Abstract
The controlled orientation of biomolecules on the sensor surface is crucial for achieving high sensitivity and accurate detection of target molecules in biosensing. FcγRI is an immune cell surface receptor for recognizing IgG-coated targets, such as opsonized pathogens or immune complexes. It plays a crucial role in T cell activation and internalization of the cargos, leading downstream signaling cascades. In this study, we repurposed the FcγRI as an analytical ligand molecule for site-oriented ADA capture, a monoclonal antibody-based biosimilar drug, on a plasmonic sensor surface and demonstrated the real-time detection of the corresponding analyte molecule, TNF-α. The study encompasses the analysis of comparative ligand behaviors on the surface, biosensor kinetics, concentration-dependent studies, and sensor specificity assays. The findings of this study suggest that FcγRI has a significant potential to serve as a universal ligand molecule for site-specific monoclonal antibody capture, and it can be used for biosensing studies, as it represents low nanomolar range affinity and excellent selectivity towards the target. However, there is still room for improvement in the surface stability and sensing response, and further studies are needed to reveal its performance on the monoclonal antibodies with various antigen binding sites and glycoforms.
Keywords: Fc gamma receptor I, Monoclonal antibody, TNF-α biosensor, Surface plasmon resonance
Graphical abstract

Highlights
-
•
FcgRI ectodomain was utilized for site-oriented ADA capture.
-
•
FcgRI ectodomain was utilized as a sensor ligand.
-
•
Tnf-a detection was achieved in the nanomolar range.
-
•
Similar kinetic performance was obtained with the Pro A ligand.
-
•
Sensor accuracy and coefficient of variation analyses were in good agreement
1. Introduction
The classical Fc receptor (FcR) family of Fc gamma receptors (FcγRs) includes several members that, depending on the receptor and the Immunoglobulin G (IgG) subclass, attach IgG molecules to the cell surface with alternating affinities. FcγRs are also divided into activating and inhibitory receptors based on the type of signal transduced. While human cells express FcγRI, FcγRIIa, FcγRIIc, FcγRIIIa, and FcγRIIIb, mice only express FcγRI, FcγRIII, and FcγRIV as activating receptors [1]. Fc gamma receptor I (FcγRI) is expressed on various immune cells and exhibits different effector activities on the target cell. It possesses three extracellular domains (D1, D2, and D3), one transmembrane, and one intracellular domain. Structural studies confirmed that IgG binding interaction occurs in the D1 and D2 domains of FcγRI and the lower hinge site of the Fragment crystallizable (Fc) region in IgG1. The binding interaction depends on the IgG subtype (IgG1, 2, 3, or 4) and the glycosylation profile of the antibodies, and FcγRI shows a nanomolar affinity towards IgG1-type monoclonal antibodies [2].
FcγRI-related studies in the literature have focused on the therapeutic effects of the interactions with the IgGs [[3], [4], [5], [6]]. A few studies reported recombinant FcR production and proposed FcγRs as IgG-capturing agents [[7], [8], [9], [10]]. For example, Brown et al. [11,12] developed an Fc-based array containing antigen-conjugated beads to capture target antibodies in a polyclonal sample solution. Then, the authors evaluated the effector functions of the specified antibodies through FcγRs on multiplex fluorescent beads. Boesch et al., on the other hand [3], recovered serum IgGs through FcγRI, FcγRIIa, and FcγRIIIa immobilized affinity columns and evaluated their effector functions among various glycan profiles and subtypes of IgGs. They reported that FcγRI lost its binding activity after the elution step, while other FcγR proteins retained their IgG binding activities. In another example, FcγRI was used for in vitro imaging of cancer biomarkers, claudin-4, mesothelin, mucin-4, and cadherin-11 [13]. Recently, our group revealed the full analytical potential of the FcγRI ectodomain as an IgG1 capture molecule and compared its performance with a well-known antibody-capturing ligand, Protein A, under various experimental conditions [14].
In the current proof of concept study, we employed FcγRI ectodomain as a ligand molecule for Adalimumab (ADA) capture on a plasmonic surface and investigated its capabilities and limitations for real-time Tumor necrosis factor-Alpha (TNF-α) detection. A graphical illustration of the suggested detection method is presented in Fig. 1. TNF-α is a cytokine that exerts its biological activity in homotrimer form by engaging Type 1 and Type 2 TNF receptors [15]. It regulates immune activities in normal cells, including intracellular pathogen responses, cytotoxicity, and local inflammation. However, its overexpression is related to inflammatory responses and may result in Crohn's disease and rheumatoid arthritis (RA). The TNF-α levels in serum have been studied as a biomarker for autoimmune diseases, monitoring of the treatment, and the alleviation effect of anti-TNF-α agents [[16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27]]. ELISA is the most preferred assay format for detecting TNF-α in serum samples. However, it takes many manual steps, and it is time-consuming. In addition to that, other disadvantages are the single-use ELISA kits and labeled secondary antibodies required for the detection. Therefore, various biosensor formats have been developed to detect TNF-α, some presented in Table S1 [17,18,21,25,26,28]. It is important to note that the standard range of circulated TNF-α levels (usually in the picomolar range) may vary depending on the assay used and the population being studied (elevated under abnormal conditions). Therefore, the reported levels of TNF-α in different studies, including the current study, should be interpreted cautiously.
Fig. 1.
A graphical summary of the suggested detection protocol.
