Abstract
Metastasis is the leading cause of cancer-related deaths. During this process, cancer cells are likely to navigate discrete tissue-tissue interfaces, enabling them to infiltrate and spread throughout the body. Three-dimensional (3D) spheroid modeling is receiving more attention due to its strengths in studying the invasive behavior of metastatic cancer cells. While microscopy is a conventional approach for investigating 3D invasion, post-invasion image analysis, which is a time-consuming process, remains a significant challenge for researchers. In this study, we presented an image processing pipeline that utilized a deep learning (DL) solution, with an encoder-decoder architecture, to assess and characterize the invasion dynamics of tumor spheroids. The developed models, equipped with feature extraction and measurement capabilities, could be successfully utilized for the automated segmentation of the invasive protrusions as well as the core region of spheroids situated within interfacial microenvironments with distinct mechanochemical factors. Our findings suggest that a combination of the spheroid culture and DL-based image analysis enable identification of time-lapse migratory patterns for tumor spheroids above matrix-substrate interfaces, thus paving the foundation for delineating the mechanism of local invasion during cancer metastasis.
Keywords: Deep learning, Spheroid, Interfacial invasion, DIC images, Image processing, Diepafitaxis
Graphical abstract

1. Introduction
Metastasis, a major cause of mortality in patients with advanced cancer, refers to the dissemination of cancer cells from a primary tumor to distant tissues and organs, where development of secondary tumors takes place [1]. Indeed, metastatic progression is a multistep process that includes a series of distinct and stepwise events: (a) local invasion (i.e., escape of cancer cells from the primary lesion and infiltration into the surrounding stroma), (b) intravasation (i.e., ingress of malignant cells into the circulatory system), (c) surviving in the bloodstream, (d) extravasation (i.e., egress of circulating malignant cells from the bloodstream into adjacent normal tissues), and (e) proliferation, colonization and establishment of overt metastases [2]. In the course of metastatic spread, though being able to navigate either as single cells or via collective means through stromal and vascular microenvironments [3], collective invasion has been suggested to be a clinically relevant mode of invasive outgrowth for the majority of solid tumors [4]. Studies have revealed that clusters of cancer cells, compared to their single-cell counterparts, are not merely more aggressive and metastatic, but also display enhanced chemoresistance [5]. In this sense, the modes of cancer cell invasion are thus related to patient prognosis and therapeutic efficacy [6]. In addition to intrinsic (e.g., genetic) factors, tumor microenvironments also play a significant role in driving the switch between different invasion modes exhibited by the cancer cells [7]. It is consequently of great interest to explore the influence of physical and chemical microenvironmental attributes on their invasive and metastatic behaviors.
The application of 3D multicellular tumor spheroids has recently made significant advancements in the fields of oncogenesis and tissue engineering, notably enhancing our understanding and progress in these areas [8]. The spheroid cultures effectively resemble in vivo avascular tissue-like cellular characteristics in morphology, signal transmission, cell-cell communication, metastasis, and mechanical stimulation when compared to monolayer cultures [9]. Although several pieces of software have been created to analyze the invasive and migratory behaviors of spheroids using microscopic data, these tools can only extract particular cell morphologies at two- or three-time points during the time-lapse observation process [[10], [11], [12], [13], [14], [15], [16], [17]]. On the other hand, researchers used to visualize the details of spheroid structures and track the status of their migration and invasion under confocal fluorescence microscopy [[18], [19], [20], [21]]. However, this technique may adversely cause tissue heating, photobleaching, and phototoxic effects [[22], [23], [24], [25]]. With both fluorescence-free and non-destructive detection advantages, differential interference contrast (DIC) microscopy instead is a much more commonly used approach to observe live cells and follow their progression dynamics in a temporal manner [[26], [27], [28]]. Besides, it enables adequately high imaging resolution that can discriminate spatial organization of spheroids and identify the contrasting aspects of their invasive and migratory patterns [29,30]. However, analyzing spheroid DIC images is a time-consuming process, and it remains challenging to automatically extract and analyze the model objectively. The supervised machine learning frameworks have contributed to major breakthroughs in achieving more precise segmentation accuracy in recent decades, and, in particular, deep convolutional neural networks (CNNs) deliver the cutting-edge solutions that enhance performance of semantic image segmentation [31]. Several deep learning (DL)-based variants (e.g., spatial pyramid pooling and encoder-decoder structure) that can exploit contextual information have been proposed for executing image segmentation [32]. The encoder-decoder networks have been shown to lend themselves to efficient computation because they consist of an encoder module that gradually decreases the feature maps and a decoder module that gradually recovers spatial information [33]. In this sense, they can capture greater semantic information and restore sharp object borders, thus showing potential applicability for analyzing spheroid behavior on DIC brightfield images.
Since the process of early tumor metastasis involves multiple phases, the ability to properly simulate tumor-stromal interactions in vivo is critical for developing superior in vitro models with pathophysiological implications [34,35]. In terms of adhesion molecule and extracellular matrix (ECM) protein profiles, a spheroid culture system itself can create an extracellular environments that are closely comparable to tumor microenvironments in vivo [36,37]. Furthermore, embedding spheroids in biomimetic ECM scaffolds, with proper levels of deposition, cross-linking, and remodeling, also paves the way to a more realistic tumor progression model [38,39]. It has been noted that tumor-induced collagen fibril re-alignment can promote early cancer cell metastasis [40]. More recently, a clinical research has shown that metastatic tumor cells preferentially navigate through the pre-existing bone cavities or gaps between adipocytes [41], which also implicates that the structural organization of the ECM may facilitate cell dispersion. As a matter of fact, Roach et al. also postulated that the discrete space between heterogeneous structures can serve as an “interfacial” track, by which cancer cells prefer migrating on a ventral two-dimensional surface [42]. Nevertheless, there still exists a controversy regarding the mechanism of how the overall interfacial stiffness and adhesive strength between interfacial structures affect cell translocation processes [43,44]. Therefore, it emphasizes the importance of establishing a reliable analytic strategy, with the existence of a robust in vitro model that replicates these parametric aspects, to characterize such unsettled issue. In this study, we proposed a DL pipeline that begins with processing of DIC images extracted from an open-source database (URL/DOI: https://doi.org/10.17632/4v625rp3cj.1), followed by using an encoder-decoder architecture to segment the core region and invasive protrusions of the spheroids as well as the detached single cells automatically, and eventually produces the parametric results. This DL-based platform could profoundly reduce the time taken to segment and track spheroids and dissociated cells on DIC images. Besides, the analytical data indicated that a matrix-substrate interfacial boundary, with higher stiffness and lower adhesive strength, led to enhancement of spheroid invasiveness and single-cell migration.
2. Materials and methods
2.1. Data acquisition
Our previous work have employed DIC microscopy to characterize the invasive behaviors of MDA-MB-231 tumor spheroids positioned above interfaces with varying stiffness and topographic properties (i.e., interfacial adhesive strengths) [45]. In short, the spheroids generated from 4-day cultures in microwells [46] were embedded within the rat tail type I collagen scaffolds over glass and polydimethylsiloxane (PDMS) substrates, with Young's moduli of 69.3 GPa and 2.85 ± 0.001 MPa, respectively. The structural design of this culture device aimed to replicate heterogeneous interfaces characterized by different stiffness levels. Besides, assorted treatments of the PDMS substrate surface by two coating agents, pluronic 127 and glutaraldehyde (GA), together with the untreated counterpart, further resulted in a range of adhesive strengths at the matrix-substrate interface. Eventually, time-lapse DIC microscopy was conducted to record the invasive outgrowth of tumor spheroids under the above-mentioned interfacial conditions. In this study, the imaging data obtained from the interfacial spheroid invasion assay were utilized for training of our DL-based automated image segmentation model and its further validation. These data were extracted from the following open-source portal (https://doi.org/10.17632/4v625rp3cj.2). The detailed experimental procedures for the results presented in Supplementary Fig. 2, specifically the 3D tumor spheroid invasion assay and melanoma spheroid interfacial invasion assay, are provided in the supplementary document.
