Abstract
BACKGROUND & AIMS:
Gaps remain in understanding the epidemiology of eosinophilic esophagitis (EoE). Our aim was to identify and validate a national cohort of individuals with EoE utilizing Veterans Health Administration (VHA) data.
METHODS:
We used two validation strategies to develop algorithms that identified adults with EoE between 1999–2020. The first validation strategy applied International Classification of Diseases (ICD) code algorithms to a base cohort of individuals who underwent esophagogastroduodenoscopy with esophageal biopsies. The second applied ICD code algorithms to a base cohort of all individuals in the VHA. For each ICD algorithm applied, a random sample of candidate EoE cases and non-EoE controls were selected and the charts manually reviewed by a blinded reviewer. Each algorithm was iteratively modified until the prespecified diagnostic accuracy endpoint (95% confidence lower bound for a positive predictive value (PPV) >88%) was achieved. We compiled individuals from each strategy’s maximum performance algorithm to construct the VA eosinophilic esophagitis cohort (VA E-O-ECHO).
RESULTS:
The maximum performance algorithm from the first validation strategy included ≥2 ICD code encounters for EoE separated by >30 days and achieved 93.3% PPV (lower bound 88.1%) for identifying true EoE cases. The maximum performance algorithm from the second validation strategy included ≥4 ICD code encounters for EoE where two codes were separated by at least 30 days, and similarly achieved 93.3% PPV (lower bound 88.1%). Combining both strategies yielded 6,637 individuals, which comprised the VA E-O-ECHO cohort.
CONCLUSIONS:
We developed and validated two highly accurate coding algorithms for EoE and established a nationwide VHA cohort of adults with EoE for future studies.
Keywords: allergy, immunology, immune-mediated, gastrointestinal
INTRODUCTION
Eosinophilic esophagitis (EoE) is a chronic allergen/immune-mediated disease of the esophagus characterized clinically by symptoms of esophageal dysfunction, including food impaction, dysphagia, heartburn, regurgitation, and non-cardiac chest pain, and histologically by eosinophilic infiltration of the esophageal epithelium.1,2 Appreciation of EoE as a distinct disease entity has increased since the late-1990s, resulting in an expansion of literature over the past two decades. Significant progress has been made in understanding the epidemiology, pathophysiology,1–13 clinical and endoscopic phenotypes, and therapeutics for EoE.3–13
There has been a steady rise in incidence and prevalence rates over the past decades. In the 1990s the incidence of EoE described in population-based studies was approximately 1 per 100,000 persons, whereas in the early 2000s the incidence increased 10-fold to approximately 10 per 100,000 persons.3–13 This rise has outpaced what would be attributable to increased awareness alone,14 suggesting that other factors are contributing to increasing incidence beyond increased case finding. Etiologic triggers and risk factors for EoE, however, remain poorly characterized.
These epidemiologic gaps stem in part from a lack of large, validated cohorts available for study.15 Veterans Health Administration (VHA) resources, including the corporate data warehouse (CDW), allow access to data compiled as part of usual health care for purposes of research including cohort studies, and contains comprehensive, longitudinal healthcare records for millions of Veterans annually. The aim of our study was to establish a first-of-its-kind validated cohort of individuals with EoE in the United States (US) utilizing VHA data to address current epidemiologic gaps in our understanding of EoE.
METHODS
Data Source and Study Population
We utilized national VHA healthcare data to identify adults ≥18 years of age diagnosed with EoE between 10/01/1999–12/31/2020. The VHA, one of the largest integrated healthcare systems in the US, includes >1,300 healthcare facilities across the US that provide longitudinal healthcare to over 9 million Veterans annually.16 Since 1999, data from millions of clinical encounters through the VHA are available to approved VHA researchers through the CDW. The CDW provides access to discrete structured and unstructured individual-level data, including demographic information, claims-based procedure and diagnostic codes, anthropometric data, medication prescription data, medical encounter notes, procedure notes, and pathology reports. CDW data are organized into longitudinal records for all Veterans enrolled in the VHA, and data are accessible and shared across all VA sites nationwide.
For this study, all data collected and queried from the CDW were maintained in the VA Informatics and Computing Infrastructure (VINCI) workspace, which can be accessed for research purposes by credentialed VHA investigators. This study was approved by the VA Institutional Review Board (#1450302).
