Summary
Alcohol-related liver disease (ALD) is one of the leading causes of liver-related death worldwide. However, roles of oral microbiota in regulating the progression of ALD remain unknown. Here, we fed mice with control or ethanol diet to establish chronic-plus-binge ALD model. 16S ribosomal DNA sequencing was performed on oral and cecum samples. We demonstrated that alcohol drinking influenced bacterial richness, microbial structure, and composition in oral samples of ethanol-fed mice compared with control mice. Alcohol consumption also remodeled relationships among oral microbes and altered functions of oral microbiota. Furthermore, oral microbiota, such as Streptococcus, Helicobacter, Alloprevotella, and Psychrobacter were closely associated with ALD parameters. Finally, we observed Sutterellaceae_uncultured, Dyella, and Gemmatimonas possibly translocated along with oral-gut axis and positively correlated with the severity of ALD. Altogether, alcohol consumption reprogramed composition and functions of oral microbiota to promote ALD progression, suggesting that oral microbes might become a new target for ALD therapy.
Subject areas: Health sciences, Medicine, Microbiology
Graphical abstract

Highlights
-
•
Alcohol intake altered oral microbial diversity, composition, and function
-
•
Alcohol drinking changes the relationships among oral microbiota
-
•
The relative abundance of oral microbiota reflects the severity of ALD
-
•
Potential oral-gut bacterial translocation was positively associated with ALD
Health sciences; Medicine; Microbiology
Introduction
Alcohol-related liver disease (ALD) is one of the most causes of mortality worldwide, with approximately half of the liver cirrhosis-induced death associated with alcohol.1,2 The spectrum of ALD covers alcoholic fatty liver, alcoholic steatohepatitis (ASH), alcoholic fibrosis, and cirrhosis.3 Besides, severe ASH may progress to alcoholic hepatitis, in which 20%–50% of patients will die within 3 months.2,3 Although ALD has been a serious threat to human health, the studies about the pathological mechanism and effective therapeutic approaches are still limited.2,3,4 Therefore, it is urgent to elucidate the pathogenesis and explore potential therapeutic strategies for ALD.
With further researches on the pathogenesis of ALD, increasing evidences have demonstrated that alcohol-associated dysbiosis of gut microbiota contributes to ALD progression by various mechanisms, including the disruption of intestinal tight junction, the microbial translocation from gut to the liver, the release of microbial toxins (like lipopolysaccharides), and the metabolism of intestinal bile acids or fatty acids.5,6,7 As the second largest microbiome after the gut microbiota, oral microbiota includes more than 700 different species of bacteria, fungi, viruses, and protozoa.8 The disturbance of oral microbiota results in the increase of pathogenic bacteria, mucosal immunity damage, and the translocation of oral microbiota to other tissues.9 In consequence, the dysbiosis of oral microbiota is not only associated with oral diseases but also has intimate relationships to systemic diseases, such as liver cirrhosis, liver cancer, pancreatic cancer, inflammatory bowel disease, and cardiovascular disease.10,11,12,13 Although heavy alcohol drinking has been reported to influence the composition of oral microbiota,14 whether oral microbiota could also take an active role in ALD progression remains unknown.
Here, we demonstrated that the impact of alcohol consumption on oral microbiota during the development of ALD. The microbial diversity was reduced and the microbial structure was changed in the oral cavity of ethanol-fed mice as compared with control mice. The relative abundance of oral Helicobacter, Janthinobacterium, Alloprevotella, Paenalcaligenes, and Psychrobacter was increased, while Lactococcus and Streptococcus was decreased following alcohol intake. In addition, we predicted the potential altered functions of oral microbiota during alcohol consumption and analyzed the correlation between the oral microbiota and ALD progression. Finally, further investigation showed that Sutterellaceae_uncultured, Dyella, and Gemmatimonas were probably translocated along with the oral-gut axis, and more importantly, their intestinal abundance was positively associated with ALD. Collectively, our findings have illustrated the critical roles of oral microbiota in promoting ALD progression.
Results
Alcohol feeding affects the microbial diversity of oral microbiota
Chronic-plus-binge model was utilized to generate mice with ALD (Figure 1A).15,16 Although alcohol feeding did not affect the body weight and food intake significantly (Figures 1B and 1C), the liver weight/body weight ratio and plasma levels of ALT and AST were markedly increased in ethanol-fed mice (Figures 1D and 1E). Histological staining revealed very obvious hepatic steatosis after alcohol diet feeding (Figure 1F). Compared with mice fed with control diet, the real-time PCR data showed the elevated mRNA expression of inflammatory cytokines, such as Il1b, Il6, Ccl2, and Tnf, in the livers of ethanol-fed mice. Gene-encoding fatty acid translocase (Cd36) was also upregulated, while Ppara, the gene responsible for fatty acid β-oxidation was decreased following alcohol dinking (Figure 1G). Taken together, alcohol feeding could induce severe hepatocytes death, hepatic steatosis, and inflammation in livers of mice.
Figure 1.
EtOH liquid diet feeding deteriorates the biochemical and histological characters to induce ALD
(A) Schematic of the mice ALD model. Mice were fed with control liquid diet for 5 days, and then diet was changed to 6% EtOH liquid diet in EtOH group for 10 days. At the 15th day, mice were gavaged EtOH or maltose dextrin, respectively. Oral samples were collected every 7:00 a.m. and 4:00 p.m. (n = 12 per group).
(B) Body weight.
(C) Food intake.
(D) Liver weight/body weight ratio.
(E) Plasma ALT and AST levels.
(F) Left: Representative images of H&E staining in liver sections. Scale bar: 100 μm. Right: Scores of steatosis.
(G) Gene expressions of inflammation and lipid metabolism in liver tissues (n = 10–12 per group). Data were presented as mean ± SEM. ∗p < 0.05. Data were analyzed by two-tailed Student’s t test except data in G were analyzed by Mann Whitney test.
