Abstract
The phenomenon of antimicrobial resistance threatens our ability to treat common infections. The clinical pipeline for new antimicrobials is pretty much dry and hence, there is a need for the development of new antimicrobial agents with low toxicities to help fight resistant microorganisms. This work aimed to design antimicrobial peptides with low toxicities using a database filtering technology and evaluate their bioactivities. The physicochemical properties of the designed peptides were explored with molecular dynamics (MD) simulations. Microbroth dilution and hemolytic assays were used to assess the peptides' antimicrobial activity and toxicity. The activity of combinations of the peptides and some standard antibiotics was tested by the checkerboard method. In general, the designed peptides had a charge of +2, chain length of 13, and hydrophobicity of 61%. The predicted secondary structures of the peptides were either extended conformations or alpha-helices, and these structures were found to fluctuate during the MD simulations, where coils, bends, and helices dominated. , of the peptides, BRG003, BRG004 and BRG002 had the greatest aggregation propensities, whereas BRG001, BRG005, and BRG006 exhibited lower aggregation propensities. The minimum inhibitory concentration (MIC) of the peptides ranged from 0.015 to >1.879 μM, with BRGP-001 exhibiting high activity against MRSA with an MIC of 15 nM. BRGP-005 and BRGP-006 exhibited synergistic effects against Escherichia coliR when used in combination with erythromycin. At the minimum hemolytic concentration, the percentage of lysed erythrocytes was lower for all the peptides in comparison to the reference peptide, indicating low hemolytic activity. The study revealed the importance of peptide self-association in the antimicrobial activity of the peptides. These peptides provide a basis for the design of potent antimicrobial peptides that can further be developed for use in antimicrobial therapy.
Keywords: Antimicrobial peptides, Peptide design, Hemolytic activity, Molecular dynamics, Database filtering technology
1. Introduction
In the 1940s, antibiotics were hailed as miracle drugs as they were able to eliminate bacteria. As the years passed, however, microorganisms began to develop resistance to these antibiotics [1]. The improper use of these antibiotics is a major contributor to antimicrobial resistance, which poses threats to human health globally [2]. Microorganisms utilize various mechanisms to make existing antimicrobials less effective. This phenomenon makes it difficult to treat diseases caused by pathogens that were once susceptible to existing antibiotics [3]. Antimicrobial resistance results in economic losses and losses of life, especially in the developing world. It has been estimated that antimicrobial-resistant infections will lead to about 10 million deaths per year by 2050 and a total gross domestic product (GDP) loss of $100.2 trillion by 2050 if appropriate actions are not taken [4]. Despite the threats posed by antimicrobial resistance, the clinical pipeline for antimicrobial drug development remains insufficient to tackle this threat. There is, therefore, an urgent need for the development of new drugs with modes of action that do not encourage resistance development.
Antimicrobial peptides (AMPs) are the major defense molecules of innate immunity in invertebrates. In vertebrates, AMPs serve as both effectors in the innate immune system and modulators in the adaptive immune system [5]. Due to their broad spectrum of activity against Gram-positive and Gram-negative bacteria, fungi, parasites, viruses, and resistant microorganisms, and their slow rate to induce resistance in bacteria, a lot of focus has shifted to the development and use of AMPs as a novel class of antibiotics [6,7]. AMPs exert their action over a wide range of mechanisms, making it difficult for microorganisms to develop resistance to them [3].
Despite their biological activities, some AMPs can be toxic and unstable as they lyse red blood cells and are also susceptible to degradation by proteases when administered orally. The antimicrobial peptide, tyrrocidine which has been shown to be effective against both Gram-negative and Gram-positive bacteria, has been found to be toxic to human red blood cells [8]. Cathelicidin LL-37 is also a potent antimicrobial peptide but susceptible to proteolytic degradation [9]. In silico technologies can be explored to overcome some of these limitations plaquing natural AMPs. These technologies consider the size, charge, hydrophobicity, amphipathicity and solubility of AMPs as crucial factors [10]. A machine-learning method based on the concept of antimicrobial activity contribution score for each amino acid has been used to design a set of antimicrobial peptides in which improved antimicrobial activity towards standard and resistant microbial strains, as well as very low hemolytic activity were achieved [11]. Strategies used in the design of antimicrobial peptides include natural template optimization through truncation or substitution, sequence shuffling and motif hybridization [12]. The activity of natural peptides can be altered by removing a series of amino acids from the peptide (truncation) or by replacing some of the amino acids with different ones (substitution). Sequence shuffling involves changing the positions of the amino acids in a sequence while motif hybridization is the combination of two different peptide motifs to form a new hybrid peptide. Other strategies utilize databases such as the antimicrobial peptide database (APD).
The APD houses natural antimicrobial peptides with varying biological activities such as antibacterial, antifungal, antiviral and anticancer [[13], [14], [15]]. The database provides an interface that allows one to sort the antimicrobial peptides based on their activity, length, charge, and hydrophobicity, and hence, provides a useful platform for classifying, decoding and designing antimicrobial peptides [13]. This led to the development of a database filtering technology (DFT) for designing new peptides with predictable activities and properties. The database filtering technology utilizes the most probable parameters that determine the properties of AMPs in a database [16]. Parameters that are considered during the design of peptides by the DFT approach include average peptide length, frequently used amino acids, net charge, and hydrophobicity. An advantage of this technique is that the designed peptide has a good chance of being antimicrobial since it utilizes the most probable parameters derived from a set of AMPs with the desired activity. This method is also cost-effective as it allows for only a few peptides to be synthesized compared to sequence shuffling or library screening. DFTamp1 was the first peptide designed using this strategy. The peptide exhibited good activity against MRSA with a minimum inhibitory concentration (MIC) of 3.1 μM [17].
In this work, we aimed to design novel antimicrobial peptides with antimicrobial activity but minimal toxicity using the database filtering technology approach. Molecular dynamics simulations were performed to evaluate the physicochemical properties of the designed peptides. Micro broth dilution and hemolytic assays were used to assess the antimicrobial activity and toxicity of the peptides. Combinations of the peptides and some standard antibiotics were tested by the checkerboard method. Three of the designed peptides were active against the strains tested with relatively low hemolytic activity. The activities of the peptides designed in this work were generally enhanced when used in combination with either of the standard antibiotics - erythromycin, cefotaxime, or chloramphenicol. These peptides provide a basis for the design of potent AMPs that can further be developed for use in antimicrobial therapy.
2. Materials and methods
2.1. Peptide design
The peptides were designed using the database filtering technology (DFT). This technology employs the most probable parameters, that is, amino acid composition, peptide hydrophobicity and net charge. In the design, the antimicrobial peptide database (APD) was sorted for peptides that are active against Gram-positive and Gram-negative bacteria, as well as fungi. Sampling of the APD was done in February 2020. Ninety-four peptides were obtained from this activity. The chain length for the peptides of interest was set to 11–15 amino acids because short peptides were preferred, as peptide length can affect the activity and toxicity of the resulting peptide. Truncated peptides such as melittin with 15 residues at its c-terminal and a shorter derivative of HP (2–20) exhibited extremely reduced hemolytic activity toward rat and human erythrocytes respectively, compared to their parent peptide [18,19]. Thus, by sticking to shorter peptides, the risk of toxicity is reduced. From the APD, the average net charge and the average length of the selected peptides were found to be +2 and 13 respectively. The frequently occurring amino acids present in the hits were isoleucine, leucine, phenylalanine, alanine, glycine, serine, and lysine. The hydrophobicities for the peptides ranged from 61 to 70%. Two and three amino acid motifs were identified and how often they occur among the peptides from the database was determined. The motifs were arranged based on the frequencies obtained, yielding six peptides (BRGP-001 – 006). BRGP-007 was designed by a random combination of amino acids other than the frequently used ones to serve as a negative control peptide. PCP-002, a positive control peptide, was selected randomly from the database [15,20].