Study Design Overview
We developed algorithms for identifying true EoE cases (Supplemental Table 1) using two different validation strategies. The first validation strategy applied International Classification of Disease (ICD) codes for EoE [530.13 (ICD-9) and K20.0 (ICD-10)] to a base cohort of individuals (n=375,794) who underwent esophagogastroduodenoscopy (EGD) with esophageal biopsies within the VHA, as determined by Current Procedural Terminology (CPT) codes for EGD and natural language processing (NLP) methods to identify esophageal biopsies. This validation strategy is hereafter referred to as the “combined claims code and NLP validation strategy.” The second validation strategy applied only ICD codes for EoE to a base cohort which included all individuals in the VHA (n=18,151,358), irrespective of being identified as having undergone an EGD within the VHA, and is, hereafter referred to as the “ICD only validation strategy.” This second validation strategy, which leverages only structured data, was conducted to ascertain a potentially more generalizable approach for identifying EoE cases within other large healthcare databases that utilize ICD codes.
Within these two primary strategies, iterative algorithms were developed and applied to the respective base cohorts (i.e. denominators) to identify candidate EoE cases (algorithm positive individuals) and non-EoE controls (algorithm negative individuals) (Figure 1). From the queried individuals, a random sample of 150 candidate EoE cases and 150 candidate non-EoE controls were mixed, and the medical charts were manually reviewed by an experienced gastroenterologist (EL) who was blinded to the algorithm diagnosis. Performance measures were computed for the respective algorithms, including the positive predictive value (PPV) of each algorithm for identifying true EoE cases among those who tested positive for the algorithm, negative predictive value (NPV) for identifying true non-EoE controls among those who tested negative for the algorithm, sensitivity for identifying algorithm positive cases among those with EoE, and specificity for identifying algorithm negative non-EoE controls among those without EoE. PPV was the performance measurement used to determine algorithm success in ascertaining true EoE diagnoses and, hence, algorithm validity. An iterative process of adjusting algorithm criteria (for example, increasing the number of ICD code encounters required) was repeated until the a priori PPV threshold was achieved for each validation approach.
Figure 1.
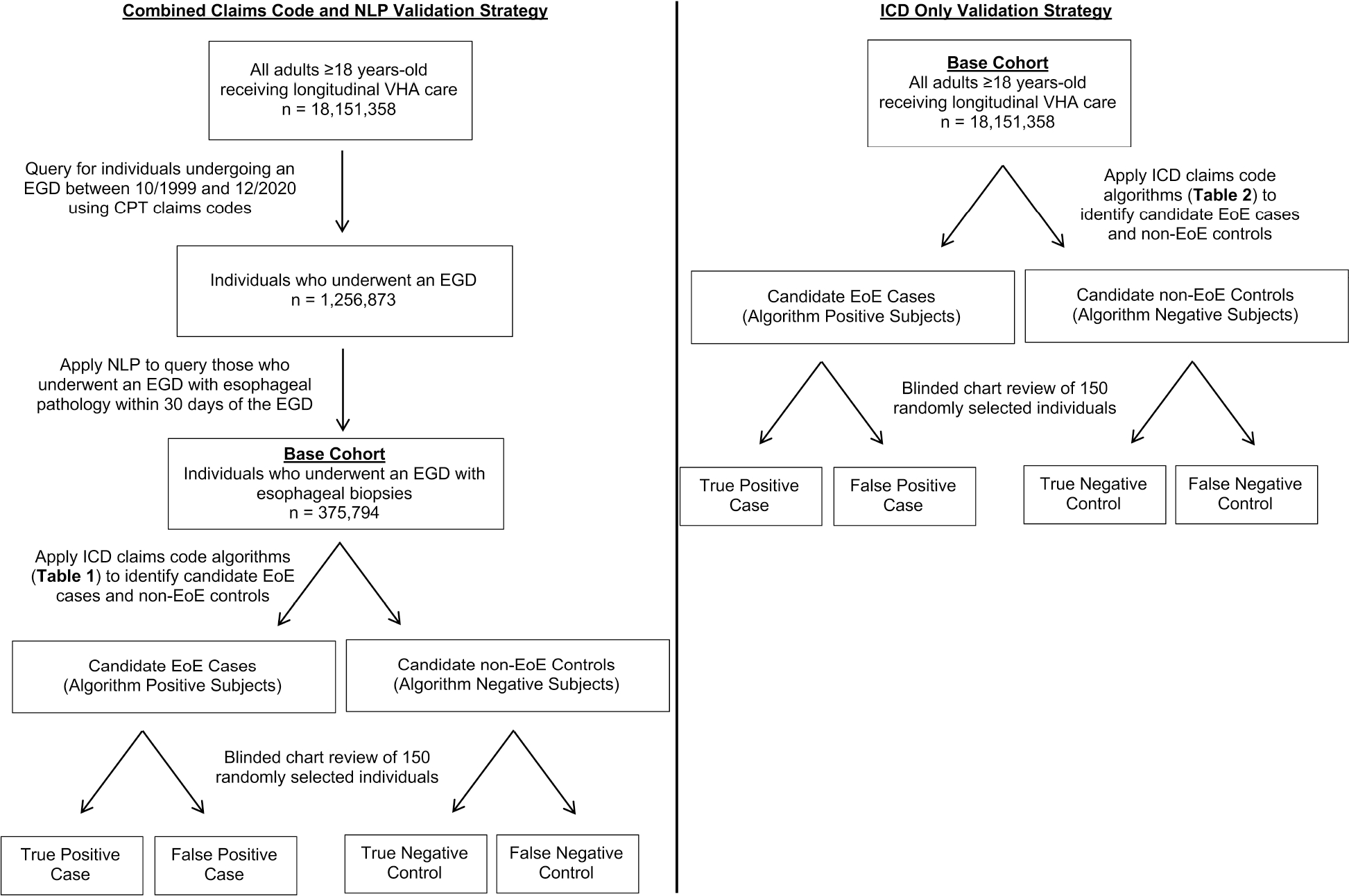
Flow diagram of the two validation strategies
Left panel shows the validation process for the combined claims code and NLP validation strategy. Right panel shows the validation process for the ICD only validation strategy.