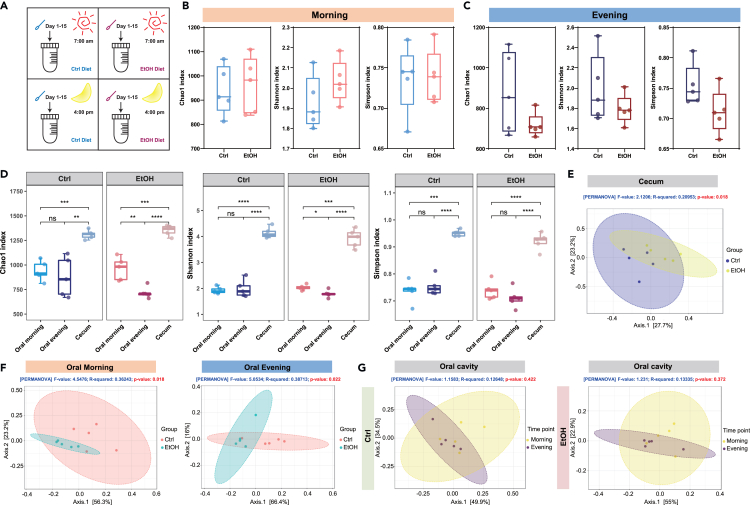
To investigate the effects of alcohol feeding on the oral microbiota and assess whether this impact will be different in the morning or in the evening, oral samples were collected daily at 7:00 a.m. and 4:00 p.m. from control and ethanol-fed mice (Figure 2A). α-diversity which represents species richness and evenness was shown by Chao 1, Shannon, and Simpson indexes. In spite of the less impact of alcohol drinking on samples collected in the morning from the oral cavity of mice (Figure 2B), oral samples collected in the evening showed obvious decrease of microbial diversity following alcohol intake (Figure 2C). We also compared the α-diversity of oral samples collected in the morning or in the evening and the cecum samples. As expected, we found that the microbial richness was quite higher in the gut microbiota than that in oral cavity of both control and ethanol groups (Figure 2D). Moreover, in mice fed with ethanol diet, the oral bacterial richness was significantly lower in samples collected in the evening than that in the morning as evidenced by Chao 1 index and Shannon index (Figure 2D), while in control diet-fed mice, the oral bacterial diversity was not changed between these two time points (Figure 2D). The compositional dissimilarity among groups was described by β-diversity. We firstly observed a significant difference in clustering between the gut microbiota of control and ethanol-fed mice (p = 0.018) (Figure 2E). Further, the β-diversity analysis revealed an obvious separation between control and ethanol diet-fed mice in oral samples collected in both the morning (p = 0.018) (Figure 2F, Left panel) and evening (p = 0.022) (Figure 2F, Right panel). Of note, we have investigated the microbial structure in oral samples collected in the morning and in the evening and found that microbiota composition in samples collected in the morning was not clustered with that in samples collected in the evening in control group (p = 0.422) (Figure 2G, Left panel) and ethanol group (p = 0.372) (Figure 2G, Right panel). Overall, alcohol feeding altered the microbial richness and structure of oral microbiota especially in samples collected in the evening.
Figure 2.
Alcohol consumption affects the microbial diversity of oral microbiota
(A) Schematic of the samples collecting.
(B and C) The α-diversity was showed by Chao 1 index, Shannon index, and Simpson index. Oral samples were collected from mice fed with control and EtOH diet in the morning (B) or in the evening (C).
(D) Comparison of the α-diversity of oral samples collected in the morning or in the evening and the cecum samples from mice fed with control and EtOH diet.
(E) β-diversity in cecum microbiota of Ctrl and EtOH groups.
(F) β-diversity of control group and EtOH group in oral samples collected in the morning (Left) and in the evening (Right).
(G) The β-diversity between samples collected in the morning or in the evening in control (Left) and ethanol-fed mice (Right). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.001.
Alcohol consumption alters the composition of oral microbiota
Since that oral samples collected in the evening showed the most significant differences in microbial diversity in control and ethanol-fed mice, we thus chose these oral samples for further analysis on microbial composition. The relative abundance of oral microbiota in phyla levels was described using bar charts (Figure 3A). We have compared the oral microbial composition between mice fed with control and alcohol diet, and observed that at the phylum level, the relative abundance of Proteobacteria was obviously elevated in ethanol-fed mice compared with control mice, while Firmicutes, Acidobacteriota, Chloroflexi, Myxococcota, and RCP2-54 was significantly decreased after alcohol feeding (Figures 3A–3C). At the level of genus, the bar charts showed the remarkable upregulation in the amount of Janthinobacterium, Psychrobacter, and Paenalcaligenes in the oral samples of ethanol-fed mice. In contrast, we found that the relative abundance of Streptococcus was significantly reduced in the oral cavity of alcohol-consumed mice, and there was also a decreased tendency in the abundance of Pseudomonas, Lactococcus, and Acinetobacter (Figures 3D–3F).
Figure 3.
Alcohol drinking changes the microbial community structures in oral cavity
(A–C) The relative abundance of oral bacterial at phylum level. (A) Top 14 phyla were presented in order of their relative abundance in the bar chart and the rest was assigned as “others”. (B) The relative abundance of Proteobacteria and Firmicutes. (C) The relative abundance of Acidobacteriota, Actinobacteriota, Bacteroidota, Chloroflexi, Verrucomicrobiota, Myxococcota, and RCP2-54 in the oral cavity. Data were presented as mean ± SEM. ∗p < 0.05.
(D–F) The relative abundance of oral microbiota at genus level. (D) Bar chart of the top 14 genera in order of their relative abundance. (E) The relative abundance of top 5 genera with abundance >5%. (F) The relative abundance of other 9 genera. Data were presented as mean ± SEM. ∗p < 0.05.
(G) The cladogram of oral microbiota in different taxonomic levels. Red and green represented the differential taxa from control and ethanol-fed mice, respectively (n = 5 per group) (Wilcoxon rank-sum test, p < 0.05).
(H) Significantly changed bacterial taxa between control and ethanol group of mice (LDA score, >2 or < −2). Data were analyzed by two-tailed Student’s t test in B, C, E, and F.
Consistently, the linear discriminant analysis effect size (LEfSe) analysis of oral microbiota in control and ethanol-fed mice also demonstrated that ethanol intake significantly increased the relative abundance of Campilobacterota and Proteobacteria at the phylum level and dramatically elevated the relative abundance of Helicobacter, Janthinobacterium, Alloprevotella, Paenalcaligenes, and Psychrobacter at the genus level, while genus of Lactococcus and Streptococcus were obviously decreased in the oral cavity following alcohol feeding (Figures 3G and 3H). Altogether, we revealed that alcohol consumption significantly upregulated the abundance of Janthinobacterium, Paenalcaligenes, and Psychrobacter and remarkably reduced the relative abundance of Streptococcus and Lactococcus in oral cavity.
