Abstract
The lack of physiologically relevant in vitro models has hampered progress in understanding human lung development and disease. Here, we describe a protocol in which human induced pluripotent stem cells (hiPSCs) undergo stepwise differentiation into definitive endoderm (>88% population) to three-dimensional (3D) lung organoids (LORGs), which contain both epithelial and mesenchymal cellular architecture and display proximal and distal airway patterning. These LORGs can maintained for more than 90 days by re-embedding in the Matrigel. We show the utility of LORGs for disease modeling and drug screening by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and treatment with antiviral drugs.
Keywords: Lung organoids, Human iPS cells, SARS infection, Lung development
Graphical abstract

Highlights
-
•
hiPSCs are first differentiated into a definitive endoderm (DE) germ layer (>88% population) by exposure to activin A, a member of the transforming growth factor β superfamily, and CHIR99021, a small molecule inhibitor of glycogen synthase kinase-3β (GSK3β) and activator of Wnt signaling.
-
•
In the next step, exposure of the DE layer to the bone morphogenic protein (BMP) inhibitor Noggin, the TGFβ inhibitor SB431542, fibroblast growth factor 4 (FGF4), and the agonist of the Sonic hedgehog pathway Smoothened agonist (SAG), leads to differentiation of DE to anterior foregut endoderm (AFE).
-
•
AFE cells are then induced to differentiate into lung progenitor cells that form aggregates (LPAs) by addition of CHIR99021, BMP4, and all-trans-retinoic acid (ATRA).
-
•
To promote LORG formation, LPAs are embedded in Matrigel and exposed to CHIR99021, ATRA, FGF10, FGF7, epidermal growth factor (EGF), vascular endothelial growth factor (VEGF), placental growth factor (PlGF), cyclic adenosine monophosphate (cAMP), dexamethasone (DEX), and 3-isobutyl-1-methylxanthine (IBMX), which results in the formation of branched 3D LORGs that express proximal and distal airway markers.
-
•
We show that the LORGs express ACE2/TMPRSS2, the cellular receptors for SARS-CoV-2, and that infection with live virus can be suppressed by treatment with known antiviral compounds, thus illustrating the potential utility of LORGs as a model system for investigation of respiratory diseases and drug screening.
1. Maintenance and passaging of hiPSCs
1.1. Timing: 45–90 min
For complete details on the use and execution of this protocol, please refer to Ref. [1].
Proper maintenance of hiPSCs in culture is essential to the success of the LORG differentiation protocol. In our hands, the optimal conditions for the maintenance of undifferentiated hiPSCs are culture in mTeSR1 medium containing Y-27632 (10 μM) in 6-well tissue culture plates coated with growth factor-reduced Matrigel basement membrane (GFR-M, 100 μg/mL, Cat No. 354230) at 37 °C in a humidified 5% CO2 incubator. Y-27632, a Rho-associated kinase (ROCK) inhibitor, is added to the medium to enhance the recovery of iPSC cells from cryopreserved stocks and increases the number of colonies and colony size. For propagation, hiPSCs from the 6-well plates, 70–80% confluent, are then split 1:6 into fresh 6-well or 24-well plates as described here.
CRITICAL: Too low confluency may result in an insufficient cell yield after DE differentiation, while too high confluency may result in incomplete differentiation into DE.
-
1.
Thaw the GFR-M stock on ice until it completely melts and dilute it to 100 μg/mL in cold DMEM/F12 medium and add 1mL/well of diluted Matrigel to a 6-well plate. Leave at 37 °C for 1 h to coat the wells.
CRITICAL: Matrigel solidifies at room temperature. Therefore, keep small aliquots of the stock at −20 °C and thaw on ice immediately before use.
-
2.
Prewarm DMEM/F12, mTeSR1, and either dispase (1 mg/mL) or Versene (1 mg/mL) in a 37 °C water bath for ∼15 min.
-
3.
Aspirate the culture medium from the hiPSC cultures, add 1 mL dispase or Versene solution to each well, and incubate for 5–10 min in a 37 °C incubator. Observe the cells under a light microscope to check for evidence that the colonies are beginning to detach from the tissue culture plate.
-
4.
Carefully aspirate the dispase/Versene reagent and gently wash (2–3 times) the still-attached colonies with prewarmed DMEM/F12.
-
5.
Add Y-27632 to the prewarmed mTeSR1 medium at a final concentration of 10 μM. Prepare tubes containing 4.5 mL of this medium (one tube per well). Using a cell scraper, gently scrape the hiPSCs from the culture plate and then transfer cells with remaining culture medium into tubes with a pipette and triturate the colonies into small aggregates using a 5 mL serological pipette.
-
6.
Take the 6- or 24-well plates containing GFR-M (step 1), aspirate the GFR-M, and add 0.5 mL/well of the triturated hiPSCs to 6-well or 24-well plates. For the 6-well plates only, add an additional 1.5 mL/well of the prewarmed mTeSR1/Y-27632 medium.
-
7.
Gently shake the plates in a side-to-side motion to evenly distribute the hiPSCs across the wells and place in a 37 °C 5% CO2 incubator.
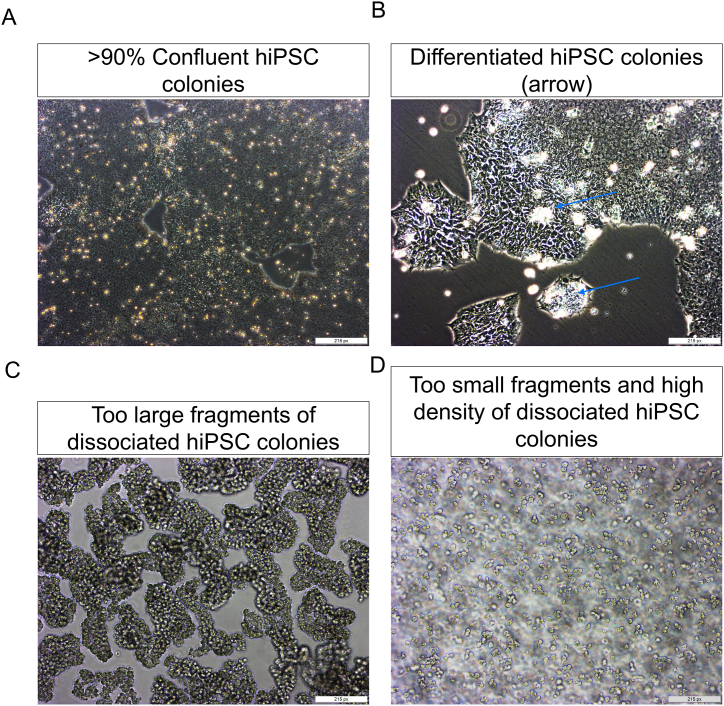
CRITICAL: hiPSCs must be evenly distributed across the well to ensure optimal colony growth before dissociation and seeding into differentiation cultures (see Troubleshooting Problem 1, Fig. 1A and B).