Combined Claims Code and NLP Validation Strategy
The first validation strategy to identify true EoE cases applied EoE ICD code algorithms to a base cohort of individuals (n=375,794) who underwent EGD with esophageal biopsies within the VHA. This base cohort defined the denominator for the combined claims code and NLP strategy. To create this base cohort, first, we applied CPT codes to query all individuals who underwent an EGD procedure during the study period (10/01/1999–12/31/2020). Using NLP, we then selected the sub-set of individuals who had esophageal biopsy results reported +/−30 days from the EGD procedure date (see Supplemental Methods for details). Creating this base cohort of individuals with esophageal biopsies on EGD allowed determination of NPV based on biopsy proven non-EoE controls.
In the algorithm development and validation stage, we applied combinations of ICD code algorithms to this base cohort. We selected a random sample of subjects who were “algorithm positive” (i.e. candidate EoE cases), along with a random sample of subjects who were “algorithm negative” (i.e. candidate non-EoE controls), to evaluate if these algorithm positive individuals satisfied the definition of a true EoE diagnosis (see “True EoE Definition” section below). A random number generator was used to select individuals at random and to mix candidate cases and controls so that the reviewer was blinded to the algorithm positive or negative status during the manual chart review process. After manual chart review, algorithm performance was assessed by calculating the PPV among candidate EoE cases. Following our previously described approach for validation of algorithms for clinical diagnoses and outcomes within large scale healthcare data,17 an a priori goal for PPV was set to a one-sided Bonferroni corrected 95% confidence lower bound for PPV of >88%. Since we also validated the algorithm for identifying non-EoE controls and estimating NPV, the Bonferroni multiple comparison adjustment was applied to adjust the confidence level. This ensured an overall confidence level of 95% for PPV and NPV estimates. Using established calculations, we determined that we would need to sample at least 150 candidate EoE cases to establish a one-sided 95% confidence lower bound for PPV >88%. This calculation is associated with an overall observed PPV ≥92.5%.17
If optimal algorithm performance was not achieved (i.e., failure to achieve a PPV associated with a one-sided 95% confidence lower bound of >88%), the algorithm was modified, and the process was repeated so that a unique set of 300 candidate individuals (150 cases/150 controls) were reviewed (blinded) for each algorithm iteration.
ICD Only Validation Strategy
For this strategy, the base cohort (i.e. denominator) comprised all individuals receiving longitudinal care within the VHA (n=18,151,358), irrespective of whether an EGD with esophageal biopsies was identified. We applied iterative ICD code algorithms to this VHA base cohort and used the same development and validation methods as described for the combined claims code and NLP strategy above (Figure 1).
Optimal algorithm performance was again defined by an a priori PPV one-sided 95% confidence lower bound of >88%. Since EoE is a rare disease and our VHA base cohort for this validation strategy included over 18 million individuals, the NPV for each algorithm would likely be 100% for the 150 randomly reviewed algorithm negative individuals. Therefore, PPV is the only performance measure reported for this validation strategy. Algorithm negative individuals, however, were still mixed with algorithm positive individuals and reviewed to maintain reviewer blinding.
True EoE Case Definition, Chart Review Process, and Diagnosis Date Validation
For both validation approaches, we adhered to established international consensus criteria for defining “true” EoE cases based on manual chart review as the reference standard. The AGREE international consensus defines EoE as the co-occurrence of “symptoms of esophageal dysfunction and at least 15 eosinophils per high-power field (or approximately 60 eosinophils per mm2) on esophageal biopsy after a comprehensive assessment of non-EoE disorders that could cause or potentially contribute to esophageal eosinophilia.”1 In the present study, any individual meeting one of the two definitions below was considered a true EoE case:
-
The reference standard diagnosis, which includes:
-
Histology demonstrating a peak count ≥15 eosinophils/high-power field on at least one esophagus biopsy.