Ethanol diet reprograms the relationships among oral microbiota
Apart from providing information about taxonomic composition of oral microbiota, we further analyzed the relationships among microbiota in mice fed with control or ethanol diet. In the oral cavity of control mice, most microbes displayed positive interactions with each other (Figures 4A and 4B, Left panels), but following alcohol feeding, we observed that a larger proportion of microbiota shifted to behave negatively correlated with other bacteria as evidenced by more blue-color area in the correlation plots (Figures 4A and 4B, Right panels). Specifically, Monoglobus was positively correlated with Lactobacillus, Aerococcus, Sutterellaceae_uncultured, Ileibacterium, Jeotgalicoccus, and Staphylococcus in control mice (Figure 4B, Left panel), while it was only positively correlated with Parabacteroides after alcohol feeding (Figure 4B, Right panel). Acidothermus and Dubosiella were both positively correlated with Candidatus_Xiphinematobacter, Sutterellaceae_uncultured, Bryobacter, and Candidatus_Solibacter, but these correlations were disappeared after alcohol feeding. These results indicated that alcohol drinking disrupted the relationships among oral microbiota. Meanwhile, we found that new correlations appeared following alcohol feeding. For example, Paenarthrobacter almost failed to correlate with other microbiota in the oral cavity of control mice, while Paenarthrobacter was inversely correlated to Candidatus_Solibacter, Parasutterella, and Bacillus following alcohol feeding. Notably, alcohol reprogramed the connections of Psychrobacter with other bacteria, making it become the busiest hub in ethanol-fed mice, while Psychrobacter had no remarkable correlations with other bacteria under the control diet feeding (Figure 4B). Collectively, alcohol consumption remodeled the relationships among microbiota in the oral cavity.
Figure 4.
Ethanol diet feeding reprograms the correlation between oral microbiota
(A) Correlation between oral bacteria under control diet (Left) or ethanol diet (Right) feeding.
(B) The significant relationships between oral microbes (p < 0.05) were selected to be shown.
Effects of alcohol on the function of oral microbiota
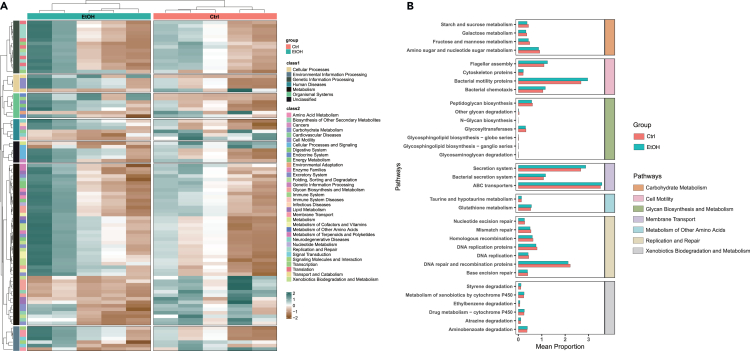
We further predicted the functions of oral microbiota in control and ethanol-fed mice, and the significant altered pathways were demonstrated in Figure 5A. More specifically, among the pathways which were decreased after alcohol feeding, a large part of them were associated with carbohydrate metabolism and glycan biosynthesis and metabolism, including pathways of galactose metabolism, glycosaminoglycan degradation, starch and sucrose metabolism, fructose and mannose metabolism, and so on (Figure 5B, orange and green panels). We also noticed that alcohol feeding decreased the DNA replication and repair pathway in ethanol-fed mice, indicating that alcohol consumption damaged the survival of bacteria (Figure 5B, yellow panel). In contrast, alcohol upregulated the pathways related to alcohol metabolism, such as glutathione metabolism and metabolism by cytochrome P450 (Figure 5B, blue and gray panels). We next revealed that alcohol feeding also increased pathways of microbial secretion system, suggesting that alcohol might promote the communication between oral microbiota and the host cells (Figure 5B, purple panel). Finally, the upregulation of pathways about bacterial motility proteins, bacterial chemotaxis, and flagellar assembly in alcohol-fed mice implied that oral microbiota might have higher capability to translocate from oral cavity to internal tissues following alcohol feeding (Figure 5B, pink panel).
Figure 5.
Effects of alcohol on the functional profile of oral microbiota
(A) The prediction of functional profiles by 16S rRNA sequencing analysis of oral microbiota in control and EtOH group of mice.
(B) The upregulated and downregulated pathways influenced by alcohol consumption.
