demonstrate that using probiotic strains including Lactobacillus spp. and Bifidobacterium spp. could improve clinical symptoms and pathological evidence in mice with colitis [6,7].
IBD is recognized by chronic inflammation of the gastrointestinal tract. Disturbed homeostasis in the intestinal cells leads to abnormal function of the innate and adaptive immune system and causes inflammation [8]. Two distinct phases are observed in patients with IBD, including remission and relapse, and predicting when relapse will occur is difficult. Although the use of various therapies could help alleviate symptoms during the relapse phase, the persistence of subclinical inflammation in the intestine leads to a return of symptoms [9]. The longer a therapeutic option is maintained in a patient with IBD, the more sustained the beneficial effect and the longer remission is achieved. It seems that probiotics can be effective for up to three weeks after the treatment is stopped [10]. Therefore, probiotics with their anti-inflammatory properties seem to be an appropriate choice to reduce inflammation during the relapse phase.
Several important immune pathways, including the Janus kinase (JAK)-signal transducer and activator of transcription (STAT), and Nuclear Factor kappa-light-chain-enhancer of activated B cells (NF-кB) have been considered as possible sources of probiotics' mechanisms of action [3]. The functions of the JAK/STAT signaling pathway lead to some outcomes that play a crucial role in autoimmune and inflammatory diseases, including IBD [11]. Besides, several studies have shown that NF-кB activation leads to the production of inflammatory cytokines, and that inhibition of NF-кB activity may play an important role in reducing inflammation [12]. Our previous studies have shown phenotypic anti-inflammatory effects of probiotics [13]. However, research on the molecular properties of probiotics is noteworthy to understand how probiotics can reduce inflammatory status. The anti-inflammatory effects of probiotic strains as preventive agents and before the induction of inflammation has been recently demonstrated [14]. Here, we aimed to investigate the efficacy of probiotics in modulating JAK/STAT and NF-кB signaling pathways to understand how probiotics might play an effective role in reducing inflammation during an inflammatory state, especially at the onset of the inflammatory phase. Thus demonstrating the beneficial effects of probiotics in patients with IBD, especially during the relapse and onset of inflammation period, could be so crucial. We also used probiotic cocktails containing four Lactobacillus spp. and three Bifidobacterium spp. and a mixture of Lactobacillus spp. and Bifidobacterium spp. (Lac/Bif) to determine which probiotic might have the best effect against inflammation.
2. Materials and methods
2.1. Bacterial strain and cell culture procedure
2.2. The in-vitro assay using HT-29 cell line was performed to evaluate the effects of probiotics on the NF-кB and JAK/STAT signaling pathways, in the present study. Probiotic strains including four Lactobacillus spp., including L. plantarum, L. rhamnosus, L. brevis, and L. reuteri, were isolated from the fecal samples of 53 volunteers of healthy individuals and three Bifidobacterium spp., including B. bifidum, B. longum, and B. infantis, that were isolated from breast milk. Also, induction of inflammation was performed by using sonicated enterotoxin-producing Escherichia coli (ETEC) and Salmonella typhimurium (ST). All these procedures along with bacterial suspensions and cell cultures have been described, previously [[13], [14], [15]]. In brief, the bacterial strains were inoculated into MRS broth containing 0.05% l-cysteine and incubated for 20 h at 37 °C. For preparing the bacterial suspension, the culture pellet of probiotic strains was collected and diluted in RPMI-1640 with 10% FBS without antibiotics to reach an Optical Density of 0.08 at 600 nm. Also, enterotoxigenic Escherichia coli (ETEC) and Salmonella typhimurium (ST), were cultured in Luria-Bertani (LB) broth (Thermo Fisher Scientific, US), followed by sonication (10 rounds, 1 min/round, and the cellular debris was centrifuged 1700 g, 15 min, 4 °C) to disrupt the cell integrity. Also, the cell culture was performed using HT-29 cells which were grown in RPMI-1640 (Thermo-Gibco, USA) supplemented with 10% fetal bovine serum (Biochrom, Berlin, Germany) and 1% penicillin-streptomycin (Sigma Aldrich, UK) to reach the final optical density concentration (OD) of 0.08 at 600 nm (1 × 108 CFU/ml). Cultures were performed at 37 °C in a 5% CO2 and 95% filtered air atmosphere. The cells were detached by 0.25% Trypsin-EDTA (Gibco, USA), washed twice with PBS, and counted. Finally, the cell suspension was centrifuged, the precipitate was diluted with RPMI-1640, and 2 × 105 cells/cm2 per well were seeded into the 6-well plates with a volume of 2 ml for 24 h. This study was performed according to the relevant guidelines and regulations, and ethical approval for the previous study was obtained from the Pasteur Institute of Iran Committee (IR.PII. REC.1398.060). In accordance with ethical research standards, all participants were required to provide their signature on a document indicating their full understanding and consent of the study's objectives and procedures prior to any data collection. Sonicated pathogen and probiotics treatments.