2.2. Characterization of the designed peptides
The secondary structures of the designed peptides were predicted and modelled with the iTASSER server [21]. I-TASSER is a hierarchical template-based method for protein structure and function prediction. For a given protein or peptide with a sequence of not less than 10 amino acids, I-TASSER identifies structural templates or super secondary structure motifs from the protein data bank (PDB) library using LOMETS - a meta-threading program that consists of multiple threading algorithms. The topology of the full-length model is then constructed. A second round of structure re-assembly is performed to refine the structural models starting from clusters obtained from SPIKER. The low free-energy conformations are further refined by full-atomic simulations using FG_MD and ModRefiner [22].
The AGGRESCAN3D (A3D) webserver was used to further predict the aggregation propensities of the peptides. A3D uses a structure-based approach which allows for the detection of spatially-adjacent aggregation from folded states. A higher score indicates a higher aggregation propensity and hence, a darker red colour. Soluble residues are coloured in gradients of blue. A lower score corresponds to higher solubility and hence, darker blue colour. Residues with A3D scores equal to or close to zero do not influence aggregation and are coloured in white [23].
2.3. Peptide molecular dynamics simulation
Molecular dynamics (MD) simulations of the peptides were performed to further study properties such as the association of the peptides. All MD simulations were performed with GROMACS package version, 2018.2 [24]. The protein database (PDB) structures of the peptides obtained from iTASSER were used as initial structures. The amber99sb-ildn force field was used to generate topology files for each peptide. The systems were neutralized by adding an appropriate number of sodium and chloride ions to each simulation box and the TIP3P water model was used to solvate the systems. The steepest descent algorithm was used to locate the closest local minimum and set some convergence criteria. Equilibration MD simulations were run to further relax system. A thermostat and barostat were used to make the temperature and pressure fluctuate around room temperature and atmospheric pressure. The molecular dynamics (MD) production simulation was set up for 100 ns. The Nos-Hoover thermostat and Parrinello-Rahman barostat were used at this point since they are more accurate in sampling the isothermal-isobaric ensemble [25].
2.4. Analyses
The trajectory files containing only protein atoms were created and visualized with VMD [26]. The energy and pressure of the simulations were checked. The root mean square deviations (RMSD), root mean square fluctuations (RMSF) and radius of gyration (Rgyr) were computed using standard GROMACS commands [27,28], and a Ramachandran plot showing Φ and Ψ angles for all residues and snapshots for the peptides were generated. The solvent-accessible surface area (SASA) was calculated by first defining the radius of a probe sphere, which is usually the van der Waals (vdW) radius of water which is rolled along the surface of the selected groups. Biomolecular processes such as folding, or aggregation can be described in terms of the molecule's free energy. The free energy was plotted along with two other parameters (RMSD and Rgyr), giving rise to a (reduced) free energy surface (FES). A python script was used to generate the FESs of the systems.
2.5. Antimicrobial activity
All the peptides used in this study, were purchased from Pepmic Company Limited, Suzhou, China. The minimum inhibitory concentrations (MICs) were determined using a standard micro broth dilution method. Microorganisms used include Escherichia coliN (standard strain), Streptococcus pyogenes, Staphylococcus aureus, Klebsiella pneumoniae, Enterococcus feacalis, and methicillin-resistant Staphylococcus aureus (MRSA), Salmonella typhi and Escherichia coliR (resistant strain). Bacterial cells (both standard and resistant strains) were grown overnight at 37 °C on nutrient agar. The overnight culture was then transferred into 0.9% normal saline to obtain a McFarland standard of 0.5. Forty microliters of this bacterial suspension were transferred into an 8 mL sterile broth. Serial dilutions of the peptides (1.878 mM–917.5 nM) were made in triplicate in microtiter plates and then 100 μL of the broth containing the bacteria was transferred into each well of the microtiter plate to make a total volume of 200 μL. The plates were incubated at 37 °C for 24 h. The MICs were determined as the lowest peptide concentration that inhibited bacterial growth using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) indicator [[29], [30], [31]].
2.6. Antibiotic modulation activity
Combinations of the six designed peptides and cefotaxime, erythromycin or chloramphenicol were tested by the checkerboard method. Briefly, two-fold serial dilution of the peptides (0.939 mM–1.834 μM) and the standard antibiotics (0.774 mM–377.5 nM for Chloramphenicol, 0.34 mM to 166 nM for Erythromycin and 0.55 mM to 267.8 nM for Cefotaxime) were made and transferred into microtiter plates. A hundred microliters of the broth containing the resistant bacteria strains (methicillin-resistant Staphylococcus aureus (MRSA), Salmonella typhiR and Escherichia coliR) were transferred into each well of the microtiter plates to make a total volume of 200 μL. The plates were incubated at 37 °C for 24 h. The new MICs of the peptides in combination with the antibiotics were determined as the lowest peptide concentration that inhibited bacterial growth using the MTT indicator. The fractional inhibitory concentration (FIC) index was calculated by the following equations:
where FIC(P) is the FIC of the peptides and FIC(A) is the FIC of the antibiotic.
Synergy was defined as FIC index ≤0.5. When the FIC index is between 0.5 and 4.0, it indicated no interaction between peptides and antibiotics. An FIC index ≥4.0 indicated antagonism between peptides and antibiotics [32]. The antibiotics used for this study were erythromycin, cefotaxime, and chloramphenicol.
2.7. Hemolytic activity
A human blood sample was centrifuged at 2000 rpm for 10 min to separate the erythrocytes from the serum. The erythrocytes were then washed 3 to 5 times with phosphate-buffered saline (PBS) at a pH of 7.40. One per cent erythrocyte was prepared as a working solution. Serial dilutions of the peptides were made. A hundred microliters of the peptide samples were added to 100 μL of 1% human erythrocytes in phosphate-buffered saline. The reaction mixture was incubated at 37 °C for 1 h in microtiter plates. Unlysed erythrocytes were removed by centrifugation. Haemoglobin release was determined spectrophotometrically at 570 nm. The minimum concentration of peptides that cause hemolysis (MHC) and per cent hemolysis caused at that concentration were determined [33].
2.8. Calculation of therapeutic index
The therapeutic index is a widely accepted parameter to represent the specificity of AMPs. It is the ratio of hemolytic activity to antimicrobial activity. Hence, larger values of the therapeutic index indicate greater specificity for prokaryotic cells [34]. For this study, the therapeutic indices were calculated as:
3. Results
3.1. Peptide design
Of the peptides obtained from the APD, the frequently occurring amino acids were isoleucine (I), leucine (L), phenylalanine (F), alanine (A), glycine (G), serine (S) and lysine (K) (Fig. 1a). The percent hydrophobicities for the peptides were between 61% and 70% (Fig. 1b) with most containing three leucine residues arranged separately (L-L-L), two isoleucine residues arranged separately (I–I) and two lysine residues either together or separately (KK or K–K). Based on these, motifs with two (IK, IL, GA, SL, KG, A) or three (GAL, FAL, GIK, SIL, K) amino acids were obtained. The motifs were then arranged based on their frequencies to obtain BRGP-001 to BRGP-006. BRGP-007 was designed at random using residues other than the frequently occurring amino acids to serve as a negative control peptide. The designed antimicrobial peptides (BRGP-001 to BRGP-006) had net charges of +2, chain length of 13 and hydrophobicity of 61%, as predicted from the APD. BRGP-007 had a net charge of +1 and hydrophobicity of 15% (Table 1).
Fig. 1.