Key resources table
| REAGENT | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-FOXJ1 | eBioscience | 14-9965-82 |
| Mouse anti-surfactant protein B (SFTPB) | Seven Hills Bioreagents | Wmab-1B9 |
| Rabbit anti-prosurfactant protein C (proSP-C) | Millipore | AB3786 |
| Rabbit anti-RAGE | Abcam | ab216329 |
| Mouse anti-Hop (E−1) | Santa Cruz | Sc-398703 |
| Rabbit anti-NKX2.1 | Abcam | ab76013 |
| Rabbit anti-SOX9 | Millipore | AB5535 |
| Rabbit anti-ECAD | Cell Signaling Technology | 3195 |
| Mouse anti-CC10 (SCGB1A1) | Santa Cruz | sc-365992 |
| Rabbit Anti-ACE2 | Novus Biologicals | NBP2-67692 |
| Rabbit anti-TMPRSS2 | Abcam | ab92323 |
| Rabbit anti-Ki67 | Millipore | AB9260 |
| Mouse anti-GFAP | Sigma-Aldrich | G3893 |
| Rabbit anti-SOX2 | Abcam | ab97959 |
| Mouse anti-β-tubulin III | Abcam | ab7751-100 |
| Chicken anti-Nestin | Abcam | ab134017 |
| Chicken anti-MAP2 | Abcam | ab5392 |
| Alexa Fluor 488, 594, and 647 conjugated secondary antibodies | Molecular Probes/Invitrogen | A11032, A11034, A11012, A11039, A11001 |
| Antifade mounting medium with DAPI (Vectashield) | Vector Laboratories | H-1200 |
| Chemicals | ||
| 1-Thioglycerol | Sigma-Aldrich | M6145 |
| l-Ascorbic acid | Tocris Bioscience | 4055 |
| Bovine serum albumin (BSA) | Fisher Scientific | BP9703–100 |
| Protease and phosphatase inhibitor cocktail | Pierce | A32955 |
| Bio-Rad DC protein assay kit | Bio-Rad | 5000112 |
| Chemiluminescence substrate | Thermo Scientific | 34580 |
| Optimal cutting temperature (OCT) compound | Tissue-Tech | 4583 |
| RIPA buffer | Teknova | R3792 |
| Falcon™ Cell Culture Inserts | BD, Falcon | 08–770 |
| β-Mercaptoethanol | Sigma-Aldrich | M7522 |
| Immun-Blot PVDF membrane | Bio-Rad Laboratories | 1620177 |
| Growth Factors | ||
| Activin A | R&D Systems | 338-AC |
| FGF10 (recombinant human fibroblast growth factor 10) | R&D Systems | 345-FG |
| FGF4 (recombinant human fibroblast growth factor 4) | R&D Systems | 7460-F4 |
| FGF7 (recombinant human fibroblast growth factor 7) | R&D Systems | 251-KG/CF |
| NOGGIN | R&D Systems | 6057 |
| Smoothened agonist (SAG) | Enzo Life Sciences | ALX-270-426-M001 |
| Epidermal growth factor (EGF) | Sigma-Aldrich | E9644 |
| Basic fibroblast growth factor (bFGF) | Sigma-Aldrich | F0291 |
| Small molecules | ||
| SB431542 (selective inhibitor of the transforming growth factor-β (TGF-β) type I receptor) | TOCRIS | 1614 |
| CHIR9902 (GSK3β inhibitor/Wnt activator) | TOCRIS | 4423 |
| Y-27632 (ROCK inhibitor) | Sigma-Aldrich | SCM075 |
| Cell Lines | ||
| Human induced pluripotent stem cells (hiPSCs) | Cell Application | hiPS11-10 |
| 293FT (HEK) | American Type Culture Collection (ATCC) | CRL-1573 |
| Calu-3 human lung epithelial cell | ATCC | HTB-55 |
| Culture Media | ||
| Advanced DMEM | Thermo Fisher Scientific | 12491015 |
| CHIR99021 | Stem cell Technologies | 72054 |
| GlutaMAX (100X) | Thermo Fisher Scientific | 35050061 |
| HEPES (1 M) | Thermo Fisher Scientific | 15630080 |
| mTeSR | Stem cell Technologies | 85850 |
| N-2 supplement | Thermo Fisher Scientific | 17502048 |
| Penicillin-streptomycin (100X) | Thermo Fisher Scientific | 15140122 |
| RPMI 1640 medium | Thermo Fisher Scientific | 11875119 |
| Matrigel basement membrane matrix (growth factor reduced) | Corning | 354230 |
| Matrigel basement membrane matrix | Corning | 354234 |
| Versene solution | Thermo Fisher Scientific | 15040066 |
| DMEM/F12 | Thermo Fisher Scientific | 11320082 |
| MEM nonessential amino acids (MEM-NEAA) | Thermo Fisher Scientific | 11140050 |
| B-27 supplement without ATRA | Thermo Fisher Scientific | 12587010 |
| B-27 supplement with ATRA | Thermo Fisher Scientific | 17504044 |
| GlutaMAX, 200 mM | Thermo Fisher Scientific | 35050061 |
| l-Glutamine | Thermo Fisher Scientific | A2916801 |
| Dispase | Thermo Fisher Scientific | 17105-0412q |
| Collagenase IV | Thermo Fisher Scientific | 17104019 |
| Fetal bovine serum (FBS) | Thermo Fisher Scientific | 10438–018 |
| Bovine serum albumin (BSA) | Fisher Scientific | BP9703-100 |
Fig. 1.
hiPSC culture and seeding for differentiation.