HT -29 cells were exposed to various bacteria, either alone or as mixtures. The constituent agents consisted of sonicated enterotoxigenic E. coli (SP-ETEC) and sonicated Salmonella typhi (SP-ST), employed as inducers of inflammation. It should be noted that the use of the sonication procedure resulted in the extraction of the inflammatory substances, including lipopolysaccharide (LPS), which cause inflammation. Lactobacillus spp. alone, Bifidobacterium spp. alone, Lactobacillus/Bifidobacterium mixture (Lac/Bif) were used as putative probiotic strains. In the present study, we aimed to evaluate the effects of using probiotic strains during the presence of inflammation. Therefore, to study the effect of probiotics during the inflammatory state, 1 ml of each strain of Lactobacillus spp., Bifidobacterium spp., and Lac/Bif were added simultaneously along with 1 ml of each of SP-ETEC and SP-ST, for 6 h. After 6 h, each well was washed twice with PBS to remove the non-adherent bacteria. These treatments were performed in duplicate and the cell culture was maintained at 37 °C and 5% CO2 for up to 48 h. Three biological repetition were used in cell culture assay. The determination of the Moment of Inertia (MOI) was executed in accordance with the previously specified methodology [16].
2.2. Cytokine assays
To evaluate the phenotypic affects of probiotic intervention on the alleviation of inflammation, an examination was conducted on cytokine production through employment of the ELISA assay (Roche Diagnostics, Germany). After SP and probiotic treatments, the supernatant of the cell culture was centrifuged at 4427 g, and the supernatant was collected to evaluate the production of pro-inflammatory cytokines, including IL-6 and IL-1β. All experiments were performed in duplicate.
2.3. RT- PCR of inflammatory signaling pathway genes
An RNA extraction kit (Roche, Germany) and the cDNA synthesis kit (Yekta Tajhiz, Iran) were used according to the manufacturer's instructions. The online website Primer Bank (http://pga.mgh.harvard.edu/primerbank) was used to select qPCR primers (Table 1). All reactions were performed in duplicate. The formula RQ = 2−ΔΔCt was used to obtain the relative gene expression in the comparative CT method [17]. The RT- PCR method was performed according to our previous study [14]. All reactions were performed in duplicate.
Table 1.
Primer sequences used in this study.
| Gene | Primer Sequence [5' > 3'] | Primer Bank ID | Product Size [bp] |
|---|---|---|---|
| STAT1 F STAT1 R |
CGGCTGAATTTCGGCACCT CAGTAACGATGAGAGGACCCT |
189458859c3 | 81 |
| STAT2 F STAT2 R |
CTGCTAGGCCGATTAACTACCC TCTGATGCAGGCTTTTTGCTG |
291219923c3 | 87 |
| STAT3 F STAT3 R |
ACCAGCAGTATAGCCGCTTC GCCACAATCCGGGCAATCT |
47080104c2 | 124 |
| STAT4 F STAT4 R |
GCTTAACAGCCTCGATTTCAAGA GAGCATGGTGTTCATTAACAGGT |
345110659c2 | 91 |
| STAT5 F STAT5 R |
CGACGGGACCTTCTTGTTG GTTCCGGGGAGTCAAACTTCC |
221316717c3 | 80 |
| STAT6 F STAT6 R |
CGAGTAGGGGAGATCCACCTT GCAGGAGTTTCTATCAAGCTGTG |
296010867c2 | 92 |
| JAK1 F JAK1 R |
CTTTGCCCTGTATGACGAGAAC ACCTCATCCGGTAGTGGAGC |
102469033c1 | 101 |
| JAK2 F JAK2 R |
ATCCACCCAACCATGTCTTCC ATTCCATGCCGATAGGCTCTG |
223671934c2 | 121 |
| JAK3 F JAK3 R |
CTGCACGTAGATGGGGTGG CACGATCAGGTTGGACTTTTCT |
189095272c2 | 78 |
| TYK2 F TYK2 R |
GAGATGCAAGCCTGATGCTAT GGTTCCCGAGGATTCATGCC |
187608614c1 | 76 |
| RIP2 F RIP2 R |
GCCCTTGGTGTAAATTACCTGC GGACATCATGCGCCACTTT |
93141034c2 | 138 |
| NEMO F NEMO R |
AAGAGCCAACTGTGTGAGATG TTCGCCCAGTACGTCCTGA |
142381344c1 | 69 |
| TIRAP F TIRAP R |
GACCCCTGGTGCAAGTACC CGACGTAGTACATGAATCGGAG |
89111123c2 | 133 |
| IRAK4 F IRAK4 R |
CTTGGATGGTACTCCACCACT AAAATTGATGCCATTAGCTGCAC |
223671887c3 | 76 |
2.4. Statistical analysis
Graphs and statistical analysis of the data were performed using SPSS (ver.25) and GraphPad Prism software (ver. 8) to compare variables of different groups. Statistically significant distinctions were evaluated between numerous groups, encompassing control (C), sonicated pathogen (SP), Lactobacillus spp., and SP that were added concurrently (P + L), Bifidobacterium spp., and SP that were added concurrently (P + B), and Lac/Bif and SP that were added concurrently (P + LB) using ordinary one-way ANOVA. P-values<0.05 were considered statistically significant. The results were presented as Standard Deviation (SD).