(a)Amino acid composition of the peptides from the antimicrobial peptide database, (b)percent hydrophobicities of the peptide from the antimicrobial peptide database (APD) and (c) secondary structures of the designed peptides as predicted by the iTASSER server.
Table 1.
Biophysical properties of the designed peptides as predicted by the antimicrobial peptide database (APD).
|
Peptide Code |
Length | Net Charge | Sequence | % Hydrophobicity | Molecular Weight | Structure |
|---|---|---|---|---|---|---|
| BRGP-001 | 13 | 2 | AILFLSLGAIKKG | 61 | 1330.672 | helix |
| BRGP-002 | 13 | 2 | FLIKSLAGAKGIL | 61 | 1330.672 | helix |
| BRGP-003 | 13 | 2 | KGGAIKSLFLILA | 61 | 1330.672 | helix |
| BRGP-004 | 13 | 2 | KGIKGALSILFAL | 61 | 1330.672 | helix |
| BRGP-005 | 13 | 2 | GIKGALKSILFAL | 61 | 1330.672 | helix |
| BRGP-006 | 13 | 2 | FALSILGALGIKK | 61 | 1330.672 | helix |
| BRGP-007 | 13 | 1 | PQQYPEMNTRRNW | 15 | 1719.904 | not helix |
3.2. Characterization and aggregation propensity
The secondary structures of the peptides predicted by iTASSER web server, ranged from extended conformations to alpha-helices. BRGP-001 and BRGP-003 exhibited extended conformations. BRGP-002, BRGP-004 and BRGP-006 took up both the extended and alpha-helical structures. For these peptides, extended conformations were observed at the termini regions of the structure. BRGP-005 was predicted to have a complete alpha-helical structure (Fig. 1c). These peptide structures were used as the initial conformations for molecular dynamics simulations and subsequently for predicting the aggregation propensities of the peptides. The A3D server predicts aggregation propensities based on the amino acid sequences. From the plots (Fig. 2), residues with a score greater than zero have a propensity to aggregate while residues with scores less than zero are soluble. Except for BRGP-007, most of the residues of the peptides had scores greater than zero, suggesting that the peptides are likely to form aggregates. BRGP-003 had the highest propensity to aggregate, followed by BRGP-004 and BRGP-002. BRGP-001, BRGP-005 and BRGP-006 have the least aggregation propensities as only a few amino acids had an A3D score >2. This ranking was based on the number of amino acids with A3D score greater than 2 for each peptide sequence.
Fig. 2.
Plots of amino acid residue and A3D score predicted with the AGGRESCAN3D web server for (a) BRGP-001, (b) BRGP-002, (c) BRGP-003, (d) BRGP-004, (e) BRGP-005, (f) BRGP-006 and (g) BRGP-007. A higher score indicates a higher aggregation propensity and hence, a darker red colour. Soluble residues are coloured in gradients of blue. A lower score corresponds to higher solubility and hence, darker blue colour. Residues with A3D scores equal to or close to zero do not influence aggregation and are coloured in white.
3.3. Molecular dynamics
RMSD was computed for the alpha carbon atoms of the peptides. This explains how different the individual conformations are to the reference structure which is usually the starting structure. As shown in Fig. 3a, the RMSD of the peptides ranged from 0.2 to 0.8 nm. Higher fluctuations were observed for BRGP-001, BRGP-002, BRGP-003 and BRGP-004 throughout the simulation period. Increased RMSD changes were observed for BRGP-005 until 40 ns where the RMSD was stable to the end of the simulation. Fluctuations in RMSD values were minimal in BRGP-006 over the simulation time. Higher fluctuations were observed for BRGP-007 for the first 30 ns, after which the fluctuations were relatively minimal till the last 5 ns when the RMSD dropped slightly. The RMSD fluctuations observed indicate that there were significant conformational changes throughout the simulation period.
Fig. 3.
(a) Root mean square deviation (RMSD), (b) Root mean square fluctuations (RMSF), (c) Radius of gyration and (d) Solvent accessible surface area obtained from the molecular simulations of the peptides.
The RMSF values of each atom give insight into the flexible regions in the peptide. RMSF values greater than 0.25 nm are characteristic of atoms or amino acid residues that belong to flexible regions [35]. High RMSF values were obtained for almost all the peptides except BRGP-006 (Fig. 3b). The fluctuations were higher in the N-termini and C-termini regions of the peptides. The peptides were generally flexible with the end termini exhibiting higher flexibility.
The radius of gyration measures the compactness of protein or peptide structure over time. Generally, a lower radius of gyration implies a more compact and stable structure. The radius of gyration for BRGP-001, BRGP-002, BRGP-003 and BRGP-004 fluctuated throughout the simulations. BRGP-004 however had the highest radius of gyration and hence was the least compact. For the first 40 ns, BRGP-005 had a high radius of gyration after which it reduced to about 0.7 nm till the end of the simulation. BRGP-006 had a lower radius of gyration compared to the rest of the peptides (Fig. 3c) and the most compact structure among the peptides.
Higher fluctuations in solvent accessible surface areas were observed for BRGP-001, BRGP-002, BRGP-003 and BRGP-004. BRGP-004, however, exhibited higher solvent accessible surface area (SASA) ranging from 17.5 to greater than 20 nm2 throughout the simulation (Fig. 3d). BRGP-005 showed high SASA around 17.5 to greater than 20 nm2 but reduced to about 15 nm2 after the first 40 ns to the end of the simulation. SASA values for BRGP-007 fluctuated between about 15 nm2 to greater than 20 nm2 throughout the simulation. This further explains the flexibility of the peptides. SASA value of approximately 15 nm2 for BRGP-006 reflects its stable, compact structure.
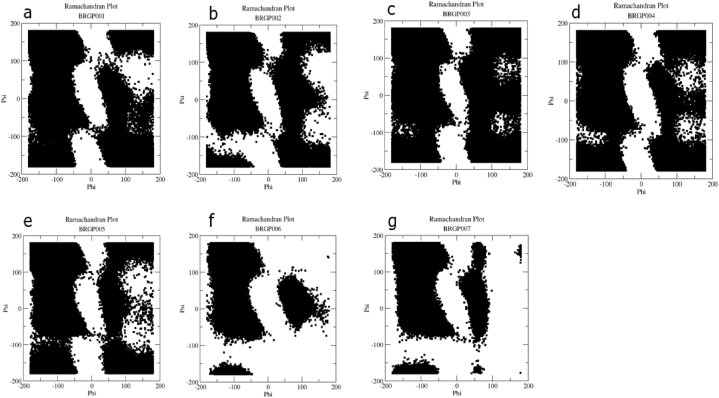
The Ramachandran plot is a plot of torsion angles of each residue of a peptide. Torsion angles determine the possible conformations of the peptide. Certain regions of the plots are characteristic of some secondary structures while others are considered forbidden. To study the conformational preferences of the peptides, Ramachandran plots for each peptide were generated. For all the peptides, data points were found in the forbidden regions which are mostly not possible for highly solvent-exposed residues. The plots (Fig. 4) are an indication of the peptides' (BRGP-001, BRGP-002, BRGP-003, BRGP-004, BRGP-005) poor solvent accessibility which is characterized by the relatively high distribution of data points in the forbidden regions. However, the low distribution of data points at the forbidden regions of BRGP-006 and BRGP-007 is indicative of their relatively high solvent accessibility. Analysis of the Ramachandran plot suggests that the distribution of the secondary structures of each of the peptides spanned between alpha helices and beta sheets.
Fig. 4.
Ramachandran plots for BRGP-001 (a), BRGP-002 (b), BRGP-003 (c), BRGP-004 (d), BRGP-005 (e), BRGP-006 (f) and BRGP-007 (g) obtained from the molecular simulations.