Many TNF-α sensing studies are based on the direct and random attachment of anti-TNF-α antibodies or anti-TNF-α aptamers to the surface via EDC/NHS coupling reaction. Since the performance of the biosensor heavily relies on the proper orientation of the capture element, the conjugation step is a significant factor in the assays. The binding interactions in the current study are assessed using a His capture method for capturing His-tagged FcγRI molecules on a standard carboxymethyl dextran (CM) chip surface. Then, the ligand antibody molecules are captured from their Fc regions through FcγRI molecules in a site-oriented manner, leaving antigen binding sites free for further analyte interactions. A Protein A immobilized sensor channel was also used to compare the data sets in parallel. Several real-time ligand positioning, biosensor performance, and selectivity studies were conducted and compared on FcγRI and Protein A immobilized surfaces.
2. Material & method
2.1. Material
N-(3-(Dimethylamino) propyl)-N′-ethyl carbodiimide hydrochloride (EDC), Sulpho-N-hydroxysuccinimide (NHS), ethanolamine hydrochloride, and CM5-type dextran chip were acquired from Cytiva (USA).
His-tagged FcγRI (NS0-derived human Fc gamma RI, >95% purity), TNF-α (E. coli-derived human TNF-alpha protein, >97% purity), and IL-1β (E. coli-derived human IL-1β, >97% purity) were purchased from R&D Systems (Minnesota, USA).
Protein A (Staphylococcus aureus, ≥95% purity), C1q (Complement component C1q from human serum, 95% purity), and thrombin (Thrombin from human plasma) was bought from Sigma Aldrich (St. Louis, MO, USA). ADA (Humira Pen, 1126059) was bought from AbbVie (Illinois, USA).
SPR running buffer, 1x HBS-EP, was prepared from 10xHBS-EP (0.1 M HEPES, 1.5 M NaCl, 0.03 M EDTA, and 0.5% v/v Surfactant P20, pH 7.4, Cytiva) solution with distilled water.
2.2. Antibody binding analysis
SPR analysis was performed with a Biacore T200 instrument (Cytiva). ADA binding responses were compared with FcγRI captured and Protein A immobilized surface. Protein A, the anti-His antibody, was immobilized on the CM5 (Cat no: 29-1496-03, Cytiva) chip using the amine coupling reaction on the second, third, and fourth flow cells for the CM5 chip.
The anti-His antibody was immobilized for the FcγRI surface with EDC/Sulpho-NHS chemistry on Flow cell 2. Anti-His surface was performed based on the manufacturer guide (His Capture kit, Cytiva). The chip surface was activated by a 1:1 mixture of EDC-NHS reagents for Protein A immobilization. Then, Protein A was diluted to 25 μg/mL in 10 mM pH 5.0 acetate buffer (Cytiva) and coupled through their primary amine groups to one flow cell. The residual activated carboxyl groups were blocked with 1 M ethanolamine-HCl (Cytiva) on the dextran matrix.
FcγRI was captured on the active flow cells for the 60 s with a 10 μL/min flow rate at 22 °C. ADA (ADA) was injected at 15 nM concentration for the 60s on blank and active flow cells. Three different concentrations (3.33 nM, 10 nM, 30 nM) of TNF-α protein samples were injected into both flow cells (active and blank) with 60 s association and 300 s dissociation with 50 μL/min flow rate at 22 °C. The surface was regenerated with 10 mM pH 1.5 glycine (Cytiva) for 60 s. Results were obtained with the double referencing method, where the presented data was subtracted from the zero-concentration sample and the blank surface.
The SPR data were presented as the mean value ± standard deviation (SD) obtained from at least three sample measurements. The kinetic parameters were calculated by Biacore Evaluation Software using the two-state binding model. KD values from affinity analysis were calculated using Biacore Evaluation Software (steady state affinity algorithm).
2.3. Concentration analysis
FcγRI was captured at 500 Response Unit (RU) on the active flow cell for 60 s with a 10 μL/min flow rate at 22 °C. ADA was injected for the 60s on active flow cells with a 500 RU final response level on FcγRI and Protein A chip surfaces. Then, TNF-α sample solutions were injected at six different concentrations (1.11 nM, 3.33 nM, 10 nM, 30 nM, 90, 270 nM) on blank, FcγRI, Protein A surfaces with 60 s association and 300 s dissociation with 50 μL/min flow rate at 22 °C. The surface was regenerated with 10 mM glycine (pH 1.5) for 60 s.
Also, TNF-α spiked samples were prepared with 0.02% BSA in 1X HBS-EP system buffer. Sample dilutions were prepared in 1.25% BSA in 1X HBS-EP. The assay setup was performed as stated above, except for the 270 nM TNF-α sample solution. Results were obtained with the double referencing method, where the presented data was subtracted from the zero-concentration sample and the blank surface [29,30].
The calibration plots were fitted by non-linear regression to the four-parameter logistic(4-PL) equation (GraphPad Prism 5.0). The limit of detection (LOD) is calculated from the three-time the SD of replicate measurements on blank samples [24]. The assay performance was evaluated based on SD, the coefficient of variation (CV%), and the accuracy/recovery values.
2.4. Specificity analysis
Four proteins (C1q, thrombin, IL-1β, and TNF-α) were prepared in 1X HBS-EP system buffer at 30 nM concentration. Sample solutions were injected on active chip surfaces (FcγRI and protein A) and blank surfaces with 60 s association and 300 s dissociation with a 50 μL/min flow rate at 22 °C.
The surface was regenerated with 10 mM glycine (pH 1.5) for 60 s. As stated earlier, Results were obtained with the double referencing method, where the presented data was subtracted from the zero-concentration sample and the blank surface.
3. Results and discussion
3.1. Analytical comparison of FcγRI and protein a for antibody and antigen binding
ADA (Humira) is an IgG1-type monoclonal antibody that interacts with TNF-α with a high affinity (KD: 10−11). Therefore, it effectively treats autoimmune diseases such as RA, Chron's, Bechet, and Psoriasis. It blocks soluble TNF-α antigens for the exaggerated inflammatory response in the tissue. ADA recruits effector functions (Complement-dependent Cell Cytotoxicity, Antibody-Dependent Cell Cytotoxicity) upon engagement with transmembrane TNF-α on the target cell membrane [31,32].