3. Results
3.1. The results of pro-inflammatory cytokines production
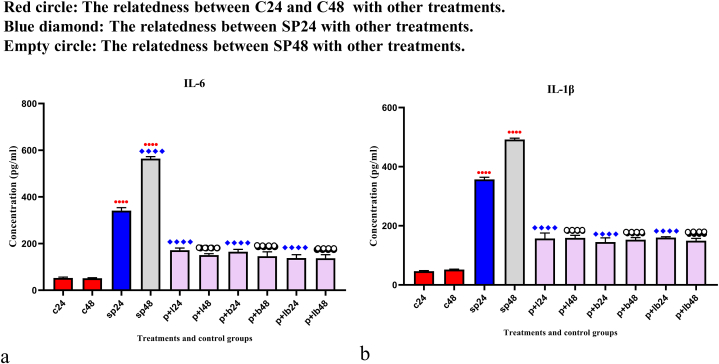
The results of pro-inflammatory cytokines production are shown in Fig. 1. Cytokine production was significantly higher after SP treatments (p < 0.0001). However, probiotic treatments (6 h after SP treatment) significantly decreased cytokine production (p < 0.0001). No significant difference was observed between Lactobacillus spp., Bifidobacterium spp. and Lac/Bif at any time point after the treatments.
Fig. 1.
Different levels of concentrations of (a) IL-6 and (b) IL-1β. Data were represented as mean SD. The number 24 and 48 refers to different time orders of HT-29 cell line treatments. C, control; SP, Sonicated Pathogen; P + L, sonicated pathogen, and Lactobacillus spp. were added simultaneously.; P + B, sonicated pathogen, and Bifidobacterium spp. were added simultaneously; P + LB, sonicated pathogen, and Lac/Bif were added simultaneously. Data were considered as statistically significant when p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001).
3.2. The results of gene expression
The efficacy of probiotics in up- or down-regulating the studied genes was examined by comparing probiotic + SP treated HT -29 cells with control cells (unexposed HT -29 cells as negative controls, i.e., C24 and C48) and HT -29 cells exposed to the sonicated pathogen as positive controls (i.e., SP24 and SP48).
3.2.1. Probiotic strains had different up- and down-regulations of STAT genes
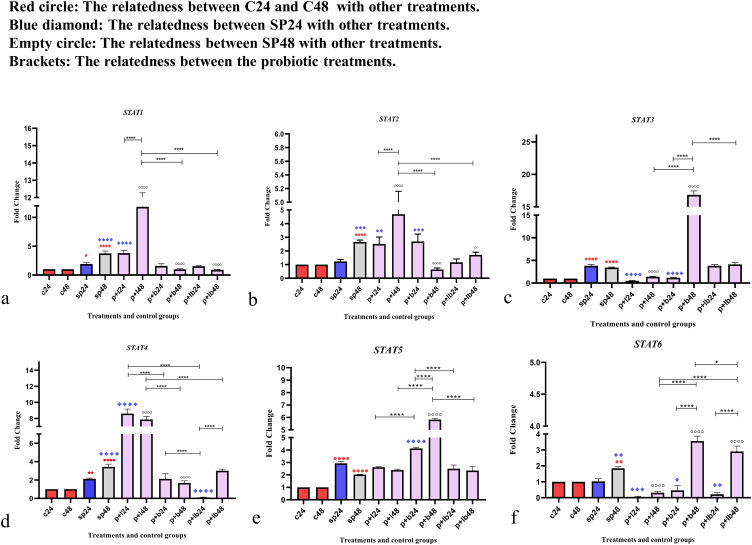
Data on STAT gene expression are shown in Fig. 2. Comparative analysis of the STAT genes showed various alters in gene expression levels. Some of the probiotic treatments significantly up-regulated the gene expression, while the others decreased the expression level. The comparative analysis of STAT gene expression between the sonicated pathogens and the negative controls showed that SP-ETEC and SP-ST could significantly increase the level of gene expression in most STAT genes.
Fig. 2.
Relative gene expression (mean fold change) of (a) STAT1, (b) STAT2, (c) STAT3, (d) STAT4, (e) STAT5, and (f) STAT6 in the different groups of treatments. Data were normalized with gapdh. Data were represented as mean SD. The number 24 and 48 refers to different time orders of HT-29 cell line treatments. C, control; SP, Sonicated Pathogen; P + L, sonicated pathogen, and Lactobacillus spp. were added simultaneously.; P + B, sonicated pathogen, and Bifidobacterium spp. were added simultaneously; P + LB, sonicated pathogen, and Lac/Bif were added simultaneously. Data were considered as statistically significant when p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001).
In STAT1, Lactobacillus spp. and Bifidobacterium spp. along with Lac/Bif had opposite effects on gene expression. Lactobacillus spp. had the strongest effect on upregulating expression after 48 h of treatment (P + L48) compared to negative and positive controls (p < 0.0001). On the other hand, in comparison to the positive control (SP48), Bifidobacterium spp. and Lac/Bif had the most significant effects on reducing the expression level, especially after 48 h of treatment (P + B48 and P + LB48) (p < 0.0001).
Comparative analysis of STAT2 gene expression revealed that Lactobacillus spp. (P + L48) could significantly increase gene expression (p < 0.0001), however, Bifidobacterium spp. at 48 h (P + B48) and Lac/Bif (P + LB48) could decrease expression level (p < 0.05). Also, concerning 48-h treatment, Bifidobacterium spp. (P + B48) had a stronger reduction effect compared to Lac/Bif (P + LB48) (p < 0.01).