2.2. Data preparation
The DIC images were pre-processed before being passed as an input to the DL model. The pixel size of these images consisted of three channels: red, green, and blue. However, the information contained in these channels was identical, so merely one of them was selected for analysis. The intensity of each pixel in the DIC image was normalized with the mean and standard deviation of the raw pixel intensities (Equation (1)). Normalization was necessary to ensure that the image pixels had a consistent data distribution.
| Equation 1 |
where I refers to the raw intensity of a certain pixel in the original DIC image, μI and σI respectively refer to the mean and standard deviation of the raw pixel intensities in this image, and In refers to the normalized intensity value of the above-specified pixel.
Annotations were required for all DIC images in order to create the ground truth masks. To accommodate the DL model, which only accepts binary masks, the ground truth masks were transferred into a binarized format of 0 and 1. Data augmentation was applied to the training datasets to enhance the data variability, allowing the model to learn more effectively and become more robust. Two genres of augmentation functions were used geometric and generic augmentation. Geometric augmentation encompassed operations such as flipping, rotation, random grid shuffle and elastic transformation. On the other hand, generic augmentation involved random resizing and cropping, random brightness contrast, sharpening and blurring [47]. Geometric augmentation functions were employed to boost the morphological variability of the spheroid, enabling the model to learn robust features related to the irregular invasion characteristics. Meanwhile, generic augmentation functions were utilized to enhance the texture variability of the spheroid. This augmentation aimed to improve the model's ability to distinguish between the spheroid and the background by capturing a wider range of texture patterns. The same augmentation function was applied for each pair of input DIC images and ground truth masks, ensuring that the input DIC image correctly aligned with its corresponding output ground truth mask.
2.3. Deep learning model architecture design
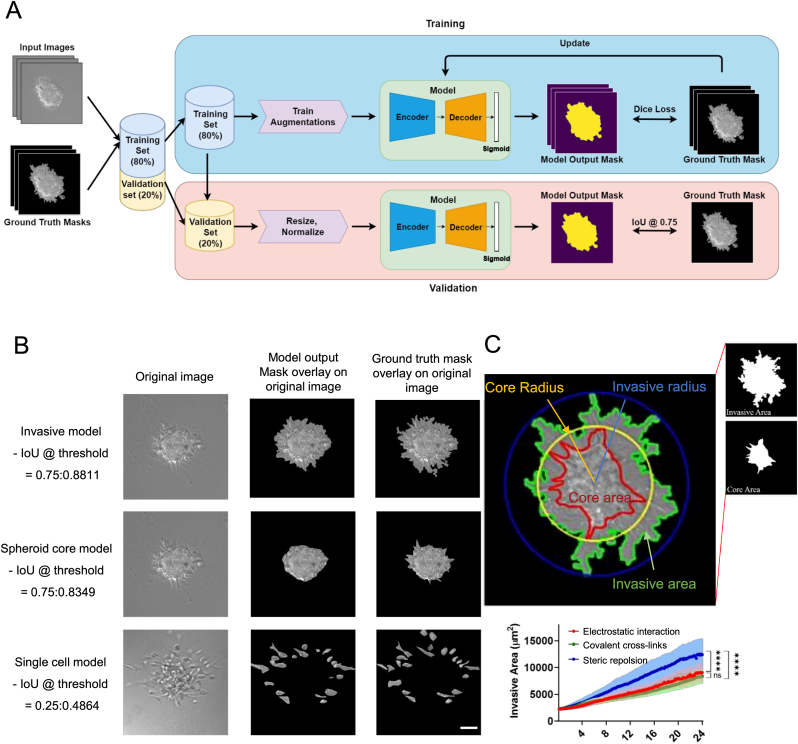
An overview of the framework was presented in Fig. 1A. For the invasive area model, EfficientNet, a convolutional neural network (CNN), was implemented in the encoder for computer vision tasks [48]. The encoder transformed the image information into a latent space and then transmitted the embeddings to the decoder. Unet++ with a squeeze-and-excitation layer was employed as the decoder model [49]. The PReLU function was employed in the encoder-decoder model as its activation function. Besides, pretrained weights from ImageNet were used for the encoder to promote the model's robustness and facilitate the training process [50]. Since the use of EfficientNet B7 and Unet++ yielded the highest IoU values for the invasive area model (Table 1), the same architecture was used for the spheroid core model.
Fig. 1.
The segmentation results of the model output for the invasive protrusions and core region of the spheroids as well as the detached single cells. (A) The comprehensive DIC image segmentation pipeline for assessing spheroid's invasiveness (B, C) The post-processing processes for quantifying variables to analyze the metastatic pattern of tumor spheroids. The scale bar represents 50 μm.
Table 1.
The mean Intersection over Union (IoU) values for the invasive area, spheroid core, and single-cell models. These values represent the mean and standard deviation across the 5-fold cross-validation results. The model with the highest IoU is indicated in bold.
| Encoder | Decoder | Intersection over Union (IoU) |
||||
|---|---|---|---|---|---|---|
| Threshold = 0.25 | Threshold = 0.35 | Threshold = 0.50 | Threshold = 0.75 | Threshold = 0.95 | ||
| Invasive model | ||||||
| EfficientNet B4 | Unet++ | 0.7577 ± 0.0181 | 0.7574 ± 0.0180 | 0.7570 ± 0.0179 | 0.7565 ± 0.0178 | 0.7555 ± 0.0177 |
| EfficientNet B6 | Unet++ | 0.7657 ± 0.0098 | 0.7659 ± 0.0095 | 0.7658 ± 0.0092 | 0.7657 ± 0.009 | 0.7651 ± 0.0087 |
| EfficientNet B7 | Unet++ | 0.7600 ± 0.0118 | 0.7599 ± 0.0118 | 0.7597 ± 0.0119 | 0.7594 ± 0.012 | 0.7588 ± 0.0123 |
| Unet++ Decoder Channel: (256, 128, 128, 64, 64) |
0.7745 ± 0.0184 | 0.7744 ± 0.0186 | 0.7742 ± 0.0188 | 0.7740 ± 0.0189 | 0.7735 ± 0.019 | |
| Spheroid core model | ||||||
| EfficientNet B7 | Unet++ Decoder Channel: (256, 128, 128, 64, 64) |
0.7497 ± 0.0002 | 0.7497 ± 0.0002 | 0.7497 ± 0.0002 | 07496 ± 0.0002 | 0.7495 ± 0.0002 |
| Single-cell model | ||||||
| EfficientNet B7 | Unet++ Decoder Channel: (256, 128, 128, 64, 64) |
0.4089 ± 0.1607 | 0.4113 ± 0.1589 | 0.3894 ± 0.1687 | 0.4065 ± 0.1698 | 0.4043 ± 0.1689 |
| RegNetY320 | Feature Pyramid Network (FPN) | 0.4779 ± 0.0556 | 0.4677 ± 0.0558 | 0.4705 ± 0.0673 | 0.4652 ± 0.0786 | 0.4622 ± 0.0725 |
**All the Unet++ decoders are configured with decoder channels of (256, 128, 64, 32, 16) unless specified otherwise.
For the single-cell DL model, the RegNetY320 and Feature Pyramid Network (FPN) were used in the encoder and decoder, respectively. RegNetY320 was equipped with streamlined networks that were both simple and fast, making them highly efficient across a wide range of computational capabilities. This design choice allowed for powerful feature extraction in various flop regimes [51]. The FPN was introduced into the single-cell model to address the inherent detection disparities between the single-cell model and the spheroid core and invasive area models. By capturing objects at various scales of feature maps, the FPN improved the detection performance, particularly in scenarios involving small objects [52].