The target antigen (TNF-α) was detected with SPR analysis by comparing two IgG1 capture ligands, FcγRI and Protein A. For this purpose, the binding interactions were analyzed based on the Antibody (ADA) and antigen (TNF-α) binding responses. The FcγRI ectodomain was captured on the anti-His antibody immobilized surface without affecting its critical binding activity (Fig. 2A). Protein A was directly conjugated with EDC/NHS chemistry to the chip surface (Fig. 2B). Immobilized Protein A and captured FcγRI levels were kept constant at 200 RU. As presented in Fig. 2C, the Protein A surface presented a 3-fold higher antibody binding response than the FcγRI surface. This result could be explained by the structure of Protein A bearing five Ig binding domains. Even though Protein A has 5 Ig-binding domains, studies showed that it could bind 2.1 to 3 IgG in solution due to the steric hindrance of IgG molecules [33]. In addition to that, amine coupling randomly immobilizes proteins through free primary amines of Protein A. Thus, antibody binding capacity is affected by both a steric hindrance and an immobilization strategy.
Fig. 2.
Comparison of FcγRI and Protein A for ADA and TNF-α binding. (A) Schematic illustration of the binding analysis with anti-His antibody surface. FcγRI was captured on an anti-His antibody immobilized surface. (B) Schematic illustration of the binding analysis with Protein A. (C) Antibody (ADA) binding performances of FcγRI and Protein A ligands. (D) Antigen (TNF-α) binding performances of FcγRI and Protein A ligands after ADA capture. All data were presented as the mean value obtained from at least three measurements. *p < 0.05, **p < 0.005.
TNF-α binding response was two-fold higher in the Protein A surface than in FcγRI (Fig. 2D). A study performed an SPR assay to show the efficiency of the anti-TNF- α agents in RA patient samples. TNF-α was immobilized with amine coupling to quantify anti-TNF-α antibody molecules within RA patient samples. They reported that the SPR assay correlated with the ELISA assay. According to the results, the TNF-α value for the control group was in the 10 to 20 RU range, and that of the RA patient group was 50–250 RU [34].
Next, we optimized critical assay parameters such as ligand concentration, TNF-α flow rate, and TNF-α concentration range. Kinetic analyses were conducted at low ligand levels (150 RU). In addition to that, the TNF-α sample solution was injected at 50 μL/min to reduce mass transport limitation on the chip surface [35]. TNF-α samples were prepared with a concentration range from 3.33 to 30 nM (3-fold dilution). The KD values of kinetic analysis obtained from the two-state binding model for both ligands were in the 0.08–0.63 nM range for ADA and TNF-α (Table 1). The steady-state affinity values were 9.54 nM and 11.9 nM for FcγRI captured and Protein A surface, respectively (Table 1). Sensorgrams obtained from these experiments are presented in Figure S1. Ogura et al. studied three anti-TNF-α agents (etanercept, infliximab, and ADA) for their binding to the membrane form of TNF-α within Jurkat cells by SPR analysis. One type of their assay was capturing anti-TNF-α agents on a Protein A immobilized chip surface. They obtained KD values for soluble TNF-α in the pM range (5.66–277 pM) for three anti-TNF-α agents [36].
Table 1.
Kinetics and affinity parameters related to TNF-α for ADA on FcγRI captured and Protein A immobilized chip surface. A standard two-state binding model was used for kinetic parameters in Biacore Evaluation Software. A standard steady-state analysis was used in Biacore Evaluation Software.
| KINETICS | |||||||
|---|---|---|---|---|---|---|---|
| Ligand | × 105 (M−1s−1) | (M−1s−1) | × 10−4 (s−1) | (s−1) | (nM) | Chi2 | kt × (108) |
| FcγRI | 15.8 ± 3.6 | 0.013 ± 0.016 | 16.45 ± 6.7 | 0.045 ± 0.05 | 0.63 ± 0.08 | 0.25 ± 0.04 | 5.55 ± 0.08 |
| Protein A | 14.1 ± 0.9 | 0.002 ± 0.002 | 0.75 ± 0.18 | 0.009 ± 0.02 | 0.008 ± 0.02 | 0.1 ± 0.02 | 2.32 ± 0.005 |
| AFFINITY | |||||||
| Ligand | (nM) | ||||||
| FcγRI | 24.1 ± 2.3 | 9.54 ± 0.6 | |||||
| Protein A | 110 ± 1.6 | 11.9 ± 0.2 | |||||
3.2. Concentration analysis
A low antigen concentration could restrict the plasmonic response (RU), resulting in an inability to offer an adequate detection window. In contrast, a larger concentration could cause a higher background response, diminishing the detection sensitivity [37]. Thus, the optimum amount of TNF-α was investigated in a concentration range, and the results are presented in Fig. 3.