(A) Representative phase contrast image of a hiPSC culture showing healthy colonies with optimal confluency before seeding. Scale bar, 200 μm. (B) Representative light microscopic image showing the optimal size of dissociated hiPSC colonies for seeding and differentiation. Scale bar, 200 μm. Inset magnification = 50 μm.
1.2. Materials and equipment
.
2. Step-by-step method details
Note: All procedures for differentiation of hiPSCs into DE, AFE, and 3D LORGs are adapted from Refs. [2,3].
3. DE induction (Days 1–4)
3.1. Timing: 4 days
In the first step, hiPSCs are induced to differentiate into DE. Physiologically, this germ layer is formed during gastrulation and gives rise to the epithelial lining of the respiratory tract and lungs and also to the digestive tract, thymus, pancreas, and liver [4].
-
1.
The protocol employs hiPSCs at about 1–2 days after splitting as described above. The cells should be at 60%–80% confluency (see Troubleshooting Problem 2, Fig. 2A–D)
-
2.
On Day 1, aspirate the mTeSR1/Y-27632 growth medium and add 2 mL or 0.5 mL per well (6- or 24-well plates) of DE differentiation medium (Day 1 medium, Table 1), supplemented with human activin A (100 ng/mL) and the GSK3β inhibitor/Wnt activator CHIR99021 (5 μM) to initiate DE differentiation.
Fig. 2.
Critical steps and troubleshooting.
(A, B) Representative images of hiPSCs that (A) are at supra optimal confluency (>90%) for dissociation before seeding and (B) have already formed colonies before seeding (blue arrows). Remove differentiated colonies carefully by scraping. Scale bars, 200 μm. (C, D) Representative images of hiPSC colonies that are too large (C) and too small and dense (D) after dissociation, both of which are suboptimal for seeding and differentiation. Scale bars, 200 μm.
Table 1.
Definitive endoderm differentiation medium.
| DAY | MEDIUM/SUPPLEMENTS (For 24- and 12-well plate use 0.5 and 1 mL medium/well, respectively, for differentiation) |
|---|---|
| 1 | RPMI 1640 + 0% FBS + activin A (100 ng/mL) + CHIR9902, (5 μM) + Y27632 (10 μM) |
| 2 | RPMI 1640 + 0.2% FBS + activin A (100 ng/mL) |
| 3–4 | RPMI 1640 + 2% FBS + activin A (100 ng/mL) |
Note: When two volumes are stated, they apply to 6-well and 24-well plates..
-
3.
On Day 2, 3, and 4, replace the medium with 2 mL or 0.5 mL of the appropriate DE differentiation medium (Day 2, Day 3, Day 4 medium, Table 1), and observe the morphology of the cells. Representative images are shown in Fig. 3A.
Fig. 3.
Differentiation of hiPSCs into DE, AFE, and LPAs.
(A, B) Representative light microscopic images showing the typical morphology of hiPSCs upon differentiation to (A) DE and (B) to AFE (Days 5–8) and LPAs (Days 9–12). Scale bars, 200 μm. (C) Representative microscopic images showing the lung progenitor aggregate (Day 12) formation. (Scale bar, 200 μm left image and 50 μm right image) (D) qRT-PCR analysis of expression of genes associated with DE (SOX17, CXCR4), AFE (FOXA1, FOXA2), and LPA (NKX2.1) in differentiating hiPSCs collected on Days 4, 8, and 12, respectively and also collected undifferentiated hiPSC for qRT-PCR analysis. Data are presented as the mean ± SEM (n = 3) and are presented as the fold change in mRNA relative to that in hiPSCs. (E) Immunofluorescence staining of LPA-associated proteins (NKX2.1, green, SOX2, red) on Day 12 of differentiation. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, blue). Scale bars, 100 μm.
Note: CHIR99021 should be removed within 24 h of initiating differentiation on Day 1.
-
4.
On Day 5, collect cell sample, extract RNA, and confirm differentiation to DE by qRT-PCR of the DE markers CXCR4, FOXA2, and SOX17 (Fig. 3D). In addition, we also extracted RNA from undifferentiated hiPSC cells to compare the genes for DE markers. Furthermore, DE formation was confirmed by flow cytometry showing >88% population (Fig. S1B).
4. AFE and LPA induction (Days 5–12)
4.1. Timing: 8 days
AFE gives rise to respiratory tissues such as the trachea and lungs [5]. Previous work established that concomitant inhibition of BMP with Noggin and of TGFβ signaling with SB431542 results in the differentiation of a highly enriched AFE population from DE [6]. At the same time, the growth factor FGF4 and the smoothened agonist (SAG) are added to the culture for differentiation into AFE..
-
5.
On Days 5, 6, 7, and 8, replace the DE differentiation medium with AFE differentiation medium (basal medium [Table 2] supplemented with HEPES buffer 1%, SB431542 10 μM, Noggin 200 ng/mL, SAG 1 μM, and FGF4 500 ng/mL).
-
6.
Check the cell morphology daily. Representative images are shown in Fig. 3B.
-
7.
On Day 8, collect a cell sample for RNA and immunofluorescence analysis to confirm AFE differentiation by qRT-PCR for expression of FOXA1, FOXA2, and NKX2.1 genes or immunofluorescence microscopy for NKX2.1, showing increase in expression during the transition from DE to AFE (Fig. 3D and E).
-
8.
We also analyzed the expression of posterior markers (CDX2.1, PAX1 and NKX2.5) as a negative control in the same sample used in step 7 and their expression is significantly low as compared to anterior markers (Fig. S1A).
-
9.
