Summary
Dysregulated lipid metabolism occurs in pathological processes characterized by cell proliferation and migration. Nonetheless, the mechanism of increased mitochondrial lipid oxidation is poorly appreciated in diabetic cardiac fibrosis, which is accompanied by enhanced fibroblast proliferation and migration. Herein, increased WTAP expression promotes cardiac fibroblast proliferation and migration, contributing to diabetic cardiac fibrosis. Knockdown of WTAP suppresses mitochondrial lipid oxidation, fibroblast proliferation and migration to ameliorate diabetic cardiac fibrosis. Mechanistically, WTAP-mediated m6A methylation of AR induced its degradation, dependent on YTHDF2. Additionally, AR directly interacts with mitochondrial lipid oxidation enzyme Decr1; overexpression of AR-suppressed Decr1-mediates mitochondrial lipid oxidation, inhibiting cardiac fibroblast proliferation and migration. Knockdown of AR produced the opposite effect. Clinically, increased WTAP and YTHDF2 levels correlate with decreased AR expression in human DCM heart tissue. We describe a mechanism wherein WTAP boosts higher mitochondrial lipid oxidation, cardiac fibroblast proliferation, and migration by enhancing AR methylation in a YTHDF2-dependent manner.
Subject areas: Biological sciences, Molecular biology, Cell biology
Graphical abstract

Highlights
-
•
Mitochondrial lipid oxidation played a key role in diabetic cardiac fibrosis
-
•
AR is involved in diabetic cardiac fibrosis and acts as a key regulator
-
•
WTAP mediated m6A methylation of AR via a YTHDF2-dependent manner
Biological sciences; Molecular biology; Cell biology
Introduction
Diabetic cardiac fibrosis remains an unresolved problem in cardiovascular diseases.1,2 Diabetic cardiac fibrosis is characterized by the proliferation and migration2 of cardiac fibroblasts (CFs), with subsequent collagen deposition.3 Such cellular proliferation and migration are affected by lipid metabolic reprogramming,4 of which enhanced mitochondrial lipid oxidation is a vital component.5,6 Indeed, proper mitochondrial lipid oxidation may enhance cell functions, including cell proliferation and migration in itself. Lipid catabolism is regulated by 2,4-dienoyl CoA reductase 1 (Decr1),7 carnitine palmitoyltransferase 1α (CPT1α),8 adipose triglyceride lipase (ATGL),9 and hormone-sensitive lipase (HSL).10 Nevertheless, enhanced mitochondrial lipid oxidation is a general aspect of diabetic cardiac fibrosis which remains poorly understood at the molecular level.
Higher mitochondrial lipid oxidation plays an important role in many diseases and is regulated by transcription factors.9,11 Of relevance to this study, androgen receptor (AR) is a transcription factor that plays a crucial role in activating/inhibiting mitochondrial lipid oxidation by targeting mitochondrial lipid oxidation enzyme.12,13 AR has been shown to control lipid metabolic remodeling in inflammation14,15 and cancers.7,16 However, whether AR regulates higher mitochondrial lipid oxidation, thereby contributing to CFs proliferation and migration, has not yet been determined.
Dysregulation of AR modifications has been linked to the epigenetic mechanism.17,18,19,20 N6-methyladenosine (m6A) is the most prevalent RNA methylation21 and is mainly mediated by m6A regulators, comprising writers, erasers, and readers.22 The core writers that catalyze m6A modification are Wilms’ tumor 1-associating protein (WTAP),23 methyltransferase-like (METTL) 14, and METTL3.24 Readers contain the YTH (YT521-B homology) protein domain and include cytoplasmic and nucleic YTH domain containing (YTHDC) 1, YTHDC2, YTH domain family (YTHDF)1, YTHDF2, and YTHDF3 proteins.25 Interestingly, WTAP-mediated m6A modification specifically cooperates with the m6A reader YTHDF2, which is essential for mRNA degradation.26 Nonetheless, whether or not AR expression as regulated by WTAP-mediated m6A modification contributes to diabetic cardiac fibrosis remains unclear.
In this study, we aimed to demonstrate that WTAP boosts higher mitochondrial lipid oxidation as well as CFs proliferation and migration by increasing AR methylation via m6A modification dependent on YTHDF2.
Results
Leptin receptor-deficient induced diabetic cardiac fibrosis, resulting in increased mitochondrial lipid oxidation and impaired heart function
We established an animal model of diabetic cardiac fibrosis in mice using leptin receptor-deficient mice (db/db), a putative model of cardiac fibrosis in type 2 diabetes (Figure 1A). After one week of acclimatization, we measured the fasting blood glucose values of mice in each group every week to ensure the successful establishment of animal models of diabetic cardiac fibrosis in mice (Figure S1A). Oil red O staining revealed a significant increase in total oil red O staining area in cardiac tissue of diabetic mice induced with fibrosis (Figures 1B and S7A). Furthermore, BODIPY staining indicated a significantly higher proportion of lipid droplets in the db/db tissue of the test group (Figures 1C and S7B), indicating excessive lipid droplet deposition.
Figure 1.
Leptin receptor-deficient induced diabetic cardiac fibrosis, resulting in increased mitochondrial lipid oxidation and impaired heart function
(A) Leptin receptor-deficient induced diabetes in mice and established an animal model of diabetic cardiac fibrosis.
(B) Lipid accumulation was evaluated by oil red O staining of mice heart sections. Scale bars: 50 μm, n = 6.
(C) Lipid accumulation was evaluated by BODIPY staining of mice heart sections. Count the quantification of stains by ImageJ software. Scale bars: 20 μm, n = 6.
(D) The expression levels of FASN, ACLY, ACCα, SREBP-1, Decr1, CPT1a, ATGL, and HSL in cardiac tissue were detected by RT-qPCR. Results were normalized to the β-actin levels in each sample and expressed relative to the untreated control, n = 6.
(E) The expression levels of FASN, ACLY, ACCα, SREBP-1, Decr1, CPT1a, ATGL, and HSL in cardiac tissue were detected by western blotting analysis. β-actin was used as an internal protein loading control. A representative sample is shown for each protein, n = 6.
(F) The expression levels of Decr1, CPT1a, and ATGL in cardiac tissue were detected by immunohistochemistry. Results were normalized to expression relative to untreated controls. Scale bars: 20 μm, n = 6.
(G) Determination of oxygen consumption rate, ROS, and MDA levels in fresh tissue samples. Results were normalized to expression relative to untreated controls, n = 6.
(H) Lipid deposition and paralipid mitochondria were observed in diabetic myocardial fibrosis mice under TEM. M represents paralipid mitochondria and L represents lipids. Scale bars: 1 μm, n = 6.
(I) Heart sections from normal and diabetic mice were co-stained with mito tracker and BODIPY. Count the quantification of individual stains separately. Scale bars: 20 μm, n = 6.
(J) Heart sections from normal and diabetic mice were co-stained with mitotracker, BODIPY, and Postn—a cardiac fibroblast marker. Count the quantification of individual stains separately. Scale bars: 50 μm, n = 6. All data are presented as mean ± SD; ∗p < 0.05,∗∗p < 0.01,∗∗∗p < 0.001.
To examine the reason for this change in lipid droplets deposition in fibrotic tissue, we used RT-qPCR and western blotting to screen a variety of enzymes related to lipid droplets synthesis and lipid droplets oxidation. The results revealed a significant increase in key mitochondrial lipid droplets oxidation enzymes (Decr1, CPT1α, ATGL, and HSL) in tissues with induced diabetic cardiac fibrosis (Figures 1D, 1E, and S7C); similar changes were demonstrated with an immunohistochemistry assay (Figures 1F and S7D). Levels of lipid droplets synthesis enzymes (FASN, ACLY, ACCα, and SREBP-1) were significantly enhanced in fibrotic tissues (Figures 1D, 1E, and S7C). This suggests that lipid droplets oxidation and lipid droplets synthesis was responsible for the observed increase in lipid droplets deposition. Yet, given the accumulation of lipid droplets, are most likely due to increased mitochondrial liposynthase-related enzymes and abnormal mitochondrial β-oxidation. Nevertheless, strikingly, we found that mitochondrial oxygen consumption, MDA, and ROS were significantly increased in fibrotic tissues (Figure 1G); moreover, we detected the lipid oxidation markers peroxisome proliferator-activated receptors (PPARs) family in the fibrotic tissue, PPARα, and PPARδ were significantly increased, while the level of PPARγ was slightly decreased (Figures S1E and S7E), indicating that accumulated lipid droplets may also simultaneously mediate enhanced mitochondrial lipid oxidation.
To get more evidence to explain this phenomenon, some characterizations were carried out. Strikingly, transmission electron microscopy revealed a significant increase in the peridroplet mitochondria associated with lipid droplets in fibrotic tissues of diabetic cardiac fibrosis mice (Figures 1H and S7F). More importantly, we found that the Mito Tracker Red probe co-stained with lipid droplets BODIPY staining in fibrotic tissues (Figures 1I and S7G). The results indicated that the adherence of mitochondrial particles to lipid droplets.
Next, we used echocardiography to monitor changes in heart function in diabetic cardiac fibrosis mice. At 20 weeks old, the db/db mice group showed impaired cardiac function in terms of significantly reduced fractional shortening and decreased ejection fraction (EF) compared with the control group (Figure S1F). At the same time, the diastolic function test E/e’ and E/A in the db/db mice group decreased significantly, showing impaired diastolic function (Figure S1F). Additionally, food intake and water intake displaying the db/db mice group were monitored and shown to be higher than that of the control group. More importantly, the body weight of db/db mice was also significantly higher than that of db/+ mice (Figures S1B–S11D).
Since increased cardiac fibrosis is closely associated with such impaired heart function,25 we validated the presence of fibrosis using histopathology and measurement of fibrosis marker expression. Moreover, Masson’s trichrome (MT) and Sirius Red staining identified significant increases in collagen deposition in the heart tissue of db/db mice (Figure S1G), which also showed increased expression of Postn, collagen I, and collagen III proteins (Figures S1H and S7H). More importantly, we found that enhanced co-staining of the Mito Tracker Red probe and BODIPY with the fibroblast marker Postn (Figures 1J and S7I), but not significantly changed of the cardiomyocyte marker cTnT in the fibrotic heart tissue (Figures S1I and S7J), indicating that mitochondria and lipid droplet-specific interactions in CFs might play a critical role in diabetic cardiac fibrosis.
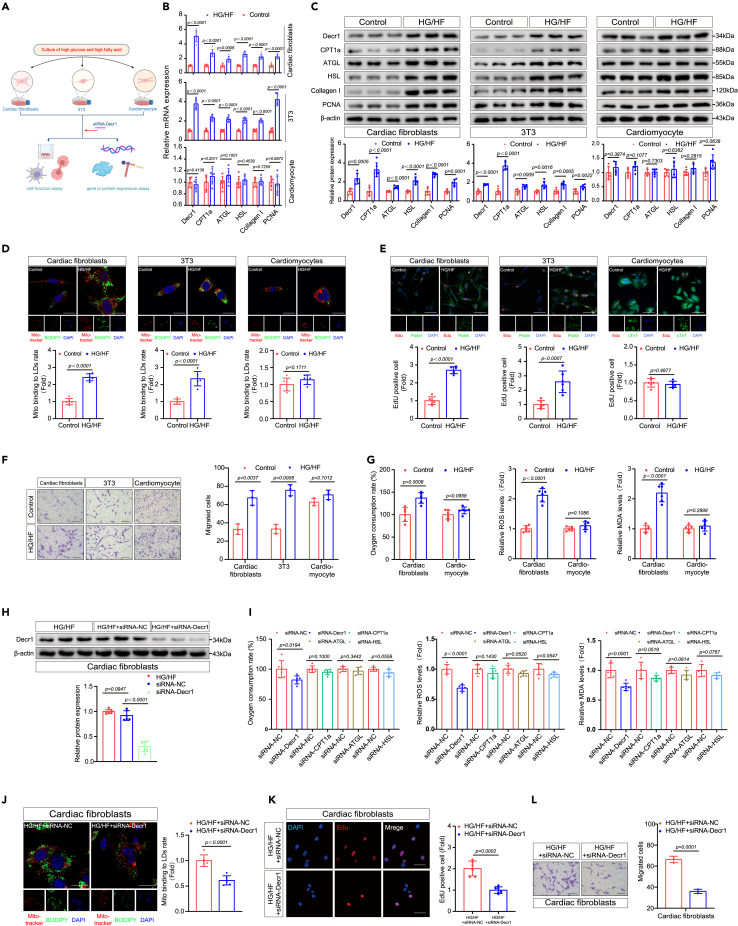
HG/HF induced enhanced mitochondrial lipid oxidation in CFs, dependent on Decr1 expression
We determined whether mitochondrial lipid oxidation is similarly augmented in vitro using primary CFs isolated from neonatal mice, cardiomyocytes, and 3T3 cells (mouse embryonic fibroblasts) (Figure 2A). First, we tested the establishment of CFs activation stimulated by HG/HF (33 mmol/L glucose, 200 μmol/L palmitate, and 200 μmol/L oleate), a putative factor that mimics type 2 diabetes in vitro.27,28,29 During cardiac fibrosis, CFs are activated and transdifferentiated into myofibroblasts, marked by enhancement of α-SMA.30 In order to determine the effect of HG/HF on promoting fibroblasts proliferation and transdifferentiation, we detected the expression changes of cell proliferation marker TGF-β1/Smad pathway31 and transdifferentiation marker α-SMA.30 Western blotting experiments showed that the expression of TGF-β1/Smad2/Smad3 increased (Figures S2A and S8A), which confirmed the significant enhancement of the proliferation of CFs under HG/HF conditions. At the same time, the significant increase of α-SMA protein level and the change of immunofluorescence staining morphology indicated that CFs had been activated and transdifferentiated into myofibroblasts (Figures S2A, S2B, S8A, and S8B). Then, RT-qPCR and western blotting were used to examine differential protein expression levels between CFs, 3T3 cells, and cardiomyocytes following exposure to HG/HF. HG/HF treatment of CFs and 3T3 cells resulted in increased levels of the mitochondrial lipid droplets oxidation enzymes Decr1, CPT1α, ATGL, and HSL (Figures 2B, 2C, and S8C), increased expression of the PCNA and collagen I mRNA and protein (Figures 2B, 2C, and S8C), and an increased proportion of mitochondrial bound to lipid droplets (Figures 2D and S8D); none of these increases were evident in cardiomyocytes. More importantly, we found that the Mito Tracker Red probe co-stained with BODIPY in fibroblasts but not significantly enhanced in cardiomyocytes. In addition, we used Edu staining to co-stain with CF marker Postn and cardiomyocyte marker cTnT to confirm the purity of the relevant cells (Figures 2E and S8E). At the same time, our results indicate that excessive lipid droplets deposition plays an important role in fibroblasts exposed to HG/HF in vitro, enhancing their proliferation rate significantly (Figures 2E and S8E). Additionally, wound healing and transwell migration assays indicated significantly increased migration activity of HG/HF-treated fibroblasts compared with that of controls, whereas cell migration activity of cardiomyocytes showed no significant change post-treatment (Figures 2F, S2C, S8F, and S8G). Enhanced mitochondrial lipid oxidation may therefore be an underlying trigger for diabetic cardiac fibrosis, which is known to arise from the proliferation and migration of CFs.