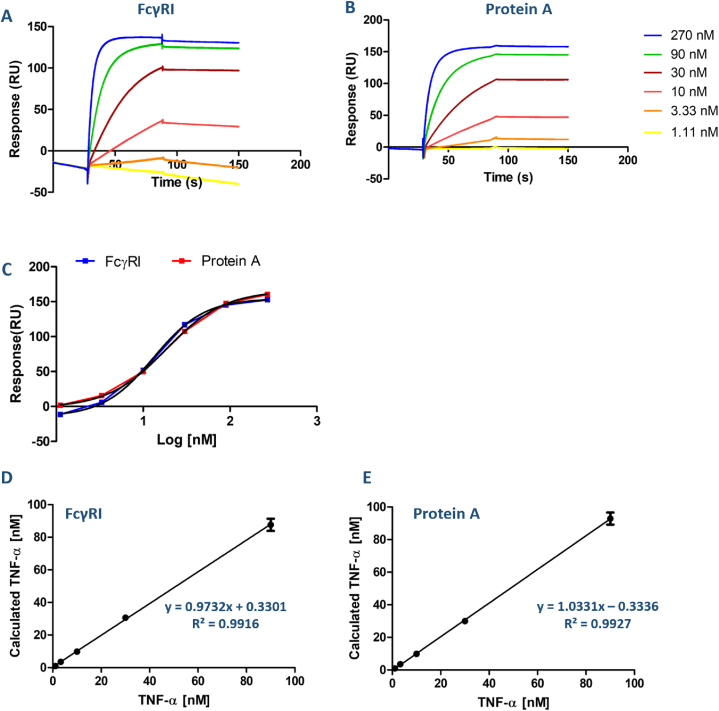
Fig. 3.
Comparison of TNF-α concentration analysis for FcγRI and Protein A ligand surfaces. (A) and (B) Representative SPR sensorgram of TNF-α binding on ADA captured surface through FcγRI and Protein A chip surfaces, respectively. TNF-α spiked in the HBS-EP buffer from lowest to highest concentration in duplicates. The surface was regenerated for each cycle with 10 mM glycine (pH 1.5) for 60 s. Results were obtained from double referencing. (C) Calibration curve of TNF-α for FcγRI and Protein A surfaces. The linearity plot of TNF-α was calculated from the output of the calibration curve analysis for FcγRI (D) and Protein A (E), respectively.
Immobilized Protein A and captured FcγRI levels were kept constant at 500 RU. ADA was injected on these surfaces at 90 nM and 74 nM concentrations for FcγRI and Protein A ligands, respectively. Antibody (ADA) binding response at this step was 500 RU for both ligand surfaces (data not shown). Then, TNF-α samples were injected from the lowest to the highest concentration. The binding response for the assays was obtained through double referencing where a zero-concentration TNF-α (HBS-EP buffer) sample and the reference channel responses were subtracted from the TNF-α sample responses.
High concentrations of TNF-α presented a curvature in the sensorgrams of both ligand surfaces, reaching a saturation of around 270 nM (Fig. 3A and B). A non-linear 4-parameter curve was obtained by plotting TNF-α concentration versus binding response (Fig. 3C). The results were analyzed with a non-linear four-parameter equation with GraphPad. The half response of the maximum response value, EC50, was found to be 13.2 nM for FcγRI and 18.4 nM for Protein A. Subsequently, the linearity was evaluated by plotting the TNF-α concentration of the sample against the calculated TNF-α concentration (Fig. 3D and E). The correlation was higher with a good correlation efficient value (R2: 0.9916 and R2: 0.9927) for FcγRI and Protein A.
LOD values were calculated as 1.93 nM for FcγRI captured surface and 0.84 nM for Protein A immobilized surface. In the SPR studies, the detection range was shown in the nM and pM range in various biological sample solutions (i.e., urine, serum, plasma) [38]. Our results were found in the nM range on both ligand surfaces. The anti-His antibody immobilized surface response decreased over time (Figure S2 and S3). This was due to the unstable baseline with anti-His antibody, so a low concentration of TNF-α sample showed a negative response when subtracted from the reference responses.
3.3. Detection of sensor accuracy and data variation
Table 2 presents the parameters such as accuracy/recovery and coefficient of variation (CV%) related to the concentration analysis of TNF-α. Parameters were evaluated based on the 80–120% accuracy/recovery and <20% CV value condition. According to these results, the FcγRI-captured surface gave a good accuracy/recovery rate and a low CV% value ranging from 3.33 to 90 nM (TNF-α). Protein A ligand surface showed a similar performance except for the highest TNF-α concentration. A biolayer interferometry-based TNF-α sensor study in the literature was performed with an aptamer-antibody pair to enhance the sensitivity of the biosensor. Biotin-labeled DNA aptamer was captured on the streptavidin chip surface then TNF-α injected into the surface. An Anti-TNF-α antibody injected aptamer-TNF-α pair on the surface for the enhancement step. They reported CV% values in the 3–8% range and concluded that their biosensor was reproducible and stable [22]. Like FcγRI-captured surface results, there was a good recovery and low standard deviations within the 0.25 nM–32 nM concentration range. LOD value was calculated as 0.0625 nM. The findings agreed with the current analysis except for the highest TNF-α concentration sample.
Table 2.
Accuracy/recovery and coefficient of variation (CV%) values related to the concentration analysis of TNF-α on different surfaces. The calculated concentration values are presented with the corresponding standard deviations (SD).
