Abstract
Activating transcription factor 6 (ATF6), one of the three sensor proteins in the endoplasmic reticulum (ER), is an important regulator of ER stress-induced apoptosis. ATF6 resides in the ER and, upon activation, is translocated to the Golgi apparatus, where it is cleaved by site-1 protease (S1P) to generate an amino-terminal cytoplasmic fragment. Although recent studies have made progress in elucidating the regulatory mechanisms of ATF6, its function during early porcine embryonic development under high-temperature (HT) stress remains unclear. In this study, zygotes were divided into four groups: control, HT, HT+ATF6 knockdown, and HT+PF (S1P inhibitor). Results showed that HT exposure induced ER stress, which increased ATF6 protein expression and led to a decrease in the blastocyst rate. Next, ATF6 expression was knocked down in HT embryos under microinjection of ATF6 double-stranded RNA (dsRNA). Results revealed that ATF6 knockdown (ATF6-KD) attenuated the increased expression of CHOP, an ER stress marker, and Ca2+ release induced by HT. In addition, ATF6-KD alleviated homeostasis dysregulation among organelles caused by HT-induced ER stress, and further reduced Golgi apparatus and mitochondrial dysfunction in HT embryos. AIFM2 is an important downstream effector of ATF6. Results showed that ATF6-KD reduced the occurrence of AIFM2-mediated embryonic apoptosis at HT. Taken together, our findings suggest that ATF6 is a crucial mediator of apoptosis during early porcine embryonic development, resulting from HT-induced ER stress and disruption of organelle homeostasis.
Keywords: ATF6, Embryo, High temperature, ER stress, Organelle homeostasis
INTRODUCTION
During oocyte maturation and preimplantation embryo development, the synthesis of new proteins by the translation of maternal mRNA is extremely important (Guzel et al., 2017; Zeng & Schultz, 2005) and various functional proteins need to be correctly folded in the endoplasmic reticulum (ER). The ER is one of the most important organelles involved in the biosynthesis of lipids, proteins, and secreted proteins (Lin et al., 2019). However, mammalian oocytes and early embryos are highly sensitive to cellular and environmental stress. Increases in environmental temperature are a major factor leading to livestock fertility decline. Studies have revealed that high-temperature (HT) seasons can reduce the viability of bovine oocytes and embryos (Al-Katanani et al., 2002), and cause ER stress in porcine embryos, eventually leading to quality decline (Lee et al., 2022).
The ER is the major organelle responsible for protein synthesis in eukaryotic cells (McAlpine & Werstuck, 2013; Tabas & Ron, 2011). Accumulation of excess unfolded or misfolded proteins in the ER lumen leads to ER stress and functional disruption, thereby activating the unfolded protein response (UPR) (Senft & Ronai, 2015). The UPR is primarily regulated by three sensors: activating transcription factor 6 (ATF6), double-stranded RNA (dsRNA)-activated protein kinase-like ER kinase (PERK), and inositol-requiring enzyme 1 (IRE1), as well as the ER chaperone immunoglobulin protein-binding protein (BiP, also known as glucose-regulated protein (GRP78)) (Ghemrawi et al., 2018; Song et al., 2018). Under normal conditions, ATF6, IRE, and PERK interact with GRP78/BiP; however, under stress conditions, misfolded proteins in the ER increase and the sensors dissociate from GRP78/BiP, thereby activating the UPR (Frakes & Dillin, 2017; Hetz & Papa, 2018). As normal cellular adaptation mechanisms, these coping responses play a key role in the development of preimplantation embryos. Therefore, UPR-associated molecules and pathways may be useful markers for diagnosing stress conditions in preimplantation embryos (Michalak & Gye, 2015).
Previous studies have revealed that loss of IRE1 is lethal, while loss of PERK induces profound metabolic disturbances in mice (Zhang et al., 2002, 2005), suggesting the involvement of these ER sensors in maintaining organelle and organismal homeostasis. However, studies using ATF6α-knockout models have not induced overt phenotypes (Wu et al., 2007; Yamamoto et al., 2007). Likewise, studies on Caenorhabditis elegans have indicated that IRE1, XBP1, and PERK/PEK1 mutants exhibit heightened sensitivity to tunicamycin, while ATF6 mutants display minimal baseline phenotypes and exhibit a normal wild-type response to tunicamycin (Bischof et al., 2008; Shen et al., 2005). These results suggest that ATF6 may play a role in maintaining ER and cellular homeostasis beyond protein folding. However, the precise role of ATF6 in early porcine embryonic development under HT conditions has not been investigated. In the present study, we explored the role of ATF6 under HT during early porcine embryonic development. Results showed that reducing the expression of ATF6, induced by HT, alleviated ER stress and Golgi and mitochondrial dysfunction in embryos, thereby reducing the occurrence of apoptosis mediated by AIFM2, leading to an increase in the blastocyst rate.