2.4. Training the deep learning model
All models were trained for 20 epochs, with a batch size of 2, and a learning rate of 2e-4. To improve training efficiency, gradient accumulation was implemented every 2 steps, resulting in backpropagation updates occurring merely once every 2 steps. This approach involved accumulating gradients over 2 steps before updating the model's weights. A weight decay of 1e-3, along with a cosine decay schedule and an AdamW optimizer, was used during the training process. Dice Loss ( ) were utilized as the loss function, as described in Equation (2).
| Equation 2 |
where pi = predicted mask, qi = ground truth mask, N = number of samples. This loss function played a crucial role in updating the model's weights. The goal of training was to minimize this loss function. Therefore, a low Dice loss indicated successful performance, as it indicated minimal discrepancy between the predicted mask and the ground truth mask.
2.5. Evaluating the deep learning model
The performance of the trained DL models was evaluated using five-fold cross-validation. Intersection over Union (IoU) in Equation (3) was used as the evaluation metric, with a tunable threshold value. The threshold value represented the confidence score of the DL models for each pixel. When the pixel value exceeded or equaled the threshold, it was set to 1, and vice versa. Five different threshold values were investigated (refer to Table 1) to evaluate the DL model's performance.
| Equation 3 |
where IoU is the intersection over union for the masks, y is the ground truth mask and is the predicted mask.
2.6. Post-processing
The predicted masks underwent a series of post-processing functions for further analysis. Various measurements, including radius, area, perimeter, and complexity were computed for both the invasive and core regions. Additionally, for single-cell analysis, the number of detached cells, aspect ratio, and distance from the spheroid core centroid to individual cells were calculated. OpenCV functions were employed to compute these variables.
The formula for calculating both core complexity and invasive complexity was:
| Equation 4 |
The formula for calculating the single-cell aspect ratio was:
| Equation 5 |
The calculated values, except for complexity, were then scaled to a micrometers using a predetermined scaling factor. The scaling factor was computed prior to image resizing:
| Equation 6 |
where IR represents the size of resized image, and IO represents the size of the original image.
2.7. Statistical analysis
Statistical significance was evaluated using a two-way ANOVA, followed by Tukey-Kramer t-test for multiple comparisons. The results were presented as means ± standard deviations. The significance level was set at P < 0.05.
3. Results
3.1. Creation and validation of DL-based models to segment the spheroid core, invasive area, and detached single cells
Table 1 represents the results of 5-fold cross-validation for IoU at different thresholds, focusing on the invasive area, spheroid core, and single cell models. The number of the encoder parameters varied across the models:19 million for EfficientNet-B4, 43 million for EfficientNet-B6, and 66 million for EfficientNet-B7. The evidence indicates that the outcomes remained largely unaffected by the different threshold values and the number of parameters, suggesting that these factors did not have a significant impact on the results.
The highest IoU for the invasive area model was EfficientNet-B7 and Unet++ with decoder channels of (256, 128, 128, 64, 64), resulting in an average IoU of 0.77. Similarly, for the spheroid core model, a comparable performance was attained with an average IoU of 0.75 (Table 1). For single-cell segmentation, the model employing RegNetY320 as the encoder and FPN as the decoder achieved an IoU of 0.48, which showed a 17.5% improvement compared to the EfficientNet-B7 and Unet++ model. Fig. 1B showcases examples of the segmented outputs from all three models. Besides, our trained models exhibited a robust parameter quantification across different environmental conditions (e.g., brightness, noises) and variations in z-stack images, as illustrated in Supplementary Fig. 1. Moreover, we conducted an evaluation of our trained model by employing breast cancer spheroids to assess their 3D invasion within collagen gel and interfacial invasion in melanoma cells at the interface between collagen matrix and glass. As depicted in Supplementary Fig. 2, despite variations in environmental conditions and spheroid size, the central region of the spheroids and the areas displaying invasive behavior consistently demonstrated remarkable performance across the majority of the experimental conditions. Our model displayed exceptional precision in accurately extracting both the spheroid core and the invasive regions. Furthermore, it successfully captured the behavior of individual cells involved in both 3D and interfacial invasions of melanoma spheroids. These results suggest the robustness of our pipeline in correctly segmenting regions of interest, making it valuable for cell morphology studies across various spheroid conditions.
3.2. Analysis of the invasive behavior exhibited by spheroids at interfaces of different stiffnesses using the trained models
Fig. 2A represents the distinct types of segmentation models utilized to automatically demarcate the contour-based boundary and coverage radius of the invasive protrusions and core region of the spheroids cultivated at collagen matrix-glass and collagen matrix-PDMS interfaces. As illustrated in Fig. 2B, there were no disparities in the parametric values of the core region between Glass and PDMS (10:1) groups within the time-lapse observation period, in terms of area, radius, perimeter, and complexity. Compared to the PDMS (10:1) group, the spheroids positioned above the collagen matrix-glass interface exhibited a significantly higher degree of invasive outgrowth, as evidenced by the invasive area, radius, and perimeter parameters, with the difference gradually becoming apparent starting after 8 h post-incubation (Fig. 2C). Although both interface groups exhibited an increasing trend in invasive complexity throughout the 24-h culture period, there was only a marginal difference observed between the two groups during the time frame from 16 h to 20 h post-incubation. Furthermore, the ratios of area, radius, perimeter between the invasive protrusions and core regions exhibited by the spheroids also demonstrated a notable disparity between the two interface stiffness conditions (Fig. 2D), suggesting that greater interface stiffness should promote tumor spheroid's invasiveness. In other words, the constituent cells within spheroids located at the collagen matrix-glass interface might display a tendency to migrate at higher velocities and travel longer distances away from spheroid's core compared to those within spheroids at the collagen matrix-PDMS interface.
Fig. 2.
The quantitative results of spheroid's invasiveness at the collagen matrix-glass and collagen matrix-PDMS interfaces (E = 69.3 GPa and 2.85 ± 0.001 MPa, separately). (A) The segmentation results of spheroids at different interface stiffness levels at 0h, 12h, and 24h post-incubation. The green line represents the contour of invasive protrusions, the red line delineates the core region, the blue circle represents the coverage radius of invasive protrusions, and the yellow circle indicates the coverage radius of the core region. (B) Analysis of the spheroid's core region within 24h. (C) Analysis of spheroid's invasive protrusions within 24h. (D) The ratios between the invasive protrusions and core regions. The scale bar corresponds to 50 μm *P < 0.05, **P < 0.01, ****P < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Analysis of spheroid's invasiveness in response to distinct interfacial adhesive strengths using the trained models
To examine the influence of interfacial adhesive strengths on a spheroid's invasiveness, the PDMS (10:1) substrates were separately subjected to two distinct surface treatment agents: pluronic F127 (referred to as Pluronic) and glutaraldehyde (GA). While Pluronic introduces steric repulsion into the interlayer space between collagen matrix and PDMS substrate, GA generates chemical cross-links within this interface region. Additionally, an uncoated PDMS substrate, referred to as the “No coating” condition, was utilized as the control, allowing for the electrostatic interaction with the collagen matrix. As depicted in Fig. 3A, the developed DL-based segmentation models were employed to accurately delineate the invasive area, core area, invasive radius, and core radius of the spheroids positioned at collagen matrix-PDMS interfaces with varying levels of interfacial strengths. Basically, the segmentation boundaries were shown to steadily enlarged over time (0h, 12h, and 24h). In terms of the area, radius, perimeter, and complexity parameters, there were no observable differences in the core region of spheroids among the above-mentioned three groups (Fig. 3B). However, the area, radius, and perimeter of invasive protrusions from spheroids in the “Pluronic” group were significantly larger than those in the “No coating” and “GA” groups in that order (Fig. 3C); this difference progressively became overt after 8h of culture. When considering the invasive complexity of spheroids, the value in the “Pluronic” group also appeared to exhibit a notable increase compared to the other two groups, with this divergence becoming evident exclusively once 20h had elapsed in the incubation process. The distribution patterns of the ratios of area, radius, perimeter, and complexity between the invasive protrusions and core regions observed in the spheroids, as depicted in Fig. 3D, were akin to those illustrated in Fig. 3C. These results indicate that modifications in spacing, achieved by adjusting the adhesive strength between the collagen matrix and PDMS substrate, impact the invasive behaviors of spheroids located within interfacial microenvironments.