| FcγRI-captured surface | ||||
|---|---|---|---|---|
| TNF-α concentration (nM) | Calculated concentration (nM) | SD | Recovery% | CV% |
| 1.11 | 0.95 | 0.12 | 85.4 | 12.2 |
| 3.33 | 3.52 | 0.10 | 105.8 | 2.86 |
| 10 | 9.83 | 0.09 | 98.3 | 0.87 |
| 30 | 30.6 | 0.26 | 102 | 0.87 |
| 90 | 87.6 | 7.51 | 97.3 | 8.57 |
| Protein A-immobilized surface | ||||
| TNF-α concentration nM) | Calculated concentration (nM) | SD | Recovery% | CV% |
| 1.11 | 0.97 | 0.04 | 87.7 | 3.96 |
| 3.33 | 3.51 | 0.13 | 105.4 | 3.66 |
| 10 | 9.9 | 0.19 | 99.0 | 1.95 |
| 30 | 29.97 | 1.19 | 99.9 | 3.98 |
| 90 | 92.87 | 7.43 | 103.2 | 8.00 |
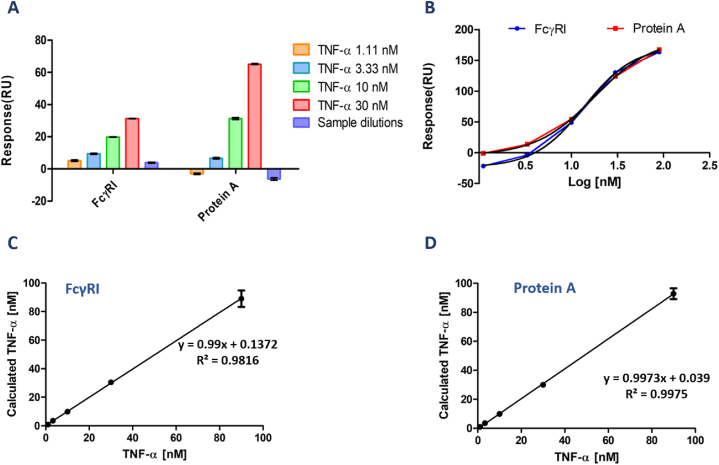
The concentration analysis was also conducted with BSA (0.02%) containing HBS-EP buffer. Antibody (ADA) capture level was kept at 500 RU for both ligand surfaces as in the HBS-EP condition. TNF-α spiking samples were prepared in 1.25% BSA containing HBS-EP buffer to mimic real serum sample conditions. TNF-α spiking concentration range was 1.11–90 nM since previous concentration results presented a high standard deviation and CV% value at 270 nM. TNF-α spiked samples were injected from lowest to highest concentration on both antibody-captured ligand surfaces. The binding response was like the HBS-EP buffer condition in FcγRI captured and Protein A immobilized surface (Fig. 4A and B). The studies with real serum samples stated that serum has a complex medium and could interact with non-specific binding to the chip surface.
Fig. 4.
TNF-α concentration analysis for FcγRI and Protein A ligands in 0.02% BSA containing HBS-EP medium. (A) Binding response graphs of TNF-α on ADA captured FcγRI and Protein A chip surfaces. TNF-α spiked samples were prepared in 1.25 %BSA containing HBS-EP buffer to mimic the real serum samples. The samples were injected from lowest to highest concentration in duplicates. The surface was regenerated for each cycle with 10 mM glycine (pH 1.5) for 60 s. Results were obtained from double referencing. (B) FcγRI and Protein A ligand surface calibration curves are fitted with a non-linear four-parameter equation on GraphPad Prism. (C) and (D) Linearity plots of TNF- α were calculated from the output of calibration curve analysis using FcγRI and Protein A surfaces, respectively.
As shown in the binding response graph, there is no non-specific binding to the chip surface. A calibration curve for TNF-α was presented in Fig. 4C and D by plotting TNF-α concentration versus binding response. The graphs were fitted with a non-linear 4-parameter curve, and the half response of the maximum response value was found to be 13.7 nM for FcγRI and 17.8 nM for Protein A. The linearity plots presented a high correlation value (R2: 0.9816 and R2: 0.9977) for FcγRI and Protein A. Table S2 shows accuracy/recovery and coefficient of variation (CV%) values related to the concentration analysis of TNF-α[NO_PRINTED_FORM]. FcγRI captured surface presented a good accuracy/recovery and low CV% value in the 3.33–90 nM TNF-α concentrations range. CV% was out of the acceptable range for CV% at the lowest TNF-α spiked sample solution.
Protein A ligand surface covered a similar concentration range regarding good accuracy/recovery and low CV% rates. Hao et al. [39] studied a graphene-based biosensing device using DNA aptamers for three cytokines (IL-6, IFN-γ, TNF- α). LOD value was found to be 611 fM in PBS for TNF-α. The study accomplished the sensitive and rapid detection of three cytokines in buffer and biofluid samples. However, the sensor surface step preparation was long and complex. Another study with a plasmonic sensor showed a LOD value at the fM level via integrating Ge2Sb2Te5 material (GST) on the gold substrate but did not present a concentration range [27]. Deng et al. [23] developed multiple cytokines (interleukin-1β, interleukin-6, and TNF-α) detection biosensors through fibers. The fibers were modified with streptavidin to capture biotinylated targets antibodies. Fluorescently conjugated secondary antibodies monitored cytokine detection. The detection range was 12.5–200 pg/mL, and LOD was 12.5 pg/mL.
3.4. Target specificity analysis
The specificity of the TNF-α binding assay was evaluated by injecting C1q, thrombin, and IL-1β protein samples. Fig. 5A and B shows that only the TNF-α sample specifically interacted with the ADA-captured surface in both ligands. The binding response of the non-specific proteins for the anti-His immobilized surface was shown as a negative response (Figure S3). That could be explained by the unstable baseline occurring due to using an Anti-His antibody immobilized surface, which was decaying throughout experiments. Also, C1q accumulated on channel one (Fc1) contributed to the negative response after double referencing subtraction. Raw data from blank samples and the Fc1 channel were presented in the supporting information file (Figure S2 and Figure S3). Results from the same specificity analysis are shown in a bar chart in Fig. 5C.
Fig. 5.
Evaluation of specificity for TNF-α binding analysis on FcγRI captured (A) and Protein A immobilized surface (B). Protein samples (TNF-α, C1q, Thrombin, IL-1β) were injected at 15 nM concentration spiked in duplicates. The surface was regenerated with 10 mM glycine (pH 1.5) for 60 s. Results were obtained from double referencing. (C) Results from the same specificity analysis are shown in a bar chart.