AND
-
A gastroenterology consultant note documenting symptoms of esophageal dysfunction, as defined above.
AND
-
No documentation of other causes of esophageal eosinophilia, such as hypereosinophilic syndrome, non-EoE eosinophil-associated gastrointestinal disorders, achalasia, Crohn’s disease, infections, connective tissue disorders, or drug hypersensitivity reactions.
OR
-
A gastroenterology consultant note documenting a diagnosis of EoE based on prior histology and endoscopy referenced in the encounter note, when endoscopy reports or esophageal histology reports were not available for review through the VHA.
If an individual met the criteria for a true EoE diagnosis as above, then they were counted as a true positive case. If an individual did not meet the criteria, then they were counted as a false positive case. A gastroenterologist (EEL) reviewed all charts with any discrepancies adjudicated by a second gastroenterologist (YC, SS).
We also validated whether the index ICD code date (i.e. the earliest occurrence of the EoE ICD-9 or -10 code) could serve as a proxy for the true date of EoE diagnosis. In order to do so, all gastroenterology-related clinical documentation, procedure reports, and histology reports were reviewed to determine the true EoE diagnosis date. If the index ICD code date was within +/− 60 days of the EoE diagnosis date determined by manual chart review, then the ICD code date was considered an appropriate approximation. If the ICD code was outside of this time interval, then it was not considered an appropriate measure of the true diagnosis date. We set an a priori minimum correlation for the index ICD date to correspond to the true diagnosis date at >88%.
Non-EoE Control Definition, Chart Review Process, and NPV Measurement
For future case-control studies, we wanted to validate a biopsy-proven non-EoE control cohort. In the combined claims code and NLP validation strategy (EGD base cohort), we used a similar approach as that used to define EoE cases. With each algorithm, a blinded review of candidate non-EoE controls was performed to estimate each algorithm’s NPV (with one-sided 95% confidence lower bound) for identifying true non-EoE status. We identified candidate non-EoE controls for each algorithm as those individuals who did not satisfy the respective EoE algorithm criteria. As described in the true EoE case section, a sample of 150 algorithm-negative subjects was randomly selected and mixed with randomly selected algorithm-positive subjects, with the reviewer (EEL) blinded to the true algorithm status. Manual chart review was conducted to assess for non-EoE status. The reference standard process for confirming a true non-EoE control was the same as for confirming a true EoE case – that is, manual chart review of all free text histology reports, EGD procedure reports, and gastroenterology consultant notes. If there was documentation of EoE that met either of the two EoE diagnostic criteria above based on AGREE international consensus, then candidate non-EoE controls were categorized as false negative controls. Otherwise, they were considered true negative controls (Figure 1).
Creating the Validated EoE Cohort: VA E-O-ECHO
EoE cases were identified from the optimal algorithm of each of the two validation strategies and combined to create the VA E-O-ECHO. Age, sex, race, ethnicity, BMI, and smoking status, which are all previously validated variables using VHA data,18,19 were recorded at the time of EoE diagnosis.
RESULTS
Algorithm Validation for Combined Claims Code and NLP Strategy
The prespecified performance endpoint (lower bound PPV >88%) was achieved after three independent validation rounds (Table 1). Algorithm 1 – at least one ICD code encounter for EoE – and algorithm 2 – two or more ICD code encounters for EoE – were applied to the EGD with esophageal biopsy base cohort (n=375,794). Both algorithms achieved suboptimal PPVs, with lower bound 53.0% and 80.2%, respectively.
Table 1.