Figure 2.
HG/HF-induced enhanced mitochondrial lipid oxidation in CFs, dependent on Decr1 expression
(A) Schematic diagram of cell culture. We used cardiac fibroblasts, 3T3, and cardiomyocytes to evaluate gene or protein expression and cell function under HG/HF conditions and after transfection with siRNA-Decr1.
(B) The expression levels of Decr1, CPT1a, ATGL, HSL, collagen I, and PCNA in different cells under HG/HF were detected by RT-qPCR. Results were normalized to the β-actin levels in each sample and expressed relative to the untreated control.
(C) The expression levels of Decr1, CPT1a, ATGL, HSL, collagen I, and PCNA in different cells under HG/HF were detected by western blotting analysis. β-actin was used as an internal protein loading control. A representative sample is shown for each protein, n = 6.
(D) Cells in the normal control group and HG/HF group were co-stained with mitotracker and BODIPY. Calculation of mitochondrial bounding to lipid droplet rate. Scale bars: 25.375 μm, n = 6.
(E) The EdU assay was performed to compare the proliferative capacity of cardiac fibroblasts, 3T3 cells, and cardiomyocytes under normal control conditions and HG/HF conditions. Scale bars: 25.375 μm, n = 6.
(F) Transwell migration assays were performed to compare the migration ability of cardiac fibroblasts, 3T3 cells, and cardiomyocytes under normal control conditions and HG/HF conditions. Scale bars: 500 μm, n = 3.
(G) Measurement of oxygen consumption rate, ROS, and MDA levels in cardiac fibroblasts and cardiomyocytes under normal control and HG/HF conditions. Results were normalized to expression relative to untreated controls, n = 6.
(H) Detection of Decr1 protein level in cardiac fibroblasts after Decr1 knockout under HG/HF condition by western blotting analysis. β-actin was used as an internal protein loading control. A representative sample is shown for each protein, n = 6.
(I) Knockdown of different lipid-related enzymes under HG/HF conditions measured oxygen consumption rate, ROS, and MDA levels in cardiac fibroblasts. Results were normalized to expression relative to untreated controls, n = 6.
(J) Cardiac fibroblasts were co-stained with mitotracker and BODIPY after Decr1 knockdown. Calculation of mitochondrial bounding to lipid droplet rate. Scale bars: 25.375 μm, n = 6.
(K) The EdU assays were performed on the proliferative capacity of cardiac fibroblasts after the Decr1 knockdown. Scale bars: 20 μm. (mean ± SD; ∗p < 0.05,∗∗p < 0.01,∗∗∗p < 0.001,n = 6).
(L) Transwell migration assays were performed to examine the migratory capacity of cardiac fibroblasts after the Decr1 knockdown. Scale bars: 500 μm, n = 3. All data are presented as mean ± SD; ∗p < 0.05,∗∗p < 0.01,∗∗∗p < 0.001.
Next, we found that HG/HF can lead to the increase of mitochondrion lipid oxidation level and mitochondrial oxygen consumption, MDA, and ROS in fibroblasts but not significantly in cardiomyocytes (Figure 2G). At the same time, we detected the expression of lipid oxidation markers PPARs family in CFs under an HG/HF environment. Similar to the expression levels in heart tissue, PPARα and PPARδ were significantly increased (Figures S2D and S8H), while the changes of PPARγ were slightly decreased (Figures S2D and S8H). We subsequently hypothesized that higher mitochondrial lipid oxidation is necessary for CFs proliferation and migration. To test this hypothesis, we inhibited Decr1, CPT1α, ATGL, and HSL expression in CFs using siRNA-mediated gene silencing (Figures 2H, S2E, and S8I). We confirmed that only Decr1 knockdown decreased Decr1 level and dampened mitochondrion lipid oxidation level and mitochondrial oxygen consumption, MDA, and ROS in CFs pretreated with HG/HF (Figure 2I), but not significantly CPT1α, ATGL, and HSL (Figure 2I). More importantly, mitochondrial bound to lipid droplets, CFs proliferation, and migration were also suppressed with Decr1 knockdown, respectively (Figures 2J–2L and S8J–S8L). In addition, Decr1 knockdown decreased lipid oxidation markers PPARα and PPARδ expression levels (Figures S2F and S8M), indicating that HG/HF-induced enhanced mitochondrial lipid oxidation in CFs, dependent on Decr1 expression.
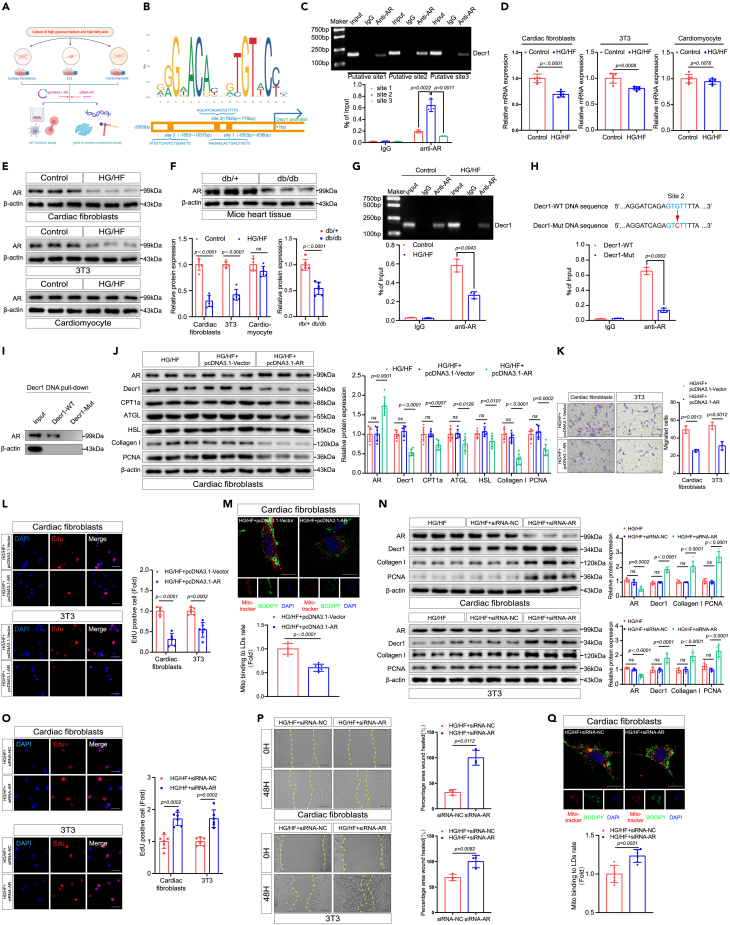
Given these findings, we investigated a possible correlation between the expression of mitochondrial lipid oxidation Decr1 and cell proliferation regulatory molecules in CFs. Given that mitochondrial lipid oxidation is regulated by transcription factors, combined with literature reports and JASPAR transcription factor analysis indicated that mitochondrial lipid oxidation enzyme Decr1 might interact with five common transcription factors, namely, AR, FOXO3, HIF-1α, Nrf2, and STAT3 (Figures 3B and S3A). To investigate which transcription factor was most closely related to Decr1-mediated mitochondrial lipid oxidation, we investigated the bind to the promoter site of Decr1 associated with transcription factors. CHIP assay confirmed that AR could bind to the promoter site of Decr1 in HG/HF-treated CFs (Figures 3C and 3G), no significant changes were observed for FOXO3, HIF-1α, Nrf2, and STAT3 binding to the promoter site of Decr1 in HG/HF-treated CFs following similar treatment (Figure S3B).
Figure 3.
Mitochondrial lipid oxidation and CFs proliferation and migration decrease with AR overexpression and increase with AR knockdown
(A) Schematic diagram of cell culture. We used cardiac fibroblasts, 3T3, and cardiomyocytes to evaluate gene or protein expression and cell function under HG/HF conditions and after transfection with pcDNA3.1-AR or siRNA-AR.
(B) Sequence motif identified from top 1,000 AR peaks and transcription factor AR-specific site on the Decr1 promoter.
(C) ChIP assay of AR binding to three different sites on Decr1. ChIP-qPCR quantification of AR binding at the Decr1 promoter site. Each group of Input was used as baseline control, n = 3.
(D) The expression levels of AR in different cells under HG/HF conditions were detected by RT-qPCR. Results were normalized to the β-actin levels in each sample and expressed relative to the untreated control, n = 6.
(E) The expression levels of AR in different cells under HG/HF conditions were detected by western blotting analysis. β-actin was used as an internal protein loading control. A representative sample is shown for each protein, n = 6.
(F) The expression levels of AR in diabetic cardiac fibrosis mice were detected by western blotting analysis. β-actin was used as an internal protein loading control. A representative sample is shown for each protein, n = 6.
(G) The predicted site 2 was selected as the site for subsequent experiments and CHIP analysis of AR on Decr1 under HG/HF conditions was performed. ChIP-qPCR quantification of AR binding to site 2 at the Decr1 promoter. Each set of inputs served as baseline control, n = 3.
(H) Predicted ARE-site 2 on wild-type Decr1 and mutated sites on the DNA promoter of mutant Decr1. ChIP-qPCR quantification of AR binding to the Decr1 promoter in wild-type and mutant-type. Each set of inputs served as baseline control, n = 3.
(I) Cardiac fibroblasts were stably transfected with plasmids containing the gene indicated by mutant Decr1 and subjected to DNA pull-down, followed by western blot analysis with anti-AR antibody, n = 3.
(J) The expression levels of AR, Decr1, CPT1a, ATGL, HSL, collagen I, and PCNA in cardiac fibroblasts after AR overexpression under HG/HF conditions were detected by western blotting analysis. β-actin was used as an internal protein loading control. Representative samples for each protein are shown n = 6.
(K) Transwell migration assays were performed to compare the migration ability of cardiac fibroblasts and 3T3 cells after AR overexpression. Scale bars: 500 μm, n = 3.
(L) The EdU assays were performed on the proliferative capacity of cardiac fibroblasts and 3T3 after AR overexpression. Scale bars: 20 μm, n = 6.
(M) Cardiac fibroblasts were co-stained with mitotracker and BODIPY after AR overexpression. Calculation of mitochondrial bounding to lipid droplet rate. Scale bars: 25.375 μm, n = 6.
(N) The expression levels of AR, Decr1, collagen I, and PCNA in cardiac fibroblasts and 3T3 after AR knockdown under HG/HF conditions were detected by western blotting analysis. β-actin was used as an internal protein loading control. Representative samples for each protein are shown n = 6.
(O) The EdU assays were performed on the proliferative capacity of cardiac fibroblasts and 3T3 after AR knockdown. Scale bars: 20 μm, n = 6.
(P) Scratch-healing assays were performed to measure the migratory capacity of cardiac fibroblasts and 3T3 after AR knockdown. Scale bars: 500 μm, n = 6.
(Q) Cardiac fibroblasts were co-stained with mitotracker and BODIPY after AR knockdown. Calculation of mitochondrial bounding to lipid droplet rate. Scale bars: 25.375 μm, n = 6. All data are presented as mean ± SD; ∗p < 0.05,∗∗p < 0.01,∗∗∗p < 0.001.
Interestingly, in a single-cell sequencing sample of mouse cardiac fibrosis through the GEO database for analysis, we found that AR was mainly expressed in CF subsets. AR was negatively correlated with the expression of Postn and collagen I (Figure S3C). Furthermore, the enrichment analysis results showed that AR was negatively correlated with mitochondrial lipid β-oxidation (Figure S3E). Given that AR can control pyroptosis32 and autophagy33 in HG/HF-treated CFs, we verified whether AR directly regulates the expression of pyroptosis and autophagy-related genes through CHIP experiments. The results showed that AR could slightly directly combine with the classic genes of pyroptosis NLRP3 and autophagy Beclin 1 in HG/HF-treated CFs (Figure S3D), indicating that Decr1 is potentially regulated by transcription factor AR. In addition, high expression of Decr1 correlated with low AR expression in HG/HF-treated CFs (Figure S3F), which was surprising given that AR has been reported to increase cell proliferation.34 In contrast, we found that overexpression of AR inhibited CF proliferation (Figures 3K, 3L, S9F, and S9G) and mitochondrial lipid oxidation (Figures 3M and S9H). Thus, we hypothesized that AR might play a role in mitochondrial lipid oxidation and resultant CFs proliferation and migration.
Mitochondrial lipid oxidation and CFs proliferation and migration decrease with AR overexpression and increase with AR knockdown
We next investigated the mechanisms underlying AR in the context of mitochondrial lipid oxidation, CFs proliferation, and migration. Indeed, we observed a decreased expression of AR in CFs and 3T3 cells treated with HG/HF in vitro (Figures 3D, 3E, and S9A) and in murine cardiac tissue induced with diabetic cardiac fibrosis (Figures 3F and S9B); no significant change was observed for corresponding AR expression in cardiomyocytes following similar treatment (Figures 3D, 3E, and S9A). Additionally, we also analyzed the trend of decreasing expression of AR in db/db diabetic mice (Figure S3G). The enrichment analysis of AR-related genes showed that AR was enriched into more negative regulation of lipoprotein lipase activity (Figure S3G).
Accumulating evidence indicates that AR plays a crucial role in controlling lipid metabolism.12 Interestingly, overexpression of AR downregulated the expression of Decr1 in CFs much more than that of CPT1α, ATGL, and HSL (Figures 3J and S9E). Notably, overexpression of AR inhibited the proliferation and migration of CFs and 3T3 cells (Figures 3K, 3L, S9F, and S9G) as well as mitochondrial lipid oxidation (Figures 3M and S9H). Furthermore, we found that the expression of PPARα and PPARδ decreased after AR overexpression (Figures S3H and S9O).