Fig. 3.
The quantitative results of the invasiveness exhibited by spheroids grown above plain and GA- and Pluronic-modified PDMS substrates. (A) The segmentation results of spheroids in response to different interfacial adhesive strengths at 0h, 12h, and 24h post-incubation. The green line represents the contour of invasive protrusions, the red line delineates the core region, the blue circle represents the coverage radius of invasive protrusions, and the yellow circle indicates the coverage radius of the core region. (B) Analysis of the spheroid's core region within 24h. (C) Analysis of spheroid's invasive protrusions within 24h. (D) The ratios between the invasive protrusions and core regions. The scale bar corresponds to 50 μm *P < 0.05, **P < 0.01, ****P < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Characterization of single-cell motility in relation to different substrate stiffness
We further developed the single-cell model and employed it to track the movement of individual cells upon detachment from spheroids at interfaces with varying stiffness levels. As depicted in Fig. 4A, the model respectively demarcated the contour-based boundary and coverage radius of the cells distributed around the spheroids located on glass and PDMS substrate surfaces at 0h, 12h, and 24h post-incubation. Regarding the level of single-cell detachment, we did not observe any significant difference between the two stiffness groups prior to 12 h; however, interface stiffness enabled enhancement of this phenomenon as the disparity progressively became noticeable, with a profound statistical significance at 24h post-incubation (Fig. 4B). Moreover, the measurement of aspect ratio possessed by detached cells revealed that substrate stiffness at the interface also implemented an influence on the morphological appearance of cells, causing them to elongate (Fig. 4C). In light of the spacing between the detached cells and spheroid center, it was observed that the glass substrate, which had higher stiffness compared to the PDMS substrate, resulted in an increment in the average distance (Fig. 4D). Taken together, these results suggest that the detached cells at stiffer interfaces are more likely to exhibit a greater migratory speed and adopt a mesenchymal-like mode of movement.
Fig. 4.
The quantitative results of utilizing the trained single-cell model to characterize the migratory behavior exhibited by detached cells above glass or PDMS substrates. (A) The segmentation results of detached cells in response to different substrate stiffness at 0h, 12h, and 24h post-incubation. The cyan line indicates the contour-based boundary of a single cell, while the purple circle demarcates its coverage radius. (B) The dynamics of cell detachment from spheroids under two stiffness conditions over a 24-h period. (C) The aspect ratio of cells detached from spheroids under two stiffness conditions at 12 and 24h post-incubation. (D) The spacing between the detached cells and spheroid center in relation to varying interface stiffness at 24h post-incubation. The scale bar represents 50 μm *P < 0.05, ***P < 0.001, ****P < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Characterization of single cell motility in relation to distinct interfacial adhesive strengths
Fig. 5A illustrates the utilization of the single-cell segmentation model to delineate the cells detached from spheroids at the interfaces with varying interfacial adhesive strengths at 0h, 12h and 24h post-culture. Despite observing an upward trend in cell detachment across all three groups over the time-lapse period, the Pluronic-coated PDMS substrate demonstrated the strongest effect in promoting this phenotypic behavior, followed by the uncoated and GA-coated PDMS substrates, respectively (Fig. 5B). With respect to the morphological characteristics, the cells detached and migrating over both of the uncoated and PDMS substrates tended to display a rounded shape, with an aspect ratio close to 1. On the flip side, the cells dislodged from spheroids present in the “Pluronic” group had a pronounced propensity to assume an elongated morphology (Fig. 5C) and migrate farther away from their central location (Fig. 5D). Overall, these findings indicate that a higher adhesive strength between the matrix and substrate corresponds to reduced cell detachment. Additionally, this adhesive interaction also physically hinder the migration of detached cells, resulting in an amoeboid-like movement.
Fig. 5.
The quantitative analysis of the migratory behavior exhibited by detached cells responding to varying interfacial adhesive strengths using the trained single-cell model. (A) The segmentation results of detached cells at 0h, 12h, and 24h post-culture. The cyan line demarcates the contour of single cells, while the purple circle indicates their coverage radius. (B) The dynamics of cell detachment from spheroids under different interfacial-adhesive-strength conditions over 24 h. (C) The aspect ratio of cells dislodged from spheroids at 12h and 24h post-incubation. (D) The spacing between the detached cells and spheroid center in response to different interfacial adhesive strengths at 24h post-culture. The scale bar corresponds to 50 μm *P < 0.05, **P < 0.01, ****P < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Both individual and collective cell migration modes play pivotal roles in the progression of many diseases, notably cancer metastasis [[53], [54], [55], [56]]. Exploring the mechanisms behind migratory and invasive behaviors of cancer cells is critical for identifying strategies to counteract metastasis. While the image-based spheroid invasion assay provides many advantages for investigating cancer cell metastasis under in vitro conditions [57,58], the concurrent image processing and analysis during the assay period still present notable challenges. Due to the extensive number of time-lapse brightfield DIC images, the manual processing and analysis of these images is a time-intensive task. In addition, the unclear differentiation between cell-cell boundaries and/or cell-ECM contacts makes it difficult to separately delineate the cells even through the assistance of electronic drawing boards or by means of manual segmentation tools [19]. By automatically learning and extracting relevant features from image data, DL-based models have the potential to improve the accuracy of semantic segmentation, thus eliminating the need for manual annotation and enabling efficient production of analytical outputs and predictions [59,60]. In this study, we utilized an encoder-decoder architecture to develop a DL-based image segmentation pipeline, which allowed us to effectively analyze time-lapse imaging data and characterize the invasive and migratory behaviors demonstrated by assorted cancer cell types (i.e., breast cancer and melanoma) under diverse conditions, without any human involvement. The inferences drawn from post-processing and analysis of the image data, facilitated by the developed DL-based automated segmentation models, align with those obtained via manual analysis in our previous report [45]. Specifically, elevated interface stiffness and diminished interfacial adhesive strength can boost the invasiveness of tumor spheroids. Furthermore, these models also showcase their effectiveness in accurately and reliably segmenting spheroids and individual cells within a 3D context, even in the presence of environmental interferences (see Supplementary Figs. 1 and 2). Taken together, the validity and robustness of our proposed DL-based models for spheroid and single-cell segmentation (see Fig. 1) have been confirmed through a series of comprehensive assessments conducted in this research.
Former studies have successfully used a combination of DL and image processing algorithms to achieve cell segmentation from DIC microscopic images, with average IoU values ranging from 0.85 to 0.9 [[61], [62], [63]]. However, the majority of these algorithms were specifically developed for segmenting round-shaped cells, resulting in contour segments characterized by smooth boundaries. In this study, the outgrowth of spheroids gave rise to formation of structures exhibiting an irregular morphology (Fig. 1B), which appeared as fine-grained objects in DIC images, thus posing greater challenges in terms of image segmentation. It is noteworthy that our proposed DL-based segmentation models for the invasive protrusions and core region of spheroids yielded satisfactory IoU results of 0.77 and 0.75, respectively. As a matter of fact, compared to the extensive datasets commonly found in computer vision domains (e.g., ImageNet) [64] that typically comprise millions of images, our study encountered the constraint of utilizing a considerably smaller training dataset consisting of merely 400 spheroid images for a DL segmentation task. However, despite this limitation, our model still demonstrated impressive performance with an IoU score of 0.77. The success in this regard could be credited to the proficient utilization of well-established standard data augmentation and transfer learning techniques. On the one hand, the implementation of data augmentation techniques, such as flipping, rotating, and random adjustments of image brightness, served to increase the variability of the training dataset, thereby bolstering the performance of the DL model. On the other hand, to leverage the benefits of pre-existing knowledge, transfer learning was applied in this study by employing well-known pre-trained DL models, namely EfficientNet, Unet++, RegNetY320, and FPN. These models were initially trained on large datasets, enabling them to acquire valuable insights into low-level features such as edges and circles. Our selection to use EfficientNet and RegNet-Y 320 as the encoders for our model was driven by their remarkable performance on the ImageNet benchmark. In our assessment, both EfficientNet-B7 and RegNet-Y320 exhibited superior accuracies compared to ResNet [65]. EfficientNet-B7 achieved an exceptional score of 86.83, while RegNet-Y320 obtained a commendable score of 80.80. Through continued training with our specific datasets, the model acquired the capacity to effectively identify and understand the distinctive features possessed by the spheroids.