In STAT3, after 48 h of treatment, Bifidobacterium spp. (P + B48) was the only probiotic that could significantly upregulate the expression level compared to the negative and positive controls (p < 0.0001), while Lactobacillus spp. decreased the expression level in both different time sequences (P + L24 and P + L48) together with Bifidobacterium spp. in the first 24 h of treatment (P + B24) (p < 0.0001).
In STAT4, it can be said that the general trend of gene expression in terms of Bifidobacterium spp. (P + B48) together with P + LB24 was downward. Actually, P + LB24 had the most reduction effect (p < 0.0001). On the other hand, Lactobacillus spp. showed the most significant increase in expression level in both time series (P + L24 and P + L48) (p < 0.0001).
Comparative analysis of STAT5 gene expression showed that Bifidobacterium spp. was the only probiotic treatment that exhibited a significant difference in the level of gene expression compared to positive controls (SP24 and SP48) and could significantly up-regulate the level of gene expression in both time series, including 24 and 48 h (P + B24, P + B24) (p < 0.0001).
In STAT6, most treatments were able to decrease gene expression, except Bifidobacterium spp. and Lac/Bif after 48 h of treatment (P + B48 and P + LB48), which were able to significantly up-regulate gene expression (p < 0.0001). In other words, the overall effect of Lactobacillus spp. was to decrease gene expression, whereas Bifidobacterium spp. and Lac/Bif decreased expression in the first 24 h of treatment but increased expression after 48 h.
3.2.2. The various results of probiotic treatments on JAK genes expression
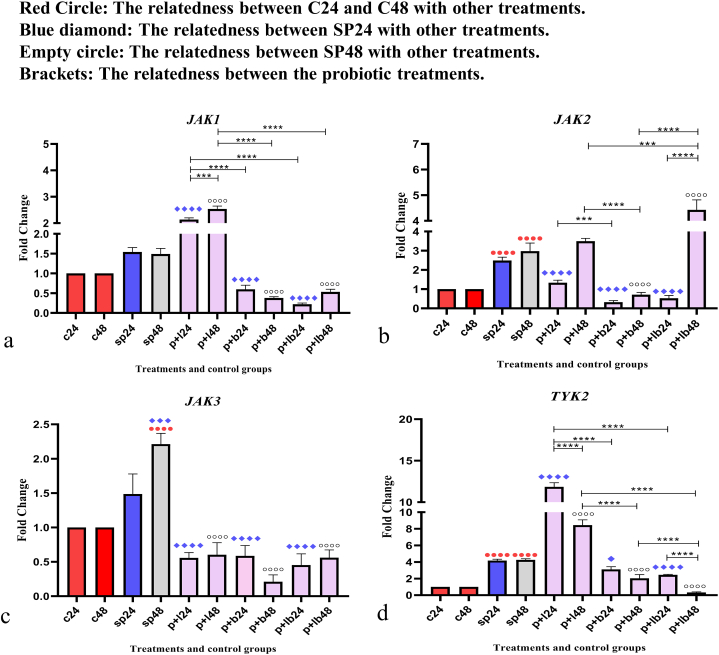
Data on JAK expressions are shown in Fig. 3. Comparative analysis of JAK gene expression between sonicated pathogens and negative control showed that SP-ETEC and SP-ST could significantly increase gene expression.
Fig. 3.
Relative gene expression [mean fold change] of (a) JAK1, (b) JAK2, (c) JAK3, and (d) TYK2 in the different groups of treatments. Data were normalized with gapdh. Data were represented as mean SD. The number 24 and 48 refers to different time orders of HT-29 cell line treatments. C, control; SP, Sonicated Pathogen; P + L, sonicated pathogen, and Lactobacillus spp. were added simultaneously.; P + B, sonicated pathogen, and Bifidobacterium spp. were added simultaneously; P + LB, sonicated pathogen, and Lac/Bif were added simultaneously. Data were considered as statistically significant when p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001).
Comparative analysis of JAK1 showed that Lactobacillus spp. up-regulated the expression level, especially after 48 h of treatment (P + L48) (p < 0.001). In contrast, Bifidobacterium spp. and Lac/Bif decrease gene expression in both time periods.
For JAK2, Lactobacillus spp. and Lac/Bif (P + L48 and P + LB48) were able to increase the expression level after 48 h of treatment, with P + LB48 having the most significant effect (p < 0.001), while the expression level decreased in the first 24 h of treatment. In contrast, Bifidobacterium spp. were able to down-regulate gene expression in both time periods (P + B24 and P + B48) (p < 0.0001).
Comparative analysis of JAK3 showed a downward trend in expression. All probiotic treatments could significantly decrease the expression level compared to positive controls (p < 0.0001).
Comparative analysis of TYK2 showed that Lactobacillus spp. up-regulated the expression level mainly after 24 h of treatment (P + L24) (p < 0.0001), while Bifidobacterium spp. and Lac/Bif were able to decrease the expression level in both time sequences (p < 0.05). It should be noted that P + LB48 had the strongest reduction effect (p < 0.0001).