In order to explore the relationship between AR mediating Decr1, we predicted and verified three sites (named site 1/2/3) where AR bound to the Decr1 promoter, and the CHIP experiment results showed that site 2 was the site where AR strongly bound to the Decr1 promoter and exerted transcriptional effects (Figure 3C). Then we selected site 2 and performed CHIP experiments in a high-glucose environment to observe the binding of AR to the Decr1 site. The results showed that AR binding to the promoter site of Decr1 was attenuated in fibroblasts under HG/HF conditions (Figures 3G and S9C), suggesting that the AR-binding site 2 on the Decr1 promoter may be a key site for AR to regulate Decr1 transcription. To verify whether site 2 on the Decr1 promoter is a key site for AR to regulate Decr1 transcription, therefore, we constructed point mutants of site 2 (mutating GTGTT to GTCTT, Figure 3H) and performed CHIP experiments and DNA pull-down experiments on the mutants to verify our conjectures. CHIP experiments and DNA pull-down experiments confirmed that the AR binding to this site on the Decr1 DNA promoter was significantly reduced after the mutation of site 2 (Figures 3H, 3I, and S9D). This indicated that AR might regulate the transcription of Decr1 through the related binding sites in the DNA promoter region of Decr1, which would indirectly regulate the process of lipid oxidation, cell proliferation, and migration in fibroblasts.
Furthermore, AR is a nuclear receptor that is activated by binding in the cytoplasm of the androgen hormone testosterone or dihydrotestosterone (DHT) and then translocated into the nucleus.35,36 We tried to validate the content of testosterone/DHT and its effect on AR and Decr1. First, the testosterone content decreased significantly in the serum of db/db mice and the CFs cultured with HG/HF (Figure S3I). At the same time, the addition of DHT to CFs significantly increased AR expression and suppressed Decr1 expression (Figures S3J and S9I). In addition, it is known that AR usually participates in the activation or repression of AR transcription by recruiting cofactors. Therefore, we explored whether AR recruits relevant cofactors to regulate the transcription of Decr1 in CFs. Interestingly, the co-immunoprecipitation (coIP) assay found that the coactivators SWI/SNF complex family members Baf60a and Baf57 were little or not recruited in CFs, while the corepressors NcoR and SMRT were significantly recruited (Figure S3K). At the same time, the coIP assay showed that the NcoR and SMRT pulled down by AR antibody in the HG/HF environment were significantly reduced (Figure S3L), which indicated that the low level of AR in the HG/HF environment might fail to recruit corepressors, resulting in increased AR-mediated Decr1 transcriptional activity and enhanced lipid oxidation, thereby promoting cell proliferation and migration. At the same time, to verify whether the NcoR/SMRT complex directly acts on the region associated with the Decr1 promoter to exert transcriptional repression, we performed ChIP experiments using the NcoR/SMRT complex antibody, and the results showed that in the HG/HF environment, the NcoR/SMRT complex bounding to the Decr1-site2 DNA fragment was significantly reduced, thereby failing to repress Decr1 transcription (Figure S3M).
Therefore, we hypothesized that the loss of AR triggers CFs proliferation, migration, and mitochondrial lipid oxidation. To evaluate the specific role of AR, this RNA was knocked down in CFs and 3T3 cells pretreated with HG/HF (Figure 3A). As a result, the expression of the fibroblast proliferation marker PCNA was promoted (Figures 3N and S9J). Furthermore, the proliferation rate and migration activity of CFs and 3T3 cells were increased in groups treated with AR-siRNA, compared with those in the siRNA-NC group, as determined by measuring EdU incorporation and conducting wound healing and transwell migration assays (Figures 3O, 3P, and S9K–S9M). Consistent with this observation, increased mitochondrial lipid oxidation in CFs treated with AR-siRNA, confirming the loss of AR triggers mitochondrial lipid oxidation (Figures 3Q and S9N). Finally, the knockdown of AR in CFs increased the expression of Decr1 (Figures 3N and S9J), PPARα, and PPARδ (Figures S3N and S9O), further indicating that the knockdown of AR in CFs promoted mitochondrial lipid oxidation.
Collectively, in vitro model mimicking diabetic cardiac fibrosis, our results demonstrated that CFs proliferation, migration, and mitochondrial lipid oxidation were significantly ameliorated by AR overexpression and aggravated by AR knockdown.
WTAP-dependent m6A upregulation suppresses AR expression
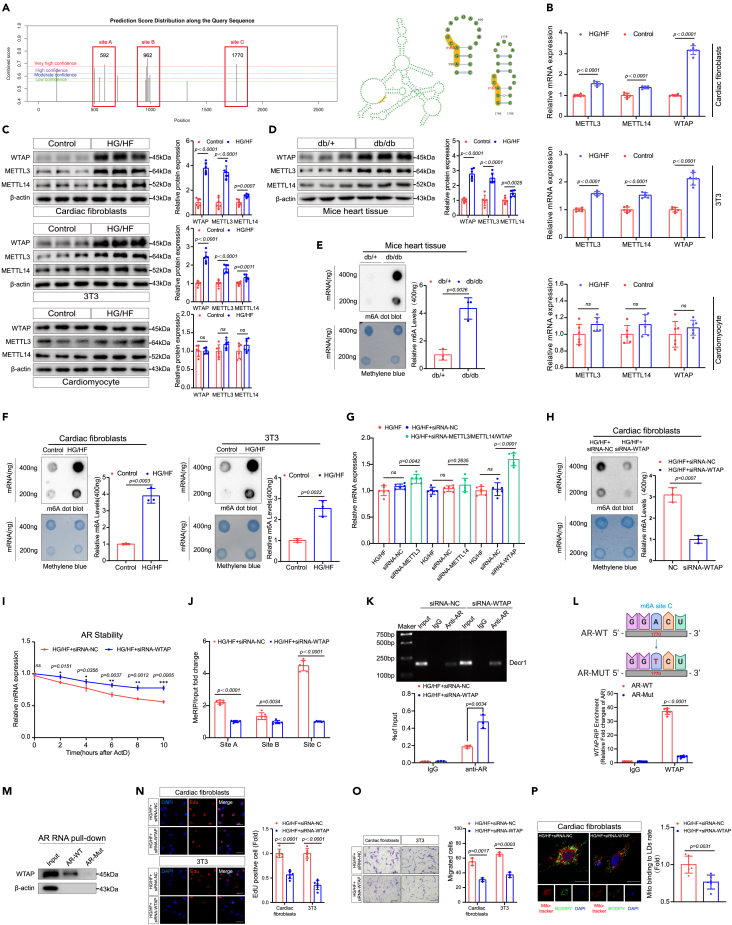
To explore the mechanism by which AR is suppressed in the HG/HF environment, we investigated whether m6A RNA modification, the most abundant epigenetic modification in eukaryotic mRNA, might be involved. By using SRAMP software (http://www.cuilab.cn/sramp), we found strong m6A peak enrichment of AR in mice (Figure 4A), indicating that AR can be regulated by m6A methylation. The expression of genes associated with m6A modifications was investigated, and our results were consistent with the bioinformatics findings. Both fibroblasts treated in vitro with HG/HF and murine heart tissue induced with diabetic cardiac fibrosis in vivo exhibited marked increases in METTL3, METTL14, and WTAP expression. In contrast, no significant enhancement was recorded for corresponding expression levels in cardiomyocytes (Figures 4B–4D, S10A, and S10B). Similarly, both the in vitro and in vivo tests demonstrated significant increases in m6A mRNA contents compared with control groups (Figures 4E, 4F, S10C, and S10D).
Figure 4.
WTAP-dependent m6A upregulation suppresses AR expression
(A) M6A-sites prediction and the sites’ confidence score for AR. M6A sites prediction by bioinformatic analysis using SRAMP software (http://www.cuilab.cn/sramp).
(B) The expression levels of METTL3, METTL14, and WTAP in different cells under HG/HF conditions were detected by RT-qPCR. Results were normalized to the β-actin levels in each sample and expressed relative to the untreated control, n = 6.
(C) The expression levels of METTL3, METTL14, and WTAP in different cells under HG/HF conditions were detected by western blotting analysis. β-actin was used as an internal protein loading control. A representative sample is shown for each protein, n = 6.
(D) The expression levels of METTL3, METTL14, and WTAP in diabetic cardiac fibrosis mice were detected by western blotting analysis. β-actin was used as an internal protein loading control. A representative sample is shown for each protein, n = 6.
(E) Global m6A levels of RNA extracted from db/db mice heart tissue were measured via m6A dot blot assays. The gray value of m6A dot blot and methylene blue were evaluated by ImageJ software and quantitative analysis of 400 ng gray value was performed, n = 3.
(F) Global m6A levels in RNA extracted from cardiac fibroblasts and 3T3 cells cultured under HG/HF were measured by m6A dot blot assay. The gray value of m6A dot blot and methylene blue were evaluated by ImageJ software and quantitative analysis of 400 ng gray value was performed, n = 3.
(G) The expression levels of AR in cardiac fibroblasts after METTL3/METTL14/WTAP knockdown were detected by RT-qPCR. Results were normalized to the β-actin levels in each sample and expressed relative to the untreated control, n = 6.
(H) Global m6A levels in RNA extracted from cardiac fibroblasts cultured under HG/HF after WTAP knockdown were measured by m6A dot blot assay. The gray value of m6A dot blot and methylene blue were evaluated by ImageJ software and quantitative analysis of 400 ng gray value was performed, n = 3.
(I) The stability of AR transcripts in ActD-treated cardiac fibroblasts after transfection with WTAP siRNA was detected by RT-qPCR, n = 3.
(J) MeRIP-qPCR analysis of m6A levels at three different sites of mRNAs from cardiac fibroblasts in siRNA negative control and siRNA-WTAP, n = 6.
(K) The predicted ARE-site 2 was selected for subsequent experiments and CHIP analysis of AR on Decr1 under HG/HF conditions after WTAP knockdown was performed. ChIP-qPCR quantification of AR binding to the Decr1 promoter. Each set of inputs served as baseline control, n = 3.
(L) Selection of m6A-site C for point mutation and relative quantification of AR by WTAP-RIP. Quantitative qPCR was analyzed by t-test. Each set of inputs served as baseline control, n = 6.
(M) RNA pull-down was used to detect the binding pull-down ability of wild-type AR to WTAP or mutant AR to WTAP, followed by western blot analysis with anti-AR antibody.
(N) The EdU assays were performed on the proliferative capacity of cardiac fibroblasts and 3T3 after WTAP knockdown. Scale bars: 20 μm, n = 6.
(O) Transwell migration assays were performed to compare the migration ability of cardiac fibroblasts and 3T3 cells after WTAP knockdown. Scale bars: 500 μm, n = 3.
(P) Cardiac fibroblasts were co-stained with mitotracker and BODIPY after WTAP knockdown. Calculation of mitochondrial bounding to lipid droplet rate. Scale bars: 25.375 μm, n = 6. All data are presented as mean ± SD; ∗p < 0.05,∗∗p < 0.01,∗∗∗p < 0.001.
Based on these results, we hypothesized that METTL3, METTL14, or WTAP might regulate AR expression. The successful knockdown of METTL3, METTL14, and WTAP were validated at both protein and mRNA levels (Figures S4A, S4B, S10E, and S10F). Surprisingly, the knockdown of WTAP promoted AR expression significantly, while the knockdown of METTL3 or METTL14 promoted AR expression to a significantly lower extent than the WTAP knockdown (Figure 4G). Correspondingly, the knockdown of WTAP significantly decreased the mRNA m6A content in CFs (Figures 4H and S10G). Crucially, sequence analysis found multiple matching m6A common sequences in AR, and we selected the three sites with the highest predicted confidence scores and named them sites A–C (Figure 4A; Table S2).
Subsequently, the RNA decay assay showed that the knockdown of WTAP remarkably enhanced the stability of AR mRNA (Figure 4I). At the same time, MeRIP-qPCR confirmed the significant m6A-mediated reduction of AR following WTAP knockdown in HG/HF-treated CFs compared with the negative control group (Figure 4J). Strikingly, the CHIP assay confirms that the knockdown of WTAP remarkably enhanced the AR mRNA, and upregulation of AR can bind to the promoter site of Decr1 and inhibit the transcription of Decr1 (Figures 4K and S10H). More importantly, the RNA decay assay confirmed that the knockdown of WTAP did not directly affect the stability of Decr1 mRNA (Figure S4C). Collectively, these results indicate that WTAP-dependent upregulation of m6A in AR suppresses AR expression in CFs, and AR regulates the expression of Decr1 in HG/HF-treated CFs by binding to the promoter site of Decr1.
After the knockdown of WTAP, the expression of total m6A on AR was detected by performing MeRIP-qPCR experiments. Surprisingly, the results showed that the predicted site C significantly reduced the expression of m6A (Figure 4J), which suggested that predicted site C might be an important site regulating AR mRNA stability. To verify whether site C could directly regulate AR mRNA, we subsequently performed a gene point mutation (we constructed an AR mutant plasmid and mutated the 1,770 position A of the AR mRNA sequence to T) at the m6A site for site C. Next, we verified whether the AR mutant is functional through decay assay. Compared with wild-type AR, AR mutant could stabilize the mRNA expression of AR (Figure S4D). Additionally, western blot results also showed that AR-Mut significantly stabilized AR mRNA and disrupted the HG/HF-induced downregulation of AR protein (Figures S4E and S10I). Finally, after determining the functionality of the AR mutants, we performed WTAP-RIP detection and RNA pull-down detection to verify the m6A function of site C respectively (Figures 4L and 4M). The RIP results showed that the expression of WTAP in the mutant type was significantly lower than that in the wild type after the site C mutation (Figure 4L). The RNA pull-down results also showed that the site C mutated AR RNA could not pull down WTAP (Figures 4M and S10J). Collectively, these results suggested that site C was an important m6A site for regulating AR mRNA stability.