Further, non-specific binding was checked for binding responses on the anti-His antibody immobilized flow cell via electrostatic interactions between analyte and ligand [40]. Anti-his immobilized two flow cells were evaluated with a 30 nM TNF-α solution. There is no non-specific binding (<4 RU response level) with an anti-His immobilized blank surface (Figure S4). A recent study was conducted to minimize non-specific binding and enhance specificity via cell membrane coating. A bio affinity membrane layer was formed red blood cell and macrophage cell membrane onto the electrode substance. They reported that TNF-α sensing was obtained from a half-diluted serum sample without non-specific binding of the serum components. LOD value was calculated as 1.6 nM in this serum sample [16]. Another study performed a charged lipid membrane coating over Protein A conjugated chip surface in the SPR method. The charged lipid membrane (ethyl phosphocholine, EPC+) prevented non-specific binding in undiluted serum samples. The assay proposed sensitively detecting cholera toxin through the site-directed IgG orientation with Protein A ligand [41]. It was reported that Protein A could bind non-specific interaction with the Fab region of antibodies and other proteins, such as host cell proteins, within the purification process of cell culture harvest [33]. Therefore, we suggested that FcγRI is a potential ligand property regarding antibody binding capacity, specificity, and antigen-sensing properties.
4. Conclusion
Site-oriented antibody capture is important to enhance the antigen-binding performance of biosensors. Antibodies are usually immobilized on surfaces via cross-linking procedures, leading to the random orientation of the molecules on a given substrate. The current study used the FcγRI ectodomain as a ligand molecule for ADA capture on a plasmonic sensor surface to detect TNF-α. Various SPR analyses were conducted in parallel with Protein A, a well-known antibody-capturing ligand, to obtain a comparative performance data set. In kinetic analysis, the KD value was calculated as 0.63 ± 0.008 nM using a two-state binding model for TNF-α. The concentration analysis of TNF-α resulted in a good recovery rate (90%–105%) and a low coefficient of variation (≤8.5 CV%) in the 3.33–90 nM concentration range for both ligand surfaces. The assay specificity was evaluated by spiking three proteins (C1q, thrombin, and IL-1β) in sample solutions. The analysis specifically responded to target TNF- α on FcγRI captured and Protein A surfaces.
FcγRI presents potential advantages and disadvantages in the current prof of the concept study. While FcγRIa has a considerable affinity for IgG1-type monomeric monoclonal antibodies and can be used as a universal antibody linker, there is still room for improvement in performance and stability. Because the FcγRI molecule used in the current study was His-tagged, we employed an anti-His antibody to capture the FcγRI ligand on the surface, leading to the ligand decay due to the weak molecular interactions between the His-tag molecules and the anti-His antibody. Alternative immobilization methods, such as biotinylating, could be explored further to achieve more stable surface immobilization and orientation of the ligand molecule. In addition, Protein A performs better than the FcγRI on the surface because it has five antibody binding sites. However, Protein A does not always bind the antibodies in a site-oriented manner, and it cannot differentiate the monomeric antibodies from aggregates; thus, it could be groundbreaking to design a new ligand with several FcγRI ectodomains, which could rival Protein A's performance in the future.
Author contribution statement
Conceived and designed the experiments: MY and EC.
Performed the experiments: EC.
Analyzed and interpreted the data: EC, MY and AK.
Contributed reagents, materials, analysis tools or data: MY and EC.
Wrote the paper: MY, EC, AK.
Data availability statement
Data included in article/supp. material/referenced in article.
The authors declare no conflict of interest.
Supplementary content related to this article has been published online at [URL].
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
E.C. acknowledges TUBITAK 2244 Industrial Ph.D. Program (Grant ID: 118C149) for her scholarship. The authors thank Assoc. Prof. Dr. Hasan Kurt (Senior Research Associate at Imperial College London) and Begum Balkan Apaydin (Personnel at Sabanci University) for useful discussions and technical assistance, respectively.