The relative abundance of oral microbiota reflects the severity of ALD
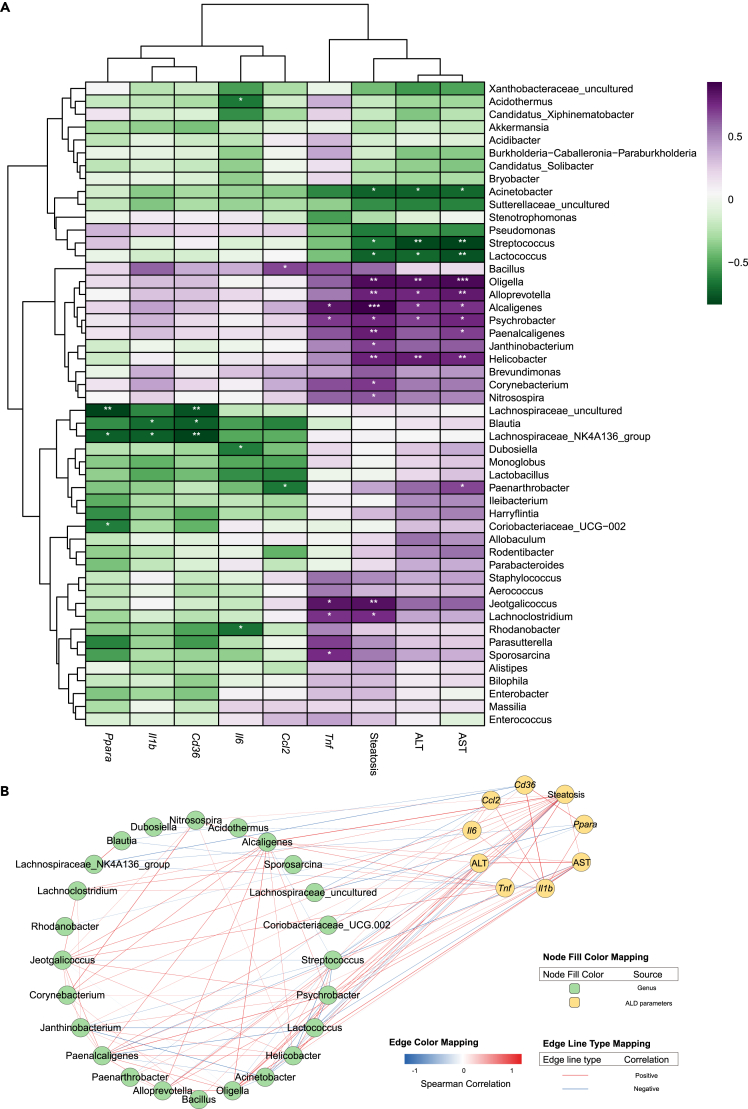
To investigate whether the change of the oral microbiota after alcohol feeding was associated with ALD progression, we analyzed the relationships of the top 50 abundant oral bacteria with ALD symptoms including ALT, AST, steatosis, and hepatic mRNA levels of inflammation and lipid metabolism (Figure 6A). Among them, 24 genera were significantly correlated with these ALD phenotypes; their Spearman correlations were described in Figure 6B. The relative abundance of Helicobacter, Oligella, Alcaligenes, Psychrobacter, and Alloprevotella was positively correlated with ALT, AST, and hepatic steatosis. In contrast, we found that the relative abundance of Streptococcus, Lactococcus, and Acinetobacter showed negative relationships with the hepatic ALT, AST, and steatosis levels (Figures 6A and 6B). Additionally, the abundance of Blautia and Lachnospiraceae_NK4A136_group was negatively correlated with the expression of Il1b in liver tissues. The alteration of Acidothermus, Rhodanobacter, and Dubosiella was revealed to be inversely correlated with hepatic Il6 expression, and the relationship of Paenarthrobacter abundance with Ccl2 mRNA level was negative as well. We also observed that the abundance of Psychrobacter, Jeotgalicoccus, and Alcaligenes had positive relationships with the hepatic Tnf level (Figures 6A and 6B). Finally, we noticed that the relative abundance of Lachnospiraceae_uncultured, Blautia, and Lachnospiraceae_NK4A136_group was dramatically negatively correlated with the expression of fatty acid translocase (Cd36) and Ppara, suggesting that this microbiota might contribute to liver steatosis (Figures 6A and 6B). Collectively, the reprogramming of oral microbiota by alcohol drinking possibly indicated the development of ALD.
Figure 6.
Oral microbiota is associated with ALD progression
(A) Heatmap of Spearman’s correlation among top 50 genera in the oral cavity and several ALD parameters. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(B) Correlation network of the significant (p < 0.05) related nodes. Colors of edge represent Spearman’s correlation coefficients, and colors of nodes for their categories.
Alcohol induces the oral-intestine translocation of microbiota
Bacterial translocation from oral cavity to other internal tissues was one of the major routes for oral microbiota to influence systemic diseases.10,11,12,13 Considering that gut microbiota has been reported to regulate ALD progression,5,6,7 we investigated whether oral bacteria could translocate to the intestine and take part in modulating ALD development. Through comparing of oral microbiota with gut microbiota in control and ethanol-fed mice, we demonstrated that the relative abundance of Sutterellaceae_uncultured, Dyella, and Gemmatimonas was decreased in oral cavity and increased in cecum after ethanol diet feeding (Figure 7A). We further revealed that the relative abundance of Sutterellaceae_uncultured, Dyella, and Gemmatimonas in the oral cavity exhibited negative correlations with alcoholic liver phenotypes including ALT, AST, steatosis, inflammation, and lipid metabolism-related genes, while in the gut, the abundance of these three bacteria was positively correlated with the previously described ALD symptoms (Figure 7B), suggesting that the shift of microbiota from oral cavity to gut might promote the ALD progression. Notably, the elevated relative abundance of Sutterellaceae_uncultured in intestine showed significant positive correlations with the hepatic levels of ALT, AST, and steatosis, as evidenced by the linear regression analysis in Figure 7C. Taken together, we found that oral microbiota possibly translocated along with the oral-gut axis to the intestine, and their enrichment in the intestine might promote the progression of ALD.
Figure 7.
Ethanol consumption induces oral-intestine translocation of oral microbiota
(A) The relative abundance of Sutterellaceae_uncultured, Dyella, and Gemmatimonas in oral and cecum samples of mice fed with control and EtOH liquid diet.
(B) Correlations among abundance of Sutterellaceae_uncultured, Dyella, Gemmatimonas, and ALD symptoms.
(C) Linear regression of the relative abundance of Sutterellaceae_uncultured with ALT, AST, and steatosis level. Data were presented as mean ± SEM. ∗p < 0.05. All data were analyzed follow the procedure of statistical analysis.
A machine learning prospective model predicts ALD development
Based on the oral microbiota which was found to be associated with alcohol drinking, we established a noninvasive risk assessment model to estimate whether mice developed ALD or not. This predictive model was created using 5 increased oral microbiota (Helicobacter, Janthinobacterium, Alloprevotella, Paenalcaligenes, and Psychrobacter) and 2 downregulated oral microbiota (Lactococcus and Streptococcus), as well as Sutterellaceae_uncultured, Dyella, and Gemmatimonas which had possibility to translocate from oral cavity to gut following alcohol drinking (Figure 8A). According to the results of 16S rRNA sequencing of oral microbiota collected in the evening from mice fed with or without alcohol drinking in this study, we calculated that this predictive model achieved a performance with the area under the receiver operating characteristic curve of 0.78 (Figure 8B), indicating that the discriminatory ability of this model was acceptable.15 However, the suitability of this model for clinical usage needs further verification, since that the samples’ number used here to establish the predictive model was very limited.
Figure 8.