There were relevant literature reports that WTAP was related to RNA methylation and cellular proliferation.37,38 We constructed WTAP knockdown models in CFs and 3T3 cells. Indeed, the knockdown of WTAP significantly decreased cell proliferation rate and migration activity (Figures 4N, 4O, S10K, and S10L). In addition, the knockdown of WTAP similarly reduced the level of mitochondrial lipid oxidation (Figures 4P, S4F, S4G, S10M, and S10N), suppressed mitochondrial oxygen consumption, MDA, and ROS (Figure S4F), and inhibited the expression of PCNA and Decr1 (Figures S4H and S10O) in CFs. More importantly, the coIP assay and ChIP assay showed that after the knockdown of WTAP, AR might recruit more corepressors to inhibit the transcription of Decr1 genes (Figures S4I and S4J).
WTAP enhances AR methylation and subsequent binding of YTHDF2 to AR suppresses its expression
Using an m6A dot blot, we showed that WTAP knockdown significantly decreased m6A levels (Figures 4H and S10G). Indeed, m6A status was dominated by the activity of m6A readers.39 A protein-protein interaction network revealed that WTAP could interact with five common m6A readers, namely, YTHDF1–3 and YTHDC1–2 (Figure 5A). To investigate which reader was most closely related to WTAP-mediated modification of AR, we investigated the expression of genes associated with m6A readers. The expression of YTHDF1 and YTHDF2 was markedly enhanced not only in HG/HF-induced CFs and 3T3 cells but also in murine heart tissue with diabetic cardiac fibrosis (Figures 5B–5D, S11A, and S11B); the expression of YTHDF3, YTHDC1, and YTHDC2 was only slightly increased in the same test groups (Figure 5B); cardiomyocytes receiving similar treatment showed no significant change in expression levels.
Figure 5.
WTAP enhances AR methylation and subsequent binding of YTHDF2 to AR suppresses its expression
(A) A protein-protein interaction network revealed that WTAP could interact with five common m6A readers, namely, YTHDF1-3 and YTHDC1-2.
(B) The expression levels of YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2 in different cells under HG/HF conditions were detected by RT-qPCR. Results were normalized to the β-actin levels in each sample and expressed relative to the untreated control, n = 6.
(C) The expression levels of YTHDF1 and YTHDF2 in different cells under HG/HF conditions were detected by western blotting analysis. β-actin was used as an internal protein loading control. A representative sample is shown for each protein, n = 6.
(D) The expression levels of YTHDF1 and YTHDF2 in diabetic cardiac fibrosis mice were detected by western blotting analysis. β-actin was used as an internal protein loading control. A representative sample is shown for each protein, n = 6.
(E) Bioinformatics analysis by SRAMP software revealed that the most common m6A motif in AR is the consistent RRACH. (where R = G or A, H = A, C, or U).
(F) MeRIP-qPCR analysis of m6A levels at three different sites of mRNAs from cardiac fibroblasts in siRNA negative control and siRNA-YTHDF2, n = 6.
(G) The expression levels of YTHDF2 and AR in different cells under HG/HF conditions after YTHDF2 knockdown were detected by western blotting analysis. β-actin was used as an internal protein loading control. A representative sample is shown for each protein, n = 6.
(H) The stability of AR transcripts in ActD-treated cardiac fibroblasts after transfection with YTHDF2 siRNA was detected by RT-qPCR, n = 3.
(I) Gene ontology enrichment analysis revealed that YTHDF2 was involved in mRNA processing, mRNA binding, and metabolism.
(J) WTAP interacts with YTHDF2 by reciprocal coIP assays in cardiac fibroblasts. Input groups were used as an internal protein loading control, n = 3.
(K) The expression levels of AR in cardiac fibroblasts under HG/HF conditions after WTAP knockdown or WTAP+YTHDF2 knockdown were detected by RT-qPCR. Results were normalized to the β-actin levels in each sample and expressed relative to the untreated control, n = 6.
(L) CHIP analysis of AR on Decr1 under HG/HF environment after YTHDF2 knockdown was performed. ChIP-qPCR quantification of AR binding to the Decr1 promoter. Each set of inputs served as baseline control, n = 3.
(M) Selection of m6A site C for point mutation and relative quantification of AR by YTHDF2-RIP. Quantitative PCR signals were analyzed by t-test. Each set of inputs served as baseline control, n = 6.
(N) RNA pull-down was used to detect the binding pull-down ability of wild-type AR to YTHDF2 or mutant AR to YYTHDF2, followed by western blot analysis with anti-AR antibody.
(O) The EdU assays were performed on the proliferative capacity of cardiac fibroblasts and 3T3 after YTHDF2 knockdown. Scale bars: 20 μm, n = 6.
(P) Cardiac fibroblasts were co-stained with mitotracker and BODIPY after YTHDF2 knockdown. Calculation of mitochondrial bounding to lipid droplet rate. Scale bars: 25.375 μm, n = 6.
(Q) Determination of oxygen consumption rate, ROS, and MDA levels in cardiac fibroblasts under HG/HF conditions after YTHDF2 knockdown. Results were normalized to expression relative to untreated controls, n = 6. All data are presented as mean ± SD; ∗p < 0.05,∗∗p < 0.01,∗∗∗p < 0.001.
Bioinformatic analysis with SRAMP software revealed that the most frequent m6A motif of AR was a consistent RRACH (where R = G or A, H = A, C, or U) (Figure 5E). More importantly, compared with an IgG control group, we could verify that YTHDF2 binds to AR in HG/HF-induced CFs, but YTHDF1 did not (Figure S5A). MeRIP-qPCR confirmed the m6A methylation of AR as recognized by YTHDF2; there was a significant reduction of the YTHDF2-mediated m6A methylation of AR mRNA following YTHDF2 knockdown (Figure 5F), but YTHDF1-knockdown did not (Figure S5A). This resulted in enhanced AR stability and increased AR expression in CFs (Figures 5G, 5H, and S11C), as determined by RNA decay assay and western blotting assay. However, AR expression did not change significantly in YTHDF1-knockdown fibroblasts (Figures S5B and S11D). Gene Ontology enrichment analysis indicated that WTAP and/or YTHDF2 were related to mRNA processing, mRNA binding, and metabolism (Figure 5I). To verify whether WTAP and YTHDF2 co-precipitate specifically in CFs cultured in an HG/HF environment, we performed a coIP assay to screen for WTAP-related m6A readers, which validated that WTAP specifically co-precipitated with YTHDF2 in CFs (Figures 5J and S11E).
Next, we detected the expression levels of AR in CFs transfected with WTAP and/or YTHDF2 siRNA. The results showed that knocking down YTHDF2 and WTAP simultaneously had a more significant effect on AR expression than knocking down WTAP alone (Figure 5K). Strikingly, the CHIP assay confirms that the knockdown of YTHDF2 remarkably enhanced the AR mRNA, and the upregulation of AR can bind to the promoter site of Decr1 (Figures 5L and S11F). More importantly, the RNA decay assay confirmed that the knockdown of YTHDF2 did not directly affect the stability of Decr1 mRNA (Figure S5C). At the same time, overexpression of AR and knockdown of AR did not significantly change the stability of Decr1 (Figure S5D). Combining with the aforementioned research, the knockdown of WTAP did not directly affect the stability of Decr1 mRNA (Figure S4C), suggesting that Decr1 expression may not be directly regulated by post-transcriptional degradation. Subsequently, we examined the expression levels of Decr1 transfected with WTAP and YTHDF2 overexpression plasmids or siRNA after overexpressing AR. The results showed that the level of Decr1 was significantly decreased after overexpression of AR, while overexpression of WTAP and YTHDF2 at the same time significantly relieved the inhibitory effect of AR on the Decr1 promoter (Figures S5E and S11G). In addition, the knockdown of WTAP and YTHDF2 while overexpressing AR further reduced the expression of Decr1 (Figures S5F and S11H). In summary, the results of RNA stability experiments and rescue experiments indicated that Decr1 is not directly regulated by WTAP/YTHDF2, and its expression changes depend on AR-mediated transcriptional regulation. Similarly, after point mutation site C, we performed YTHDF2-RIP experiments and RNA pull-down experiments to verify the altered binding of YTHDF2 to AR mRNA. Consistently, the RIP results showed that the expression of YTHDF2 using anti-AR in the mutant type was significantly lower than that in the wild type (Figure 5M). The RNA pull-down results also showed that the m6A site C on AR was mutated, the mutated AR RNA could not pull down YTHDF2 (Figures 5N and S11I). In short, these results also suggested that site C was an important m6A site for AR mRNA stability.
Indeed, the knockdown of YTHDF2 significantly reduced CFs proliferation and migration abilities (Figures 5O, S5G, S11J, and S11K), decreased mitochondrial lipid oxidation in CFs (Figures 5P, S5H, S11L, and S11M), suppressed mitochondrial oxygen consumption, MDA, ROS (Figure 5Q) and decreased the expression levels of Decr1 and PCNA (Figures S5I and S11N). In addition, to explore the effect of knocking down YTHDF2 on AR recruitment cofactor-mediated Decr1 transcription, coIP assay, and ChIP assay showed that after knocking down YTHDF2, the corepressors binding to AR were significantly increased, thereby inhibiting the transcription of Decr1 and reducing the level of lipid oxidation (Figures S5J and S5K).
Importantly, AR is activated and functions upon binding to its ligand, we verified the role of DHT in the WTAP/YTHDF2 pathway. RT-qPCR results showed that after overexpression of WTAP and YTHDF2, the expression of AR was significantly decreased while the expression of Decr1 was increased (Figure S5L). Meanwhile, after overexpressing WTAP and YTHDF2 and adding DHT, the expression of AR was restored to some extent, and the expression of Decr1 was suppressed (Figure S5L). In fact, overexpression of WTAP and YTHDF2 significantly increased CFs mitochondrial lipid oxidation (Figures S5M and S11O) and proliferation ability (Figures S5N and S11P). However, the supplementation of DHT inhibited the mitochondrial lipid oxidation (Figures S5M and S11O) and proliferation (Figures S5N and S11P) of CFs to a certain extent.
Collectively, our data revealed that WTAP facilitates CFs proliferation, migration, and mitochondrial lipid oxidation by increasing AR methylation in an m6A-YTHDF2-dependent manner.
Knockdown of WTAP/YTHDF2 ameliorated Db/db mice diabetic cardiac fibrosis by increasing AR expression
At the age of 16 weeks, db/db mice with diabetic cardiac fibrosis were injected with either LV3-WTAP lentiviral vectors to introduce shRNA for WTAP knockdown or LV3-YTHDF2 lentiviral vectors to similarly induce knockdown of YTHDF2 (Figure 6A). Body weight and fasting blood glucose did not differ between the treatment and control groups (Figures S6C, S6D, S6G, and S6H), although the food and water intake slightly decreased in LV3-WTAP and LV3-YTHDF2 treated mice at 16–20 weeks (Figures S6A, S6B, S6E, and S6F). The knockdown of WTAP and YTHDF2 substantially decreased the expression levels of Decr1, Postn, and collagen I (Figures 6B–6D, S12A, and S12B). The expression of WTAP and YTHDF2 was markedly reduced in LV3-WTAP- and LV3-YTHDF2-treated mice compared with mice treated with the vector (Figures S12C and S12D). We observed significantly reduced mRNA m6A content in the diabetic cardiac fibrosis tissues of the LV3-WTAP-treated group compared with that of the vector groups (Figures 6F and S12F). Consistent with the aforementioned results, MT and Sirius red staining showed that LV3-WTAP- and LV3-YTHDF2-treated mice had markedly reduced collagen deposition and fibrosis compared to those treated with the vector (Figures 6E and S12E). Moreover, after evaluating LV3-WTAP- and LV3-YTHDF2-treated mice by echocardiography, we observed that their cardiac function was ameliorated (Figure 6G). Furthermore, we found decreased mitochondrial lipid oxidation (Figures 6H and S12G) and mitochondrial oxygen consumption, MDA, and ROS (Figure S12H) in mice with diabetic cardiac fibrosis following WTAP and YTHDF2 knockdown. Further validation by immunofluorescence staining found that both WTAP knockdown (Figures 6I and S12I) and YTHDF2 knockdown (Figures 6J and S12J) significantly increased the expression of AR in mice with diabetic cardiac fibrosis, which was consistent with that the expression of AR in western blot and RT-qPCR (Figures 6B, 6C, and S12A).
Figure 6.
Knockdown of WTAP/YTHDF2 ameliorated Db/db mice diabetic cardiac fibrosis by increasing AR expression
(A) An animal model of lentivirus targeting cardiac fibroblasts injected from the tail vein in db/db mice.
(B) The expression levels of AR, Decr1, Postn, and collagen I in the heart tissue of LV3-WTAP-injected or LV3-YTHDF2-injected mice were detected by RT-qPCR. Results were normalized to the β-actin levels in each sample and expressed relative to the untreated control, n = 6.
(C) The expression levels of AR, Decr1, Postn, and collagen I in the heart tissue of LV3-WTAP-injected or LV3-YTHDF2-injected mice were detected by western blotting analysis. β-actin was used as an internal protein loading control. A representative sample is shown for each protein, n = 6.
(D) The expression levels of Decr1, collagen I, and Postn in the heart tissue of LV3-WTAP-injected or LV3-YTHDF2-injected mice were detected by immunohistochemistry. Results were normalized to expression relative to untreated controls. Scale bars: 20 μm, n = 6.
(E) Sirius Red and Masson trichrome staining of heart sections from LV3-WTAP, LV3-YTHDF2, and LV3-Vector-injected mice were performed. Semi-quantitative analysis of Masson trichrome and Sirius red staining was performed by using ImageJ software. Scale bars: 20 μm, n = 6.
(F) Global m6A levels in RNA extracted from LV3-WTAP/LV3-Vector lentivirus-injected hearts were measured by m6A dot blot assay. The gray value of m6A dot blot and methylene blue were evaluated by ImageJ software and quantitative analysis of 400 ng gray value was performed, n = 3.
(G) Representative M-mode recordings of echocardiography from LV3-Vector and LV3-WTAP/LV3-YTHDF2. Quantitative analysis of ejection fraction (EF), fractional shortening (FS), E/e’, or E/A in each group of lentivirus-injected mice was determined by echocardiography, n = 6.
(H) Heart sections from LV3-Vector and LV3-WTAP/YTHDF2 mice were co-stained with mitotracker and BODIPY. Count the quantification of individual stains separately. Scale bars: 20 μm, n = 6.