3.2.3. Probiotic treatments lead to a downward trend in inflammatory genes expression
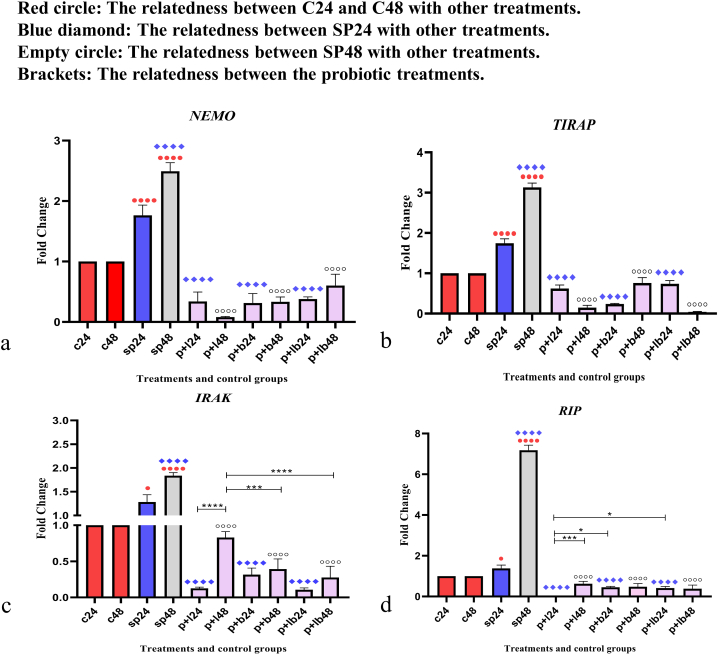
Inflammatory gene expression data are shown in Fig. 4. Comparative analysis of inflammatory gene expression, including NEMO, TIRAP, IRAK, and RIP between sonicated pathogens and negative controls showed that SP-ETEC and SP-ST could significantly increase gene expression, especially after 48 h (p < 0.0001). A downward trend in gene expression was observed for all studied genes and all probiotic treatments down-regulated inflammatory genes (p < 0.001). Although all probiotics had a reducing effect, the extent varied somewhat. For example, Lactobacillus spp. had the weakest reducing effect concerning IRAK expression level (P + L48) (p < 0.0001), whereas Lactobacillus spp. had almost the strongest reducing effect on the RIP gene in the 24-h treatment (P + L24) (p < 0.05).
Fig. 4.
Relative gene expression [mean fold change] of (a) NEMO, (b) TIRAP, (c) IRAK, and (d) RIP in the different groups of treatments. Data were normalized with gapdh. Data were represented as mean SD. The number 24 and 48 refers to different time orders of HT-29 cell line treatments. C, control; SP, Sonicated Pathogen; P + L, sonicated pathogen, and Lactobacillus spp. were added simultaneously.; P + B, sonicated pathogen, and Bifidobacterium spp. were added simultaneously; P + LB, sonicated pathogen, and Lac/Bif were added simultaneously. Data were considered as statistically significant when p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001).
4. Discussion
As a relapsing disease, IBD is usually recognized between the ages of 15 and 25, however, most treatments are used to control symptoms, and patients usually need to continuously use various treatment options [18]. Subtypes of IBD, including ulcerative colitis, may have a more severe course and require more aggressive strategies, including surgery, to reduce severity [19]. According to various studies, the medical cost of IBD is $4 billion (USD) per year, especially if the patient requires hospitalization [20]. Therefore, the implementation of the experimental procedure to improve the effective treatment options with the least side effects may be necessary to improve the quality life of IBD patients. As mentioned above, immune system overactivity in IBD along with suppression of regulatory T cells (Tregs) has been demonstrated in IBD, and probiotics may play a critical role in modulating the signaling pathways involved in the immune system and progression of IBD [18]. Thus, in the present study, we aimed to investigate the precise molecular mechanisms of probiotics in three formats Lactobacillus spp., Bifidobacterium spp, and Lac/Bif (mixed form of Lactobacillus spp. and Bifidobacterium spp.) to find out, first, whether probiotics can influence the signaling pathway and, second, which probiotic version has the best effects. We also used sonicated pathogens as inflammatory triggers and probiotics simultaneously to elucidate the effects of probiotics on the occurrence of inflammation and to understand the efficacy of taking probiotics even in the early stages of IBD detection.
In the present study, comparative analysis of JAK/STAT genes revealed differential changes in gene expression. The general trend for almost all JAK/STAT genes was downward. One of the remarkable results is that Lactobacillus spp. and Bifidobacterium spp. seem to have opposite effects on most JAK/STAT genes. The results showed that, Lactobacillus spp. could significantly upregulate the expression level of STAT1, STAT2, STAT4, JAK1, JAK2, and TYK2, while, Bifidobacterium spp. and Lac/Bif almost had a significant reduction effect. For STAT3, STAT5, and STAT6 Bifidobacterium spp. together with Lac/Bif increased gene expression levels, especially compared to negative controls and SP48, while Lactobacillus spp. was able to downregulate the expression levels, especially of STAT3 and STAT6.