Validation Algorithms and Performance Measurements Using the Combined Claims Code and NLP Validation Strategy
| PPV Assessment | NPV Assessment | Sensitivity/Specificity Assessment | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Algorithms | Algorithm Description | True Positive Cases | Sample Size Reviewed | PPV (Lower Bound)+ | Total Individuals Meeting Algorithm Criteria from the National VHA | Cases Where Index ICD Code Date Matched True EoE Diagnosis Date (%) | True Negative Controls | Sample Size Reviewed | NPV (Lower Bound)+ | Sensitivity (Lower Bound^) | Specificity (Lower Bound^) |
| Algorithm 1 | A single ICD9 or ICD10 code for EoE AND a CPT code for EGD with NLP confirming esophageal biopsies | 92 | 150 | 61.3 (53.0) | 8,815 | 83 (90.2) | 150 | 150 | 100 (97.6) | 100 (97.6) | 72.1 (65.5) |
| Algorithm 2 | Two or more ICD9 and/or ICD10 codes for EoE AND a CPT code for EGD with NLP confirming esophageal biopsies | 130 | 150 | 86.7 (80.2) | 6,286 | 121 (93.1) | 150 | 150 | 100 (97.6) | 100 (97.6) | 88.2 (82.4) |
| Algorithm 3 | Two or more ICD9 and/or ICD10 codes for EoE separated by at least 30 days AND a CPT code for EGD with NLP confirming esophageal biopsies | 140 | 150 | 93.3 (88.1) | 5,417 | 131 (93.6) | 150 | 150 | 100 (97.6) | 100 (97.6) | 93.8 (88.8) |
CPT = Current Procedural Terminology; EoE = eosinophilic esophagitis; ICD = International Classification of Diseases; PPV = positive predictive value; NPV = negative predictive value; VHA = Veterans Health Administration
95% one-sided confidence lower bound based on exact binomial test with Bonferroni correction for two performance measures, PPV and NPV
95% one-sided confidence lower bound based on exact binomial test with Bonferroni correction for two performance measures, Sensitivity and Specificity
Algorithm 3, which queried any individual in the EGD base cohort with two or more ICD code encounters for EoE separated by at least 30 days, achieved PPV 93.3% (lower bound 88.1%), and was thus selected as the optimal validated algorithm for the first strategy. Of note, for this algorithm, 99% of the true EoE cases reviewed met the reference standard diagnosis based on AGREE international consensus criteria (Supplemental Table 2a).
Algorithm Validation for ICD Only Strategy
At the time of analysis, the total number of individuals receiving VHA care during the 1999–2020 study period was 18,151,358 (“VHA base cohort”). Applying algorithms to this VHA base cohort, the prespecified performance endpoint (lower bound PPV >88%) was achieved after four independent validation rounds (Table 2). Algorithm 1 – at least one ICD code encounter for EoE, algorithm 2 – two or more ICD code encounters for EoE separated by at least 30 days, and algorithm 3 – three or more ICD code encounters for EoE, where two encounter codes were separated by at least 30 days – achieved suboptimal PPVs, with lower bound PPV 56.5%, 65.5%, and 75.6%, respectively.
Table 2.
Validation Algorithms and Performance Measurements Using the ICD Only Validation Strategy
| PPV Assessment | ||||||
|---|---|---|---|---|---|---|
| Algorithms | Algorithm Description | True Positive Cases | Sample Size Reviewed | PPV (Lower Bound)+ | Total Individuals Meeting Algorithm Criteria from the National VHA | Cases Where Index ICD Code Date Matched True EoE Diagnosis Date (%) |
| Algorithm 1 | A single ICD9 or ICD10 code | 97 | 150 | 64.7 (56.5) | 12,131 | 87 (89.7) |
| Algorithm 2 | Two or more ICD9 and/or ICD10 codes* | 110 | 150 | 73.3 (65.5) | 7,564 | 97 (88.2) |
| Algorithm 3 | Three or more ICD9 and/or ICD10 codes* | 124 | 150 | 82.7 (75.6) | 6,324 | 113 (91.1) |
| Algorithm 4 | Four or more ICD9 and/or ICD10 codes* | 140 | 150 | 93.3 (88.1) | 5,396 | 124 (88.6) |
EoE = eosinophilic esophagitis; ICD = International Classification of Diseases; PPV = positive predictive value; VHA = Veterans Health Administration
At least 2 code encounters were separated by 30 days
95% one-sided confidence lower bound based on exact binomial test with Bonferroni correction for one performance measure
Algorithm 4, which queried any individual with four or more ICD code encounters for EoE where two encounter codes were separated by at least 30 days, achieved PPV 93.3% (lower bound 88.1%), and was thus selected as the optimal validated algorithm for the second strategy. For this algorithm, 97% of the true EoE cases reviewed met the reference standard diagnosis based on AGREE international consensus criteria (Supplemental Table 2b).
Analysis of Algorithm NPV, Sensitivity and Specificity in Combined Claims Code and NLP Strategy
For the combined claims code and NLP validation strategy, NPV for each of the separate algorithms was 100% (one-sided 95% confidence lower bound of 97.6%). Sensitivity and specificity of each algorithm are provided in Table 1.
Validation of Index ICD code to Predict true EoE Diagnosis Date
During manual chart review, the index ICD code correlated with initial EoE diagnosis 88.2% of the time or greater for all algorithms tested, independent of the validation strategy (Table 1 and 2), justifying this encounter date as an acceptable proxy for true EoE diagnosis date in VHA.