(I and J) AR expression in cardiac sections from LV3-Vector or LV3-WTAP/YTHDF2 mice was detected by tissue immunofluorescence. Scale bars: 20 μm, n = 6. All data are presented as mean ± SD; ∗p < 0.05,∗∗p < 0.01,∗∗∗p < 0.001.
Crucially, similar to HG/HF cultured cells, db/db mice after 4 weeks of DHT feeding showed significantly increased expression of AR and decreased expression levels of Decr1, fibrosis markers collagen I, and Postn (Figures S6I, S6J, S12K, and S12L). In addition, we examined the expression of WTAP and YTHDF2 in the hearts of STZ-induced type 1 diabetic cardiac fibrosis mice. The results showed that the expressions of WTAP and YTHDF2 were significantly increased in STZ-induced diabetic cardiac fibrosis mouse hearts (Figures S6K and S12M). More importantly, the expression of WTAP and YTHDF2 decreased in the treatment group mice injected with LV3-WTAP and LV3-YTHDF2, and the cardiac fibrosis was alleviated to a certain extent (Figures S6L, S6M, S12N and S12O).
Downregulated AR expression correlates with upregulated WTAP, YTHDF2, and Decr1 expression in human DCM patients
We examined AR levels, WTAP and YTHDF2 expression, m6A mRNA methylation, expression of mitochondrion lipid oxidation markers and fibrosis-related markers, and cardiac function in human patients with DCM (Figure 7A). First, we compared AR expression between the heart tissues of DCM patients and those of patients with control (Figures 7C, 7D, S13B, and S13C). The expression of WTAP and YTHDF2 was significantly higher in the heart tissues of DCM patients than those of patients with control (Figures 7C, 7D, 7F, S13B, and S13C), according to the result of immunofluorescence assays (Figures 7I and S13F). Accordingly, and in line with our previous observations, we also detected significantly higher m6A mRNA content in the heart tissues of DCM patients than in those of control patients (Figures 7B and S13A). Notably, MT and Sirius Red staining indicated thick and highly disorganized collagen content in DCM patient heart tissues; these histological alterations were confirmed with H&E staining (Figures 7E and S13D). Next, both the mRNA and protein expression of fibrosis markers (collagen I, Postn, and α-SMA) and the expression of mitochondrial lipid oxidation marker (Decr1) were increased in patients with DCM (Figures 7C, 7D, 7F, S13B, and S13C). We also observed impaired cardiac function in the heart tissues of DCM patients, measured as significantly reduced fractional shortening and decreased EF (Table S1). Lastly, mitochondrial lipid droplets deposition and mitochondrion lipid oxidation was significantly increased in DCM patient samples than in those of control patients (Figures 7G, 7H, S13E, and S13G).
Figure 7.
Downregulated AR expression correlates with upregulated WTAP, YTHDF2, and Decr1 expression in human DCM patients
(A) Flow chart of obtaining human myocardial tissue samples.
(B) Global m6A levels in RNA extracted from Control healthy and DCM patients' myocardial tissue were measured by m6A dot blot assay. The gray value of m6A dot blot and methylene blue were evaluated by ImageJ software and quantitative analysis of 400 ng gray value was performed, n = 6.
(C) The expression levels of WTAP, YTHDF2, AR, Decr1, Postn, collagen I, and α-SMA in the heart tissue of Control healthy and DCM patients were detected by western blotting analysis. β-actin was used as an internal protein loading control. A representative sample is shown for each protein, n = 6.
(D) The expression levels of WTAP, YTHDF2, AR, Decr1, Postn, collagen I, and α-SMA in the heart tissue of Control healthy and DCM patients were detected by RT-qPCR. Results were normalized to the β-actin levels in each sample and expressed relative to the untreated control, n = 6.
(E) H&E, Sirius Red, and Masson trichrome staining of heart sections from Control healthy and DCM patients were performed. Semi-quantitative analysis of H&E, Masson trichrome, and Sirius red staining was performed by using ImageJ software. Scale bars: 20 μm, n = 6.
(F) The expression levels of WTAP, YTHDF2, Decr1, and Postn in the control healthy group and DCM patients group were detected by immunohistochemistry. Results were normalized to expression relative to untreated controls. Scale bars: 20 μm, n = 6.
(G) Lipid deposition and paralipid mitochondria were observed in the control healthy group and DCM patients group under TEM. M represents paralipid mitochondria and L represents lipids. Scale bars: 1 μm, n = 6.
(H) Heart sections from the control healthy group and DCM patients group were co-stained with mitotracker, BODIPY, and Postn—a cardiac fibroblast marker. Count the quantification of individual stains separately. Scale bars: 50 μm, n = 6.
(I) WTAP/YTHDF2 expression in cardiac sections from the control healthy group and DCM patients group was detected by tissue immunofluorescence. Scale bars: 20 μm, n = 6. All data are presented as mean ± SD; ∗p < 0.05,∗∗p < 0.01,∗∗∗p < 0.001.
Discussion
Excessive mitochondrial lipid oxidation occurs in several diseases, such as liver injury,40 neurodegeneration,41 and cancers.42 An increasing number of studies suggest that excessive mitochondrial bound to lipid droplets may also cause adverse effects on the cardiovascular system.43,44,45 Nonetheless, we still lack a comprehensive understanding of the underlying mechanisms of this phenomenon. In this study, we combine evidence for such a mechanism by investigating cells cultured in vitro, animal models, and clinical samples; thereby, we demonstrate that WTAP boosts mitochondrial lipid oxidation, CFs proliferation, and fibroblast migration by increasing AR m6A methylation that is dependent on the YTHDF2 binding protein (Graphic abstract).
Several of our findings support this hypothesis. Firstly, WTAP and YTHDF2 expression were significantly elevated in the fibrotic heart tissue of both diabetic humans and mice. The upregulation of WTAP was furthermore associated with increased m6A methylation of RNA in these tissues. This corresponds with previous research suggesting that WTAP and m6A modification of RNA may be necessary for cardiac fibrosis.46
Secondly, the m6A RNA modification involves a crucial step in controlling cellular processes such as proliferation, migration,37,38 and mitochondrial lipid oxidation.47 Indeed, a link between m6A regulation and cardiac fibrosis has been demonstrated.46,48,49,50 Based on our aforementioned findings, the knockdown of WTAP or YTHDF2 ameliorated diabetic cardiac fibrosis induced in mice decreased collagen deposition, decreased the expression of fibroblast marker Postn and mitochondrion lipid oxidation marker Decr1, and suppressed mitochondrial lipid oxidation and fibrosis. This suggests that WTAP and YTHDF2 are intimately associated with mitochondrial lipid oxidation, cell proliferation, and cell migration.
Thirdly, there is an increasing body of evidence demonstrating that AR regulates cell proliferation, cell migration, and mitochondrion lipid oxidation. Different functions of AR regulating cells have been reported. For instance, AR indirectly regulated pyroptosis32 and autophagy33,51 in CFs, which was consistent with our ChIP assay results in this paper. Strikingly, in mice with ISO-induced cardiac fibrosis, AR regulated cardiac fibrosis by mediating glycolytic reprogramming50 and indicated that fundamental differences in AR-mediated glycolytic reprogramming and enhanced lipid metabolism were disease-specific. Nonetheless, more importantly, we report that AR directly acts on the key promoter region of the lipid oxidation gene Decr1, and then regulates the enhancement of lipid oxidation and cell proliferation in CFs. In addition, AR has been shown to be regulated by epigenetic factors.20,52 Our results showed that CFs, stimulated to differentiate with HG/HF, displayed increased mRNA and protein levels of WTAP and YTHDF2, respectively, with a corresponding decrease in AR expression. In our bioinformatics analysis, mutant assay, and MeRIP-qPCR assay confirmation, AR was modified by the WTAP-related m6A methylation. Knockdown of WTAP resulted in a significant reduction of m6A modification of AR, leading to increased AR expression. The m6A readers consistently recognize m6A sites, resulting in different destinies of mRNA, including molecular degradation.21 YTHDF2, an m6A reader, induces the targeted degradation of a tumor suppressor protein to accelerate tumor progression, with this regulatory function being catalyzed by the formation of m6A by WTAP.37,53 We provided evidence for direct binding between YTHDF2 and WTAP. Strikingly, the knockdown of YTHDF2 and WTAP simultaneously had a more significant effect on AR expression than knocking down WTAP alone. MeRIP-qPCR confirms that m6A methylation of AR was specifically recognized by YTHDF2; following YTHDF2 knockdown, there was a significant reduction of m6A modification of AR, which promoted AR expression. These results further support the theory that WTAP catalyzes m6A methylation of AR, with subsequent binding of YTHDF2 to AR suppressing the expression of the latter.
Fourth, we demonstrated that the augmentation of mitochondrial lipid oxidation is necessary to affect CFs proliferation and migration, similar to the findings reported for chronic cholestatic liver disease.54 We discovered that the fibrotic heart tissue of humans and mice showed increased expression of Decr1, Postn, and collagen I. Crucially, overexpression of AR strongly decreased the expression of both PCNA and Decr1 in CFs, suppressing the proliferation and migration of these fibroblasts, as well as mitochondrial lipid oxidation; the knockdown of AR produced opposite results. Mechanistically, the CHIP assay confirms that AR can bind to the promoter site of Decr1. Mechanistically, the CHIP assay and co-IP assay confirmed that AR can bind to the promoter site of Decr1 and recruit related corepressors to regulate the transcription of Decr1. In the HG/HF environment, the number of corepressors recruited was reduced by AR binding to the response element of Decr1, which led to enhanced Decr1 transcription and increased lipid oxidation, and increased proliferation of CFs. Additionally, our reported results are consistent with a report in prostate cancer, Decr1 expression is notably more abundant in AR-negative cells (PC3) than in AR-expressing cells. In VCaP cells overexpressing AR, AR mediates repression of Decr1 transcription through Decr1 promoter region-specific binding sites.55 Nevertheless, another report showed that the Decr1 was consistently upregulated in ARI-resistant cells when compared to WT LNCaP and AR silencing did not affect Decr1 protein levels in any of the LNCaP derivatives.7 Therefore, the effect of AR on mitochondrial lipid oxidation and Decr1 might be due to the distinctness of cancer cells and CFs.
The findings of our current study defined the mechanism underlying the role of enhanced mitochondrial lipid oxidation in CFs proliferation, migration, and fibrosis. This study demonstrates that WTAP boosts mitochondrial lipid oxidation, CFs proliferation, and fibroblast migration by increasing AR methylation in an m6A-YTHDF2-dependent manner. Overall, our research offers a better understanding of the roles of mitochondrial lipid oxidation and m6A regulation in diabetic cardiac fibrosis.
Limitations of the study
Although our study does much mechanistic research, the genetic roles of WTAP and AR remain to be determined. WTAP has been shown in the literature to promote cardiac injury through ischemia/reperfusion of cardiomyocytes.56 Higher expression of WTAP promoted the death of human cardiomyocytes (AC16), which was consistent with the disease outcome in our report. Both the death of cardiomyocytes and the activation and proliferation of CFs aggravated the process of cardiac fibrosis, and this might be due to the different functions of different species of cells in the heart. In addition, the molecular mechanisms of cardiac fibrosis development in WTAP−/− mice and YTHDF2−/− mice is beyond the scope of this first report. Constructing gene knockout mice for further verification will be one of the subjects of our future research.