Alcohol drinking remodels oral microbiota to modulate ALD progression
(A) Bar plot of feature importance in the random forest model. Blue and pink bar: microbiota with decreased or increased abundance, respectively, in the oral samples collected in the evening following alcohol drinking. Yellow bar: bacteria which might shift along with the oral-gut axis to the intestine.
(B) Performance of the machine learning model discriminating mice fed with control or ethanol diet.
(C) The schematic diagram summarized our findings that alcohol feeding altered the bacterial diversities and compositions of oral microbiota in ethanol-fed mice as compared with control mice. The relationships among oral microbiota were also changed following alcohol feeding. Notably, the alterations of microbiota in the oral cavity were significantly correlated with ALD progression after ethanol intake. We further identified that three genera of bacteria, Sutterellaceae_uncultured, Dyella, and Gemmatimonas, might shift along the oral-gut axis to the intestine to promote the development of ALD.
Discussion
ALD contributes markedly to the global morbidity and mortality of liver diseases.2,16 Given the complexity of ALD pathogenesis, the clinical diagnostic and therapeutic strategies are still limited.6 Increasing evidences have showed the importance of oral microbiota in systemic diseases;17,18 however, the detailed connection of oral microbiota with ALD has not been well described.14,19 Therefore, here we performed 16S rRNA sequencing on the oral microbiota of chronic-plus-binge-induced ALD mice to investigate the changes of oral microbiota forced by alcohol intake. We demonstrated that alcohol feeding altered the microbial diversities and compositions as well as functions of oral microbiota in ethanol-fed mice as compared with control mice. The relationships among oral microbiota were also changed following alcohol feeding. Notably, we observed the significant correlations between the alterations of oral microbiota and ALD progression after ethanol intake. We further identified that three genera of bacteria, Sutterellaceae_uncultured, Dyella, and Gemmatimonas, might shift along with the oral-gut axis to the intestine to promote the development of ALD (Figure 8C). Consistent with our findings, several studies also demonstrated that human participants with the long-term alcohol drinking habit or alcohol dependence had the altered oral microbiota compositions and metabolisms.14,19,20 Although there is no report up to today about the roles of oral microbiota in ALD patients in clinics, our results and the previously mentioned studies indeed shed new light on the potential important functions of oral microbiota in regulating ALD progression in human patients, which merits further investigation.
Since evidences showed that saliva microbiome was influenced by the diurnal rhythm in patient with alcohol dependence,20 we thus collected oral samples from control and ethanol-fed mice at 7:00 a.m. and 4:00 p.m. daily to investigate the differences of oral microbiota in the morning and in the evening. In our study, we observed a slight increase of α-diversity after alcohol feeding in oral samples collected in the morning and significant decrease of microbiota diversity in oral samples collected in the evening, whereas previous report on saliva microbiota in people with alcohol drinking showed an obvious increase in microbial richness at 7:00 a.m. or at 3:00 p.m.20 This discrepancy might be due to the fact that experiment on human samples involved much more variables, including diet, smoking, diseases, and other confounding factors. However, our investigation on mice was able to exhibit a more direct impact of alcohol feeding on oral microbiota. Further, although the effects of alcohol intake on oral microbiota were not obviously different between samples collected in the morning and in the evening according to the analyses about microbial structure, we still found that alcohol had affected much more on oral samples collected in the evening, indicating that the changes of alcohol intake on oral microbiota were indeed influenced by diurnal rhythm in some degree.20
In this study, we observed significant increases in the relative abundance of Janthinobacterium and Helicobacter after alcohol feeding in the oral cavity, while the abundance of Streptococcus was obviously decreased. It was reported that oral Helicobacter may promote periodontal disease, and then induce the release of oral inflammatory cytokines.21 Moreover, the relationship between periodontal disease and liver diseases has also been revealed.9 These might be consistent with our findings that the elevated Helicobacter in the oral cavity was positively correlated with ALD parameters. In addition, Streptococcus is the dominant species of oral cavity, whose abundance may be related to the pH level of oral cavity.22 The abundance of certain acid-nonresistant Streptococcus might be decreased due to the influence of organic acids produced by alcohol metabolism.22 Further, evidences showed that Janthinobacterium played important roles in proteolysis, lipolysis, and saccharolysis,23 while we have no clear clue about the function of Janthinobacterium in the oral cavity, which might need further investigations.
The dysbiosis of oral microbiota could influence the systemic diseases, such as pancreatic cancer, liver cirrhosis, and cardiovascular disease.10,13,18 Pathogenic bacteria and toxin metabolite in oral cavity may transfer to the systemic circulation via damaged oral mucosa.24 Oral microbiota could also translocate to the gut and increase the permeability of the intestinal mucosa to invade to other tissues.24 In this study, we observed that oral microbiota Sutterellaceae_uncultured, Dyella, and Gemmatimonas translocated to the intestine after alcohol feeding. One of the possibilities to explain the promotion of ALD progression by oral microbiota was that these oral microbiota or their metabolites might further shift from gut to liver, since evidence showed that the breach of the intestinal mucosal barrier by alcohol facilitated gut microbiota translocation to liver.7,24 Nevertheless, all these findings indicated that the oral-gut axis might play an important role in the ALD progression.
Currently, liver biopsy is the golden standard of clinical diagnosis of ALD, which is invasive and has the risk of complications and sampling error.25 Developing novel noninvasive diagnostic methods is thus urgently needed. Increasing evidence showed that oral microbiota could be biomarkers for the clinical diagnosis in several diseases, such as hepatocellular carcinoma26 and colorectal carcinogenesis.27 In this study, we established a machine learning model to predict ALD in mice; although the sample numbers were very limited, the model might still find its way into the prediction of ALD in clinical samples in the near future.
Collectively, our study demonstrated that the microbial structure and compositions as well as functions of oral microbiota could be regulated by alcohol drinking. We also discovered that the relationships among microbes in the oral cavity were reprogrammed with alcohol consumption. More importantly, the identified oral microbiota which translocated to the gut could be potential culprits for the progression of ALD. All these findings indicated that oral microbiota might be served as novel noninvasive biomarkers and therapeutic targets for ALD treatment.