MATERIALS AND METHODS
Unless otherwise indicated, all chemicals were purchased from Millipore Sigma (Burlington, Massachusetts, USA), and all experiments were conducted on a heating table adjusted to 38.5 °C.
Ethics statement
This study was performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of Chungbuk National University Laboratory Animal Center, Cheongju, Korea. All surgeries involving porcine embryos were performed according to the committee guidelines.
Porcine oocyte collection and in vitro maturation
Porcine ovaries obtained from a local slaughterhouse were sent to the laboratory in saline at 37 °C, supplemented with 75 mg/mL penicillin G and 50 mg/mL streptomycin sulfate. Cumulus-oocyte complexes (COCs) were obtained from follicles (3–6 mm in diameter) via aspiration using an 18-gauge needle attached to a 10 mL disposable syringe, with COCs showing at least three layers of dense cumulus cells and an evenly granulated ooplasm selected for experiments. The COCs were transferred to in vitro maturation (IVM) medium (TCM-199 (11150-059; Gibco, USA) supplemented with 10% (v/v) porcine follicular fluid, 0.1 g/L sodium pyruvate, 10 ng/mL epidermal growth factor, 0.6 mmol/L L-cysteine, 10 IU/mL luteinizing hormone, and 10 IU/mL follicle-stimulating hormone) and covered with mineral oil. The plates were cultured in a humidified 5% CO2 incubator at 38.5 °C for 44 h.
Parthenogenetic activation and in vitro culture
The COCs were placed in 1 mg/mL hyaluronidase via repeated pipetting to remove the cumulus cells, with the denuded oocytes then passed through two direct-current pulses of 120 V for 60 µs (FL201, Nepa gene, Japan) in 297 mmol/L mannitol containing 0.05 mmol/L MgSO4, 0.1 mmol/L CaCl2, 0.5 mmol/L mmol/L HEPES, and 0.01% polyvinyl alcohol (PVA, w/V) (pH 7.2). The oocytes were subsequently cultured for 3 h in bicarbonate-buffered porcine zygote medium 5 (PZM-5) containing 4 mg/mL bovine serum albumin (BSA) and 7.5 µg/mL cytochalasin B to suppress pseudo extrusion of the second polar body. The activated oocytes were thoroughly washed using in vitro culture (IVC) medium (PZM-5 supplemented with 4 mg/mL BSA). The activated oocytes were divided into four groups: (a) control group (control), (b) HT group, (c) HT group with ATF6-KD (HT+ATF6-KD), and (d) HT group with PF inhibitor (PF 429242 dihydrochloride) (HT+PF). Each group was cultured in IVC medium covered with mineral oil for 7 days at 38.5 °C in a humid environment containing 5% CO2, among which the HT, HT+ATF6-KD, and HT+PF groups were cultured at 39.5 °C or 40.5 °C.
Preparation and microinjection of ATF6 dsRNA
To prepare ATF6 dsRNA for knockdown experiments, ATF6 was amplified using a pair of specific primers containing the T7 promoter sequence at the 5ʹ end (ATF6 dsRNA: forward, TAA TAC GAC TCA CTA TAG GGG TTA CTT CCA GCA GCA CCC A; reverse, TAA TAC GAC TCA CTA TAG GGC CTG TTC CAA TAT ACT CAT AGG TCC). The purified polymerase chain reaction (PCR) products were used to synthesize dsRNA using a MEGAscript T7 Kit (AM1333; Thermo Fisher Scientific, USA) according to the manufacturer’s instructions (Niu et al., 2021; Zhou et al., 2021). After in vitro transcription, dsRNA was treated with DNase I and RNase A to remove the DNA template and any single-stranded RNA, with the purified dsRNA then dissolved in RNase-free water. Concentration of dsRNA were determined by measuring the optical density at 260 nm (Nanodrop, Thermo Fisher, Deutsch, Germany), followed by adjustment to a final concentration of 1 µg/µL. The dsRNA aliquots were stored at −80 °C until use.
For the knockdown experiments, ATF6 dsRNA was microinjected into the cytoplasm of oocytes using Femto-Jet electronic microinjectors (Eppendorf AG, Germany) and a Diaphot ECLIPSE TE300 microscope (Nikon, Japan) equipped with MM0-202N hydraulic three-dimensional micromanipulators (Narishige, Japan) following parthenogenetic activation and cytochalasin B treatment for 3 h. After microinjection, the embryos were cultured in PZM-5. As a negative control, water was microinjected into the zygotes under the same conditions.