Footnotes
Declaration of interest's statement.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e19469.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Macri C., Morgan H., Villadangos J.A., Mintern J.D. Regulation of dendritic cell function by fc-γ-receptors and the neonatal Fc receptor. Mol. Immunol. 2021;139:193–201. doi: 10.1016/j.molimm.2021.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Lu J., Ellsworth J.L., Hamacher N., Oak S.W., Sun P.D. Crystal structure of Fc gamma receptor I and its implication in high affinity gamma -Immunoglobulin binding. J. Biol. Chem. 2011;286:40608–40613. doi: 10.1074/jbc.M111.257550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boesch A.W., Kappel J.H., Mahan A.E., Chu T.H., Crowley A.R., Osei-Owusu N.Y., Alter G., Ackerman M.E. Enrichment of high affinity subclasses and glycoforms from serum-derived IgG using FcγRs as affinity ligands. Biotechnol. Bioeng. 2018;115:1265–1278. doi: 10.1002/bit.26545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorion-Thibaudeau J., Raymond C., Lattová E., Perreault H., Durocher Y., De Crescenzo G. Towards the development of a surface plasmon resonance assay to evaluate the glycosylation pattern of monoclonal antibodies using the extracellular domains of CD16a and CD64. J. Immunol. Methods. 2014;408:24–34. doi: 10.1016/j.jim.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Jefferis R., Lund J. Interaction sites on human IgG-fc for FcγR: current models. Immunol. Lett. 2002;82:57–65. doi: 10.1016/S0165-2478(02)00019-6. [DOI] [PubMed] [Google Scholar]
- 6.Thomann M., Schlothauer T., Dashivets T., Malik S., Avenal C., Bulau P., Rüger P., Reusch D. glycoengineering of IgG1 and its effect on Fc receptor binding and ADCC activity. PLoS One. 2015;10:1–16. doi: 10.1371/journal.pone.0134949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asaoka Y., Hatayama K., Tsumoto K., Tomita M., Ide T. Engineering of recombinant human fcγ receptor i by directed evolution. Protein Eng. Des. Sel. 2012;25:835–842. doi: 10.1093/protein/gzs053. [DOI] [PubMed] [Google Scholar]
- 8.Hatayama K., Asaoka Y., Hoya M., Ide T. Effective expression of soluble aglycosylated recombinant human fcγ receptor i by low translational efficiency in Escherichia coli. Appl. Microbiol. Biotechnol. 2012;94:1051–1059. doi: 10.1007/s00253-012-3902-x. [DOI] [PubMed] [Google Scholar]
- 9.Jung S.T., Kang T.H., Georgiou G. Efficient expression and purification of human aglycosylated fcγ receptors in Escherichia coli. Biotechnol. Bioeng. 2010;107:21–30. doi: 10.1002/bit.22785. [DOI] [PubMed] [Google Scholar]
- 10.Kiyoshi M., Caaveiro J.M.M., Tada M., Tamura H., Tanaka T., Terao Y., Morante K., Harazono A., Hashii N., Shibata H., et al. Assessing the heterogeneity of the fc-glycan of a therapeutic antibody using an engineered FcγReceptor IIIa-immobilized column. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-22199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown E.P., Dowell K.G., Boesch A.W., Normandin E., Mahan A.E., Chu T., Barouch D.H., Bailey-Kellogg C., Alter G., Ackerman M.E. Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J. Immunol. Methods. 2017;443:33–44. doi: 10.1016/j.jim.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown E.P., Weiner J.A., Lin S., Natarajan H., Normandin E., Barouch D.H., Alter G., Sarzotti-Kelsoe M., Ackerman M.E. Optimization and qualification of an Fc array assay for assessments of antibodies against HIV-1/SIV. J. Immunol. Methods. 2018;455:24–33. doi: 10.1016/j.jim.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C., Galloway J.F., Lee K.H., Searson P.C. Universal antibody conjugation to nanoparticles using the fcγ receptor i (FcγRI): quantitative profiling of membrane biomarkers. Bioconjug Chem. 2014;25:1893–1901. doi: 10.1021/bc5003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capkin E., Kurt H., Gurel B., Bicak D., Akgun Bas S., Daglikoca D.E., Yuce M. Characterization of FcγRIa (CD64) as a ligand molecule for site-specific IgG1 capture: a side-by-side Comparison with protein A. Langmuir. 2022;38:14623–14634. doi: 10.1021/acs.langmuir.2c02022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horiuchi T., Mitoma H., Harashima S.I., Tsukamoto H., Shimoda T. Transmembrane TNF-α: structure, function and interaction with anti-TNF agents. Rheumatology. 2010;49:1215–1228. doi: 10.1093/rheumatology/keq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vargas E., Zhang F., Ben Hassine A., Ruiz-Valdepeñas Montiel V., Mundaca-Uribe R., Nandhakumar P., He P., Guo Z., Zhou Z., Fang R.H., et al. Using cell membranes as recognition layers to construct ultrasensitive and selective bioelectronic affinity sensors. J. Am. Chem. Soc. 2022;144:17700–17708. doi: 10.1021/jacs.2c07956. [DOI] [PubMed] [Google Scholar]
- 17.Pruna R., Baraket A., Bonhommé A., Zine N., Errachid A., López M. Novel nanostructured indium tin oxide electrode for electrochemical immunosensors: suitability for the detection of TNF-α. Electrochim. Acta. 2018;283:1632–1639. doi: 10.1016/j.electacta.2018.07.066. [DOI] [Google Scholar]
- 18.Martinez-Perdiguero J., Retolaza A., Bujanda L., Merino S. Surface plasmon resonance immunoassay for the detection of the TNFα biomarker in human serum. Talanta. 2014;119:492–497. doi: 10.1016/j.talanta.2013.11.063. [DOI] [PubMed] [Google Scholar]
- 19.Li H., Li X., Chen L., Li B., Dong H., Liu H., Yang X., Ueda H., Dong J. Quench-release-based fluorescent immunosensor for the rapid detection of tumor necrosis factor α. ACS Omega. 2021;6:31009–31016. doi: 10.1021/acsomega.1c03941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim B.Y., Lee H.B., Lee N.E. A durable, stretchable, and disposable electrochemical biosensor on three-dimensional micro-patterned stretchable substrate. Sens Actuators B Chem. 2019;283:312–320. doi: 10.1016/j.snb.2018.12.045. [DOI] [Google Scholar]