Finally, although gene silencing with siRNA may have at least one allele functioning due to limitations in the gene silencing efficiency of siRNA, the fact of gene silencing and regulated changes in related downstream genes is visible, and it does not affect the reliability of our conclusions. Due to the advancement of related technologies, works on constructing CRISPR-CAS9-sgRNA-related plasmids will be considered in the future. In addition, all m6A sites related to AR cannot be verified, which is limited. Relatedly, we only validated the relationship between AR and canonical effector molecules, and the role of AR as a powerful transcription factor beyond the regulation of non-lipid metabolism could not be fully verified. In the development of diabetic cardiac fibrosis, AR-mediated other processes that regulate proliferation or death will become one of the topics of our future research.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CPT1a antibody | Proteintech | Cat#:15184-1-AP; RRID:AB_2084676 |
| anti-ATGL antibody | Proteintech | Cat#:55190-1-AP; RRID:AB_11182818 |
| anti-HSL antibody | Proteintech | Cat#:17333-1-AP; RRID:AB_2878386 |
| anti-FASN antibody | Proteintech | Cat#:10624-2-AP; RRID:AB_2100801 |
| anti-ACLY antibody | Proteintech | Cat#:15421-1-AP; RRID:AB_2223741 |
| anti-ACCα antibody | Proteintech | Cat#:21923-1-AP; RRID:AB_11042445 |
| anti-SREBP-1 antibody | Proteintech | Cat#:14088-1-AP; RRID:AB_2255217 |
| Anti-DECR1 rabbit polyclonal antibody | Sangon Biotech | Cat#:D161613; |
| anti-Postn antibody | Proteintech | Cat#:19899-1-AP; RRID:AB_10732682 |
| anti-Postn antibody | Proteintech | Cat# 66491-1-Ig; RRID:AB_2881856 |
| anti-cTnT antibody | Proteintech | Cat#:15513-1-AP; RRID:AB_2206563 |
| anti-PCNA antibody | Proteintech | Cat#:10205-2-AP; RRID:AB_2160330 |
| anti-AR antibody | Proteintech | Cat#:22089-1-AP; RRID:AB_11182176 |
| anti-AR antibody | Proteintech | Cat#:66747-1-Ig; RRID:AB_2882094 |
| anti-METTL3 antibody | Proteintech | Cat#:15073-1-AP; RRID:AB_2142033 |
| anti-METTL14 antibody | Proteintech | Cat#:26158-1-AP; RRID:AB_2800447 |
| anti-WTAP antibody | Proteintech | Cat#:10200-1-AP; RRID:AB_2216349 |
| anti-YTHDF1 antibody | Proteintech | Cat#:66745-1-Ig; RRID:AB_2882093 |
| anti-YTHDF2 antibody | Proteintech | Cat#:24744-1-AP; RRID:AB_2687435 |
| anti-SMARCD1(Baf60a) antibody | Proteintech | Cat#:10998-2-AP; RRID:AB_2192133 |
| anti-SMARCE1(Baf57) antibody | Proteintech | Cat#:10814-1-AP; RRID:AB_2192174 |
| anti-NCOR1 antibody | Proteintech | Cat#:20018-1-AP; RRID:AB_10665360 |
| anti-NCOR2(SMRT) antibody | Thermo Fisher | Cat#:PA1-843; RRID:AB_2149135 |
| anti-PPARα antibody | Proteintech | Cat#:66826-1-Ig; RRID:AB_2882169 |
| anti-PPARδ antibody | Proteintech | Cat#:60193-1-Ig; RRID:AB_10896827 |
| anti-PPARγ antibody | Proteintech | Cat#:16643-1-AP; RRID:AB_10596794 |
| anti-Collagen Type I antibody | Proteintech | Cat#:14695-1-AP; RRID:AB_2082037 |
| anti-Collagen Type I antibody | Proteintech | Cat#:67288-1-Ig; RRID:AB_2882554 |
| anti-TGF-β1 antibody | Proteintech | Cat#:21898-1-AP; RRID:AB_2811115 |
| anti-SMAD2 antibody | Proteintech | Cat#:12570-1-AP; RRID:AB_2193037 |
| anti-SMAD3 antibody | Proteintech | Cat#:66516-1-Ig; RRID:AB_2881879 |
| anti-SMAD7 antibody | Proteintech | Cat#:25840-1-AP; RRID:AB_2848137 |
| anti-α-SMA antibody | Proteintech | Cat#:14395-1-AP; RRID:AB_2223009 |
| anti-Mouse IgG | Proteintech | Cat#:B900620; RRID:AB_2883054 |
| anti-Rabbit IgG | Proteintech | Cat#:30000-0-AP; RRID:AB_2819035 |
| anti-β-actin antibody | Affinity | Cat#:AF7018; RRID:AB_2839420 |
| Goat anti-Mouse IgG (H + L) HRP | Bioworld | Cat#:BS12478; RRID:AB_2773727 |
| Goat anti-Rabbit IgG (H + L) HRP | Bioworld | Cat#:BS13278; RRID:AB_2773728 |
| Goat anti-Rabbit IgG/Alexa Fluor® 594 | ZSGB-BIO | Cat#:ZF-0516; RRID:AB_2936330 |
| Goat anti-Mouse IgG/Alexa Fluor® 594 | ZSGB-BIO | Cat#:ZF-0513; RRID:AB_2892725 |
| Goat Anti-Rabbit IgG (H + L) Fluor647-conjugated | Affinity | Cat#:S0013; RRID:AB_2844801 |
| Goat Anti-Mouse IgG (H + L) Fluor647-conjugated | Affinity | Cat#:S0014; RRID:AB_2844802 |
| Goat anti-Rabbit IgG/Alexa Fluor® 488 | ZSGB-BIO | Cat#:ZF-0511; RRID:AB_2864279 |
| Goat anti-Mouse IgG/Alexa Fluor® 488 | ZSGB-BIO | Cat#:ZF-0514; |
| Chemicals, peptides, and recombinant proteins | ||
| Oil Red O kit | Solarbio | Cat#:G1261 |
| Modified Masson Trichrome Staining Kit. | Solarbio | Cat#:G1346 |
| Modified hematoxylin and eosin (HE) staining kit. | Solarbio | Cat#:G1121 |
| Modified Sirius Red Staining Kit. | Solarbio | Cat#:G1472 |
| BODIPY 493/503 | Invitrogen | Cat#:D3922 |
| Oxygen Species Assay Kit | Beyotime | Cat#:S0033S |
| Lipid Oxidation (MDA) Detection Kit | Beyotime | Cat#:S0131S |
| Oxygen Consumption Assay Kit | Bestbio | Cat#:BB-48211 |
| DCFDA/H2DCFDA - Cellular ROS Assay Kit | Abcam | Cat#:ab113851 |
| BCA kit | Beyotime | Cat#:P0012 |
| T(Testosterone) ELISA Kit | Sangon Biotech | Cat#:D751045 |
| PrimeScript™ RT Master Mix | TaKaRa | Cat#:RR036B |
| SYBR Premix Ex Taq II | TaKaRa | Cat#:740703 |
| Co-IP kit | Absin | Cat#:abs955 |
| MeRIP kit | Bersinbio | Cat#:Bes5203-1 |
| SimpleChIP® Plus Enzymatic Chromatin IP Kit (Agarose Beads) | Cell Signaling Technology | Cat#:#9004S |
| Magnetic RNA-Protein Pull down Kit | Thermo Scientific | Cat#:20164 |
| Edu assay kit | Ribobio | Cat#:R11053.10 |
| Actinomycin D | Glpbio | Cat#:GC16866 |
| Streptozocin | Solarbio | Cat#:S8050 |
| TRIzol | Invitrogen | Cat#:15596018 |
| MitoTracker™ Red | Invitrogen | Cat#:M7512 |
| PEI-Transferrinfection Kit | Invitrogen | Cat#:BMS1003-A |
| Antifade Mounting Medium with DAPI | Beyotime | Cat#:P0131 |
| Deposited data | ||
| RNA high throughput sequencing data | This paper | GEO:GSE202418 |
| Single-cell sequencing data | This paper | GEO:GSE120064 |
| Experimental models: Cell lines | ||
| 3T3 | CCTCC | Cat# GDC0030, RRID:CVCL_0594 |
| H9C2 | CCTCC | Cat# GDC0606; RRID:CVCL_0286 |
| Software and algorithms | ||
| SPSS 23.0 | IBM SPSS | https://www.ibm.com/docs/zh/spss-statistics/ |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/ |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hui Tao (taohui@ahmu.edu.cn).
Materials availability
Reagents and methods in this study will be made available by the lead contact upon request.
Experimental model and study participant details
Animal experiments and models
All animal experimental protocols complied with the “Guidelines for the Use of Laboratory Animals” of the National Institutes of Health and were approved by the Second Affiliated Hospital of Anhui Medical University and the Ethics Committee of Anhui Medical University.
Leptin receptor-deficient (db/db, male, 6 weeks old) mice and lean control mice (db/+, male, 6 weeks old) were obtained from Hangzhou Ziyuan Laboratory Animal Technology Co., Ltd. (Zhejiang, China) and housed in a specific pathogen-free facility (4 per cage) with a 12-h light-dark cycle and access to food and water ad libitum. Weekly measurements of blood glucose and recording of food and water intake were conducted for all mice. At 16 weeks old, db/db mice in the empty virus vector group were injected with an empty virus vector through the tail vein. The LV3-WTAP and LV3-YTHDF2 groups of db/db mice were injected with lentiviral vectors containing shRNA-WTAP and shRNA-YTHDF2 through the tail vein, respectively. The DHT group was orally administered 20 mg/kg DHT daily for four weeks. C57BL/6 mice were injected with STZ at a concentration of 100 mg/kg or an equivalent volume of normal saline on an empty stomach at 8 weeks of age, and LV3-WTAP or YTHDF2 lentivirus was injected through the tail vein at 16 weeks of age. After four weeks (20 weeks old), all mice were weighed and blood glucose was measured. Echocardiographic measurements were performed after atropine injection and anesthesia with isoflurane. Subsequently, cervical dislocation was performed, and blood and heart samples were collected. Cardiac tissue sections were snap-frozen in liquid nitrogen (−80°C) or fixed in 10% formalin for histology. Blood samples were collected in EDTA tubes for plasma separation, which was stored at −80°C.
Isolation and culture of primary cardiac fibroblasts
Primary CFs were isolated from neonatal C57BL/6 mice hearts (1–3 days old) and cultured. Hearts were isolated and washed in pre-cooled PBS. After mincing, complete digestion was performed using trypsin (0.16%, Beyotime, Shanghai, China) and type II collagenase (0.67%, Biofrox) at 37°C for 30 min. Digestion was halted by adding DMEM medium containing 5% fetal bovine serum (FBS), followed by centrifugation at 1200 r/min. The cells were resuspended in DMEM medium with 5% FBS (5.5 mM glucose) and incubated in a 37°C, 5% CO2 incubator for 2 h. Based on the principle of differential adhesion of CFs to cardiomyocytes, adherent cells within 2 h were considered primary fibroblasts. The purity of primary CFs, as detected by vimentin and Periostin, was positive. Upon reaching 80% confluency, cells were passaged at a 1:1 ratio, and the second passage was used for genetic and functional experiments.
Cell culture and treatments
H9C2(rat cardiomyocytes) and 3T3(mouse embryonic fibroblasts.), which were provided by the Cell Bank of the Type Culture Collection, Chinese Academy of Sciences (Shanghai, China), were placed in DMEM (Hyclone) containing 10% FBS (Gibco, USA), 100 U/ml penicillin and 100 μg/mL streptomycin (Beyotime) in a humid environment with 5% CO2 at 37°C. In addition, primary CFs, 3T3, and cardiomyocytes were placed in low glucose medium (5.5 mmol/L glucose and 27.5 mmol/L mannitol) and high glucose + palmitic acid + oleic acid medium (33 mmol/L glucose, 200 μmol/L palmitate, and 200 μmol/L oleate) for 24 h to simulate type 2 diabetes in vitro. The DHT group was treated with 10nM DHT for 24 h at the same time as HG/HF treatment.
Human samples
Samples from 15 patients with DCM and 13 non-diabetic healthy control subjects were obtained from the Second Affiliated Hospital of Anhui Medical University. The study received approval from the Ethics Committee of Anhui Medical University and the Second Affiliated Hospital of Anhui Medical University. Inclusion criteria for DCM patients were: (a) diagnosed with diabetes, (b) no other causes of decreased left ventricular function, (c) no history of hypertension, myocardial infarction, or suspected coronary heart disease, (d) normal liver and kidney function, and blood lipids. The control group’s inclusion criteria were: (a) normal blood lipids, blood pressure, and blood glucose, (b) normal cardiac function and pulmonary arterial pressure, and (c) matching age and gender with the DCM group.
Cardiac tissue was obtained during heart valve replacement surgery or cardiac catheterization after obtaining written consent from the patients. The samples were stored at −80°C for subsequent experiments. The research procedures were performed by the Declaration of Helsinki and the Declaration of Istanbul.
Method details
Postn-promoter non-myocyte shRNA-WTAP and shRNA-YTHDF2 lentivirus construction
Non-myocyte lentivirus was generated with a periostin promoter (Postn_x0002_promoter) added before the shRNA-WTAP sequence and shRNA-YTHDF2 sequence to specifically target periostin+ cells.
Construction of mutants
Lentiviral constructs expressing mutant Decr1 (GTGTT to GTCTT in Decr1 DNA promoter region) and mutant AR (GGACU to GGTCU in m6A motif of AR mRNA CDS region) were obtained from Generalbiol (Anhui, China) and used to establish CFs. Cells were infected with lentivirus for 24 h and subsequently selected with puromycin after 72 h.
Echocardiography to assess left ventricular systolic and diastolic function
All grouped mice underwent transthoracic echocardiography at 20 weeks old to evaluate cardiac function. Mice were inhaled and anesthetized with 2 percent isoflurane and transthoracic echocardiography was performed while maintaining their heart rates at 400–500 beats per minute. (Vevo 2100 Imaging System; VisualSonics, Canada). Heart rate left ventricular end-systolic diameter (LVIDd), left ventricular end-diastolic diameter (LVIDd), percent shortening (FS), and EF were measured in M-mode at the mid-papillary level to assess systolic and diastolic function. Doppler signals of mitral inflow velocity and mitral annular velocity were obtained to calculate E/e‘ and E/A for assessment of diastolic function.
Oil red O staining
Cardiac tissues were OCT cryo-embedded sections using standard techniques, and cryosections were made for Oil Red O staining analysis. Oil Red O staining was performed according to the staining procedures in the Oil Red O kit (Solarbio).
Histopathology
Heart tissues were fixed in 10% formalin for 24 h and subsequently embedded and sectioned. After deparaffinization, serial sections were collected for hematoxylin-eosin (HE), Sirius red, and MT staining.
TEM
Samples were prepared for transmission electron microscope as reported previously[50] and viewed with a transmission electron microscope (TALOS L120C, ThermoScientific).
Immunohistochemistry
Briefly, heart tissue sections were subjected to a 30-min incubation in a 65°C incubator, followed by deparaffinization with xylene and rehydration using graded ethanol. Subsequently, the tissue was treated with 3% hydrogen peroxide for 15 min, and antigen retrieval was performed with 0.01M citrate buffer (pH = 6.0) for 30 min. After blocking with 5% BSA, the sections were incubated overnight at 4°C with the corresponding primary antibodies. Following the washing steps, different types of secondary antibodies were applied and incubated at room temperature for 1 h. Streptavidin-HRP conjugate and diaminobenzidine (DAB; Dako) were utilized for visualization. Nuclei were stained with hematoxylin, and the differential protein expression was examined using a binocular microscope.
Immunofluorescence
Cardiac tissue sections on glass slides were first permeabilized in 0.5% Triton X-100 after fixation in 4% paraformaldehyde. Blocking was done using 5% BSA. The heart tissue was then incubated overnight at 4°C with the primary antibody (1:200). After washing, the corresponding secondary antibody was incubated for 1 h at room temperature. BODIPY and mitotrack staining were performed as described. Nuclei were counterstained with DAPI. Samples were observed using a laser confocal microscope (DMI4000B, Leica).
BODIPY and MitoTracker red staining
Sections were prepared as described above for Immunofluorescence. After autoclaving fixation and blocking with 5% BSA, the sections were stained with 0.2 μg/mL BODIPY 493/503 (Invitrogen, California) for 30 min. Nuclei were counterstained with DAPI for 20 min, and images were obtained using a confocal microscope. MitoTracker Red (Invitrogen, California) staining followed the manufacturer’s instructions.
Plasmid, siRNA, and transfection
The plasmid and siRNA were transfected in serum-free DMEM using the PEI-Transferrinfection Kit (Thermofisher), following the manufacturer’s instructions. After 24 h of transfection, relevant experiments were conducted. The transfection efficiency was evaluated using RT-qPCR and Western blotting. See Table S3 for the related plasmid and siRNA sequences.
Western blot analysis
Cell lysates and cardiac tissue homogenates were prepared in RIPA lysate containing protease and phosphatase inhibitors (Sigma, USA). After centrifugation at 13,200 rpm for 30 min at 4°C, the supernatant containing total protein lysate was collected. The lysate was then incubated with SDS sample buffer at 100°C for 10 min. Protein separation was achieved through SDS-PAGE using a reducing 6–12% gradient gel with 20–30 μg of protein. The protein was transferred to a PVDF membrane, and the membrane was incubated with the primary antibody overnight at 4°C with gentle shaking. Subsequently, HRP-conjugated secondary antibodies (Bioworld, Nanjing, China) were incubated at 1:5000-1:10,000 dilutions for 1 h at room temperature. Antibody detection was performed using an ECL substrate.