Given that our model architecture comprises of only an encoder and decoder, it can be easily modified for performance enhancement if the suggested combination of EfficientNet and Unet++ does not yield satisfactory results. Modifications can be made based on the code available at [https://github.com/qubvel/segmentation_models.pytorch]. In contrast to previous works [61,62] that employed a combination of DL and watershed algorithms to perform semi-automatic segmentation, our pipeline distinguishes itself as a completely automated segmentation solution. While the Watershed algorithm has been proposed as a simpler alternative for spheroid segmentation, it relies heavily on precise determination of the region of interest (ROI) center as a marker for boundary detection, making it prone to over-segmentation when the ROI center is inaccurate [66]. In addition, the generic algorithm for spheroid segmentation requires fewer data and resources for training, but it necessitates various configurations to accommodate changes in the nature of spheroid images [67]. In comparison, our proposed pipeline for spheroid segmentation offers a straightforward, adaptable and efficient solution that eliminates the demand for customized configurations when dealing with images captured under varying conditions.
However, it is important to acknowledge that the current limitation of this pipeline lies in the manual annotation requirement, which can be a time-consuming process. In recent advancements, Self-Supervised Learning (SSL) models have emerged as a promising approach to circumvent the manual annotation issue in DL tasks [68,69]. SSL models have the capability to learn from the data itself without the need for explicit annotations, and they have demonstrated comparable performance to other DL models. In the future, it would be worthwhile to explore the feasibility of SSL models in the context of spheroid segmentation tasks.
The majority of researches have unraveled the effects of surface stiffness and matrix-surface adhesive strength on cell migration in traditional monolayer cultures [[70], [71], [72], [73]]. In this manuscript, we employed our DL-based automated segmentation pipeline to perform post-processing and analysis of image datasets that were previously manually segmented in our previous study [45], These datasets were used to investigate the impact of interface stiffness and interfacial adhesive strength on invasiveness of tumor spheroids. Our automated platform outperforms the manual approach in terms of efficiency, as it effectively reduces the time required for data processing and analysis. On the one hand, it offers a wealth of quantitative metrics, including area, radius, perimeter, and complexity, which greatly contribute to the comprehensive characterization of spheroid invasiveness throughout the duration of the time-lapse observation. On the other hand, it can delineate single motile cells detached from the observed spheroids, and automatically calculate their aspect ratio and migratory distance. Even so, both segmentation measures yielded comparable conclusions regarding enhancement of spheroid invasiveness and single-cell migration in interfacial microenvironments with higher mechanical stiffness and weaker matrix-surface adhesive strength. It merits emphasizing that we have introduced the term “diepafitaxis” to put forth a plausible mechanism that elucidates the distinct interface-mediated tropism observed in tumor spheroids situated at matrix-surface boundaries [45]. Despite the hypothetical explanation provided for this “tropistic behavior”, we previously asserted that the stiffness and topographic features of the interface play crucial roles in determining the spreading dynamics of tumor spheroids. Based on the results illustrated in Fig. 2, the spheroids exhibited a significantly higher invasive capacity, as evidenced by the indicated parameters, when being situated above glass substrate, which possess a considerably greater Young's modulus in comparison to PDMS. Likewise, migratory cells showed much greater detachment from spheroids located on stiffer glass substrates, resulting in increased speed and distance of movement. Moreover, they tended to display a larger aspect ratio (i.e., mesenchymal morphology) compared to those positioned on softer PDMS substrates (Fig. 4). From the viewpoint of geographic features, the levels of matrix-substrate interactions arising from different substrate surface modifications also had considerable impacts on spheroid outgrowth, single-cell detachment, and migration speed and morphology of the detached single cells (Fig. 5). Collectively, these results suggest that tissue-tissue boundaries characterized by greater stiffness and weaker interactions might play a significant role in promoting cancer invasion and metastasis.
Advances in development of DL-based algorithms from artificial neural networks display excellent prospects in mining intricate biological and physiological data to extract features and identify patterns [74]. The utilization of DL-based image segmentation has gained significant popularity in clinical settings for the analysis of medical images, including magnetic resonance imaging (MRI), radiography, X-rays, tomography and ultrasound, mammography. This approach plays a crucial in the diagnosis and treatment of cancer and other diseases [[75], [76], [77], [78], [79], [80]]. As mentioned above, metastasis is the leading cause of cancer-related mortality. In the course of metastasis, cancer cells frequently traverse complex tissue microenvironments, where they encounter diverse ECM microstructures at various interfaces. To circumvent metastasis, it is crucial to enhance our comprehension of how mechanical properties affect interfacial cancer cell invasion. DIC microscopy provides a facile way for time-lapse observation of cancer cell-microenvironment interactions in a non-destructive and fluorescence-free manner. By leveraging its advantages, we can gain valuable insights into the dynamic interplay between cancer cells and their surrounding microenvironment, shedding light on potential strategies to inhibit their invasive behavior. However, researchers still face significant challenges in post-invasion image analysis. In this paper, we suggest employing DL encoder-decoder models to discern the effects of different interfacial influences on spheroid invasion dynamics. In comparison to the prior assessment conducted by Nakhjavani et al. [81], our DL-based automated segmentation technique can enable detection of alterations in the invasion patterns of MDA-MB-231 breast cancer spheroids in 2.5 D (i.e., interfacial) or 3D conditions during the early hours of incubation. To broaden the segmentation capability for various types of cancer, it may be necessary to expand this DL-based pipeline by encompassing additional cell types in the future. Even so, it is worth highlighting that our proposed framework may showcase promise for pre-clinical utility in expediting evaluation of the metastatic behavior of cancer cells under physiologically-relevant in vitro conditions, while its further advancement, by incorporating with multi-drug screening platforms (e.g., phenotypic response surfaces) [82,83], can also promote, in clinical practice, identification and development of novel synergistic personalized cocktails against cancer metastasis. What's more, while our DL-based automated segmentation pipeline has been developed specifically for DIC microscopic images to differentiate spheroids and detached single cells under interfacial conditions, its applicability can be extended to other imaging modalities commonly used in clinical settings (e.g., brightfield microscopy, confocal fluorescence microscopy, and MRI, after training the models on representative data acquired from the specific imaging modality of interest.
5. Conclusion
In conclusion, we have successfully employed a DL-based architecture to develop models for segmenting time-lapse DIC images, especially targeting the invasive protrusions and core region of spheroids as well as detached single cells. The adoption of this innovative approach represents a paradigm shift in the efficient processing of post-invasion spheroid images, achieving outstanding prediction accuracy while minimizing the time investment. Through the use of the aforementioned models, we successfully obtained detailed insights into the interfacial invasive and migratory behaviors displayed by breast cancer and melanoma cells. Notably, these analyses were carried out in an entirely automated manner, eliminating any requirement for human involvement. The findings presented in this manuscript were consistent with the conclusions drawn through manual assessments in our prior report. This consistent agreement reaffirms the reliability and effectiveness of our developed models in accurately capturing and quantifying essential image features. Moreover, it emphasizes the promising potential of DL-based approaches as valuable tools for image analysis in the field of tumor research.