On Days 9, 10, 11, and 12, replace the AFE differentiation medium with LPA medium (AFE differentiation medium supplemented with BMP4 - 10 ng/mL, CHIR99021 - 3 μM, and ATRA - 0.1 μM). CHIR99021 activates Wnt-signaling by binding secreted Wnt-protein to its receptor and extremely potent glycogen synthase kinase (GSK) 3 inhibitor, inhibiting both GSK3β and GSK3α. Retinoic acid signaling play critical role in balancing lung epithelial progenitor cell growth and differentiation [7](Ng-Blichfeldt et al., 2018). Prior studies provide evidence that Wnt + BMP signaling (in the presence of RA) is necessary to specify lung progenitors from AFE, whereas FGF and BMP signaling promotes specification to thyroid [[8], [9], [10]], thus we added the CHIR99021, BMP4 and ATRA to specify the AFE into lung progenitors.
-
10.
Monitor the cell morphology daily between Days 8 and 12. Representative images are shown in Fig. 3B and C.
-
11.
On Day 12, collect a cell sample for RNA extraction and immunofluorescence analysis and confirm LPA formation by qRT-PCR and immunofluorescence microscopy analysis of expression of SOX2 and NKX2.1, which are highly expressed in LPAs (Fig. 3C–E and S1A).
Table 2.
Basal medium.
| REAGENT | FINAL CONCENTRATION | AMOUNT for 500 mL |
|---|---|---|
| Iscove's Modified Dulbecco's Medium (IMDM) | n/a | 375 mL |
| Ham's F12 | n/a | 125 mL |
| N2 supplement | 1X | 5 mL |
| B27 supplement (without ATRA) | 1X | 10 mL |
| l-Glutamine | 2 mM | 5 mL |
| Ascorbic acid | 50 μg/mL | 500 μL |
| Monothioglycerol | 0.4 μM | 500 μL |
| BSA (10% stock) | 0.05% | 2.5 mL |
| Penicillin-streptomycin (5000 U/mL) | 1X | 5 mL |
n/a, not applicable.
5. Formation of 3D LORGs and re-embedding in matrigel (Day 12–60)
5.1. Timing: ∼48 days
In the next steps of the protocol, LPAs are mixed with Matrigel (Cat No 354234) and allowed to form 3D LORGs, consisting of an airway-like structure with distal and proximal patterning of mesenchymal and alveolar epithelial cells.
-
12.
Thaw GFR-M on ice.
-
13.
Cut the tip off of a P1000 pipette filter tip and gently triturate the LPAs from the Day 12 plates.
-
14.
Transfer the LPA suspension to from each well into separate 1.5 mL tubes and allow to stand at room temperature for 5–10 min for the aggregates to settle. Alternatively, the tubes can be gently centrifuged at 200RPM for about 10 s.
CRITICAL: Handle the cell aggregates gently to avoid disrupting and forming a single cell suspension.
-
15.
Remove the medium from the tubes without disturbing the LPA pellet, add 200 μL of cold GFR-M avoiding bubbles, and transfer the LPA/Matrigel mixture to Transwell membrane inserts. Alternatively, pipette ∼25 μL aliquots of the LPA/GFR-M mixture onto squares of sterile parafilm. Place the Transwell inserts or parafilm squares in a 37 °C incubator for 20–30 min to allow the LPA-containing GFR-M to solidify.
-
16.
Once Matrigel embedded cells solidify, add the fresh LPA medium in lower chamber of trans well insert or carefully add medium on sterile film to drop the Matrigel aggregate into 24-well plate which float in the medium.
6. CRITICAL: The LPA/GFR-M mixture should contain no more than 10–15 aggregates per 200 μL of GFR-M
6.1. CRITICAL: Keep the matrigel cool at all steps preceding the 37 °C incubator to avoid solidification
-
17.
On Days 13 and 14, replace the medium with fresh LPA medium/Matrigel.
-
18.
On Day 15, add 1 mL of LORG culture medium (consisting of basal medium, Table 2 supplemented with FGF10 100 ng/mL, FGF7 10 ng/mL, CHIR99021 3 μM, EGF 10 ng/mL, and Y-27632 10 μM) to the lower chamber of the Transwell and replace the insert. Change the medium every other day from Day 15 to Day 23. If using parafilm based method, then only carefully remove the medium from side of well without disturbing Matrigel aggregate and add fresh medium.
-
19.
On Day 24, replace the medium in the lower chamber with 1 mL LORG branching medium (consisting of basal medium, Table 2 supplemented with FGF10 100 ng/mL, FGF7 10 ng/mL, CHIR99021 3 μM, EGF 10 ng/mL, ATRA 50 nM, VEGF/PIGF 10 ng/mL). Change the medium every other day from Day 24 to Day 32.
-
20.
Monitor the cultures for evidence of branching. On Day 33, replace the medium with 1 mL LORG maturation medium (consisting of LORG branching medium supplemented with cAMP 100 μM, DEX 50 nM, and IBMX 100 μM). Change the medium every other day from Day 33 to Day 40.
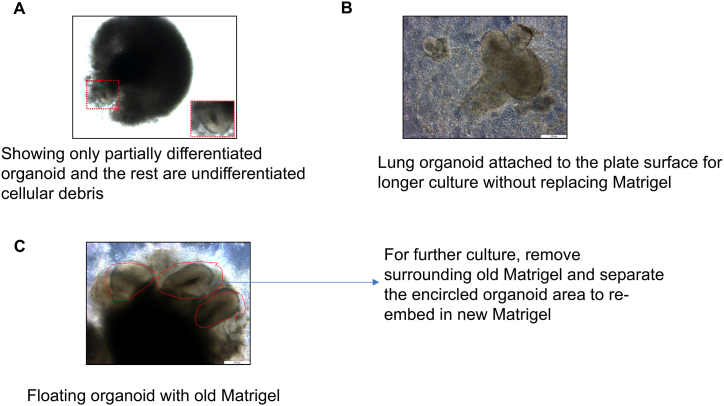
Note: If LPAs have not differentiated (as indicated by images in Fig. 3B) or cultures show evidence of cell death or the LORG/Matrigel droplet appears to have adhered to the culture well the cells should be re-embedded as described below (before the scheduled 2-week interval, if necessary).
7. Passaging and re-embedding of LORGs
7.1. Time: 1–2 h
From Day 40 onwards, the LORG cultures are monitored and LORGs are re-embedded in GFR-M every 2 weeks (or earlier if necessary) to continue branching and maturation..