The secondary structural changes of the peptides were calculated throughout the simulation period. Secondary structures common among the peptides were coil, bend, turn, alpha-helix, and 3-helix. There was a high fluctuation in the secondary structure of BRGP-001 (Fig. 5a) throughout the simulation. However, the coil conformation was prominent specifically at the C-terminus around 5 ns to the end of the simulation. Similarly, some fluctuations were observed for BRGP-002 (Fig. 5b) throughout the simulation. In addition to the coil conformation observed at the C-terminus, the turn conformation was also prominent for some residues at different simulation times. For BRGP-003 (Fig. 5c), a 3-helix conformation was observed from Phe 9 to Leu 12 between 5 ns and 45 and then from 55 ns to the end of the simulation. Between 45 ns and 75 ns, a β-bridge was observed between Gly 2 and Phe 11 for BRGP-004 (Fig. 5d). Between 35 ns and 45 ns, BRGP-005 (Fig. 5e) exhibited β-sheet structure at Ile 2, Lys 3 and Leu 10, Phe 11; followed by the formation of β-bridge at Ile 2 and Phe 11 to the end of the simulation period. Throughout the simulation period, a stable α-helix was formed from Leu 3 to Leu 9 with the intermittent formation of a turn at Leu 3, Ser 4 and Leu 9, Gly 10 for BRGP-006 (Fig. 5f). A similar effect was observed for BRGP-007 (Fig. 5g) from Gln 3 to Met 7 between 5 ns and 95 ns. The structural analysis further reveals the stability and compactness of the secondary structures of BRGP-006 and BRGP-007.
Fig. 5.
Secondary structure patterns for (a) BRGP-001, (b) BRGP-002, (c) BRGP-003, (d) BRGP-004, (e) BRGP-005, (f) BRGP-006 and (g) BRGP-007 predicted from DSSP after molecular dynamics simulations.
To determine the aggregation propensities of the peptides, the free energy surface (FES) for each system was computed using the RMSD and radius of gyration as order parameters. For each peptide (Fig. 6), the different intensities of the FES represent the energy state of the peptides with different secondary structures. The energy minima were dominated by extended secondary structures or unfolded states for BRGP-002 and BRGP-005, partially structured for BRGP-003, BRGP-006 and BRGP-007 as well as a compact structure for BRGP-001. Two energy minima were obtained for BRGP-004 which were dominated by an extended structure and a partially structured secondary structure. Aggregation mostly occurs from unfolded or unstructured states [36]. For structured states, aggregation occurs from the partially folded intermediates as observed for BRGP-006, BRGP-007 and BRGP-001. These structures may either result in amyloid fibrils or amorphous aggregates. The free energy surfaces suggest that the peptides understudy have the propensity of forming aggregates.
Fig. 6.
The free energy surface for (a) BRGP-001, (b) BRGP-002, (c) BRGP-003, (d) BRGP-004, (e) BRGP-005, (f) BRGP-006 and (g) BRGP-007 computed using the RMSD and radius of gyration as order parameters. The different intensities of the FES represent the energy state of the peptides with different conformational structures.
3.4. Determination of the activity and toxicity of the designed peptides
The minimum inhibitory concentration (MIC) of the antimicrobial peptides ranged from 0.015 mM to >1.879 mM (Table 2). Generally, most of the microorganisms were less susceptible to the peptides. Of all the peptides tested, BRGP-001, BRGP-005 and BRGP-006 were active against E. coliN, S. pyogenes, S. aureus, K. pneumoniae, E. feacalis, MRSA, S. typhi R and E. coli R. BRGP-001 however, had higher activity against MRSA with an MIC of 0.015 mM. The MIC of BRGP-001, BRGP-005 and BRGP-006 were comparable to PCP-002 (Temporin-1P, a known antimicrobial peptide from the APD), a positive control peptide (MIC ranged from 0.228 to >1.825 mM) used in this experiment. Results from the hemolytic activity (Table 3) indicate that the peptides were less toxic in comparison to PCP-002; the positive control peptide. At the MHC, the percentage of lysed RBCs was lower for all the peptides in comparison to PCP-002.
Table 2.
Minimum inhibitory concentration; MIC (mM).
| Peptides | E. coliN | S. pyogenes | S. aureus | K. pneumoniae | S. typhiN | MRSA | S. typhiR | E. coliR |
|---|---|---|---|---|---|---|---|---|
| BRGP-001 | >1.879 | >1.879 | 0.117 | 0.117 | >1.879 | 0.015 | >1.879 | >1.879 |
| BRGP-002 | >1.879 | >1.879 | >1.879 | >1.879 | >1.879 | >1.879 | >1.879 | >1.879 |
| BRGP-003 | >1.879 | >1.879 | >1.879 | >1.879 | >1.879 | >1.879 | >1.879 | >1.879 |
| BRGP-004 | >1.879 | >1.879 | >1.879 | >1.879 | >1.879 | >1.879 | >1.879 | >1.879 |
| BRGP-005 | 0.470 | 0.470 | >1.879 | >1.879 | >1.879 | >1.879 | 0.470 | 0.235 |
| BRGP-006 | 0.470 | 0.470 | 0.939 | 1.879 | >1.879 | 0.235 | 0.235 | 0.470 |
| BRGP-007 | >1.454 | >1.454 | >1.454 | >1.454 | >1.454 | >1.454 | >1.454 | >1.454 |
| PCP-002 | 0.913 | 0.456 | 0.456 | 0.456 | 1.825 | 0.228 | 1.825 | 0.913 |
All experiments were performed in triplicates.
Table 3.
Hemolytic activity of the antimicrobial peptides.
|
Sample |
MHC (mM) | % Hemolysis | MIC (mM) range | Therapeutic Indices |
|---|---|---|---|---|
| BRGP-001 | 1.127 | 17.48 | 0.015–0.117 | 75.15–9.63 |
| BRGP-002 | 1.879 | 2.87 | Nd | – |
| BRGP-003 | 1.127 | 8.80 | Nd | – |
| BRGP-004 | 1.503 | 27.90 | Nd | – |
| BRGP-005 | 0.188 | 13.81 | 0.235–0.470 | 0.80–0.40 |
| BRGP-006 | 1.503 | 16.34 | 0.235–1.879 | 6.40–0.80 |
| BRGP-007 | 1.454 | 7.58 | Nd | – |
| PCP-002 | 0.183 | 28.02 | 0.228–13.825 | 0.80–0.10 |
The FIC indices calculated for BRGP-001, BRGP-005 and BRGP-006 indicate that there was no interaction between the peptide in combination with the antibiotics used. BRGP-005 and BRGP-006 exhibited a synergistic effect against E. coliR when used in combination with erythromycin (Table 4, Table 6). BRGP-002, BRGP-003 and BRGP-004 showed good activity (defined by the reduction in MICs from >1.879 mM to MICs ranging from 0.007 to 0.939 mM) when used in combination with cefotaxime and chloramphenicol (Table 5). However, there was no significant change in the MICs of cefotaxime and chloramphenicol when used in combination with BRGP-002, BRGP-003 and BRGP-004 (Table 7, Table 8). Similarly, the antimicrobial activity of BRGP-002 was not enhanced in the presence of cefotaxime against E. coli. The combination of BRGP-002 with erythromycin resulted in an enhanced activity against MRSA (with MIC of 0.007 mM) but poor activity against S. typhi and E. coli (MIC >0.939 mM). Similarly, poor activity was observed against S. typhi when BRGP-003 was combined with erythromycin (MIC >0.939 mM). This combination (BRGP-003 with erythromycin) however, resulted in an enhanced activity against MRSA and E. coli with MICs of 0.004 mM and 0.470 mM respectively (Table 5). The combination of BRGP-004 and erythromycin resulted in an enhanced activity against the strains tested.