Quantitative real-time reverse transcription (RT-qPCR)
Total RNA from tissues and cells was extracted using 1 mL TRIzol Reagent. After quantification at 260 nm, samples were used for qPCR with β-actin as an internal reference. The relative expression levels of each mRNA were calculated using the 2ˆ-ΔΔCt method. Primers can be found in Table S3.
Determination of oxygen consumption rate in isolated heart tissue and cells
Heart tissue was rapidly dissected, rinsed in PBS, and minced before being placed in a 96-well plate for in vitro OCR determination. The tissue OCR was measured using an oxygen consumption assay kit (Bestio, China) as per the manufacturer’s instructions. Fluorescence measurements were taken at 37°C with excitation at 460 nm and emission at 600 nm, using a microplate reader. Measurements were recorded every 3 min for a total of 10 times, and the stable plateau period was selected and calculated based on the OCR formula.
Similarly, cells (0.5∗105 cells/well) were pretreated and seeded in 96-well plates for cell OCR measurement. A cell-free group was included as a control. The OCR was measured and calculated in the same manner as described above.
MDA (Malondialdehyde) content detection
Tissue or cell homogenates were collected using RIPA and centrifuged at 13200 r/min at 4°C for 30 min. The supernatant was collected, and the protein concentration was determined using a BCA protein detection kit for MDA quantification. MDA content in tissues and cells was determined using a lipid oxidation (MDA) detection kit (Beyotime) following the manufacturer’s protocol. Each sample was measured at least twice, and the average value was recorded.
Cardiac tissue and cellular ROS assay
ROS levels in cardiac tissues and cells were assessed using a reactive oxygen species detection kit (Beyotime) following the manufacturer’s instructions. Briefly, fresh heart tissue was dissected and washed in pre-chilled PBS, followed by preparation of a single-cell suspension in DMEM (Free-FBS). The suspension was then incubated for 20 min in a 37°C, 5% CO2 incubator. After washing with DMEM (Free-FBS), the fluorescence intensity was measured using a fluorescent microplate reader (excitation wavelength 488 nm, emission wavelength 525 nm). ROS levels in cells were measured following the kit instructions.
5-Ethynyl-2′-deoxyuridine (EdU) staining
Cells were seeded in 24-well plates at 1 × 105 cells per well and incubated with EdU labeling reagent (diluted 1:1000) for 3 h. After fixing with 4% formaldehyde for 1 h at room temperature, cells were permeabilized with 0.5% Triton X-100 for 15 min, followed by staining with 1× Apollo reaction buffer (300 μL) to detect EdU. Subsequently, cells were incubated overnight with the corresponding concentration of primary antibody and then with fluorescent secondary antibodies. Nuclei were stained with Hoechst 33342, and fluorescence was observed and captured using a confocal microscope. EdU-positive cells were quantified using ImageJ software, and differences in Edu percentages were analyzed using t-tests.
Transwell migration assay
Cells were seeded in the upper chamber of a matrigel-free 24-well plate. The lower chamber was filled with 20% FBS-DMEM, while the upper chamber contained Free-DMEM. After 24 h of incubation, cells were fixed with paraformaldehyde and stained with 0.5% crystal violet. Migrated cells in the upper chamber were removed using a cotton swab. Five randomly selected fields of view (x200) were photographed with an inverted microscope. The experiment was conducted three times, and cell counts were analyzed using ImageJ software.
Cell scratches experiment (wound healing experiment)
Briefly, cells were seeded in 6-well plates and treated with 10 μg/mL mitomycin C (Sigma) 24 h post-transfection to inhibit cell division. Scratch wounds were created using plastic pipette tips (100μL). Images were captured at 0h, 24h, and 48h after scratching using a 20× objective lens (Leica). Each experiment was performed in triplicate.
CHIP assays and Chip-qPCR
ChIP assays were conducted using the Simple ChIP Plus Enzymatic Chromatin IP Kit (CST) following the protocol. Briefly, cells were treated with 1% formaldehyde for cross-linking. The chromatin was then sonicated to obtain fragmented genomic DNA. Non-precipitated chromatin served as a positive control (input). The remaining chromatin was incubated with relevant antibodies, and the DNA was purified for PCR using ChIP-specific primers (Table S3). The reaction products were analyzed by agarose gel electrophoresis (2%) and subjected to qPCR for relative quantification.
M6A dot blot assay
Total RNA isolation and extraction from cardiac tissue or cells followed the same procedure as mentioned above. Denatured total RNA (400 ng, 200 ng) was spotted onto Amersham Hybond-N+ membranes (Biosharp, China) and crosslinked by UV for 45 min. After blocking with 5% nonfat milk (Biorad) for 1 h, the membranes were incubated overnight at 4°C with the m6A antibody (1:5000, Proteintech, #202003). Subsequently, HRP-conjugated mouse anti-rabbit IgG (1:1000, Proteintech, China) was applied to the membranes for 2 h at room temperature. The immune complexes were visualized using Immobilon Western Chemiluminescent HRP Substrate (Millipore) and captured with an ECL imaging system (Bio-Rad). Relative m6A levels were quantified through grayscale intensity analysis using ImageJ. As an internal reference control, 0.02% methylene blue (Biotechnology, China) staining was performed for 2 h after UV cross-linking, and the corresponding images were recorded. The blue dot scan indicated the amount of input RNA.
Co-immunoprecipitation
Different groups of CFs were lysed using an IP/CO-IP kit (Absin, China). Follow the instructions of the IP/CO-IP kit. The extracted protein was subjected to Western blot experiment.
RNA stability assay
For the actinomycin D RNA stability assay, pretreated cells of different groups were seeded in 6-well plates at 1 × 10 6 cells/well. Actinomycin D (5 μL/mL) was added at the indicated time points so that the time of actinomycin D treatment was varied. The total RNA extraction method was as described above, and then RT-qPCR was performed to detect the stability of AR/Decr1.
MeRIP and MeRIP-qPCR
M6A modifications of individual genes were determined using a MeRIP-qPCR assay. Total RNA was extracted using Trizol reagent (Invitrogen, California), and mRNA was purified using the RiboMinus Eukaryote Kit v2 (A15020, Invitrogen) to remove ribosomal RNA. The RNA was subsequently fragmented into approximately 100-nucleotide fragments using RNA Fragmentation Reagent (AM8740, Invitrogen). Purified RNA was then incubated with a specific anti-m6A antibody for 1 h at 4°C, followed by overnight mixing with pre-washed Pierce Protein A/G Magnetic Beads in immunoprecipitation buffer at 4°C. The m6A antibody was digested with proteinase K digestion buffer, and the immunoprecipitated m6A RNA was reverse transcribed to cDNA using the PrimeScript RT kit (Takara Bio, JAPAN) following the manufacturer’s instructions. Primers for MeRIP-qPCR are listed in Table S3. Real-time PCR was performed after m6A-IP to quantify m6A methylation changes in the target gene. The MeRIP-qPCR results for each gene were then calculated using the ΔΔCt method.
RNA pull-down
RNA pull-down assays were performed using the Magnetic RNA-Protein Pull-down Kit (Thermo Scientific) following the manufacturer’s instructions. Briefly, desthiobiotinylated cytidine bisphosphate was attached to the 3′ ends of full-length sense and antisense RNA AR sequences. Subsequently, 50 pmol of labeled RNA was incubated with 50 μL of streptavidin magnetic beads in RNA capture buffer (20 mM Tris [pH 7.5], 1 M NaCl, 1 mM EDTA) for 20 min at room temperature. After equilibrating the RNA-bound beads with protein-RNA binding buffer (0.2 M Tris [pH 7.5], 0.5 M NaCl, 20 mM MgCl2, 1% Tween 20), cell lysates were added to the beads and incubated for 2 h at 4°C on a rotator. The beads were then washed twice and incubated with elution buffer, and the supernatants were subjected to Western blotting assays.
DNA pull-down
A DNA fragment containing a wild-type or mutant Decr1-binding site was PCR-amplified using a 5'-biotin-labeled forward primer (primer sequences in Appendix Table S3). Biotinylated probes (1.5 μg) were mixed with cell lysates in 400 μL of a binding buffer (25 mM HEPES, pH 7.9, 50 mM NaCl, 1 mM MgCl2, 1 mM DTT, 2 μg polydI-dC) for 2 h at 4°C. Subsequently, 20 μL of streptavidin-agarose beads (Invitrogen) were added and incubated for an additional 1 h. The streptavidin-agarose beads were washed five times with the binding buffer, and then the SDS-sample buffer was added. The complexes were subjected to SDS-PAGE, followed by immunoblotting with anti-AR and anti-β-actin.
The data source of bioinformatics analysis
RNA high throughput sequencing data GSE202418 and single-cell transcriptome files GSE120064 were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/).
Single-cell RNA sequencing (scRNA-seq) analysis
Single-cell sequencing data GSE12006457 was analyzed in R (Seurat (V4.1.1) package),58 and cell-filtering parameters are nFeature_RNA> 200, nFeature_RNA< 5000. Clusters were identified using the FindClusters and FindNeighbors functions in Seurat with resolution = 0.8 and 2D visualization by UMAP. The Nebulosa (v1.6.0) package59 performed weighted kernel density estimation of gene expression using the plot_density function with default parameters. Ar’s differential expression genes (DEGs) were identified using the FindMarkers function based on Ar> 1 compared with Ar≤ 1, logfc. threshold = 0.4, min.pct = 0. Downregulated DEGs filtered by logfc< −0.4, p value< 0.05.
Protein-protein interaction (PPI) networks
Mouse PPI networks were collected from geneMANIA60 (http://genemania.org/).
Enrichment analysis
Reactome or GO analysis using the website EnrichR61 (https://maayanlab.cloud/Enrichr/) or ClueGO62 v2.5.8+ CluePedia v1.5.8 plugin on Cytoscape v3.9.1 with corrected p < 0.05 were considered significantly enriched. Ar correlated genes in GSE202418 were using Pearson correlation, the |Pearson r| > 0.8 was considered to show a robust correlation.
Quantification and statistical analysis
We used GraphPad Prism 8 or SPSS 23.0 software for statistical analysis. We used Student’s t test and analysis of variance (ANOVA) to compare the different groups and paired t-test was used to compare pre-and post-treatment groups. p values less than 0.05 were considered statistically significant. All data are presented as mean ± SD.
Acknowledgments
This project was supported by the National Natural Science Foundation of China (82170236, 81700212), Key research and development projects of Anhui Province (202104j07020037), Excellent Youth Research Project in University of Anhui Province (2023AH030116), Translational medicine research project of Anhui Province (2021zhyx-C61), Excellent Top Talents Program of Anhui Province Universities (gxyqZD2022023), National Natural Science Foundation Incubation Program of the Second Affiliated Hospital of Anhui Medical University (2020GMFY02), and Postgraduate Innovation Research and Practice Program of Anhui Medical University (YJS20230083, YSJ20230082). The graphical abstract was created using biorender.com.
Author contributions
H.T., J.Y.Z., Y.Z., and J.J.Y. conceived the project and designed the experiments. K.S., H.S., B.T., and Y.Z. performed experiments and data analyses. L.C.L., Z.Y.L., R.L., and J.J.Y. assisted with the data analysis. H.T. and J.Y.Z. wrote the manuscript, with all authors contributing to providing feedback.
Declaration of interests
The authors declare no conflict of interest.
Published: September 15, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107931.
Contributor Information
Jing-Jing Yang, Email: yangjingjing@ahmu.edu.cn.
Ye Zhang, Email: zhangy@ahmu.edu.cn.
Jian-Yuan Zhao, Email: zhaojy@vip.163.com.
Hui Tao, Email: taohui@ahmu.edu.cn.