Limitations of the study
The first limitation of the current study is that the findings observed in mice were not confirmed in the oral samples collected from the patients with ALD, so it is not sure whether the oral microbiota in humans has similar alteration following alcohol drinking, which deserves further investigation. The second limitation is that the mice numbers used to establish the machine learning model to predict ALD were very small, and the model was not tested in larger cohort, so the predictive power of this model also needs further verification in the future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Raw data | This paper | SRA: PRJNA943134 |
| Critical commercial assays | ||
| ALT kit | Nanjing Jiancheng Bioengineering Institute | Cat#C009-2-1 |
| AST kit | Nanjing Jiancheng Bioengineering Institute | Cat#C010-2-1 |
| TIANamp Stool DNA Kit | TIANGEN | Cat#DP328 |
| Control liquid diet | Bio-Serv | Cat#F1259SP |
| EtOH liquid diet | Bio-Serv | Cat#F1258SP |
| Experimental models: Organisms/strains | ||
| C57BL/6J mice | Beijing Vital River Laboratory Animal Technology Company | N/A |
| Software and algorithms | ||
| Prism 9 | GraphPad | https://www.graphpad.com/ |
| R package | R CRAN | https://www.r-project.org/ |
| USEARCH v10.0.240 | Edgar28 | http://www.drive5.com/usearch |
| VSEARCH v2.13.6 | Rognes et al.29 | https://github.com/torognes/vsearch |
| Cytoscape v. 3.9.1. | Cytoscape Consortium | https://cytoscape.org/ |
Resource availability
Lead contact
Further information and requests for any resources should be directed to and will be fulfilled by the lead contact, Lirui Wang (wanglirui@nju.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Mice
Female C57BL/6J mice were obtained from Beijing Vital River Laboratory Animal Technology Company (Beijing, China). For ALD mice model, 10-12 week-old female mice were fed with control liquid diet for 5 days, followed by 10 days 6% EtOH liquid diet.30 Mice from control group were fed with control liquid diet for the total of 15 days. At the 7:00-9:00 a.m. in the final day, mice were gavage with 31.5% ethanol (5 g/kg) or 45% maltose dextrin and sacrificed after 9 h.30 Oral samples were collected every 7:00 a.m. in the morning and 4:00 p.m. in the evening. All animals were maintained under a specific pathogen-free (SPF) room with a constant temperature (25°C) and 12:12-h light/dark cycle. All animal procedures were approved by the institutional Animal Care and Use Committee of China Pharmaceutical University.
Method details
Plasma ALT and AST measurement
Blood from inferior vena cava was mixed with 0.5 M EDTA and centrifuged at 4°C, 10000 ×g for 5 min to collect plasma. ALT and AST levels were measured according to the manufacturer’s protocols (Nanjing Jiancheng Bioengineering Institute).31,32 Briefly, 20 μL of substrate solution was mixed with 5 μL plasma and incubated at 37°C for 20 min. The mixture then added 20 μL of phenylhydrazine solution and incubated at 37°C for 30 min. Next, 200 μL 0.4 M NaOH was added to each well, and the absorbance at 510 nm was detect after 15 min standing at RT. For control wells, plasma was added after the first step of incubation, and the concentration of ALT and AST were calculated from standard curves.
Histology
The fixing and embedding of liver tissues and the H&E staining with sections were performed according to our previous studies.31,33 Briefly, liver tissues were fixed with formalin, embedded in paraffin and sectioned in 5 μm. Liver sections were deparaffinized by xylene and concentration gradients of ethyl alcohol (100%, 95% and 70%). Hematoxylin and Eosin solution (Servicebio) were used for H&E staining. The score of steatosis showed the percentage of steatosis in liver sections with H&E staining.
Real-time PCR
The detailed procedure of real-time PCR was described in our previous studies.31,33 Total RNA of liver tissues was extracted with RNAiso Plus (Takara), and then dissolved with DEPC-treated water (Sangon Biotech). HiScript III 1st Strand cDNA Synthesis Kit (Vazyme) was used to obtain cDNA. Real-time PCR was performed using ChamQ SYBR qPCR Master Mix (Vazyme) for 40 cycles of 10 s at 95°C and 30 s at 60°C on an ABI StepOnePlus instrument. Primer sequences for each gene were shown in the Table S1. Relative expressions of target genes were calculated using ΔΔCT and normalized to 18S as internal controls.
Genomic DNA extraction
The oral samples were collected using oral swabs by rubbing the oral cavity of mice for three times clockwise and scraping the inside and outside of the mice teeth. The oral swabs after sampling were rinsed in 500 μL Buffer SA (TIANamp Stool DNA Kit) for 50 times. The genomic DNA from oral samples and cecum samples were extracted by TIANamp Stool DNA Kit (DP328; TIANGEN). All the procedures were followed by the manufacturer’s protocols.
16S rRNA sequencing and analysis
The isolated genomic DNA from mice oral cavity and cecum were used for 16S rRNA sequencing and analysis. The 16S ribosomal RNA (rRNA) V4 region was amplified and sequenced by the Illumina NovaSeq platform (Illumina, San Diego, CA) in Novogene Technology Co, Ltd (Beijing, China) to evaluate microbiota diversity. The analysis of raw sequencing data were described as before.31,34 Briefly, the sequences were denoised in amplicon sequence variants (ASV) by UNOISE3 based on USEARCH28 v10.0.240 (http://www.drive5.com/usearch) and VSEARCH29 v2.13.6 (https://github.com/torognes/vsearch). The sequences were then aligned to SILVA138 database in order to calculate taxonomy table. α-diversity and β-diversity were calculated with USEARCH (https://drive5.com/usearch/manual/pipe_diversity.html) and online tools (https://www.microbiomeanalyst.ca/). The R package ‘edgeR’35 was utilized to find differential genus. Then LEfSe was performed online (http://huttenhower.sph.harvard.edu/galaxy) and the R package edgeR was obtained for identifying differential taxons.35,36 The correlation analyses between microbiota and ALD phenotypes were performed using corr.test() function of psych package v.2.2.9 in R and visualized via corrplot and pheatmap package v.1.0.12. The significant correlations between oral microbiota and disease parameters were visualized by cytoscape v. 3.9.1. The raw data have been deposited in the National Center for Biotechnology Information Sequence Read Archive database (accession no. PRJNA943134).