Table 4.
FIC index of peptides in combination with Cefotaxime, Erythromycin and Chloramphenicol.
| Sample | Cefotaxime |
Erythromycin |
Chloramphenicol |
||||||
|---|---|---|---|---|---|---|---|---|---|
| S. typhiR | MRSA | E. coliR | S. typhiR | MRSA | E. coliR | S. typhiR | MRSA | E. coliR | |
| BRGP-001 | – | 4.40 | – | – | 0.81 | – | – | 2.063 | – |
| BRGP-002 | – | – | – | – | – | – | – | – | – |
| BRGP-003 | – | – | – | – | – | – | – | – | – |
| BRGP-004 | – | – | – | – | – | – | – | – | – |
| BRGP-005 | 0.75 | – | 2.25 | 0.63 | – | 0.37 | 1.032 | – | 1.25 |
| BRGP-006 | 1.00 | 0.75 | 2.50 | 1.50 | 1.030 | 0.25 | 0.53 | 0.75 | 0.75 |
Table 6.
FIC index of Cefotaxime, Erythromycin and Chloramphenicol in combination with peptides.
| Sample | Cefotaxime |
Erythromycin |
Chloramphenicol |
||||||
|---|---|---|---|---|---|---|---|---|---|
| S. typhiR | MRSA | E. coliR | S. typhiR | MRSA | E. coliR | S. typhiR | MRSA | E. coliR | |
| BRGP-001 | 0.50 | 0.50 | – | 0.50 | 0.54 | 0.12 | 0.50 | 0.063 | 0.25 |
| BRGP-002 | 1.00 | 0.50 | – | – | 1.00 | – | 1.00 | 1.00 | 0.50 |
| BRGP-003 | 1.00 | 0.50 | 0.50 | – | 0.54 | 1.00 | 1.00 | 0.50 | 1.00 |
| BRGP-004 | 1.00 | 0.50 | 0.50 | 0.50 | 0.54 | 0.12 | 1.00 | 0.50 | 0.50 |
| BRGP-005 | 0.50 | 0.50 | 0.25 | 0.50 | 1.00 | 0.12 | 1.00 | 0.50 | 0.25 |
| BRGP-006 | 0.50 | 0.50 | 0.50 | 1.00 | 1.00 | 0.12 | 0.50 | 0.25 | 0.25 |
Table 5.
MICs (mM) of Peptides in the presence of Cefotaxime, Erythromycin and Chloramphenicol.
| Sample | Cefotaxime |
Erythromycin |
Chloramphenicol |
||||||
|---|---|---|---|---|---|---|---|---|---|
| S. typhiR | MRSA | E. coliR | S. typhiR | MRSA | E. coliR | S. typhiR | MRSA | E. coliR | |
| BRGP-001 | 0.117 | 0.059 | >0.939 | 0.059 | 0.004 | 0.059 | 0.007 | 0.029 | 0.235 |
| BRGP-002 | 0.235 | 0.059 | >0.939 | >0.939 | 0.007 | >0.939 | 0.015 | 0.470 | 0.470 |
| BRGP-003 | 0.235 | 0.059 | 0.939 | >0.939 | 0.004 | 0.470 | 0.015 | 0.235 | 0.939 |
| BRGP-004 | 0.235 | 0.059 | 0.939 | 0.059 | 0.004 | 0.059 | 0.015 | 0.253 | 0.470 |
| BRGP-005 | 0.117 | 0.059 | 0.470 | 0.059 | 0.007 | 0.059 | 0.015 | 0.253 | 0.235 |
| BRGP-006 | 0.117 | 0.059 | 0.939 | 0.117 | 0.007 | 0.059 | 0.007 | 0.117 | 0.235 |
Table 7.
MICs (mM) of antibiotic alone.
| Sample | S. typhi | MRSA | E. coli |
|---|---|---|---|
| Erythromycin | 0.0213 | 0.0013 | 0.0852 |
| Chloramphenicol | 0.0060 | 0.1934 | 0.3868 |
| Cefotaxime | 0.0686 | 0.0343 | 5.4890 |
Table 8.
MICs (mM) of Cefotaxime, Erythromycin and Chloramphenicol in the presence of the Peptides.
| Sample | Cefotaxime |
Erythromycin |
Chloramphenicol |
||||||
|---|---|---|---|---|---|---|---|---|---|
| S. typhiR | MRSA | E. coliR | S. typhiR | MRSA | E. coliR | S. typhiR | MRSA | E. coliR | |
| BRGP-001 | 0.0343 | 0.0172 | >2.7445 | 0.0106 | 0.0007 | 0.0106 | 0.0030 | 0.0121 | 0.0967 |
| BRGP-002 | 0.0686 | 0.0172 | >2.7445 | >0.1703 | 0.0013 | >0.1703 | 0.0060 | 0.1934 | 0.1934 |
| BRGP-003 | 0.0686 | 0.0172 | 2.7445 | >0.1703 | 0.0007 | 0.0852 | 0.0060 | 0.0967 | 0.3868 |
| BRGP-004 | 0.0686 | 0.0172 | 2.7445 | 0.0106 | 0.0007 | 0.0106 | 0.0060 | 0.0967 | 0.1934 |
| BRGP-005 | 0.0343 | 0.0172 | 1.3722 | 0.0106 | 0.0013 | 0.0106 | 0.0060 | 0.0967 | 0.0967 |
| BRGP-006 | 0.0343 | 0.0172 | 2.7445 | 0.0213 | 0.0013 | 0.0106 | 0.0030 | 0.0484 | 0.0967 |
4. Discussion
The database filtering technology was used to design 6 peptides (BRGP-001 to BRGP-006), with physicochemical parameters such as hydrophobicity, net charge, chain length, amphipathicity as well as secondary structure being the main factors considered. These parameters are critical for a peptide's antimicrobial and hemolytic activity. Peptide length, for instance, affects the mode of action and the hemolytic activity of AMPs. Seven to eight amino acids are needed to form an amphipathic structure [8]. AMPs that interact with bacterial membranes do so in a variety of ways – Barrel Stave, toroidal pore, or the carpet models. AMPs that induce barrel stave pores usually have a minimum amino acid length of approximately 22 residues for α-helical structures and 8 residues for β-sheets [37]. Alamethicin (a 20-residue fungal peptide that folds into an amphipathic α-helix [38]), pardaxin (a 33-residue peptide, composed of two alpha helical segments [34]) and protegrins (16–18 residues, characterized by a beta sheet secondary structure [39]) are AMPs that function by forming barrel stave channels in bacteria membranes. Other peptides also exhibit antimicrobial activity despite their amino acid composition and chain length through the carpet model at high concentrations due to their amphipathic nature. Cecropin (30- to 40-residue alpha-helical peptides [40]), indolicidin (a 13 residue with extended conformation [41]), aurein 1.2 (a 13 residue that rearrange into α-helix [42]) and LL-37 (a 37-residue, amphipathic, helical peptide [43]) are examples of AMPs that act by the carpet model. Although the mode of action of the peptides was not determined in this study, the length of the designed peptides suggests an activity through the carpet model. In the carpet model, the AMPs lie parallel to the lipid bilayer till a threshold concentration is reached to cover the surface of the membrane. This leads to unfavorable interactions on the membrane surface. Eventually, the membrane integrity is lost, producing a detergent-like effect that later disintegrates the membrane by forming micelles. The final collapse of the membrane bilayer structure into micelles is also known as the detergent-like model. Most peptides exhibit antimicrobial activity despite their amino acid composition and chain length. These AMPs mostly act through the carpet model at high concentrations due to their amphipathic nature [37,44]. A shortened melittin with 15 residues at its c-terminal and a shorter derivative of HP (2–20) exhibited extremely reduced hemolytic activity toward rat and human erythrocytes respectively, compared to their parent peptide [45,46]. Hence, the length of the designed peptides could contribute to their relatively low hemolytic activity.