National Cohort of Individuals with Eosinophilic Esophagitis
Applying the validated optimal performance algorithm from the combined claims code and NLP validation strategy (algorithm 3) to VHA nationwide data identified 5,417 individuals diagnosed with EoE between 10/1999–12/2020. Applying the validated algorithm (algorithm 4) from the ICD only validation strategy identified 5,396 total individuals diagnosed with EoE between 10/1999–12/2020. There were 1,241 unique patients identified from algorithm 4 of the ICD only strategy which were not identified in our combined claims code and NLP validation strategy. After merging individuals from both validation strategies and removing duplicates (n=4,176), the VA E-O-ECHO comprised of 6,637 total EoE cases. The diagnoses of EoE in VA E-O-ECHO increased each year from 2008 to 2018 (Figure 2) and spanned the entire US (Figure 3). The median age at EoE diagnosis was 45 years (Quartile1 (Q1)–Quartile3 (Q3): 35–57) (Table 3). Among those with EoE, 6,046 (91.1%) were male and 6,032 (90.9%) were non-Hispanic. The vast majority were White [n=5,614 (84.6%)], followed by Black [559 (8.4%)], American Indian or Alaska Native [39 (0.6%)], Asian [39 (0.6%)], and Native Hawaiian or Other Pacific Islander [38 (0.6%)]. Median BMI was 29.3 kg/m2 (Q1–Q3: 26.3–32.9 kg/m2). Smoking exposure (i.e. current or former smokers) was present in 2,509 (37.8%).
Figure 2.

VA E-O-ECHO EoE diagnoses (n=6,637) by year from 1999–2020
Percentage is reflective of the VA E-O-ECHO total cohort.
Figure 3.

VA E-O-ECHO EoE diagnoses (n=6,637) across US states/territories from 1999–2020
Percentage is reflective of the VA E-O-ECHO total cohort.
Table 3.
VA E-O-ECHO Baseline Characteristics from 1999 to 2020 (n=6,637)
| Variable | |
|---|---|
| Age, median (Q1-Q3) | 45 (35 – 57) |
| Sex, n (%) | |
| Male | 6,046 (91.1) |
| Female | 591 (8.9) |
| Ethnicity, n (%) | |
| Hispanic | 391 (5.9) |
| Non-Hispanic | 6,032 (90.9) |
| Unknown | 214 (3.2) |
| Race, n (%) ^ | |
| American Indian or Alaska Native | 39 (0.6) |
| Asian | 39 (0.6) |
| Black | 559 (8.4) |
| Native Hawaiian or Other Pacific Islander | 38 (0.6) |
| White | 5,614 (84.6) |
| Unknown | 348 (5.2) |
| BMI, median (Q1-Q3) | 29.3 (26.3 – 32.9) |
| Smoking Status, n (%) | |
| Current | 1,215 (18.3) |
| Former | 1,294 (19.5) |
| Never | 3,406 (51.3) |
| Unknown | 722 (10.9) |
DISCUSSION
Using VHA national data, we applied two rigorous, stepwise approaches to develop and then validate algorithms to identify cases of EoE with a very high degree of accuracy. Using our validated algorithms, we were able to create a large, population-based cohort of 6,637 individuals with EoE (termed the VA E-O-ECHO) to be used in future research. There has been growing interest in studying EoE in the Veteran population,6,20 and the VHA provides a unique opportunity to study the epidemiology of EoE as Veterans often live or travel through multiple locations during their service appointment and, as such, may encounter various environmental exposures.
Our US-based cohort of individuals with EoE was developed utilizing a systematic validation scheme, a critical feature of a reliable dataset. During our algorithm development and validation process, we found that using a single ICD code for EoE was suboptimal for identifying true EoE diagnoses, regardless of the base cohort used to query cases, and resulted in a PPV <65%. These results suggest that identifying EoE cases using this criterion is insufficient for identifying a high percentage of true EoE cases due to false positives. Thus, prior epidemiologic studies that utilize ICD-based codes for EoE case ascertainment without validation could be subject to bias6,7,12 (publications summarized in Supplemental Table 3). We also were able to perform validation using manual chart review as the reference (gold) standard since we had access to patient-level data, which are not available for many US-based claims databases.
Using our combined claims code and NLP validation strategy, we identified a combination of structured administrative codes and NLP that achieved high PPV (93.3%) for identifying true EoE cases: a CPT code encounter for EGD, documentation of esophageal biopsies within 30 days of an EGD as identified by NLP, and two or more ICD code encounters for EoE separated by at least 30 days. This validated algorithm was likely successful for two main reasons. First, EGD is required to obtain esophageal biopsies and confirm a histopathologic diagnosis. Accordingly, we incorporated a claims code (CPT) requirement for EGD and then used NLP to confirm that esophageal biopsies were taken during that encounter. Secondly, we required two ICD encounter codes and ensured these encounter dates were separated by at least 30 days. Requiring multiple ICD encounters likely reduced ICD-related miscoding, which was prevalent in our review of single ICD code encounters. Moreover, multiple ICD codes 30 days apart suggests that there was longitudinal care of EoE in the VHA.