-
21.
If images show poorly differentiated organoid with cellular debris (Fig. 6A), old Matrigel with attached organoid (Fig. 6B) and healthy floating organoids (Fig. 6C), remove the old Matrigel and re-embed organoids in new Matrigel for further culture.
-
22.
Under a dissecting microscope, use a P1000 pipette tip with the tip cut off to gently pick up the LORG/GFR-M droplet and transfer it to a Petri dish. Use a scalpel and 27-gauge needle to gently remove the old Matrigel and undifferentiated cellular debris. For transwell, we carefully detach all the Matrigel containing organoid with sterile spatula from the wall side of trans well and transfer with 1 mL pipette tip (cut to widen the tip mouth) to a 10 cm Petri-dish containing medium. Under dissecting microscope, carefully remove the old Matrigel and undifferentiated cellular debris with scalpel and 27-gauge needle. Collect the rounded structure containing lung progenitor cells as shown in Fig. 6C. To avoid damage to organoid during removal of old Matrigel, carefully dissect the organoid. In addition, if there is a minor damage, it will not affect the branching and differentiation because they also maintain the lung progenitor cells.
-
23.
Transfer the healthy LORG to a 1.5 mL tube, allow to settle, and remove the remaining medium.
-
24.
Add 200 μL of fresh Matrigel to the LORG and use a P200 pipette tip with the tip cut off to gently mix avoiding bubbles. Pipette 40–50 μL of the mixture onto sterile parafilm squares and place in a 37 °C incubator for 15–30 min to allow the LORG/Matrigel mixture to solidify.
-
25.
After solidification, carefully transfer the droplets to a 24-well plate (one per well) and add 500 μL of LORG maturation medium to each well.
Fig. 6.
Passaging and re-embedding of organoids.
(A) Representative phase contrast image showing failed Lung organoid differentiation and cell death. Scale bars, 200 μm (B) Microscopic image showing lung organoid attached to plate surface because of longer culture without changing the Matrigel. Scale bars, 200 μm (C) Representative phase contrast image showing well differentiated Lung organoid that requires replacing old Matrigel and passaging the organoid into new Matrigel. Scale bars, 200 μm.
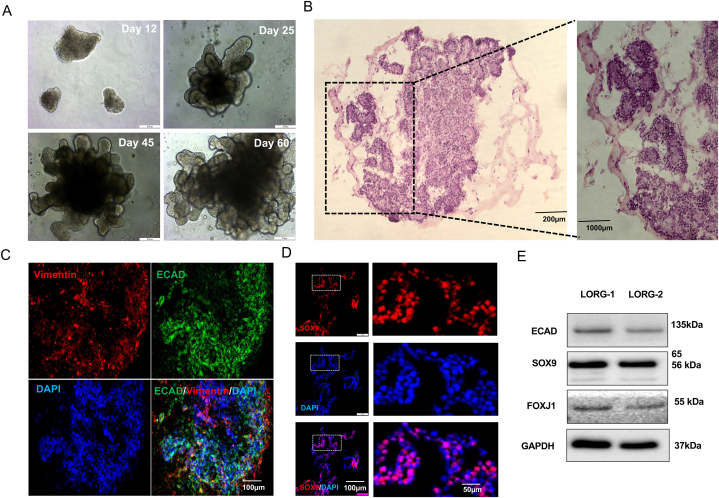
The LORGs can be maintained by repeating steps 20–23 every 2 weeks, but the epithelium is lost over time and cell death increases. Therefore, the LORGs are best used for experiments between ∼ Days 50 and 90. LORG showing the branching pattern at different developmental stages (Fig. 4A). We also analyzed intestinal (LGR5, DCAMKL1, BMPR1a) and thyroid (FOXE1, PAX8) markers in lung organoid (Fig. S4B) and we couldn't detect expression of these markers, except low expression of PAX8.
Fig. 4.
Formation and characterization of 3D LORGs.
(A) Representative phase-contrast images of lung organoids (LORGs) differentiation at indicated days of protocol. Scale bar, 200 μm (B) H&E staining of 60D LORG showing alveolar-like morphology. Scale bars, 200 μm and 100 μm (C) Confocal fluorescence microscopy of a LORG stained at Day 60 for the proximal epithelial marker E-cadherin (ECAD, green) and the mesenchymal cell-type marker vimentin (VIM, red). Nuclei were stained with DAPI (blue). Scale bars, 100 μm. (D) Immunofluorescence images of a LORG stained for the distal epithelial marker SOX9 (green). Nuclei were stained with DAPI (blue). Scale bars, 100 μm. (E) Immunoblot analysis of LORG lysate showing the intensity of protein level in two individual LORG organoids for epithelial protein (ECAD, band intensities 0.452 and 0.345), ciliated protein (FOXJ1, band intensities 0.231 and 0.211) and lung progenitor markers (SOX9, band intensities 1.45 and 1.323) as compared with internal control GAPDH, band intensities 2.92 and 3.18, n = 2 organoids).
8. Immunohistochemical analysis of LORGs
8.1. Timing: ∼2–3 days
-
26.
Collect the LORGs from culture gently with P1000 pipette tip with the tip cut off, place in a 1.5 mL tube, and wash by adding 1 mL phosphate-buffered saline (PBS). Allow to settle and remove the residual Matrigel by adding 500 μL of cell recovery solution and incubating at room temperature for ∼30 min.
-
27.
Remove the cell recovery solution and wash with PBS after organoids have settled down (Settling time is around 10min and no need to spin down). Wash the LORGs with PBS and fix by adding 500 μL 4% paraformaldehyde and incubating for 1 h at 4 °C.
-
28.
Wash the LORGS again with PBS and dehydrate by adding 500 μL 30% sucrose solution followed by incubation overnight at 4 °C. Remove the sucrose, wash the LORGs with PBS and embed in OCT compound or paraffin wax using standard procedures for sectioning Tiwari et al., 2021.
-
29.
Cut 20 μm sections from the LORG blocks and perform antigen retrieval by citrate buffer.
-
30.