The predicted secondary structures of the peptides ranged from the extended conformation to the alpha-helix as predicted by the iTASSER webserver. MD simulation over a 100 ns period was used to further investigate the stability of these secondary structures. Results from MD simulations of the peptides revealed that the secondary structures of the peptides were very unstable. This is evidenced by the fact that RMSD, RMSF, and SASA values obtained were high for almost all the peptides, except BRGP-006. Higher SASA values mean that the peptides are not accessible to solvent molecules while lower values indicate the compactness of the peptide's structure. High values of RMSD, RMSF and SASA are indicators of structural instability; instability refers to rapid changes of the physical state (secondary structure) of the peptides without any change in the chemical composition. Disrupted peptide folding or decreased peptide stability can result in partially unfolded or misfolded peptides which ultimately leads to the formation of aggregates [47]. Conformational stability is therefore a major factor in peptide aggregation. Based on the results for the free energy surface, it is possible, that the peptides will likely form aggregates (either amorphous or amyloid fibrils) at high concentrations. The partially folded or extended conformations of the designed peptides corresponding to the energy minima as well as their intrinsic higher hydrophobicities further confirm the aggregation propensities of the peptides.
The FES for BRGP-002, BRGP-003, BRGP-004 and BRGP-005 corresponded to an extended conformation suggesting that these peptides have a propensity of forming aggregates. Peptides in the extended conformation are prone to aggregation, as shown for the model peptide, GNNQQNY. GNNQQNY is a polar heptapeptide from the N-terminal prion-determining region of the yeast prion protein Sup35. Using replica exchange molecular dynamics (REMD) to survey the free-energy landscape of the peptide, it was revealed that the entropic force favors an extended structure for the GNNQQNY monomer, which is a sign of its β-propensity, and thus suggesting the formation of amyloid fibrils [48]. The study of Ding and coworkers revealed that conversion of alpha-helix to beta-sheets with exposed hydrophobic surfaces also results in the formation of aggregates [49]. Some peptides self-assemble into highly structured amyloid fibrils, forming highly regular beta-strand conformations [36]. Other peptides also form amorphous aggregates which contains no ordered intermolecular interactions. In our study, there was a conversion of alpha-helical structures to the extended conformations for BRGP-002, BRGP-004 and BRGP-005 according to FES (Fig. 6). This suggests that the formation of amorphous aggregates were favored. The free energy surface for BRGP-003 corresponded to a beta-strand conformation suggesting that BRGP-003 is capable of forming a highly structured amyloid fibril. It has been reported that hydrophobic interactions are more effective in protein aggregation [50]. Hence, the conformational stability and the relatively high hydrophobicities of the peptides contribute to their aggregation propensities.
Among the many design strategies used in peptide design, the database filtering technology is preferred because it allows one to design AMPs with the desired activity in mind since it utilizes the most probable parameters of a group of active peptides. For this work, peptides were designed to have activity against Gram-positive and Gram-negative bacteria. The MICs of the three peptides with some activity (BRGP-001, BRGP-005 and BRGP-006) ranged from 0.015 mM to 1.879 mM (Table 2).
Antimicrobial activity and hemolytic activities of AMPs are affected by physicochemical parameters used in their design. These parameters are intimately related such that altering one parameter can result in changes to other parameters. Hydrophobicity is crucial for biological activity, especially for AMPs that target bacterial cell membranes as the hydrophobic regions of the peptides interact with membrane lipids which subsequently results in pore formation. Generally, increasing hydrophobicity increases antimicrobial activity. However, a study on 26-residue amphipathic peptides revealed that increasing hydrophobicity beyond a threshold specific to the peptide under study increases hemolytic activity [34]. Furthermore, increased hydrophobicity is also associated with higher aggregation propensities or self-association. Self-association is an important parameter for antimicrobial activity. A study on the relationship between intermolecular interaction strength and antimicrobial activity revealed that increasing aggregation in magainin II (MGN) and cecropin A-melittin (CAM) resulted in reduced antimicrobial activity [51]. Hence, AMPs with strong self-association propensities have a reduced ability to penetrate the cytoplasmic membrane to kill target cells [52]. The distribution of positive charges at the termini of AMPs eliminates aggregation beyond dimer formation as established by a study which showed that, positive charge distribution does not necessarily increase the antimicrobial activity of peptides [53]. However, a reduction in the hemolytic activity of peptides with the same hydrophobic core was observed at low concentrations.
The AMPs understudy are similar to each other in terms of their physicochemical properties. Differences are only realized in the arrangement of the amino acid residues and hence, the distribution of lysine (positively charged) residues. It was observed that BRGP-001 and BRGP-006 which had the two lysine residues positioned around the C-termini regions, exhibited relatively low hemolytic activity, and this is reflected in their high therapeutic indices (75.15–9.63 and 6.40–0.80). The other peptides exhibited hemolytic activity only at high concentrations (1.879 mM, 1.127 mM and 1.503 mM for BRGP-002, BRGP-003 and BRGP-004 respectively). BRGP-005 was an exception. It exhibited the highest hemolytic activity even at a low concentration of 0.188 mM and hence, had low therapeutic indices against the studied microbes with values ranging from 0.80 to 0.40. Again, BRGP-002, BRGP-003 and BRGP-004 with high aggregation propensities showed no activity at the concentrations tested. BRGP-001, BRGP-005 and BRGP-006 with relatively low aggregation propensities exhibited some activity against the organisms tested. Hence, the antimicrobial activity and hemolytic activity observed for the AMPs understudy could be attributed to their aggregation propensities and the distribution of positive charges on the peptides.
The designed peptides’ antimicrobial activity was improved when combined with either cefotaxime, erythromycin, or chloramphenicol. This phenomenon is characteristic of most AMPs. Combining AMPs with conventional antibiotics is a strategy to enhance the treatment of diseases caused by multidrug-resistant strains and reduce the toxicity of drugs. Various studies have demonstrated the synergistic effect of some cationic AMPs in combination with some conventional antibiotics. Both the D- and the l-amino isoforms of bombinin exhibited synergistic activity with ampicillin whereas BHL-bombinin showed an additive action with ampicillin against S. aureus [54]. The combination of the hybrid peptide, HPMA (HP(2–9)-MA(1–12)) with cefotaxime, ciprofloxacin or erythromycin resulted in a synergistic effect against some strains of A. baumannii. CAME (CA(1–8)-ME(1–12)) also exhibited a similar effect when combined with cefotaxime or erythromycin. HPME (HP(2–9)-ME(1–12)) on the other hand exhibited an additive and an indifferent effect when combined with cefotaxime but showed partial synergy when combined with erythromycin [55]. Cefotaxime exhibited additive effects with all four peptides. The generic membrane disrupting activity of AMPs increases access to bacterial cells for conventional antibiotics such as cefotaxime, meropenem, erythromycin and the β-lactam antibiotics and exerts a synergistic effect. Some studies have also shown that antibacterial drugs with similar mechanisms exert synergistic effects more readily. However, the antimicrobial peptide, DP7, exhibited a strong synergistic effect with azithromycin and vancomycin; two antibiotics with extremely different modes of action. The peptide, however, showed almost no synergy with gentamycin whose mode of action is similar to that of azithromycin [56]. Antagonism may be the result of the different modes of action of a given peptide and the conventional antibiotics. When combined, these mechanisms may interfere with each other, leading to reduced efficacy or in some cases, even neutralization of the antibacterial properties of one or both agents. Again, it is possible that peptides and antibiotics may compete for binding sites on bacterial cells or interact with each other, leading to reduced uptake or penetration into the bacteria. This interference can hinder their ability to disrupt bacterial processes effectively.