These observations have been confirmed by other reports. According to various reports, STATs, including STAT1 and STAT3, may play an important role in triggering inflammation and, together with NF-κB signaling pathways, activate the progression of IBD [21,22]. In addition, studies have shown that Th1 and Th17 are associated with STAT1 and STAT3. Since dysregulation of Th1 and Th17 may lead to the development of IBD, the use of agents with inhibitory effects on STAT1 and STAT3 may target Th1 and Th17 and be useful to prevent IBD [23]. Some of the reports also focused on other components of STATs, including STAT4, and showed the crucial role of STAT4 in autoimmune diseases such as autoimmune encephalomyelitis, rheumatoid arthritis, and IBD. They demonstrated that increased expression and activation of the IL-12-induced STAT4 signaling was generally found in the mucosa of patients with UC [24]. Regarding STAT6, studies have shown that activation of STAT6 was observed in ulcerative colitis [25]. Therefore, it seems that finding an inhibitory agent to suppress STAT4 and STAT6 may play a crucial role in the treatment of IBD. However, it appears that the different components of STATs sometimes may play different roles in controlling IBD. For example, activation of STAT3, as mentioned above, plays different roles in IBD progression. On the contrary, according to Zundler et al. STAT3 might promote IL-10-dependent Treg function and thus have effects on Treg activity and improve inflammatory status in IBD patients [26]. As shown by the current study, the inactivation of STATs could be helpful in controlling IBD. However, activation of STATs might also play a crucial role in controlling of IBD. Thus, it could be concluded that STATs have different roles that sometimes seem to be opposite.
Concerning JAK expression, as could be seen in Fig. 2, Bifidobacterium spp. and Lac/Bif almost downregulated the expression of JAK genes, while, Lactobacillus spp. increased the expression level, especially for JAK1, JAK2, and TYK2. According to various studies that were conducted to introduce different treatments for IBD, key cytokines in the pathogenesis of IBD are targeted by Janus kinase (JAK) inhibitors [27]. Indeed, probiotics act as JAK inhibitors here and thus could play a crucial role in controlling IBD. In this case, Bifidobacterium spp. was able to decrease gene expression in all JAK genes. However, JAKs are associated with anti-inflammatory cytokines, including IL -4 and IL-10, so increasing gene expression may have some anti-inflammatory effects [28].
The current study also examined the effect of probiotics on the NF-кB pathway. The role of pathogens in causing inflammation was completely clear, as the expression of inflammatory genes was clearly observed after exposure to pathogens. However, probiotic treatments had a remarkable reducing effect. All probiotic versions had a reducing effect on the expression of inflammatory genes. A reduction to zero was observed for the genes TIRAP and RIP. Since these genes play an important role in inflammatory signaling pathways, a reduction in their expression could lead to a reduction in the production of pro-inflammatory cytokines [29].
On the other hand, the ELISA assay also showed that using probiotics simultaneously with inflammatory agents could significantly decrease the production of the pro-inflammatory cytokines, including IL-6 and IL-1β. According to other reports, various cytokines, including IL-1α, IL-1β, IL-2, IL-6, TNF-α, and IFNγ are associated with the development and progression of ulcerative colitis and Crohn's disease as two types of IBD. Indeed, the production of inflammatory cytokines has such a great effects on the development of the IBD that blocking the function of these cytokines could be considered as one of the most important therapeutic options to control IBD [30,31]. According to our results, probiotics as safe agents with the least side effects could be an appropriate option to control the symptoms of IBD.
It is worth mentioning that our study may have certain limitations that should be taken into consideration. As mentioned in the Material and Methods section, the duration of probiotic administration was up to 48 h. It seems that a longer administration of probiotics would lead to clearer and more meaningful results on the anti-inflammatory and immunomodulatory effects of our probiotic strains. Furthermore, by adding another treatment group in which probiotic treatments were performed alone and without the use of sonicated pathogens, we were able to show the usefulness of using probiotic strains even in a healthy situation without the presence of an inflammatory status.
5. Conclusion
The present study provides remarkable evidence for the effect of probiotics on immune system modulation and anti-inflammatory activity even in the presence of inflammation. Our previous study has shown that the use of probiotic strains, especially Lactobacillus spp. prior to inflammation induction could have remarkable anti-inflammatory effects [14]. Here, we used three different formats of probiotic treatment to determine which format might have the best anti-inflammatory effect during inflammation. Considering the idea that reducing the expression of JAK/STAT along with NF-кB might have a better effect on controlling inflammation, it seems that Bifidobacterium spp. with the greatest reduction in effect on JAK/STAT and NF-кB was the most effective probiotic during the presence of inflammation in the current study. Also, the comparison of the data between in vivo and phenotypic results with the current molecular examination showed the high probability of clinical relevance of probiotic strains in reducing inflammation. As mentioned earlier, IBD is a relapsing disease and the current study was able to confirm the hypothesis that probiotics may play a crucial role at the onset of inflammation. The utilization of probiotics that have minimal adverse reactions may prove to be beneficial in enhancing the overall health of patients. It is imperative to discover the most effortless approach to enhancing the quality of life of individuals with IBD, thereby emphasizing the crucial significance of such probiotics.
Declarations
Author contribution statement
Mahdi Rohani: Conceived and designed the experiments; Wrote the paper.
Mohammad R. Pourshafie: Conceived and designed the experiments.
Shadi Aghamohammad: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Seyedeh Tina Miri and Saeideh Najafi: Performed the experiments.
Amin Sepehr: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Data availability statement
Data will be made available on request.
Additional information
No additional information is available for this paper.