While the combined claims code and NLP approach was accurate, the ICD only algorithm applied to the full VHA base cohort achieved similar accuracy but only when four or more ICD code encounters (where two encounter codes were separated by at least 30 days) was used. Indeed, this algorithm yielded 93.3% PPV for true EoE cases and may be generalizable to other healthcare databases where access to EGD and biopsy reports, among other patient-level data, may not be readily available. This approach supports the hypothesis that multiple ICD code encounters reduce ICD-related miscoding, and that multiple ICD code encounters may be required if the goal is to construct an accurate disease-based cohort.
The VA E-O-ECHO will allow us and other investigators to address important gaps in knowledge pertaining to EoE epidemiology and disease pathogenesis. A unique feature of a Veteran-based cohort is the ability to evaluate etiologic risk factors which may be enriched in a VA cohort but may not be present in a civilian population, and future studies are planned to investigate these. Other epidemiologic studies can also be done such as identifying factors associated with treatment outcomes and generating hypotheses about the mechanisms of EoE development.
There are several novel aspects and strengths of our study. First, in order to develop this first-of-its-kind cohort, we used a rigorous, previously established systematic approach to algorithm application and chart review.17,21 We also used two validation strategies, which were both accurate in identifying EoE diagnoses and in improving EoE case ascertainment. It is notable that during manual chart review of randomly selected candidate cases and controls, EoE status was able to be determined for all reviewed individuals. Additionally, for our validated algorithms, >97% of true EoE cases satisfied AGREE international consensus criteria (criteria which require both histologic and clinical data available in the chart). We also found that the index ICD encounter code date was reflective of the true EoE diagnosis date >88% of the time, and therefore may serve as a proxy for EoE diagnosis date in future longitudinal studies where incident cases of EoE are required. Lastly, it is important to note that as Veterans continue to enroll in the VHA and care is updated, we will be able to apply our validated algorithm to current VHA data and update the VA E-O-ECHO with incident cases. As this nationally representative cohort expands, it will yield higher statistical power for analyses.
There are some limitations that should be considered when interpreting our approach. First, even though the second (ICD only) approach might be generalizable to other healthcare systems, it is at this time unclear whether our validated algorithms can be used to identify EoE patients in other databases. Additionally, our validated cohort is representative of a VHA patient population with predominance of older White males and may limit generalizability to the entire US population. That said, since EoE predominantly affects males,1 having a large proportion of males in our cohort may help advance understanding of EoE epidemiology, natural history, and therapeutic considerations. Lastly, our validated algorithms might not capture those individuals who only underwent EGD outside of the VHA and who were subsequently diagnosed with EoE.
In conclusion, we developed and validated two highly accurate coding algorithms to identify cases of EoE in the VHA. Using these algorithms we identified a nationwide cohort of 6,637 adults with EoE, termed the VA E-O-ECHO, which is one of the largest established to date in the US and the first VHA cohort. We plan to utilize the VA E-O-ECHO to address unmet knowledge gaps in EoE epidemiology in future studies.
Supplementary Material
What you need to know.
BACKGROUND:
The incidence of eosinophilic esophagitis (EoE) is rising, and is outpacing what would be expected by increased disease-related awareness alone. Accordingly, it is critical to generate large, validated cohorts for impactful epidemiologic and outcomes studies of EoE.
FINDINGS:
Using Veterans Health Administration (VHA) national data, we applied two rigorous, stepwise strategies to develop and then validate claims-based algorithms to identify cases of EoE with a very high degree of accuracy (both with PPV 93%).
IMPLICATIONS FOR PATIENT CARE:
We successfully created a large, population-based cohort of 6,637 individuals with EoE (termed the VA E-O-ECHO) to help address unmet knowledge gaps in EoE epidemiology in future anticipated studies.