Credit author statement
Conceptualization, Y.L.K, J.H.T, S.N.S. and T.Y.T.; Methodology, T.K.N.N, T.K.M.N, S.J.Y., Q.D.N, T.Y.T. and S.N.S.; Investigation, T.K.N.N, S.J.Y.,Q.D.N, T.Y.T. and S.N.S.; Writing – Original Draft, T.K.N.N and S.J.Y.; Writing – Review & Editing, B.H.M, S.N.S. and T.Y.T.; Funding Acquisition, T.Y.T.; Resources, T.K.N.N., S.J.Y., T.K.M.N, T.Y.T. and S.N.S.; Supervision, S.N.S. and T.Y.T.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This research was funded by the National Science and Technology Council (NSTC), Taiwan, and the Young Scholar Program (MOST 111-2740-B-006-002, MOST 111-2636-B-006-010-, NSTC 112-2321-B-006-021-, NSTC 112-2636-B-006-001- and NSTC 112-2740-B-006-002- to T.Y.T.). This research was also supported in part by the Higher Education Sprout Project, Ministry of Education to the Headquarters of University Advancement and Southeast Asia and Taiwan Universities (SATU) at National Cheng Kung University. The authors are grateful for the support from the Core Research Laboratory, College of Medicine, National Cheng Kung University.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.mtbio.2023.100820.
Contributor Information
Shier Nee Saw, Email: sawsn@um.edu.my.
Ting-Yuan Tu, Email: tingyuan@mail.ncku.edu.tw.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Data availability
Data will be made available on request.
References
- 1.Fazilaty H. Assessing reproducibility of the core findings in cancer research. iScience. 2022 doi: 10.1016/j.isci.2022.105125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turajlic S., Swanton C. Metastasis as an evolutionary process. Science. 2016;352:169–175. doi: 10.1126/science.aaf2784. [DOI] [PubMed] [Google Scholar]
- 3.Ha N.-H., Faraji F., Hunter K.W. Drug Deliv.; 2013. Mechanisms of Metastasis, Cancer Target; pp. 435–458. [Google Scholar]
- 4.Leggett S.E., Nelson C.M. Biomarkers Tumor Microenviron. Springer; 2022. Tissue architecture in cancer initiation and progression; pp. 91–107. [Google Scholar]
- 5.Klein C.A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer. 2009 doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 6.Karrar H.R. Clinical aspects of breast cancer. World Fam. Med. 2022;20:136–142. [Google Scholar]
- 7.Klein C.A. Cancer progression and the invisible phase of metastatic colonization. Nat. Rev. Cancer. 2020;20:681–694. doi: 10.1038/s41568-020-00300-6. [DOI] [PubMed] [Google Scholar]
- 8.Weiswald L.B., Bellet D., Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia. 2015;17:1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X., Farach-Carson M.C., Jia X., Xian Xua X.J., Farach-Carsonb Mary C., Xu X., Farach-Carson M.C., Jia X. Three-dimensional in vitro tumor models for cancer research and drug evaluation. Biotechnol. Adv. 2014;32:1256–1268. doi: 10.1016/j.biotechadv.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blacher S., Erpicum C., Lenoir B., Paupert J., Moraes G., Ormenese S., Bullinger E., Noel A. Cell invasion in the spheroid sprouting assay: a spatial organisation analysis adaptable to cell behaviour. PLoS One. 2014;9:1–10. doi: 10.1371/journal.pone.0097019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W., Wong C., Vosburgh E., Levine A.J., Foran D.J., Xu E.Y. High-throughput image analysis of tumor spheroids: a user-friendly software application to measure the size of spheroids automatically and accurately. J. Vis. Exp. 2014:1–10. doi: 10.3791/51639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Härmä V., Schukov H.P., Happonen A., Ahonen I., Virtanen J., Siitari H., Åkerfelt M., Lötjönen J., Nees M. Quantification of dynamic morphological drug responses in 3D organotypic cell cultures by automated image analysis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cisneros Castillo L.R., Oancea A.D., Stüllein C., Régnier-Vigouroux A. A novel computer-assisted approach to evaluate multicellular tumor spheroid invasion assay. Sci. Rep. 2016;6:1–16. doi: 10.1038/srep35099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirai K., Kato H., Imai Y., Shibuta M., Kanie K., Kato R. The importance of scoring recognition fitness in spheroid morphological analysis for robust label-free quality evaluation. Regen. Ther. 2020;14:205–214. doi: 10.1016/j.reth.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ducker M., Millar V., Ebner D., Szele F.G. A semi-automated and scalable 3D spheroid assay to study neuroblast migration. Stem Cell Rep. 2020;15:789–802. doi: 10.1016/j.stemcr.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grexa I., Diosdi A., Harmati M., Kriston A., Moshkov N., Buzas K., Pietiäinen V., Koos K., Horvath P. SpheroidPicker for automated 3D cell culture manipulation using deep learning. Sci. Rep. 2021;11:1–11. doi: 10.1038/s41598-021-94217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z., Ma N., Sun X., Li Q., Zeng Y., Chen F., Sun S., Xu J., Zhang J., Ye H., Ge J., Zhang Z., Cui X., Leong K., Chen Y., Gu Z. Automated evaluation of tumor spheroid behavior in 3D culture using deep learning-based recognition. Biomaterials. 2021;272 doi: 10.1016/j.biomaterials.2021.120770. [DOI] [PubMed] [Google Scholar]
- 18.Hou Y., Konen J., Brat D.J., Marcus A.I., Cooper L.A.D. TASI: a software tool for spatial-temporal quantification of tumor spheroid dynamics. Sci. Rep. 2018;8:1–9. doi: 10.1038/s41598-018-25337-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen C., Miura T., Voleti V., Yamaguchi K., Tsutsumi M., Yamamoto K., Otomo K., Fujie Y., Teramoto T., Ishihara T., Aoki K., Nemoto T., Hillman E.M.C., Kimura K.D. 3DeeCellTracker, a deep learning-based pipeline for segmenting and tracking cells in 3D time lapse images. Elife. 2021;10 doi: 10.7554/eLife.59187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alsehli H., Mosis F., Thompson C., Hamrud E., Wiseman E., Gentleman E., Danovi D. An integrated pipeline for high-throughput screening and profiling of spheroids using simple live image analysis of frame to frame variations. Methods. 2021;190:33–43. doi: 10.1016/j.ymeth.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulligan J.A., Ling L., Leartprapun N., Fischbach C., Adie S.G. Computational 4D-OCM for label-free imaging of collective cell invasion and force-mediated deformations in collagen. Sci. Rep. 2021;11:1–13. doi: 10.1038/s41598-021-81470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alzeeb G., Dubreuil M., Arzur D., Rivet S., Corcos L., Le Grand Y., Le Jossic-Corcos C. Gastric cancer multicellular spheroid analysis by two-photon microscopy. Biomed. Opt Express. 2022;13:3120–3130. doi: 10.1364/BOE.450518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazzari G., Vinciguerra D., Balasso A., Nicolas V., Goudin N., Garfa-Traore M., Fehér A., Dinnyés A., Nicolas J., Couvreur P., Mura S. Light sheet fluorescence microscopy versus confocal microscopy: in quest of a suitable tool to assess drug and nanomedicine penetration into multicellular tumor spheroids. Eur. J. Pharm. Biopharm. 2019;142:195–203. doi: 10.1016/j.ejpb.2019.06.019. [DOI] [PubMed] [Google Scholar]
- 24.Santi P.A. Light sheet fluorescence microscopy: a review. J. Histochem. Cytochem. 2011;59:129–138. doi: 10.1369/0022155410394857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang H., Liu Y., Li Y. 2022 7th Int. Conf. Intell. Comput. Signal Process. 2022. Fluorescence image denoising based on self-supervised deep learning; pp. 86–90. [DOI] [Google Scholar]