In this study, a strong synergistic effect was observed for the combination of BRGP-006 and erythromycin. A similar effect was observed for BRGP-005 in combination with erythromycin. However, the effect observed was not significant (as FIC indices indicated no interaction) when the two peptides were combined with either cefotaxime or chloramphenicol. The combination of erythromycin with either BRGP-005 or BRGP-006 could be a possible strategy for treating diseases caused by E. coli resistant strain. The antimicrobial activities of BRGP-002, BRGP-003 and BRGP-004 were enhanced greatly by the conventional antibiotics (cefotaxime, erythromycin, and chloramphenicol) used in this study.
The high hydrophobicities and the extended conformations of the peptides under study resulted in their ability to self-associate, which impacted the antimicrobial and hemolytic activity of the peptides. Although specific peptides with significant effects were not identified, this study provided valuable insights into peptide design and also highlighted the importance of considering self-association in the design of potent antimicrobial peptides. Hence, future studies may focus on tuning the hydrophobicities in an attempt to enhance the activity of the peptides.
5. Conclusion
In this study, we successfully designed several antimicrobial peptides using the database filtering technology. The designed peptides had a charge of +2 and a hydrophobicity of 61%, with 13 amino acid residues in each peptide. Prediction of the secondary structures of the peptides suggested that the peptides were mainly alpha-helices or extended conformations. In general, the peptides exhibited a high propensity towards aggregation, with BRGP-003, BRGP-004 and BRGP-002 being the most prone to aggregation. BRGP-001, BRGP-005 and BRGP-006 displayed very good activities against both Gram-positive and Gram-negative bacteria. The designed peptides generally exhibited low hemolytic activity. The antimicrobial activity of the peptides was enhanced when combined with either cefotaxime, erythromycin, or chloramphenicol. Results from MD simulations suggested that peptide self-association has significant effects on the antimicrobial activity and toxicity of the peptides. Hence, this parameter should be considered in designing potent antimicrobial peptides. The work gives more insight into the design of antimicrobial peptides using the database filtering technology. Future studies may focus on tuning the physicochemical parameters, particularly, the hydrophobicity and net charge of the peptides to obtain new peptides that can serve as leads for the development of new antibiotics.
Author contribution statement
Margaret Amerley Amarh: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Michael Konney Laryea, PhD: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Lawrence Sheringham Borquaye, PhD: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Data availability statement
Data included in article/supp. material/referenced in article.
Additional information
No additional information is available for this paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors are very grateful to the Center for High Performance Computing, Cape Town, South Africa who granted generous access to the Lengau cluster for the MD simulations.
References
- 1.Aslam B., et al. Antibiotic resistance: a rundown of a global crisis. Infect. Drug Resist. 2018;11:1645. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stephens L.J., Werrett M.V., Sedgwick A.C., Bull S.D., Andrews P.C. Antimicrobial innovation: a current update and perspective on the antibiotic drug development pipeline. Future Med. Chem. 2020;12(22):2035–2065. doi: 10.4155/fmc-2020-0225. [DOI] [PubMed] [Google Scholar]
- 3.Koo H.B., Seo J. Antimicrobial peptides under clinical investigation. Pept. Sci. 2019;111(5) [Google Scholar]
- 4.Chokshi A., Sifri Z., Cennimo D., Horng H. Global contributors to antibiotic resistance. J. Global Infect. Dis. 2019;11(1):36. doi: 10.4103/jgid.jgid_110_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang G. Cabi; 2017. Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies. [Google Scholar]
- 6.Browne K., et al. A new era of antibiotics: the clinical potential of antimicrobial peptides. Int. J. Mol. Sci. 2020;21(19):7047. doi: 10.3390/ijms21197047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta BBA-Biomembr. 2009;1788(8):1687–1692. doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Bahar A.A., Ren D. Antimicrobial peptides. Pharmaceuticals. 2013;6(12):1543–1575. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sieprawska-Lupa M., et al. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 2004;48(12):4673–4679. doi: 10.1128/AAC.48.12.4673-4679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakare O.O., Fadaka A.O., Klein A., Pretorius A. Dietary effects of antimicrobial peptides in therapeutics. Life. 2020;13(1):78–91. [Google Scholar]
- 11.Wu X., et al. In vitro and in vivo activities of antimicrobial peptides developed using an amino acid-based activity prediction method. Antimicrob. Agents Chemother. 2014;58(9):5342–5349. doi: 10.1128/AAC.02823-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loose C., Jensen K., Rigoutsos I., Stephanopoulos G. A linguistic model for the rational design of antimicrobial peptides. Nature. Oct. 2006;443(7113) doi: 10.1038/nature05233. Art. no. [DOI] [PubMed] [Google Scholar]
- 13.Wang G. The antimicrobial peptide database provides a platform for decoding the design principles of naturally occurring antimicrobial peptides. Protein Sci. 2020;29(1):8–18. doi: 10.1002/pro.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G., Li X., Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37(suppl_1):D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G., Li X., Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44(D1):D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hazam P.K., Goyal R., Ramakrishnan V. Peptide based antimicrobials: design strategies and therapeutic potential. Prog. Biophys. Mol. Biol. 2019;142:10–22. doi: 10.1016/j.pbiomolbio.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Mishra B., Wang G. Ab initio design of potent anti-MRSA peptides based on database filtering technology. J. Am. Chem. Soc. 2012;134(30):12426–12429. doi: 10.1021/ja305644e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subbalakshmi C., Nagaraj R., Sitaram N. Biological activities of C-terminal 15-residue synthetic fragment of melittin: design of an analog with improved antibacterial activity. FEBS Lett. 1999;448(1):62–66. doi: 10.1016/S0014-5793(99)00328-2. [DOI] [PubMed] [Google Scholar]
- 19.Park Y., Park S.-C., Park H.-K., Shin S.Y., Kim Y., Hahm K.-S. Structure-activity relationship of HP (2-20) analog peptide: enhanced antimicrobial activity by N-terminal random coil region deletion. Biopolymers. 2007;85(4):392–406. doi: 10.1002/bip. [DOI] [PubMed] [Google Scholar]
- 20.Bobde S.S., Alsaab F.M., Wang G., Van Hoek M.L. Ab initio designed antimicrobial peptides against gram-negative bacteria. Front. Microbiol. 2021:3460. doi: 10.3389/fmicb.2021.715246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasu E.N., Mensah J.K., Borquaye L.S. Computer-aided design of proline-rich antimicrobial peptides based on the chemophysical properties of a peptide isolated from Olivancillaria hiatula. J. Biomol. Struct. Dyn. Oct. 2022:1–22. doi: 10.1080/07391102.2022.2131626. [DOI] [PubMed] [Google Scholar]
- 22.Roy A., Kucukural A., Zhang Y. “I-TASSER: a unified platform for automated protein structure and function prediction,”. Nat. Protoc. Apr. 2010;5(4) doi: 10.1038/nprot.2010.5. Art. no. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zambrano R., Jamroz M., Szczasiuk A., Pujols J., Kmiecik S., Ventura S. AGGRESCAN3D (A3D): server for prediction of aggregation properties of protein structures. Nucleic Acids Res. 2015;43(W1) doi: 10.1093/nar/gkv359. W306–W313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kutzner C., Páll S., Fechner M., Esztermann A., de Groot B.L., Grubmüller H. More bang for your buck: improved use of GPU nodes for GROMACS 2018. J. Comput. Chem. 2019;40(27):2418–2431. doi: 10.1002/jcc.26011. [DOI] [PubMed] [Google Scholar]