Supplemental information
Data and code availability
-
•
The raw sequencing data analyzed in this study are stored in the GEO database. These data can be obtained at GEO: GSE202418; GSE120064. Accession numbers are listed in the key resources table. All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Frangogiannis N.G. Cardiac fibrosis. Cardiovasc. Res. 2021;117:1450–1488. doi: 10.1093/cvr/cvaa324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ritchie R.H., Abel E.D. Basic Mechanisms of Diabetic Heart Disease. Circ. Res. 2020;126:1501–1525. doi: 10.1161/CIRCRESAHA.120.315913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuleta I., Frangogiannis N.G. Fibrosis of the diabetic heart: Clinical significance, molecular mechanisms, and therapeutic opportunities. Adv. Drug Deliv. Rev. 2021;176 doi: 10.1016/j.addr.2021.113904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishiga M., Horie T., Kuwabara Y., Nagao K., Baba O., Nakao T., Nishino T., Hakuno D., Nakashima Y., Nishi H., et al. MicroRNA-33 Controls Adaptive Fibrotic Response in the Remodeling Heart by Preserving Lipid Raft Cholesterol. Circ. Res. 2017;120:835–847. doi: 10.1161/CIRCRESAHA.116.309528. [DOI] [PubMed] [Google Scholar]
- 5.Barrera G., Pizzimenti S., Dianzani M.U. Lipid peroxidation: control of cell proliferation, cell differentiation and cell death. Mol. Aspect. Med. 2008;29:1–8. doi: 10.1016/j.mam.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Zhao R., Cao L., Gu W.J., Li L., Chen Z.Z., Xiang J., Zhou Z.Y., Xu B., Zang W.D., Zhou X.Y., et al. Gestational palmitic acid suppresses embryonic GATA-binding protein 4 signaling and causes congenital heart disease. Cell Rep. Med. 2023;4 doi: 10.1016/j.xcrm.2023.100953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blomme A., Ford C.A., Mui E., Patel R., Ntala C., Jamieson L.E., Planque M., McGregor G.H., Peixoto P., Hervouet E., et al. 2,4-dienoyl-CoA reductase regulates lipid homeostasis in treatment-resistant prostate cancer. Nat. Commun. 2020;11:2508. doi: 10.1038/s41467-020-16126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miguel V., Tituaña J., Herrero J.I., Herrero L., Serra D., Cuevas P., Barbas C., Puyol D.R., Márquez-Expósito L., Ruiz-Ortega M., et al. Renal tubule Cpt1a overexpression protects from kidney fibrosis by restoring mitochondrial homeostasis. J. Clin. Invest. 2021;131 doi: 10.1172/JCI140695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haemmerle G., Moustafa T., Woelkart G., Büttner S., Schmidt A., van de Weijer T., Hesselink M., Jaeger D., Kienesberger P.C., Zierler K., et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-α and PGC-1. Nat. Med. 2011;17:1076–1085. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zechner R., Zimmermann R., Eichmann T.O., Kohlwein S.D., Haemmerle G., Lass A., Madeo F. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metabol. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun N., Shen C., Zhang L., Wu X., Yu Y., Yang X., Yang C., Zhong C., Gao Z., Miao W., et al. Hepatic Krüppel-like factor 16 (KLF16) targets PPARα to improve steatohepatitis and insulin resistance. Gut. 2021;70:2183–2195. doi: 10.1136/gutjnl-2020-321774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heemers H.V., Tindall D.J. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr. Rev. 2007;28:778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- 13.Li D., Zhou W., Pang J., Tang Q., Zhong B., Shen C., Xiao L., Hou T. A magic drug target: Androgen receptor. Med. Res. Rev. 2019;39:1485–1514. doi: 10.1002/med.21558. [DOI] [PubMed] [Google Scholar]
- 14.de Bono J.S., Guo C., Gurel B., De Marzo A.M., Sfanos K.S., Mani R.S., Gil J., Drake C.G., Alimonti A. Prostate carcinogenesis: inflammatory storms. Nat. Rev. Cancer. 2020;20:455–469. doi: 10.1038/s41568-020-0267-9. [DOI] [PubMed] [Google Scholar]
- 15.Vom Steeg L.G., Dhakal S., Woldetsadik Y.A., Park H.S., Mulka K.R., Reilly E.C., Topham D.J., Klein S.L. Androgen receptor signaling in the lungs mitigates inflammation and improves the outcome of influenza in mice. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerratana L., Basile D., Buono G., De Placido S., Giuliano M., Minichillo S., Coinu A., Martorana F., De Santo I., Del Mastro L., et al. Androgen receptor in triple negative breast cancer: A potential target for the targetless subtype. Cancer Treat Rev. 2018;68:102–110. doi: 10.1016/j.ctrv.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Fernandes R.C., Leach D.A., Bevan C.L. Epigenetic Coregulation of Androgen Receptor Signaling. Adv. Exp. Med. Biol. 2022;1390:277–293. doi: 10.1007/978-3-031-11836-4_16. [DOI] [PubMed] [Google Scholar]
- 18.Lu X., Fong K.W., Gritsina G., Wang F., Baca S.C., Brea L.T., Berchuck J.E., Spisak S., Ross J., Morrissey C., et al. HOXB13 suppresses de novo lipogenesis through HDAC3-mediated epigenetic reprogramming in prostate cancer. Nat. Genet. 2022;54:670–683. doi: 10.1038/s41588-022-01045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh N., Ramnarine V.R., Song J.H., Pandey R., Padi S.K.R., Nouri M., Olive V., Kobelev M., Okumura K., McCarthy D., et al. The long noncoding RNA H19 regulates tumor plasticity in neuroendocrine prostate cancer. Nat. Commun. 2021;12:7349. doi: 10.1038/s41467-021-26901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C., Jin J., Yang Y., Sun H., Wu L., Shen M., Hong X., Li W., Lu L., Cao D., et al. Androgen receptor-mediated CD8(+) T cell stemness programs drive sex differences in antitumor immunity. Immunity. 2022;55:1747. doi: 10.1016/j.immuni.2022.07.016. [DOI] [PubMed] [Google Scholar]
- 21.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y., Dominissini D., Rechavi G., He C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 23.Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., Cheng T., Gao M., Shu X., Ma H., et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer K.D., Jaffrey S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaccara S., Jaffrey S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell. 2020;181:1582–1595.e18. doi: 10.1016/j.cell.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., Adhikari S., Shi Y., Lv Y., Chen Y.S., et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue F., Cheng J., Liu Y., Cheng C., Zhang M., Sui W., Chen W., Hao P., Zhang Y., Zhang C. Cardiomyocyte-specific knockout of ADAM17 ameliorates left ventricular remodeling and function in diabetic cardiomyopathy of mice. Signal Transduct. Targeted Ther. 2022;7:259. doi: 10.1038/s41392-022-01054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M., Sui W., Xing Y., Cheng J., Cheng C., Xue F., Zhang J., Wang X., Zhang C., Hao P., Zhang Y. Angiotensin IV attenuates diabetic cardiomyopathy via suppressing FoxO1-induced excessive autophagy, apoptosis and fibrosis. Theranostics. 2021;11:8624–8639. doi: 10.7150/thno.48561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y., Cao Y., Zheng R., Xiong Z., Zhu Z., Gao F., Man W., Duan Y., Lin J., Zhang X., et al. Fibroblast-specific activation of Rnd3 protects against cardiac remodeling in diabetic cardiomyopathy via suppression of Notch and TGF-β signaling. Theranostics. 2022;12:7250–7266. doi: 10.7150/thno.77043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Y., Hu C., Song Q., Li Y., Da X., Yu Y., Li H., Clark I.M., Chen Q., Wang Q.K. ADAMTS16 activates latent TGF-β, accentuating fibrosis and dysfunction of the pressure-overloaded heart. Cardiovasc. Res. 2020;116:956–969. doi: 10.1093/cvr/cvz187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castelino F.V., Bain G., Pace V.A., Black K.E., George L., Probst C.K., Goulet L., Lafyatis R., Tager A.M. An Autotaxin/Lysophosphatidic Acid/Interleukin-6 Amplification Loop Drives Scleroderma Fibrosis. Arthritis Rheumatol. 2016;68:2964–2974. doi: 10.1002/art.39797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi P., Zhao X.D., Shi K.H., Ding X.S., Tao H. MiR-21-3p triggers cardiac fibroblasts pyroptosis in diabetic cardiac fibrosis via inhibiting androgen receptor. Exp. Cell Res. 2021;399 doi: 10.1016/j.yexcr.2020.112464. [DOI] [PubMed] [Google Scholar]
- 33.Tao H., Shi P., Xuan H.Y., Ding X.S. DNA methyltransferase-1 inactivation of androgen receptor axis triggers homocysteine induced cardiac fibroblast autophagy in diabetic cardiac fibrosis. Arch. Biochem. Biophys. 2020;692 doi: 10.1016/j.abb.2020.108521. [DOI] [PubMed] [Google Scholar]
- 34.Lamhamedi-Cherradi S.E., Maitituoheti M., Menegaz B.A., Krishnan S., Vetter A.M., Camacho P., Wu C.C., Beird H.C., Porter R.W., Ingram D.R., et al. The androgen receptor is a therapeutic target in desmoplastic small round cell sarcoma. Nat. Commun. 2022;13:3057. doi: 10.1038/s41467-022-30710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., Horn J.L., Banda K., Goodman A.Z., Lim Y., Jana S., Arora S., Germanos A.A., Wen L., Hardin W.R., et al. The androgen receptor regulates a druggable translational regulon in advanced prostate cancer. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aaw4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heath E.I., Hillman D.W., Vaishampayan U., Sheng S., Sarkar F., Harper F., Gaskins M., Pitot H.C., Tan W., Ivy S.P., et al. A phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with hormone-refractory metastatic prostate cancer. Clin. Cancer Res. 2008;14:7940–7946. doi: 10.1158/1078-0432.CCR-08-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y., Peng C., Chen J., Chen D., Yang B., He B., Hu W., Zhang Y., Liu H., Dai L., et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer. 2019;18:127. doi: 10.1186/s12943-019-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho S., Lee G., Pickering B.F., Jang C., Park J.H., He L., Mathur L., Kim S.S., Jung S., Tang H.W., et al. mTORC1 promotes cell growth via m(6)A-dependent mRNA degradation. Mol. Cell. 2021;81:2064–2075.e8. doi: 10.1016/j.molcel.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 40.Mansouri A., Gattolliat C.H., Asselah T. Mitochondrial Dysfunction and Signaling in Chronic Liver Diseases. Gastroenterology. 2018;155:629–647. doi: 10.1053/j.gastro.2018.06.083. [DOI] [PubMed] [Google Scholar]
- 41.Angelova P.R., Esteras N., Abramov A.Y. Mitochondria and lipid peroxidation in the mechanism of neurodegeneration: Finding ways for prevention. Med. Res. Rev. 2021;41:770–784. doi: 10.1002/med.21712. [DOI] [PubMed] [Google Scholar]
- 42.Su P., Wang Q., Bi E., Ma X., Liu L., Yang M., Qian J., Yi Q. Enhanced Lipid Accumulation and Metabolism Are Required for the Differentiation and Activation of Tumor-Associated Macrophages. Cancer Res. 2020;80:1438–1450. doi: 10.1158/0008-5472.CAN-19-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benador I.Y., Veliova M., Liesa M., Shirihai O.S. Mitochondria Bound to Lipid Droplets: Where Mitochondrial Dynamics Regulate Lipid Storage and Utilization. Cell Metabol. 2019;29:827–835. doi: 10.1016/j.cmet.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menendez-Montes I., Abdisalaam S., Xiao F., Lam N.T., Mukherjee S., Szweda L.I., Asaithamby A., Sadek H.A. Mitochondrial fatty acid utilization increases chromatin oxidative stress in cardiomyocytes. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2101674118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vazquez E.J., Berthiaume J.M., Kamath V., Achike O., Buchanan E., Montano M.M., Chandler M.P., Miyagi M., Rosca M.G. Mitochondrial complex I defect and increased fatty acid oxidation enhance protein lysine acetylation in the diabetic heart. Cardiovasc. Res. 2015;107:453–465. doi: 10.1093/cvr/cvv183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang B., Jiang H., Dong Z., Sun A., Ge J. The critical roles of m6A modification in metabolic abnormality and cardiovascular diseases. Genes Dis. 2021;8:746–758. doi: 10.1016/j.gendis.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z., Yu G.L., Zhu X., Peng T.H., Lv Y.C. Critical roles of FTO-mediated mRNA m6A demethylation in regulating adipogenesis and lipid metabolism: Implications in lipid metabolic disorders. Genes Dis. 2022;9:51–61. doi: 10.1016/j.gendis.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathiyalagan P., Adamiak M., Mayourian J., Sassi Y., Liang Y., Agarwal N., Jha D., Zhang S., Kohlbrenner E., Chepurko E., et al. FTO-Dependent N(6)-Methyladenosine Regulates Cardiac Function During Remodeling and Repair. Circulation. 2019;139:518–532. doi: 10.1161/CIRCULATIONAHA.118.033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng T., Liu M., Hu L., Guo D., Wang D., Qi B., Ren G., Hu C., Zhang F., Chun H.J., et al. LncRNA Airn alleviates diabetic cardiac fibrosis by inhibiting activation of cardiac fibroblasts via a m6A-IMP2-p53 axis. Biol. Direct. 2022;17:32. doi: 10.1186/s13062-022-00346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y., Song K., Tu B., Sun H., Ding J.F., Luo Y., Sha J.M., Li R., Zhang Y., Zhao J.Y., Tao H. METTL3 boosts glycolysis and cardiac fibroblast proliferation by increasing AR methylation. Int. J. Biol. Macromol. 2022;223:899–915. doi: 10.1016/j.ijbiomac.2022.11.042. [DOI] [PubMed] [Google Scholar]
- 51.Jia Z., Wang K., Zhang Y., Duan Y., Xiao K., Liu S., Ding X. Icariin Ameliorates Diabetic Renal Tubulointerstitial Fibrosis by Restoring Autophagy via Regulation of the miR-192-5p/GLP-1R Pathway. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.720387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davies A., Nouruzi S., Ganguli D., Namekawa T., Thaper D., Linder S., Karaoğlanoğlu F., Omur M.E., Kim S., Kobelev M., et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat. Cell Biol. 2021;23:1023–1034. doi: 10.1038/s41556-021-00743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei W., Sun J., Zhang H., Xiao X., Huang C., Wang L., Zhong H., Jiang Y., Zhang X., Jiang G. Circ0008399 Interaction with WTAP Promotes Assembly and Activity of the m(6)A Methyltransferase Complex and Promotes Cisplatin Resistance in Bladder Cancer. Cancer Res. 2021;81:6142–6156. doi: 10.1158/0008-5472.CAN-21-1518. [DOI] [PubMed] [Google Scholar]
- 54.Moustafa T., Fickert P., Magnes C., Guelly C., Thueringer A., Frank S., Kratky D., Sattler W., Reicher H., Sinner F., et al. Alterations in lipid metabolism mediate inflammation, fibrosis, and proliferation in a mouse model of chronic cholestatic liver injury. Gastroenterology. 2012;142:140–151.e12. doi: 10.1053/j.gastro.2011.09.051. [DOI] [PubMed] [Google Scholar]
- 55.Nassar Z.D., Mah C.Y., Dehairs J., Burvenich I.J., Irani S., Centenera M.M., Helm M., Shrestha R.K., Moldovan M., Don A.S., et al. Human DECR1 is an androgen-repressed survival factor that regulates PUFA oxidation to protect prostate tumor cells from ferroptosis. Elife. 2020;9 doi: 10.7554/eLife.54166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J., Zhang J., Ma Y., Zeng Y., Lu C., Yang F., Jiang N., Zhang X., Wang Y., Xu Y., et al. WTAP promotes myocardial ischemia/reperfusion injury by increasing endoplasmic reticulum stress via regulating m6A modification of ATF4 mRNA. Aging. 2021;13:11135–11149. doi: 10.18632/aging.202770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ren Z., Yu P., Li D., Li Z., Liao Y., Wang Y., Zhou B., Wang L. Single-Cell Reconstruction of Progression Trajectory Reveals Intervention Principles in Pathological Cardiac Hypertrophy. Circulation. 2020;141:1704–1719. doi: 10.1161/CIRCULATIONAHA.119.043053. [DOI] [PubMed] [Google Scholar]
- 58.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alquicira-Hernandez J., Powell J.E. Nebulosa recovers single cell gene expression signals by kernel density estimation. Bioinformatics. 2021;37:2485–2487. doi: 10.1093/bioinformatics/btab003. [DOI] [PubMed] [Google Scholar]
- 60.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'Ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw sequencing data analyzed in this study are stored in the GEO database. These data can be obtained at GEO: GSE202418; GSE120064. Accession numbers are listed in the key resources table. All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.