- 26.Stephens D.J., Allan V.J. Light microscopy techniques for live cell imaging. Science. 2003;300:82–86. doi: 10.1126/science.1082160. [DOI] [PubMed] [Google Scholar]
- 27.Ziv N., Schiller J. Differential interference contrast (DIC) imaging of living cells. CSH Protoc. 2007;2007:prot4787. doi: 10.1101/pdb.prot4787. [DOI] [PubMed] [Google Scholar]
- 28.Moscato Z., Jaiswal D., Dixit K., Langanis C.J., Claffey K.P., Hoshino K. In: Rasooly A., Baker H., Ossandon M.R., editors. vol. 2. Springer US; New York, NY: 2022. Label-free morphological phenotyping of in vitro 3D microtumors; pp. 31–46. (BT - Biomed. Eng. Technol.). [DOI] [PubMed] [Google Scholar]
- 29.Wang G., Fang N. P.M.B.T.-M. In E. Conn (Ed.), Imaging Spectrosc. Anal. Living Cells. Academic Press; 2012. Chapter four - detecting and tracking nonfluorescent nanoparticle probes in live cells; pp. 83–108. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., Yang Q., He S., Wang R., Luo H. ACS Photonics; 2022. Computing Metasurfaces Enabled Broad-Band Vectorial Differential Interference Contrast Microscopy. [DOI] [Google Scholar]
- 31.Seo H., Badiei Khuzani M., Vasudevan V., Huang C., Ren H., Xiao R., Jia X., Xing L. Machine learning techniques for biomedical image segmentation: an overview of technical aspects and introduction to state-of-art applications. Med. Phys. 2020;47:e148. doi: 10.1002/mp.13649. –e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Gross L., Li Z., Li X., Fan X., Qi W. Automatic building extraction on high-resolution remote sensing imagery using deep convolutional encoder-decoder with spatial pyramid pooling. IEEE Access. 2019;7:128774–128786. doi: 10.1109/ACCESS.2019.2940527. [DOI] [Google Scholar]
- 33.Chen L.C., Zhu Y., Papandreou G., Schroff F., Adam H. Encoder-decoder with atrous separable convolution for semantic image segmentation. Lect. Notes Comput. Sci. (Including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics). 11211 LNCS. 2018:833–851. doi: 10.1007/978-3-030-01234-2_49. [DOI] [Google Scholar]
- 34.Habanjar O., Diab-Assaf M., Caldefie-Chezet F., Delort L. 3D cell culture systems: tumor application, advantages, and disadvantages. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khawar I.A., Ghosh T., Park J.K., Kuh H.-J. Tumor spheroid-based microtumor models for preclinical evaluation of anticancer nanomedicines. J. Pharm. Investig. 2021;51:541–553. doi: 10.1007/s40005-021-00534-y. [DOI] [Google Scholar]
- 36.Jessup J.M., Brown D., Fitzgerald W., Ford R.D., Nachman A., Goodwin T.J., Spaulding G. Induction of carcinoembryonic antigen expression in a three-dimensional culture system. In Vitro Cell. Dev. Biol. Anim. 1997 doi: 10.1007/s11626-997-0005-6. [DOI] [PubMed] [Google Scholar]
- 37.Redmond J., McCarthy H., Buchanan P., Levingstone T.J., Dunne N.J. Advances in biofabrication techniques for collagen-based 3D in vitro culture models for breast cancer research. Mater. Sci. Eng. C. 2021;122 doi: 10.1016/j.msec.2021.111944. [DOI] [PubMed] [Google Scholar]
- 38.Carey S.P., Starchenko A., McGregor A.L., Reinhart-King C.A. Leading malignant cells initiate collective epithelial cell invasion in a three-dimensional heterotypic tumor spheroid model. Clin. Exp. Metastasis. 2013;30:615–630. doi: 10.1007/s10585-013-9565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Micalet A., Moeendarbary E., Cheema U. 3D in vitro models for investigating the role of stiffness in cancer invasion. ACS Biomater. Sci. Eng. 2021 doi: 10.1021/acsbiomaterials.0c01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conklin M.W., Eickhoff J.C., Riching K.M., Pehlke C.A., Eliceiri K.W., Provenzano P.P., Friedl A., Keely P.J. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 2011 doi: 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dumas J.-F., Brisson L. Interaction between adipose tissue and cancer cells: role for cancer progression. Cancer Metastasis Rev. 2021;40:31–46. doi: 10.1007/s10555-020-09934-2. [DOI] [PubMed] [Google Scholar]
- 42.Roach P., Parker T., Gadegaard N., Alexander M.R. Surface strategies for control of neuronal cell adhesion: a review. Surf. Sci. Rep. 2010;65:145–173. doi: 10.1016/j.surfrep.2010.07.001. [DOI] [Google Scholar]
- 43.Rape A.D., Kumar S. A composite hydrogel platform for the dissection of tumor cell migration at tissue interfaces. Biomaterials. 2014;35:8846–8853. doi: 10.1016/j.biomaterials.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merino-Casallo F., Gomez-Benito M.J., Hervas-Raluy S., Garcia-Aznar J.M. Unravelling cell migration: defining movement from the cell surface. Cell Adh. Migr. 2022;16:25–64. doi: 10.1080/19336918.2022.2055520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao B.-H., Nguyen Thi K.M., Tang M.-J., Kamm R.D., Tu T.-Y. The interface stiffness and topographic feature dictate interfacial invasiveness of cancer spheroids. Biofabrication. 2023;15 doi: 10.1088/1758-5090/acaa00. [DOI] [PubMed] [Google Scholar]
- 46.Wu K., Kuo C., Tu T. A highly reproducible micro U‐well array plate facilitating high‐throughput tumor spheroid culture and drug assessment. Glob. Challenges. 2021;5 doi: 10.1002/gch2.202000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buslaev A., Iglovikov V.I., Khvedchenya E., Parinov A., Druzhinin M., Kalinin A.A. Albumentations: fast and flexible image augmentations. Inf. 2020;11 doi: 10.3390/info11020125. [DOI] [Google Scholar]
- 48.Tan M., Le Q.V., EfficientNet . 36th Int. Conf. Mach. Learn. ICML 2019. 2019-June. 2019. Rethinking model scaling for convolutional neural networks; pp. 10691–10700. [Google Scholar]
- 49.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. UNet++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imag. 2020;39:1856–1867. doi: 10.1109/TMI.2019.2959609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hendrycks D., Lee K., Mazeika M. Using pre-training can improve model robustness and uncertainty, 36th. Int. Conf. Mach. Learn. ICML 2019. 2019-June. 2019:4815–4826. [Google Scholar]
- 51.Radosavovic I., Kosaraju R.P., Girshick R., He K., Dollár P. Designing network design spaces. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recogn. 2020:10425–10433. doi: 10.1109/CVPR42600.2020.01044. [DOI] [Google Scholar]