Real-time quantitative PCR (RT-qPCR)
Thirty embryos were collected from each stage in each group, and mRNA was extracted using a Dynabeads mRNA Direct Purification Kit (61012; Thermo Fisher Scientific, USA) according to the manufacturer’s instructions (Zhou et al., 2020). The cDNA was obtained via reverse transcription of mRNA using oligo(dT) 20 primers and SuperScript III reverse transcriptase (Thermo Fisher Scientific, USA). RT-qPCR was performed using a WizPure qPCR Master Kit at a final volume of 20 μL per reaction containing 1 μL of each forward and reverse primer, 10 μL of SYBR Green, and 2 μL of cDNA template. Amplification was performed at 95 °C for 3 min, followed by 40 cycles at 95 °C for 15 s, 60 °C for 25 s, 72 °C for 10 s, and a final extension at 72 °C for 5 min. The target and housekeeping genes were ATF6 (forward, AAT GGA TCA CTG AAG CGG CA; reverse, CCT GTT CCA ATA TAC TCA TAG GTC C) and 18S (forward, CGC GGT TCT ATT TTG TTG GT; reverse, AGT CGG CAT CGT TTA TGG TC), respectively. The mRNA quantification data were analyzed using the 2-ΔΔCt method (Schmittgen & Livak, 2008).
Immunofluorescence staining and confocal microscopy
For immunofluorescence staining, live embryos were immediately fixed in 3.7% paraformaldehyde for 1 h and permeabilized with phosphate-buffered saline (PBS)/PVA containing 0.5% Triton X-100 for 1 h at room temperature. Embryos were then transferred to blocking buffer (PBS containing 1% BSA and 0.01% Triton X-100) and blocked for 1 h at room temperature or overnight at 4 °C. Embryos were then incubated at 4 °C overnight with primary antibodies against ATF6 (1:100; Novus Biologicals, NBP1-40256, USA), AIFM2 (1:100; Proteintech, 20886-1-AP, China), and giantin (1:100; Abcam, ab80864, UK). After washing three times in PBS/PVA (3 min each time), the embryos were transferred to a secondary antibody mixture containing goat anti-rabbit Alexa Fluor 488 (Life Technologies, A10040, USA) or goat anti-mouse Alexa Fluor 568 (Life Technologies, A21202, USA) (1:200), and incubated for 1 h at room temperature. The sections were then stained with Hoechst 33342 (bis-benzyl-imine H33342 trihydrochloride; 1:2000; Sigma Life Science, USA) for 15 min, washed three times with PBS/PVA, mounted on slides, and analyzed using a confocal microscope (Zeiss LSM 710 META, Germany). Images were processed using Zen (v8.0; Zeiss, Germany).
Measurement of Ca2+
Embryos were incubated at 38.5 °C for 1 h in IVC medium containing Fluo-4-AM (5 µmol/L, Invitrogen, USA). After washing three times with fresh IVC, the embryos were transferred to medium containing Rhod-2-AM (2 µmol/L, Invitrogen, USA), then incubated at 38.5 °C for 1 h. Fluorescence signals were captured using an epifluorescence microscope after three additional washes.
MitoTracker staining
Blastocysts were incubated with 500 nmol/L MitoTracker Red CMXRos (Invitrogen, Cat # M7512, USA) in IVC at 38.5 °C for 30 min. After washing three times with fresh culture medium, the embryos were fixed in 3.7% paraformaldehyde at room temperature for 1 h, then stained with 10 µg/mL Hoechst 33342 for 10 min, washed three times with PBS/PVA, mounted onto slides, and examined under a confocal microscope (Zeiss LSM 710 META, Germany).
Mitochondrial membrane potential (MMP) assay
Embryos were incubated for 30 min at 38.5 °C under 5% CO2 with 2.5 µmol/L 5,5′,6,6′-tetrachloro-1,1′,3,3′- tetraethylbenzimidazolylcarbocyanine iodide (JC-1) (M34152; Thermo Fisher, USA) in PZM-5. MMP was calculated as the ratio of red fluorescence, corresponding to activated mitochondria (J-aggregates), to green fluorescence, corresponding to less-activated mitochondria (J-monomers). Embryos were stained with 10 µg/mL Hoechst 33342 for 10 min, washed three times with PBS/PVA, mounted on slides, and examined under a scanning laser confocal microscope (Zeiss LSM 710 META, Germany).