- 25.Copps J., Murphy R.F., Lovas S. In: Peptide-Based Drug Design. Otvos L., editor. Humana Press; Totowa, NJ: 2008. Molecular dynamics simulations of peptides; pp. 115–126. (In Methods in Molecular BiologyTM). [DOI] [Google Scholar]
- 26.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. Feb. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 27.Boakye A., Gasu E.N., Mensah J.O., Borquaye L.S. Computational studies on potential small molecule inhibitors of Leishmania pteridine reductase 1. J. Biomol. Struct. Dyn. Jan. 2023;0(0):1–14. doi: 10.1080/07391102.2023.2166119. [DOI] [PubMed] [Google Scholar]
- 28.Mensah J.O., et al. Computational studies provide a molecular basis for the quorum sensing inhibitory action of compounds from dioon spinulosum dyer ex eichler. ChemistrySelect. 2023;8(1) doi: 10.1002/slct.202203773. [DOI] [Google Scholar]
- 29.Gasu E.N., Ahor H.S., Borquaye L.S. “Peptide extract from olivancillaria hiatula exhibits broad-spectrum antibacterial activity,”. BioMed Res. Int. Dec. 2018;2018 doi: 10.1155/2018/6010572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasu E.N., Ahor H.S., Borquaye L.S. Peptide mix from olivancillaria hiatula interferes with cell-to-cell communication in Pseudomonas aeruginosa. BioMed Res. Int. Sep. 2019 doi: 10.1155/2019/5313918. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mensah C.N., et al. N-alkylimidazole derivatives as potential inhibitors of quorum sensing in Pseudomonas aeruginosa. Heliyon. Dec. 2022;8(12) doi: 10.1016/j.heliyon.2022.e12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trabelsi A., El Kaibi M.A., Abbassi A., Horchani A., Chekir-Ghedira L., Ghedira K. Phytochemical study and antibacterial and antibiotic modulation activity of Punica granatum (pomegranate) leaves. Scientifica. 2020;2020 doi: 10.1155/2020/8271203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mant C.T., et al. De Novo designed amphipathic α-helical antimicrobial peptides incorporating dab and dap residues on the polar face to treat the gram-negative pathogen, Acinetobacter baumannii. J. Med. Chem. 2019;62(7):3354–3366. doi: 10.1021/acs.jmedchem.8b01785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y., Guarnieri M.T., Vasil A.I., Vasil M.L., Mant C.T., Hodges R.S. Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007;51(4):1398–1406. doi: 10.1128/AAC.00925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhuri A. Comparative analysis of non structural protein 1 of SARS-CoV2 with SARS-CoV1 and MERS-CoV: an in silico study. J. Mol. Struct. 2021;1243 doi: 10.1016/j.molstruc.2021.130854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zapadka K.L., Becher F.J., Gomes dos Santos A.L., Jackson S.E. Factors affecting the physical stability (aggregation) of peptide therapeutics. Interface Focus. 2017;7(6) doi: 10.1098/rsfs.2017.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar P., Kizhakkedathu J.N., Straus S.K. Antimicrobial peptides: diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules. 2018;8(1):4. doi: 10.3390/biom8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wimley W.C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010;5(10):905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J., et al. Development of protegrins for the treatment and prevention of oral mucositis: structure–activity relationships of synthetic protegrin analogues. Pept. Sci. 2000;55(1):88–98. doi: 10.1002/1097-0282(2000)55:1<88::AID-BIP80>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 40.Andrä J., Berninghausen O., Leippe M. Cecropins, antibacterial peptides from insects and mammals, are potently fungicidal against Candida albicans. Med. Microbiol. Immunol. 2001;189(3):169–173. doi: 10.1007/s430-001-8025-x. [DOI] [PubMed] [Google Scholar]
- 41.Rozek A., Friedrich C.L., Hancock R.E. Structure of the bovine antimicrobial peptide indolicidin bound to dodecylphosphocholine and sodium dodecyl sulfate micelles. Biochemistry. 2000;39(51):15765–15774. [PubMed] [Google Scholar]
- 42.Fernandez D.I., Le Brun A.P., Whitwell T.C., Sani M.-A., James M., Separovic F. The antimicrobial peptide aurein 1.2 disrupts model membranes via the carpet mechanism. Phys. Chem. Chem. Phys. 2012;14(45):15739–15751. doi: 10.1039/c2cp43099a. [DOI] [PubMed] [Google Scholar]
- 43.Dürr U.H., Sudheendra U.S., Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim. Biophys. Acta BBA-Biomembr. 2006;1758(9):1408–1425. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 44.Shai Y. Mode of action of membrane active antimicrobial peptides. Pept. Sci. 2002;66(4):236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 45.Subbalakshmi C., Nagaraj R., Sitaram N. Biological activities of C-terminal 15-residue synthetic fragment of melittin: design of an analog with improved antibacterial activity. FEBS Lett. 1999;448(1):62–66. doi: 10.1016/s0014-5793(99)00328-2. [DOI] [PubMed] [Google Scholar]
- 46.Park Y., Park S.-C., Park H.-K., Shin S.Y., Kim Y., Hahm K.-S. Structure-activity relationship of HP (2–20) analog peptide: enhanced antimicrobial activity by N-terminal random coil region deletion. Pept. Sci. 2007;88(2):199–207. doi: 10.1002/bip.20679. [DOI] [PubMed] [Google Scholar]
- 47.Housmans J.A., Wu G., Schymkowitz J., Rousseau F. FEBS J.; 2021. A Guide to Studying Protein Aggregation. [DOI] [PubMed] [Google Scholar]
- 48.Strodel B., Whittleston C.S., Wales D.J. Thermodynamics and kinetics of aggregation for the GNNQQNY peptide. J. Am. Chem. Soc. 2007;129(51):16005–16014. doi: 10.1021/ja075346p. [DOI] [PubMed] [Google Scholar]
- 49.Ding F., LaRocque J.J., Dokholyan N.V. Direct observation of protein folding, aggregation, and a prion-like conformational conversion. J. Biol. Chem. 2005;280(48):40235–40240. doi: 10.1074/jbc.M506372200. [DOI] [PubMed] [Google Scholar]
- 50.Calamai M., Taddei N., Stefani M., Ramponi G., Chiti F. Relative influence of hydrophobicity and net charge in the aggregation of two homologous proteins. Biochemistry. Dec. 2003;42(51):15078–15083. doi: 10.1021/bi030135s. [DOI] [PubMed] [Google Scholar]
- 51.Zou R., Zhu X., Tu Y., Wu J., Landry M.P. Activity of antimicrobial peptide aggregates decreases with increased cell membrane embedding free energy cost. Biochemistry. 2018;57(18):2606–2610. doi: 10.1021/acs.biochem.8b00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y., Huang J., Chen Y. “Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein Cell. 2010;1:143–152. doi: 10.1007/s13238-010-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin L.M., Edwards M.A., Li J., Yip C.M., Deber C.M. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J. Biol. Chem. 2012;287(10):7738–7745. doi: 10.1074/jbc.M111.303602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang J., Zhou M., Wu Y., Chen T., Shaw C., Wang L. The synergistic antimicrobial effects of novel bombinin and bombinin H peptides from the skin secretion of Bombina orientalis. Biosci. Rep. 2017;37(5) doi: 10.1042/BSR20170967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gopal R., et al. Synergistic effects and antibiofilm properties of chimeric peptides against multidrug-resistant acinetobacter baumannii strains. Antimicrob. Agents Chemother. Mar. 2014;58(3):1622–1629. doi: 10.1128/AAC.02473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu X., et al. Synergistic effects of antimicrobial peptide DP7 combined with antibiotics against multidrug-resistant bacteria. Drug Des. Dev. Ther. 2017;11:939. doi: 10.2147/DDDT.S107195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. material/referenced in article.