Using standard tissue immunostaining techniques such as H & E staining and label the LORGs with antibodies against the desired cell markers, including SFTPC, SOX9, SOX2, HTII-280, P63, ECAD and vimentin and immunoblotting (Fig. 4, Fig. 5, Fig. 7E, Figs. S5–A).
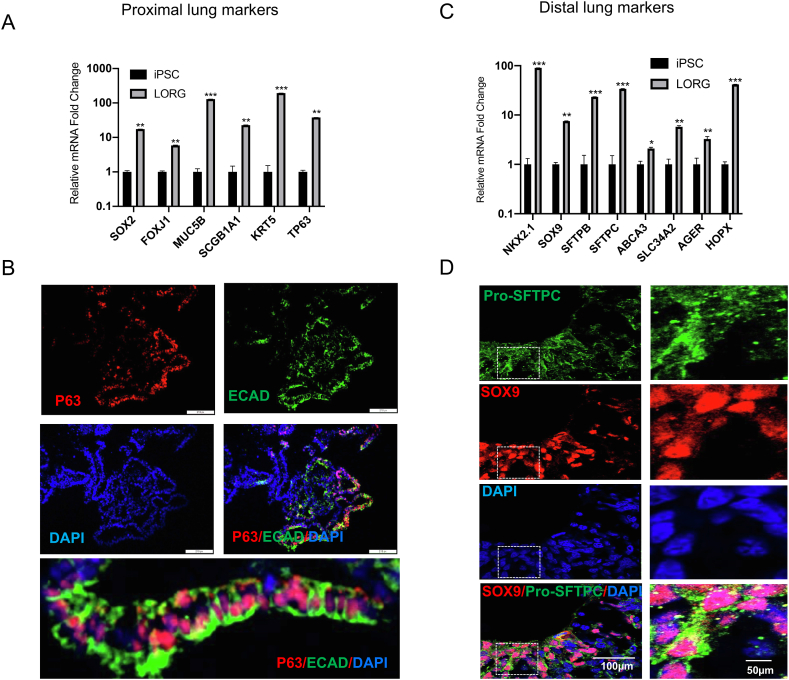
Fig. 5.
Expression of proximal and distal lung markers in LORGs.
(A) qRT-PCR analysis of the proximal lung genes SOX2, FOXJ1, MUC5B, SCGB1A1, KRT5, and TP63 in hiPSCs or in LORGs collected on Day 60. Data are the mean ± SEM (n = 3) and are presented as the fold change in mRNA level relative to that in hiPSCs.*P < 0.05, **P < 0.01, ***P < 0.001. (B) Immunofluorescence confocal microscopy of a LORG stained for the basal cell marker P63 (red) and the epithelial marker E-cadherin (ECAD, green). Nuclei were stained with DAPI (blue). Scale bars, 100 μm. (C) qRT-PCR analysis of the distal lung genes NKX2.1, SOX9, SFTPB, SFTPC, ABCA3, SLC34A2, AGER, and HOPX in hiPSCs or in LORGs collected on Day 60. Data are the mean ± SEM (n = 3) and are presented as the fold change in mRNA level relative to that in hiPSCs.*P < 0.05, **P < 0.01, ***P < 0.001. (D) Immunofluorescence confocal microscopy of a LORG stained for the distal progenitor cell marker SOX9 (red) and ATII cell-type marker Pro-SFTPC (green). Nuclei were stained with DAPI (blue). Scale bars, 100 μm.
Fig. 7.
LORGs as a model system for investigation of viral infection and screening of antiviral drugs.
(A) qRT-PCR analysis of the SARS-CoV-2 receptors ACE2 and TMPRSS2 in hiPSCs or in LORGs collected on Day 60. Data are the mean ± SEM (n = 3) and are presented as the fold change in mRNA level relative to that in hiPSCs.*P < 0.05, **P < 0.01. (B) Immunoblot analysis of ACE2 (band intensities 0.321, 0.245) and TMPRSS2 (band intensities 0.421 and 0.398) protein expression in two independent LORGs collected on Day 60. GAPDH (band intensities 2.92 and 3.18) was probed as a loading control. (C) Immunofluorescence staining of LORGs for ACE2 (red) and the ATII cell-type marker SFTPC (green). Nuclei were stained with DAPI (blue). Scale bars, 100 μm (D) Phase contrast (upper left) and fluorescence microscopy images of a LORG at 24 h after infection with a GFP-tagged SARS-CoV-2 pseudovirus (green) at a MOI of 2. Upper right and lower left panels show GFP signals of pseudovirus and merged GFP signals with organoid, respectively. Lower right panel shows an enlarged portion of the lower left panel. Scale bars, 200 μm, except lower right, 50 μm. (E) Immunofluorescence staining of a LORG for the SARS-CoV-2 nucleocapsid protein (CoV-2-NCP, red) and the lung ATII cell marker HT280 (green) at 96 h after infection with SARS-CoV-2 USA-WA1/2020 virus at a MOI of 2. Nuclei were stained with DAPI. Scale bars, 100 μm. Magnification of enlarged merged image = 50 μm. (F) Luciferase activity in Day 60 LORGs pretreated with vehicle (red) or the TMPRSS2 inhibitor camostat mesylate (10 μM, blue) for 2 h, infected with SARS-CoV-2-luciferase pseudovirus for 2 h, and then washed and incubated in camostat-containing medium for an additional 22 h. Luciferase activity was measured at 24 h post-infection. Data are shown as the mean ± SEM of n = 3 independently cultured LORGs. **P < 0.01. (G) Luciferase activity in Day 60 LORGs pretreated with ethanol or 25-hydroxy cholesterol (25HC, 5 μM) for 16 h, infected with SARS-CoV-2-luciferase pseudovirus at MOI of 0.1 for 2 h, and then washed and incubated in 25HC-containing medium for an additional 22 h. Luciferase activity was measured at 24 h post-infection. Data are shown as the mean ± SEM of 3 LORGs. ****P < 0.0001. Data are taken from Fig. 5D in (Wang et al., 2020). H) LORGsLORG were infected with SARS-CoV-2 USA-WA1/2020 virus at MOI = 2 for 2, 48 and 72 h and after respective post-infection, qRT-PCR showing increased replication of SARS-CoV-2. N = 3, mean ± SEM **P < 0.01, ***P < 0.001 by Student's t-test (taken from Fig. 2D, Tiwari et al., 2021) (I, J) qRT-PCR analysis of SARS-CoV-2 nucleocapsid mRNA expression in LORGs pretreated with the protease inhibitor E24 (5 μM) for 2 h, infected with SARS-CoV-2 USA-WA1/2020 at a MOI of 1 for 2 h, and then washed and incubated in E24-containing medium for an additional 48 h. Data are shown as the mean ± SEM (n = 3 LORGs) and are presented as the fold change in mRNA level relative to that in DMSO-treated LORGs. *P < 0.05.