Function prediction and data analysis
The ASV sequences analyzed by USEARCH using relevant procedures described in 16S rRNA sequencing and analysis, were further annotated with Greengenes database: https://greengenes.secondgenome.com. v.13_5.37 Then the annotation results were adopted to predict metagenomic functions via PICRUST,38 a suite implanted on a Galaxy website (http://huttenhower.sph.harvard.edu/galaxy/). KEGG pathways abundance of specific samples was extracted. R packages ‘edgeR’35 and ‘limma’39 were used to filter low-abundance features and find the differential ones with a threshold of adj.P.Val <0.05, which is a p value adjusted by Benjamini – Hochberg method.40 After scaling by row, the scaled matrix was obtained for plotting heatmap via R package ‘ComplexHeatmap’ v. 2.15.1.41,42 The visualization was used via ggplot2 v.3.4.1.
Establishment of machine learning model
The establishment of machine learning prospective model was described as before.43 The machine learning model was built using 10 microbial features. The randomForest package in R was used for model construction. A 10-fold cross-validation was utilized to evaluate the model’s performance. The pROC package was employed to generate the ROC curve, which assessed the discriminatory ability of this model.
Quantification and statistical analysis
Statistical analysis
In this study, data were presented as mean ± SEM and graphed using GraphPad PRISM 9. Two-tailed Student’s t test and Mann-Whitney test were used for statistical analysis of the two-group comparisons; PerMANOVA test was used for β-diversity analysis. For the relative abundance of differential bacteria in control and ethanol group at the genus level, Wilcoxon rank sum tests and Paired samples t-test were utilized depending on whether the abundance of bacteria apply to normal distribution. ∗p value < 0.05, ∗∗p value < 0.01, ∗∗∗p value < 0.001, ∗∗∗∗p value < 0.001 were considered statistically significant.
Acknowledgments
We thank Daolong Chen and Qianyu Zhang in our lab for assistance in sample collection. This study was supported by the “Double First-Class” University Project (CPU2018GF10 and CPU2018GY31 to L.W.).
Author contributions
C.P. was responsible for experiments design, data acquisition and analysis, and manuscript writing; C.L. was responsible for data analysis; W.J. was responsible for performing experiments; D.Z. was responsible for data analysis in Figure 5; X.C. and X.Z. assisted in performing experiments; M.Y. assisted in data analysis in Figure 2D; L.W. was responsible for the study concept and design, study supervision, data analysis, and manuscript writing.
Declaration of interests
The authors report no conflict of interest.
Published: September 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107977.
Supplemental information
Data and code availability
-
•
16S rRNA sequencing data have been deposited at National Center for Biotechnology Information Sequence Read Archive database and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This article contains no original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Simon L., Souza-Smith F.M., Molina P.E. Alcohol-Associated Tissue Injury: Current Views on Pathophysiological Mechanisms. Annu. Rev. Physiol. 2022;84:87–112. doi: 10.1146/annurev-physiol-060821-014008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seitz H.K., Bataller R., Cortez-Pinto H., Gao B., Gual A., Lackner C., Mathurin P., Mueller S., Szabo G., Tsukamoto H. Alcoholic liver disease. Nat. Rev. Dis. Primers. 2018;4:16. doi: 10.1038/s41572-018-0014-7. [DOI] [PubMed] [Google Scholar]
- 3.Avila M.A., Dufour J.F., Gerbes A.L., Zoulim F., Bataller R., Burra P., Cortez-Pinto H., Gao B., Gilmore I., Mathurin P., et al. Recent advances in alcohol-related liver disease (ALD): summary of a Gut round table meeting. Gut. 2020;69:764–780. doi: 10.1136/gutjnl-2019-319720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singal A.K., Bataller R., Ahn J., Kamath P.S., Shah V.H. ACG Clinical Guideline: Alcoholic Liver Disease. Am. J. Gastroenterol. 2018;113:175–194. doi: 10.1038/ajg.2017.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang R., Tang R., Li B., Ma X., Schnabl B., Tilg H. Gut microbiome, liver immunology, and liver diseases. Cell. Mol. Immunol. 2021;18:4–17. doi: 10.1038/s41423-020-00592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bajaj J.S. Alcohol, liver disease and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019;16:235–246. doi: 10.1038/s41575-018-0099-1. [DOI] [PubMed] [Google Scholar]
- 7.Wang L., Fouts D.E., Stärkel P., Hartmann P., Chen P., Llorente C., DePew J., Moncera K., Ho S.B., Brenner D.A., et al. Intestinal REG3 Lectins Protect against Alcoholic Steatohepatitis by Reducing Mucosa-Associated Microbiota and Preventing Bacterial Translocation. Cell Host Microbe. 2016;19:227–239. doi: 10.1016/j.chom.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters B.A., Wu J., Hayes R.B., Ahn J. The oral fungal mycobiome: characteristics and relation to periodontitis in a pilot study. BMC Microbiol. 2017;17:157. doi: 10.1186/s12866-017-1064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acharya C., Sahingur S.E., Bajaj J.S. Microbiota, cirrhosis, and the emerging oral-gut-liver axis. JCI insight. 2017;2:e94416. doi: 10.1172/jci.insight.94416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohammed H., Varoni E.M., Cochis A., Cordaro M., Gallenzi P., Patini R., Staderini E., Lajolo C., Rimondini L., Rocchetti V. Oral Dysbiosis in Pancreatic Cancer and Liver Cirrhosis: A Review of the Literature. Biomedicines. 2018;6:115. doi: 10.3390/biomedicines6040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D., Xi W., Zhang Z., Ren L., Deng C., Chen J., Sun C., Zhang N., Xu J. Oral microbial community analysis of the patients in the progression of liver cancer. Microb. Pathog. 2020;149:104479. doi: 10.1016/j.micpath.2020.104479. [DOI] [PubMed] [Google Scholar]
- 12.Read E., Curtis M.A., Neves J.F. The role of oral bacteria in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2021;18:731–742. doi: 10.1038/s41575-021-00488-4. [DOI] [PubMed] [Google Scholar]