Acknowledgments
Grant Support:
T32 NIH Grant 5T32DK007202-44 (Ghosh, PI); NIH K23 DK125266 (Yadlapati, PI); NIH/NCI 5 R37 CA 222866-02 (Gupta, PI); VA ICX002027A (Shah, PI); AGA Research Scholar Award (Shah, PI); IK2BX004648 (Choksi, PI)
Abbreviations:
- CDW
Corporate Data Warehouse
- CPT
Current Procedural Terminology
- EGD
esophagogastroduodenoscopy
- EoE
eosinophilic esophagitis
- ICD
International Classification of Diseases
- NPV
negative predictive value
- PPV
positive predictive value
- VA E-O-ECHO
Veterans Affairs Eosinophilic Esophagitis Cohort
- VHA
Veterans Health Administration
- VINCI
VA Informatics and Computing Infrastructure
Footnotes
Disclosures:
EEL: None
QS: None
RY: Consultant: Medtronic (Institutional), Ironwood Pharmaceuticals (Institutional), Phathom Pharmaceuticals, StatDataLink Research support: Ironwood Pharmaceuticals; Advisory Board with Stock Options: RJS Mediagnostix
ESD: None
SA: None
LL: None
SG: None
YAC: None
SCS: Consultant: Phathom Pharmaceuticals, RedHill Biopharma
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Dellon ES et al. Updated International Consensus Diagnostic Criteria for Eosinophilic Esophagitis: Proceedings of the AGREE Conference. Gastroenterology. 2018. Oct;155(4):1022–1033.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Straumann A, Katzka DA. Diagnosis and Treatment of Eosinophilic Esophagitis. Gastroenterology. 2018. Jan;154(2):346–359. [DOI] [PubMed] [Google Scholar]
- 3.Prasad GA et al. Epidemiology of eosinophilic esophagitis over three decades in Olmsted County, Minnesota. Clin Gastroenterol Hepatol. 2009. Oct;7(10):1055–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Syed AA et al. The rising incidence of eosinophilic oesophagitis is associated with increasing biopsy rates: a population-based study. Aliment Pharmacol Ther. 2012. Nov;36(10):950–8. [DOI] [PubMed] [Google Scholar]
- 5.Hruz P et al. Swiss EoE study group. Escalating incidence of eosinophilic esophagitis: a 20-year prospective, population-based study in Olten County, Switzerland. J Allergy Clin Immunol. 2011. Dec;128(6):1349–1350.e5. [DOI] [PubMed] [Google Scholar]
- 6.Ally MR et al. Prevalence of eosinophilic esophagitis in a United States military health-care population. Dis Esophagus. 2015. Aug-Sep;28(6):505–11. [DOI] [PubMed] [Google Scholar]
- 7.van Rhijn BD et al. Rapidly increasing incidence of eosinophilic esophagitis in a large cohort. Neurogastroenterol Motil. 2013. Jan;25(1):47–52.e5. [DOI] [PubMed] [Google Scholar]
- 8.Arias Á, Lucendo AJ. Prevalence of eosinophilic oesophagitis in adult patients in a central region of Spain. Eur J Gastroenterol Hepatol. 2013. Feb;25(2):208–12. [DOI] [PubMed] [Google Scholar]
- 9.Dellon ES et al. Development and validation of a registry-based definition of eosinophilic esophagitis in Denmark. World J Gastroenterol. 2013. Jan 28;19(4):503–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dellon ES et al. Prevalence of eosinophilic esophagitis in the United States. Clin Gastroenterol Hepatol. 2014. Apr;12(4):589–96.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giriens B et al. Escalating incidence of eosinophilic esophagitis in Canton of Vaud, Switzerland, 1993–2013: a population-based study. Allergy. 2015. Dec;70(12):1633–9. [DOI] [PubMed] [Google Scholar]
- 12.Mansoor E, Cooper GS. The 2010–2015 Prevalence of Eosinophilic Esophagitis in the USA: A Population-Based Study. Dig Dis Sci. 2016. Oct;61(10):2928–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molina-Infante J et al. Rising incidence and prevalence of adult eosinophilic esophagitis in midwestern Spain (2007–2016). United European Gastroenterol J. 2018. Feb;6(1):29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dellon ES et al. The increasing incidence and prevalence of eosinophilic oesophagitis outpaces changes in endoscopic and biopsy practice: national population-based estimates from Denmark. Aliment Pharmacol Ther. 2015. Apr;41(7):662–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rybnicek DA et al. Administrative coding is specific, but not sensitive, for identifying eosinophilic esophagitis. Dis Esophagus. 2014. Nov-Dec;27(8):703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.U.S. Department of Veterans Affairs. Veterans health administration. https://www.va.gov/health/aboutvha.asp. Accessed on 10/1/2022.
- 17.Liu L et al. A strategy for validation of variables derived from large-scale electronic health record data. J Biomed Inform. 2021Sep;121:103879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noel PH et al. VHA Corporate Data Warehouse height and weight data: opportunities and challenges for health services research. J Rehabil Res Dev. 2010;47:739–750. [DOI] [PubMed] [Google Scholar]
- 19.McGinnis KA et al. Validating smoking data from the Veteran’s Affairs Health Factors dataset, an electronic data source. Nicotine Tob Res. 2011; 13: 1233–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trovato A et al. The Impact of Obesity on the Fibrostenosis Progression of Eosinophilic Esophagitis in a U.S. Veterans Cohort. Dysphagia. 2022. Sep 8. [DOI] [PubMed] [Google Scholar]
- 21.Low EE et al. Development and Validation of a National US Achalasia Cohort: The Veterans Affairs Achalasia Cohort (VA-AC). Clin Gastroenterol Hepatol. 2022. Sep 6:S1542-3565(22)00826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