8.2. Application of LORGs for modeling viral infection and antiviral drug testing
Note: For additional details about infection and sample processing see Tiwari et al0.2021..
LORGs formed using various protocols have been shown to express receptors for various viruses in the proximal and distal airway cell types and are thus useful for investigating not only the mechanism of viral pathogenesis but also the efficacy and mechanisms of action of candidate antiviral drugs [1,11]. We recently employed the LORG protocol described here to investigate SARS-CoV-2 infection and to assess the activity of antiviral drugs [1]. Some of our findings are briefly summarized here to illustrate the utility of the LORG model.
We first asked whether LORGs express the SARS-CoV-2 host co-receptors ACE2 and TMPRSS2. Indeed, qRT-PCR analysis, Western blot analysis, and immunofluorescence staining of LORGs confirmed that the expression of both receptors was equally as robust in Day 60 LORGs as compared to starting hiPSCs (Fig. 7A–C, Figs. S5–B). Moreover, immunofluorescence staining of ACE2 and SFTPC, a marker of SARS-CoV-2 receptor and Alveolar type II (ATII) cells, showed coincident staining, suggesting that SARS-CoV-2 may be able to infect ATII cells in LORGs.
To test this, we infected LORGs with a GFP-tagged SARS-CoV-2 pseudovirus at a multiplicity of infection of 2. GFP fluorescence, indicative of productive SARS-CoV-2 infection, was evident throughout fluorescence imaging of infected LORGs (Fig. 7D).
Moreover, when we infected LORGs with the live SARS-CoV-2 isolate USA-WA1/2020 and co-immunostained for the SARS-CoV-2 nucleocapsid protein (NCP) and HTII-280 a biomarker of lung ATII cells, we observed an overlapping staining pattern (Fig. 7E), consistent with the known ability of SARS-CoV-2 to infect ATII cells in humans [12,13].
Next, to assess whether LORGs might be a useful drug screening model system, we examined the efficacy of several known antiviral compounds, including camostat mesylate, a TMPRSS2 inhibitor [14,15]; 25-hydroxycholesterol (25HC), a naturally occurring antimicrobial product induced by Toll-like receptor engagement [16,17]; and E24, an inhibitor of SARS-CoV-2 protease.
LORGs were preincubated with camostat (10 μM) for 2 h and then infected with SARS-CoV-2-luciferase pseudovirus. Two hours later, the virus-containing supernatant was removed, and the cells were incubated for an additional 22 h in camostat-containing medium before the cells were lysed and luciferase activity was measured. As shown in Fig. 7F, camostat efficiently (∼2–3-fold) and significantly inhibited SARS-CoV-2 infection of LORGs. Similar experiments with LORGs treated with 5 μM 25HC and then infected with SARS-CoV-2-luciferase pseudovirus showed an even more impressive suppression of SARS-CoV-2 infection by 25 HC (Fig. 7G).
Finally, we assessed the effects of treatment with E24 (5 μM) on LORG infection with live SARS-CoV-2 isolate USA-WA1/2020. qRT-PCR analysis of infected cells demonstrated significant suppression of SARS-CoV-2 NCP expression in supernatant and cellular RNA, confirming the antiviral activity of E24 (Fig. 7H, I, J).
These data highlight the utility of the LORG model system for investigation of the molecular events underlying respiratory viral infection as well as for drug screening. The LORG model will undoubtedly also prove useful for investigating the cellular and molecular pathways that regulate early lung development and cell fate specification [[18], [19], [20]].
Expected outcomes.
-
•
hiPSC cultures should be 60–80% confluent after 1–2 days of plating before DE induction. During DE induction, daily medium exchange removes dead cells and maintains healthy adherent cells, as indicated by expression of SOX17 and CXCR4 by Day 4 (Fig. 3A, D).
-
•
By Day 7–9 of AFE induction, a heterogenous population of adherent cells appears with some floating spheroids, which were termed “foregut spheroids” by Miller et al., 2019 to differentiate them from the loosely adherent AFE aggregates (i.e., LPAs) that appear around Day 12 (Fig. 3B and C).
-
•
Analysis of gene expression should confirm the presence of AFE expressing FOXA1, FOXA2, and NKX2.1 by Day 8, and of LPAs expressing high levels of SOX2 and NKX2.1 by Day 12 (Fig. 3D–E and S1A).
-
•
The AFE/LPA cultures contain both lung (NKX2.1) and gastric (SOX2+) progenitor cells (Fig. 3E), as is seen during early embryonic development in humans.
-
•
Earlier protocols for differentiation of LORGs from AFE/LPAs (e.g., sorting of NKX2.1+ cells) resulted in enrichment of ATII-like cells but lacked mesenchymal cells. Thus, our protocol improves upon these methods by yielding LORGs that contain both epithelial and mesenchymal cell populations (Fig. 4A–D, E).
-
•
LORGs express proximal lung markers (SOX2, FOXJ1, MUC5B, KRT5, and TP63) and distal lung markers (SOX9, SFTPB, SFTPC, AGER, ABCA3, and HOPX) (Fig. 5A–D). Additionally, LORGs show strong expression of SOX9 (Fig. 5C, D), a transcription factor required for distal lung epithelial cell formation.
-
•
As noted above, the LORGs produced by this protocol express ACE2 and TMPRSS2 receptors and can be infected with SARS-CoV-2, reminiscent of the human lung (Fig. 7A–C), particularly the ATII cells (Fig. 7E).
-
•
Finally, the LORG model system has utility for antiviral drug candidate testing, as shown here for camostat, 25HC, and E24 (Fig. 7F–J).