- 13.Koren O., Spor A., Felin J., Fåk F., Stombaugh J., Tremaroli V., Behre C.J., Knight R., Fagerberg B., Ley R.E., Bäckhed F. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA. 2011;108:4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan X., Peters B.A., Jacobs E.J., Gapstur S.M., Purdue M.P., Freedman N.D., Alekseyenko A.V., Wu J., Yang L., Pei Z., et al. Drinking alcohol is associated with variation in the human oral microbiome in a large study of American adults. Microbiome. 2018;6:59. doi: 10.1186/s40168-018-0448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandrekar J.N. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol. 2010;5:1315–1316. doi: 10.1097/JTO.0b013e3181ec173d. [DOI] [PubMed] [Google Scholar]
- 16.Dang K., Hirode G., Singal A.K., Sundaram V., Wong R.J. Alcoholic Liver Disease Epidemiology in the United States: A Retrospective Analysis of 3 US Databases. Am. J. Gastroenterol. 2020;115:96–104. doi: 10.14309/ajg.0000000000000380. [DOI] [PubMed] [Google Scholar]
- 17.Dong J., Li Y., Xiao H., Zhang S., Wang B., Wang H., Li Y., Fan S., Cui M. Oral microbiota affects the efficacy and prognosis of radiotherapy for colorectal cancer in mouse models. Cell Rep. 2021;37:109886. doi: 10.1016/j.celrep.2021.109886. [DOI] [PubMed] [Google Scholar]
- 18.Herremans K.M., Riner A.N., Cameron M.E., McKinley K.L., Triplett E.W., Hughes S.J., Trevino J.G. The oral microbiome, pancreatic cancer and human diversity in the age of precision medicine. Microbiome. 2022;10:93. doi: 10.1186/s40168-022-01262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao Y., Tong X.T., Jia Y.J., Liu Q.Y., Wu Y.X., Xue W.Q., He Y.Q., Wang T.M., Zheng X.H., Zheng M.Q., Jia W.H. The Effects of Alcohol Drinking on Oral Microbiota in the Chinese Population. Int. J. Environ. Res. Public Health. 2022;19:5729. doi: 10.3390/ijerph19095729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Zhao K., Chen J., Ni Z., Yu Z., Hu L., Qin Y., Zhao J., Peng W., Lu L., et al. Diurnal changes of the oral microbiome in patients with alcohol dependence. Front. Cell. Infect. Microbiol. 2022;12:1068908. doi: 10.3389/fcimb.2022.1068908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L., Chen X., Ren B., Zhou X., Cheng L. Helicobacter pylori in the Oral Cavity: Current Evidence and Potential Survival Strategies. Int. J. Mol. Sci. 2022;23:13646. doi: 10.3390/ijms232113646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abranches J., Zeng L., Kajfasz J.K., Palmer S.R., Chakraborty B., Wen Z.T., Richards V.P., Brady L.J., Lemos J.A. Biology of Oral Streptococci. Microbiol. Spectr. 2018;6 doi: 10.1128/microbiolspec.GPP3-0042-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chernogor L., Bakhvalova K., Belikova A., Belikov S. Isolation and Properties of the Bacterial Strain Janthinobacterium sp. Microorganisms. 2022;10:1071. doi: 10.3390/microorganisms10051071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng X., Cheng L., You Y., Tang C., Ren B., Li Y., Xu X., Zhou X. Oral microbiota in human systematic diseases. Int. J. Oral Sci. 2022;14:14. doi: 10.1038/s41368-022-00163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bedossa P., Dargère D., Paradis V. Sampling variability of liver fibrosis in chronic hepatitis C. Hepatology. 2003;38:1449–1457. doi: 10.1016/j.hep.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 26.Rao B.C., Lou J.M., Wang W.J., Li A., Cui G.Y., Yu Z.J., Ren Z.G. Human microbiome is a diagnostic biomarker in hepatocellular carcinoma. Hepatobiliary Pancreat. Dis. Int. 2020;19:109–115. doi: 10.1016/j.hbpd.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Zhang S., Kong C., Yang Y., Cai S., Li X., Cai G., Ma Y. Human oral microbiome dysbiosis as a novel non-invasive biomarker in detection of colorectal cancer. Theranostics. 2020;10:11595–11606. doi: 10.7150/thno.49515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 29.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertola A., Mathews S., Ki S.H., Wang H., Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat. Protoc. 2013;8:627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian M., Liu J., Zhao D., Cai P., Pan C., Jia W., Gao Y., Zhang Y., Zhang N., Zhang Y., et al. Aryl Hydrocarbon Receptor Deficiency in Intestinal Epithelial Cells Aggravates Alcohol-Related Liver Disease. Cell. Mol. Gastroenterol. Hepatol. 2022;13:233–256. doi: 10.1016/j.jcmgh.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian M., Hu H., Yao Y., Zhao D., Wang S., Pan C., Duan X., Gao Y., Liu J., Zhang Y., et al. Coordinated changes of gut microbiome and lipidome differentiates nonalcoholic steatohepatitis (NASH) from isolated steatosis. Liver Int. 2020;40:622–637. doi: 10.1111/liv.14316. [DOI] [PubMed] [Google Scholar]
- 33.Wang L., Mazagova M., Pan C., Yang S., Brandl K., Liu J., Reilly S.M., Wang Y., Miao Z., Loomba R., et al. YIPF6 controls sorting of FGF21 into COPII vesicles and promotes obesity. Proc. Natl. Acad. Sci. USA. 2019;116:15184–15193. doi: 10.1073/pnas.1904360116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y., Zhao D., Qian M., Liu J., Pan C., Zhang X., Duan X., Zhang Y., Jia W., Wang L. Amlodipine, an anti-hypertensive drug, alleviates non-alcoholic fatty liver disease by modulating gut microbiota. Br. J. Pharmacol. 2022;179:2054–2077. doi: 10.1111/bph.15768. [DOI] [PubMed] [Google Scholar]
- 35.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langille M.G.I., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Clemente J.C., Burkepile D.E., Vega Thurber R.L., Knight R., et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 41.Gu Z. Complex heatmap visualization. iMeta. 2022;1 doi: 10.1002/imt2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 43.Leung H., Long X., Ni Y., Qian L., Nychas E., Siliceo S.L., Pohl D., Hanhineva K., Liu Y., Xu A., et al. Risk assessment with gut microbiome and metabolite markers in NAFLD development. Sci. Transl. Med. 2022;14:eabk0855. doi: 10.1126/scitranslmed.abk0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
16S rRNA sequencing data have been deposited at National Center for Biotechnology Information Sequence Read Archive database and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This article contains no original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.