Ethics approval and consent to participate
This study was established following the Declaration of Helsinki and approved by the ethics committee of Pasteur Institute of Iran (IR.PII.REC.1398.060). Signed informed consent was obtained from all participants.
Consent for publication
Not applicable.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Acknowledgments
The authors would like to thank Pasteur Institute of Iran as funding agency (with grant number 1694) and Dr. Sana Eybpoosh in the Department of Epidemiology and Biostatistics of Pasteur Institute of Iran for her help and support.
Contributor Information
Shadi Aghamohammad, Email: shadi.aghamohammad@yahoo.com.
Amin Sepehr, Email: aminsepehr20@gmail.com.
Seyedeh Tina Miri, Email: tina.miri@yahoo.com.
Saeideh Najafi, Email: saeideh.njf@gmail.com.
Mohammad R. Pourshafie, Email: pour62@yahoo.com.
Mahdi Rohani, Email: kia.rohani@gmail.com.
References
- 1.Presti I., D’orazio G., Labra M., La Ferla B., Mezzasalma V., Bizzaro G., et al. Evaluation of the probiotic properties of new Lactobacillus and Bifidobacterium strains and their in vitro effect. Appl. Microbiol. Biotechnol. 2015;99(13):5613–5626. doi: 10.1007/s00253-015-6482-8. [DOI] [PubMed] [Google Scholar]
- 2.Abdelhamid A.G., El-Masry S.S., El-Dougdoug N.K. Probiotic Lactobacillus and Bifidobacterium strains possess safety characteristics, antiviral activities and host adherence factors revealed by genome mining. EPMA J. 2019;10(4):337–350. doi: 10.1007/s13167-019-00184-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacPherson C.W., Shastri P., Mathieu O., Tompkins T.A., Burguière P. Genome-wide immune modulation of TLR3-mediated inflammation in intestinal epithelial cells differs between single and multi-strain probiotic combination. PLoS One. 2017;12(1) doi: 10.1371/journal.pone.0169847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang S.W., Chang C.H., Tai T.F., Chang T.C. Comparison of the β-glucuronidase assay and the conventional method for identification of Escherichia coli on eosin-methylene blue agar. J. Food Protect. 1997;60(1):6–9. doi: 10.4315/0362-028x-60.1.6. [DOI] [PubMed] [Google Scholar]
- 5.Abraham B.P., Quigley E.M. Probiotics in inflammatory bowel disease. Gastroenterol. Clin. 2017;46(4):769–782. doi: 10.1016/j.gtc.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Toumi R., Abdelouhab K., Rafa H., Soufli I., Raissi-Kerboua D., Djeraba Z., et al. Beneficial role of the probiotic mixture Ultrabiotique on maintaining the integrity of intestinal mucosal barrier in DSS-induced experimental colitis. Immunopharmacol. Immunotoxicol. 2013;35(3):403–409. doi: 10.3109/08923973.2013.790413. [DOI] [PubMed] [Google Scholar]
- 7.Toumi R., Soufli I., Rafa H., Belkhelfa M., Biad A., Touil-Boukoffa C. Probiotic bacteria lactobacillus and bifidobacterium attenuate inflammation in dextran sulfate sodium-induced experimental colitis in mice. Int. J. Immunopathol. Pharmacol. 2014;27(4):615–627. doi: 10.1177/039463201402700418. [DOI] [PubMed] [Google Scholar]
- 8.Owaga E., Hsieh R.-H., Mugendi B., Masuku S., Shih C.-K., Chang J.-S. Th17 cells as potential probiotic therapeutic targets in inflammatory bowel diseases. Int. J. Mol. Sci. 2015;16(9):20841–20858. doi: 10.3390/ijms160920841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liverani E., Scaioli E., Digby R.J., Bellanova M., Belluzzi A. How to predict clinical relapse in inflammatory bowel disease patients. World J. Gastroenterol. 2016;22(3):1017. doi: 10.3748/wjg.v22.i3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morelli L., Pellegrino P. A critical evaluation of the factors affecting the survival and persistence of beneficial bacteria in healthy adults. Benef. Microbes. 2021 Aug 30;12(4):15–25. doi: 10.3920/BM2021.0017. PubMed PMID: 34323162. Epub 2021/07/30. eng. [DOI] [PubMed] [Google Scholar]
- 11.Kim T.-W., Shin J.-S., Chung K.-S., Lee Y.-G., Baek N.-I., Lee K.-T. Anti-inflammatory mechanisms of koreanaside A, a lignan isolated from the flower of forsythia koreana, against LPS-induced macrophage activation and DSS-induced colitis mice: the crucial role of AP-1, NF-κB, and JAK/STAT signaling. Cells. 2019;8(10):1163. doi: 10.3390/cells8101163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dejban P., Nikravangolsefid N., Chamanara M., Dehpour A., Rashidian A. The role of medicinal products in the treatment of inflammatory bowel diseases (IBD) through inhibition of TLR4/NF‐kappaB pathway. Phytother Res. 2021;35(2):835–845. doi: 10.1002/ptr.6866. [DOI] [PubMed] [Google Scholar]