8.3. Advantages and disadvantages
Our aim of this protocol is to create whole lung organoid model containing both bronchial/proximal and alveolar/distal cells (Fig. 5A–D) for better modeling the lung development and infection mechanisms. Advantages and disadvantages of the protocol is outlined below.
-
1
Current protocol is based on human iPSC derived cells which is easy to get and culture while obtaining bronchial tissue is difficult from patient due to ethical concern.
-
2.
Current protocol showing complete lung organoids formation which express markers for both bronchial airway cells (SOX2, FOXJ1, KRT5 and TP63) and alveolar cells (SOX9, SFTPC) (Fig. 5A–D), while bronchial epithelial cell culture not showing distal organoid markers.
-
3.
Bronchial epithelial cell culture in interface air-liquid condition has more physiological relevance due to mimicking similar to human bronchial airway structure compared to current protocol. We could adapt the current protocol to culture as air-liquid condition during trans well culture in future.
-
4.
In addition, current protocol shows branching morphogenesis and lung mesenchymal cells. Further by re-embedding, we can culture for longer duration without the loss of cellular integrity, while tissue obtained bronchial epithelial cells in interface air-liquid conditions for longer culture is difficult. (Included in text at page 11)
8.4. Quantification and statistical analysis (optional)
The data presented here are expressed as the means ± standard error (SEM) of three independent biological replicates. Differences between group means was analyzed with by unpaired two-tailed Student's t-test. A P value < 0.05 was considered significant.
8.5. Limitations
-
•
In this protocol, we have not established the transplantation of these lung organoids into mice for further understanding the vascularization and mature cellular architecture …
-
•
We have tested this protocol using only two hiPSC lines (iPSC-11 and iPSC-12).
-
•
The LORGs do not appear to contain pulmonary vasculature.
-
•
The LORGs are not terminally differentiated.
-
•
The LORGs contain random branching and the pattern of mesenchyme is undefined. (Mesenchymal cell type determining the shape and size of the lung, so here mentioned the pattern of mesenchyme)
9. Troubleshooting
9.1. Problem 1
Compromised pluripotency state of hiPSCs during culture.
9.2. Potential solution
Remove differentiated cells by scraping and colonies should be well separated from each other as shown in Fig. 1, Fig. 2B.
9.3. Problem 2
Failure of DE to form AFE/LPAs.
9.4. Potential solution
9.5. Problem 3
Many LPAs fail to differentiate into LORGs and cell death is extensive.
9.6. Potential solution
Remove the viable LORGs and re-embed them in fresh LORG medium/GFR-M to rescue and continue the culture.
10. Problem 4
LORGs may adhere to the plates and fail to grow properly.
10.1. Potential solution
-
•
Re-embed the LORGs in fresh LORG medium/GFR-M.
-
•
Handle GFR-M carefully and place at 4 °C only for the indicates times.
Resource availability
We require three subheadings in this section (lead contact, materials availability, and data and code availability). The guidelines include instructions and examples.
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Tariq M. Rana (trana@health.ucsd.edu).
Materials availability
This study did not create new unique materials or chemicals.
Data and code availability
This study did not generate any unique datasets or code.
Funding details
This work was supported in part by institutional funds and grants from the National Institutes of Health (CA177322, DA039562, DA049524, and AI125103).
Author contributions
S·K.T. designed and performed experiments, analyzed the data, and wrote the manuscript. T.M.R. conceived the overall project and experimental design, participated in data analysis and interpretation, and co-wrote the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Dr. F. Furnari for the hiPSC lines and members of the Rana lab for helpful discussions and advice. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e19601.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Tiwari S.K., et al. Revealing tissue-specific SARS-CoV-2 infection and host responses using human stem cell-derived lung and cerebral organoids. Stem Cell Rep. 2021;16:437–445. doi: 10.1016/j.stemcr.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leibel S.L., et al. Reversal of surfactant protein B deficiency in patient specific human induced pluripotent stem cell derived lung organoids by gene therapy. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-49696-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller A.J., et al. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 2019;14:518–540. doi: 10.1038/s41596-018-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Amour K.A., et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 5.Zorn A.M., Wells J.M. Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 2009;25:221–251. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green M.D., et al. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat. Biotechnol. 2011;29:267–272. doi: 10.1038/nbt.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng-Blichfeldt J-P., Schrik A., Kortekaas R., Noordhoek J.A., Heijink I.H., Hiemstra P.S., Stolk J, Königshoff M., Gosens R. Retinoic acid signaling balances adult distal lung epithelial progenitor cell growth and differentiation. EBioMedicine. 2018;36:461–474. doi: 10.1016/j.ebiom.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serra M., et al. Pluripotent stem cell differentiation reveals distinct developmental pathways regulating lung- versus thyroid-lineage specification. Development. 2017;144:3879–3893. doi: 10.1242/dev.150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varghese B., Ling Z., Ren X. Reconstructing the pulmonary niche with stem cells: a lung story. Stem Cell Res. Ther. 2022;13:161. doi: 10.1186/s13287-022-02830-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W., et al. Human lung organoid: models for respiratory biology and diseases. Dev. Biol. 2023;494:26–34. doi: 10.1016/j.ydbio.2022.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Han Y., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salahudeen A.A., et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature. 2020;588:670–675. doi: 10.1038/s41586-020-3014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katsura H., et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell. 2020;27:890–904 e8. doi: 10.1016/j.stem.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393 e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M., et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021;65 doi: 10.1016/j.ebiom.2021.103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zang R., et al. Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 2020;117:32105–32113. doi: 10.1073/pnas.2012197117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S., et al. Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020;39 doi: 10.15252/embj.2020106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fatehullah A., Tan S.H., Barker N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 19.Nikolic M.Z., Rawlins E.L. Lung organoids and their use to study cell-cell interaction. Curr Pathobiol Rep. 2017;5:223–231. doi: 10.1007/s40139-017-0137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCulley D., Wienhold M., Sun X. The pulmonary mesenchyme directs lung development. Curr. Opin. Genet. Dev. 2015;32:98–105. doi: 10.1016/j.gde.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.