- 52.Li H., Liu B., Zhang Y., Fu C., Han X., Du L., Gao W., Chen Y., Liu X., Wang Y., Wang T., Ma G., Lei B. 3D IFPN: improved feature pyramid network for automatic segmentation of gastric tumor. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.618496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain S., Cachoux V.M.L., Narayana G.H.N.S., de Beco S., D'Alessandro J., Cellerin V., Chen T., Heuzé M.L., Marcq P., Mège R.-M., Kabla A.J., Lim C.T., Ladoux B. The role of single-cell mechanical behaviour and polarity in driving collective cell migration. Nat. Phys. 2020;16:802–809. doi: 10.1038/s41567-020-0875-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reffay M., Petitjean L., Coscoy S., Grasland-Mongrain E., Amblard F., Buguin A., Silberzan P. Orientation and polarity in collectively migrating cell structures: statics and dynamics. Biophys. J. 2011;100:2566–2575. doi: 10.1016/j.bpj.2011.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lintz M., Muñoz A., Reinhart-King C.A. The mechanics of single cell and collective migration of tumor cells. J. Biomech. Eng. 2017;139 doi: 10.1115/1.4035121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.George M., Bullo F., Campàs O. Connecting individual to collective cell migration. Sci. Rep. 2017;7:9720. doi: 10.1038/s41598-017-10069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shabalina E.Y., Skorova E.Y., Chudakova D.A., Anikin V.B., V Reshetov I., Mynbaev O.A., V Petersen E. The matrix-dependent 3D spheroid model of the migration of non-small cell lung cancer: a step towards a rapid automated screening. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.610407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mei J., Böhland C., Geiger A., Baur I., Berner K., Heuer S., Liu X., Mataite L., Melo-Narváez M.C., Özkaya E., Rupp A., Siebenwirth C., Thoma F., Kling M.F., Friedl A.A. Development of a model for fibroblast-led collective migration from breast cancer cell spheroids to study radiation effects on invasiveness. Radiat. Oncol. 2021;16:159. doi: 10.1186/s13014-021-01883-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jang J., Wang C., Zhang X., Choi H.J., Pan X., Lin B., Yu Y., Whittle C., Ryan M., Chen Y., Lee K. A deep learning-based segmentation pipeline for profiling cellular morphodynamics using multiple types of live cell microscopy. Cell Reports Methods. 2021;1 doi: 10.1016/j.crmeth.2021.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Din N.U., Yu J. Training a deep learning model for single-cell segmentation without manual annotation. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-03299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W., Taft D.A., Chen Y.-J., Zhang J., Wallace C.T., Xu M., Watkins S.C., Xing J. Learn to segment single cells with deep distance estimator and deep cell detector. Comput. Biol. Med. 2019;108:133–141. doi: 10.1016/j.compbiomed.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lux F., Matula P. 2019 IEEE 16th Int. Symp. Biomed. Imaging. ISBI 2019; 2019. DIC image segmentation of dense cell populations by combining deep learning and watershed; pp. 236–239. [DOI] [Google Scholar]
- 63.Adhikary S., Seth S., Das S., Naskar T.K., Barui A., Maity S.P. Feature assisted cervical cancer screening through DIC cell images. Biocybern. Biomed. Eng. 2021;41:1162–1181. doi: 10.1016/j.bbe.2021.07.005. [DOI] [Google Scholar]
- 64.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., Berg A.C., Fei-Fei L. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115:211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 65.He K., Zhang X., Ren S., Sun J. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 66.Kornilov A.S., V Safonov I. An overview of watershed algorithm implementations in open source libraries. J. Imaging. 2018;4:123. [Google Scholar]
- 67.Lacalle D., Castro-Abril H.A., Randelovic T., Domínguez C., Heras J., Mata E., Mata G., Méndez Y., Pascual V., Ochoa I. SpheroidJ: an open-source set of tools for spheroid segmentation. Comput. Methods Progr. Biomed. 2021;200 doi: 10.1016/j.cmpb.2020.105837. [DOI] [PubMed] [Google Scholar]
- 68.Caron M., Touvron H., Misra I., Jégou H., Mairal J., Bojanowski P., Joulin A. Proc. IEEE/CVF Int. Conf. Comput. Vis. 2021. Emerging properties in self-supervised vision transformers; pp. 9650–9660. [Google Scholar]
- 69.Bardes A., Ponce J., LeCun Y. Vicreg: variance-invariance-covariance regularization for self-supervised learning, 10th International Conference on Learning Representations. ICLR. 2022;2022 [Google Scholar]
- 70.Vazquez K., Saraswathibhatla A., Notbohm J. Effect of substrate stiffness on friction in collective cell migration. Sci. Rep. 2022;12:1–13. doi: 10.1038/s41598-022-06504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia-Gonzalez D., Muñoz-Barrutia A. Computational insights into the influence of substrate stiffness on collective cell migration. Extrem. Mech. Lett. 2020;40 doi: 10.1016/j.eml.2020.100928. [DOI] [Google Scholar]
- 72.Yu Y., Ren L.-J., Liu X.-Y., Gong X.-B., Yao W. Effects of substrate stiffness on mast cell migration. Eur. J. Cell Biol. 2021;100 doi: 10.1016/j.ejcb.2021.151178. [DOI] [PubMed] [Google Scholar]
- 73.Califano J.P., Reinhart-King C.A. Substrate stiffness and cell area predict cellular traction stresses in single cells and cells in contact. Cell. Mol. Bioeng. 2010;3:68–75. doi: 10.1007/s12195-010-0102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao C., Liu F., Tan H., Song D., Shu W., Li W., Zhou Y., Bo X., Xie Z. Deep learning and its applications in biomedicine, genomics. Proteomics Bioinforma. 2018;16:17–32. doi: 10.1016/j.gpb.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karabatak M., Ince M.C. An expert system for detection of breast cancer based on association rules and neural network. Expert Syst. Appl. 2009;36:3465–3469. [Google Scholar]
- 76.Bejnordi B.E., Veta M., Van Diest P.J., Van Ginneken B., Karssemeijer N., Litjens G., Van Der Laak J.A.W.M., Hermsen M., Manson Q.F., Balkenhol M. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kang J., Schwartz R., Flickinger J., Beriwal S. Machine learning approaches for predicting radiation therapy outcomes: a clinician's perspective. Int. J. Radiat. Oncol. Biol. Phys. 2015;93:1127–1135. doi: 10.1016/j.ijrobp.2015.07.2286. [DOI] [PubMed] [Google Scholar]
- 78.Übeyli E.D. Implementing automated diagnostic systems for breast cancer detection. Expert Syst. Appl. 2007;33:1054–1062. doi: 10.1016/j.eswa.2006.08.005. [DOI] [Google Scholar]
- 79.Di Ieva A., Russo C., Liu S., Jian A., Bai M.Y., Qian Y., Magnussen J.S. Application of deep learning for automatic segmentation of brain tumors on magnetic resonance imaging: a heuristic approach in the clinical scenario. Neuroradiology. 2021;63:1253–1262. doi: 10.1007/s00234-021-02649-3. [DOI] [PubMed] [Google Scholar]
- 80.Ahn S.H., Yeo A.U., Kim K.H., Kim C., Goh Y., Cho S., Lee S.B., Lim Y.K., Kim H., Shin D., Kim T., Kim T.H., Youn S.H., Oh E.S., Jeong J.H. Comparative clinical evaluation of atlas and deep-learning-based auto-segmentation of organ structures in liver cancer. Radiat. Oncol. 2019;14:213. doi: 10.1186/s13014-019-1392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakhjavani M., Palethorpe H., Tomita Y., Smith E., Price T., Yool A., Pei J., Townsend, Hardingham J. Stereoselective anti-cancer activities of ginsenoside Rg3 on triple negative breast cancer cell models. Pharmaceuticals. 2019;12:117. doi: 10.3390/ph12030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang H.-Y., Goudar V.S., Hung Y.-C., Ouyang C.-H., Rashid M.B.M.A., Juo L.-Y., Wu J.-K., Cheng Y.-W., Wei P.-L., Tzeng H.-E., Jiang J.-K., Chow E.K.-H., Yang C.-Y., Ho C.-M., Yen Y., Tseng F.-G. In vitro study on AI-PRS enabled precision cocktail drugs design for treating human colorectal carcinoma. Adv. Ther. 2023;6 doi: 10.1002/adtp.202200298. [DOI] [Google Scholar]
- 83.Al-Shyoukh I., Yu F., Feng J., Yan K., Dubinett S., Ho C.-M., Shamma J.S., Sun R. Systematic quantitative characterization of cellular responses induced by multiple signals. BMC Syst. Biol. 2011;5:88. doi: 10.1186/1752-0509-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.