- 13.Rohani M., Noohi N., Talebi M., Katouli M., Pourshafie M.R. Highly heterogeneous probiotic Lactobacillus species in healthy Iranians with low functional activities. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0144467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aghamohammad S., Sepehr A., Miri S.T., Najafi S., Rohani M., Pourshafiea M.R. The effects of the probiotic cocktail on modulation of the NF-kB and JAK/STAT signaling pathways involved in the inflammatory response in bowel disease model. BMC Immunol. 2022;23(1):1–10. doi: 10.1186/s12865-022-00484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eshaghi M., Bibalan M.H., Rohani M., Esghaei M., Douraghi M., Talebi M., et al. Bifidobacterium obtained from mother's milk and their infant stool; A comparative genotyping and antibacterial analysis. Microb. Pathog. 2017;111:94–98. doi: 10.1016/j.micpath.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Ghanavati R., Asadollahi P., Shapourabadi M.B., Razavi S., Talebi M., Rohani M. Inhibitory effects of Lactobacilli cocktail on HT-29 colon carcinoma cells growth and modulation of the Notch and Wnt/β-catenin signaling pathways. Microb. Pathog. 2020;139 doi: 10.1016/j.micpath.2019.103829. [DOI] [PubMed] [Google Scholar]
- 17.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008 2008/06/01;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 18.Philippe D., Favre L., Foata F., Adolfsson O., Perruisseau-Carrier G., Vidal K., et al. Bifidobacterium lactis attenuates onset of inflammation in a murine model of colitis. World J. Gastroenterol.: WJG. 2011;17(4):459. doi: 10.3748/wjg.v17.i4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fornaro R., Caratto M., Barbruni G., Fornaro F., Salerno A., Giovinazzo D., et al. Surgical and medical treatment in patients with acute severe ulcerative colitis. Journal of digestive diseases. 2015;16(10):558–567. doi: 10.1111/1751-2980.12278. [DOI] [PubMed] [Google Scholar]
- 20.Constantin J., Atanasov P., Wirth D., Borsi A. Indirect costs associated with ulcerative colitis: a systematic literature review of real-world data. BMC Gastroenterol. 2019;19(1):1–10. doi: 10.1186/s12876-019-1095-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang M., Zhong G., Zhu Y., Wang L., He Y., Sun Q., et al. Retardant effect of dihydroartemisinin on ulcerative colitis in a JAK2/STAT3-dependent manner. Acta Biochim. Biophys. Sin. 2021;53(9):1113–1123. doi: 10.1093/abbs/gmab097. [DOI] [PubMed] [Google Scholar]
- 22.Xie F., Zhang H., Zheng C., Shen X.-F. Costunolide improved dextran sulfate sodium-induced acute ulcerative colitis in mice through NF-κB, STAT1/3, and Akt signaling pathways. Int. Immunopharm. 2020;84 doi: 10.1016/j.intimp.2020.106567. [DOI] [PubMed] [Google Scholar]
- 23.Tao F., Qian C., Guo W., Luo Q., Xu Q., Sun Y. Inhibition of Th1/Th17 responses via suppression of STAT1 and STAT3 activation contributes to the amelioration of murine experimental colitis by a natural flavonoid glucoside icariin. Biochem. Pharmacol. 2013;85(6):798–807. doi: 10.1016/j.bcp.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Glas J., Seiderer J., Nagy M., Fries C., Beigel F., Weidinger M., et al. Evidence for STAT4 as a common autoimmune gene: rs7574865 is associated with colonic Crohn's disease and early disease onset. PLoS One. 2010;5(4) doi: 10.1371/journal.pone.0010373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosen M.J., Frey M.R., Washington K.M., Chaturvedi R., Kuhnhein L.A., Matta P., et al. STAT6 activation in ulcerative colitis: a new target for prevention of IL-13-induced colon epithelial cell dysfunction. Inflamm. Bowel Dis. 2011;17(11):2224–2234. doi: 10.1002/ibd.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zundler S., Neurath M.F. Integrating immunologic signaling networks: the JAK/STAT pathway in colitis and colitis-associated cancer. Vaccines. 2016;4(1):5. doi: 10.3390/vaccines4010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Vries L., Wildenberg M., De Jonge W., D'Haens G. The future of Janus kinase inhibitors in inflammatory bowel disease. Journal of Crohn's and Colitis. 2017;11(7):885–893. doi: 10.1093/ecco-jcc/jjx003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busch-Dienstfertig M., González-Rodríguez S. IL-4, JAK-STAT signaling, and pain. JAK-STAT. 2013;2(4) doi: 10.4161/jkst.27638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao L., Li R., Chen X., Xue Y., Liu D. Neougonin A inhibits lipopolysaccharide-induced inflammatory responses via downregulation of the NF-kB signaling pathway in RAW 264.7 macrophages. Inflammation. 2016;39(6):1939–1948. doi: 10.1007/s10753-016-0429-9. [DOI] [PubMed] [Google Scholar]
- 30.Tigari P., Janadri S., Madhu K., Taj N. Evaluation of anti-inflammatory effect of wedelolactone on indomethacin induced colitis in rats: involvement of IL-6/STAT3 pathway. Biointerface Res Appl Chem. 2021;12:2813–2825. [Google Scholar]
- 31.Marafini I., Sedda S., Dinallo V., Monteleone G. Inflammatory cytokines: from discoveries to therapies in IBD. Expet Opin. Biol. Ther. 2019;19(11):1207–1217. doi: 10.1080/14712598.2019.1652267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.