- 21.Ghosh S., Datta D., Chaudhry S., Dutta M., Stroscio M.A. Rapid detection of tumor necrosis factor-alpha using quantum dot-based optical aptasensor. IEEE Trans. NanoBioscience. 2018;17:417–423. doi: 10.1109/TNB.2018.2852261. [DOI] [PubMed] [Google Scholar]
- 22.Gao S., Cheng Y., Zhang S., Zheng X., Wu J. A biolayer interferometry-based, aptamer–antibody receptor pair biosensor for real-time, sensitive, and specific detection of the disease biomarker TNF-α. Chem. Eng. J. 2022;433 doi: 10.1016/j.cej.2021.133268. [DOI] [Google Scholar]
- 23.Deng F., Qiao L., Li Y. A fluorescent immunosensor on optical fibre for the multiplex detection of proinflammatory cytokines. Sens Biosensing Res. 2022;37:0–5. doi: 10.1016/j.sbsr.2022.100501. [DOI] [Google Scholar]
- 24.Baydemir G., Bettazzi F., Palchetti I., Voccia D. Strategies for the development of an electrochemical bioassay for TNF-alpha detection by using a non-immunoglobulin bioreceptor. Talanta. 2016;151:141–147. doi: 10.1016/j.talanta.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 25.Barhoumi L., Baraket A., Bellagambi F.G., Karanasiou G.S., Ali M. Ben, Fotiadis D.I., Bausells J., Zine N., Sigaud M., Errachid A. A novel chronoamperometric immunosensor for rapid detection of TNF-Α in human saliva. Sens Actuators B Chem. 2018;266:477–484. doi: 10.1016/j.snb.2018.03.135. [DOI] [Google Scholar]
- 26.Arya S.K., Estrela P. Electrochemical immunosensor for tumor necrosis factor-alpha detection in undiluted serum. Methods. 2017;116:125–131. doi: 10.1016/j.ymeth.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., Zeng S., Crunteanu A., Xie Z., Humbert G., Ma L., Wei Y., Brunel A., Bessette B., Orlianges J.C., et al. Targeted sub-attomole cancer biomarker detection based on phase singularity 2D nanomaterial-enhanced plasmonic biosensor. Nano-Micro Lett. 2021;13:1–11. doi: 10.1007/s40820-021-00613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yagati A.K., Lee M.H., Min J. Electrochemical immunosensor for highly sensitive and quantitative detection of tumor necrosis factor-α in human serum. Bioelectrochemistry. 2018;122:93–102. doi: 10.1016/j.bioelechem.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Gürel B., Çapkın E., Parlar A., Özkan A., Çorbacıoğlu M., Dağlikoca D.E., Yüce M. Optimized methods for analytical and functional Comparison of biosimilar MAb drugs: a case study for avastin, mvasi, and zirabev. Sci. Pharm. 2022;90:36. doi: 10.3390/SCIPHARM90020036. Page 36 2022, 90. [DOI] [Google Scholar]
- 30.Gurel B., Berksoz M., Capkin E., Parlar A., Pala M.C., Ozkan A., Capan Y. Daglikoca, D.E.; yuce, M. Structural and functional analysis of CEX fractions collected from a novel Avastin® biosimilar candidate and its innovator. A Comparative Study. Pharmaceutics. 2022;14:1571. doi: 10.3390/pharmaceutics14081571. 10.3390/PHARMACEUTICS14081571 2022, 14, 1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang D., Lee A., Shin H., Song H., Park J., Kang T., Lee S., Yang S. The role of tumor necrosis factor alpha (TNF- α) in autoimmune disease and current TNF- α inhibitors in therapeutics. Int. J. Mol. Sci. 2021;22:1–16. doi: 10.3390/ijms22052719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tebbey P.W., Varga A., Naill M., Clewell J., Venema J. Consistency of quality attributes for the glycosylated monoclonal antibody Humira® (Adalimumab) mAbs. 2015;7:805–811. doi: 10.1080/19420862.2015.1073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghose S., Hubbard B., Cramer S.M. Binding capacity differences for antibodies and fc‐fusion proteins on protein A chromatographic materials. Biotech Bioengineering. 2006;96:768–779. doi: 10.1002/bit.21044. [DOI] [PubMed] [Google Scholar]
- 34.Bustos R.H., Zapata C., Esteban E., García J.C., Jáuregui E., Jaimes D. Label-free quantification of anti-TNF-α in patients treated with Adalimumab using an optical biosensor. Sensors. 2018;18 doi: 10.3390/s18030691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W., Thiemann S., Chen Q. Utility of SPR technology in biotherapeutic development : qualification for intended use. Anal. Biochem. 2022;654 doi: 10.1016/j.ab.2022.114804. [DOI] [PubMed] [Google Scholar]
- 36.Ogura T., Tanaka Y., Toyoda H. Whole cell-based surface plasmon resonance measurement to assess binding of anti-TNF agents to transmembrane target. Anal. Biochem. 2016;508:73–77. doi: 10.1016/j.ab.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 37.Masson J.F. Surface plasmon resonance clinical biosensors for medical diagnostics. ACS Sens. 2017;2:16–30. doi: 10.1021/acssensors.6b00763. [DOI] [PubMed] [Google Scholar]
- 38.Zhu H., Lu Y., Xia J., Liu Y., Chen J., Lee J., Koh K., Chen H. Aptamer-assisted protein orientation on silver magnetic nanoparticles: application to sensitive leukocyte cell-derived chemotaxin 2 surface plasmon resonance sensors. Anal. Chem. 2022;94:2109–2118. doi: 10.1021/acs.analchem.1c04448. [DOI] [PubMed] [Google Scholar]
- 39.Hao Z., Luo Y., Huang C., Wang Z., Song G., Pan Y., Zhao X., Liu S. An intelligent graphene‐based biosensing device for cytokine storm syndrome biomarkers detection in human biofluids. Small. 2021;17:1–11. doi: 10.1002/smll.202101508. [DOI] [PubMed] [Google Scholar]
- 40.Arney J.W., Weeks K.M. RNA − ligand interactions quantified by surface plasmon resonance with reference subtraction. Biochemistry. 2022;61(15):1625–1632. doi: 10.1021/acs.biochem.2c00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mckeating K.S., Hinman S.S., Rais N.A., Zhou Z., Cheng Q. Antifouling lipid membranes over protein a for orientation-controlled immunosensing in undiluted serum and plasma. ACS Sens. 2019;4:1774–1782. doi: 10.1021/acssensors.9b00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.
The authors declare no conflict of interest.
Supplementary content related to this article has been published online at [URL].