TUNEL assay
The blastocysts were fixed in 3.7% (w/v) paraformaldehyde for 1 h at room temperature and permeabilized by incubation in 0.5% (v/v) Triton X-100 for 1 h at room temperature. The embryos were incubated with fluorescein-conjugated dUTP and terminal deoxynucleotidyl transferase enzyme (1:20, In Situ Cell Death Detection Kit; Roche, Germany) for 2 h at room temperature, then washed three times with PBS/PVA. The embryos were treated with Hoechst 33342 for 10 min, thrice washed with PBS/PVA, mounted onto glass slides, and imaged using a confocal microscope (LSM 710, Zeiss, Germany).
Western blotting
We collected at least 70 embryos per group in 20 µL of ice-cold Laemmli sample buffer (sodium dodecyl sulfate (SDS) sample buffer containing 2-mercaptoethanol) heated at 95 °C for 10 min and stored at –20 °C. Based on previous study (Zhou et al., 2022), proteins were separated by molecular weight using SDS-polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to polyvinylidene fluoride membranes. The membranes were blocked in Tris-buffered saline (TBS) containing 0.1% Tween 20 and 5% skim milk for 1 h at room temperature, then incubated overnight at 4 °C with ATF6 (1:1 000; Cat # 24169-1-AP, Proteintech, China) or GAPDH (1:1 000, 5174S; Cell Signaling Technology, USA) antibodies. Subsequently, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (IgG) or goat anti-mouse IgG (1:20 000; Santa Cruz Biotechnology, USA) secondary antibodies for 1 h at room temperature. Finally, the membranes were exposed to the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, USA). Band intensity values were analyzed using ImageJ (version 1.53, National Institutes of Health, USA).
Statistical analysis
Each experiment was repeated at least three times, with representative figures shown. Statistical comparisons were performed using an independent-sample t-test. Statistical significance was set at P<0.05. Fluorescence intensity of the control group was arbitrarily set to 1 and fluorescence intensities of the other groups were expressed relative to the control group. Statistical analysis was performed using GraphPad Prism (v5.0; GraphPad, USA).
RESULTS
Effects of HT on ATF6 expression and localization
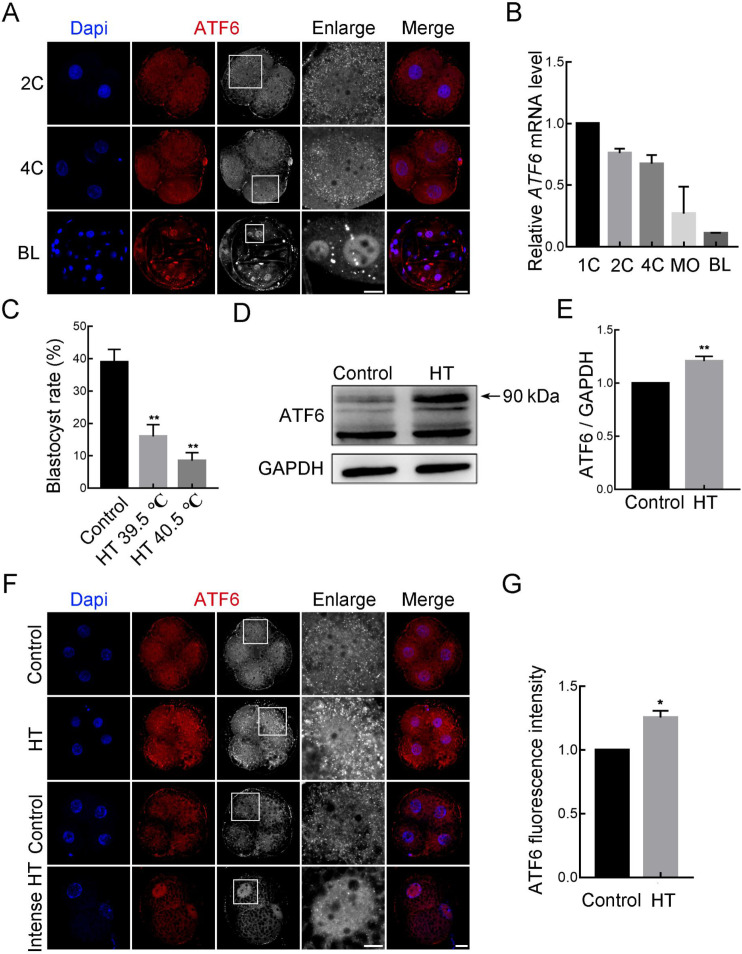
To determine the function of ATF6 during early porcine embryonic development, we detected its localization and expression patterns at different stages using immunofluorescence staining and RT-qPCR. As shown in Figure 1A and B, we cultured embryos to the 1-cell (1C), 2-cell (2C), 4-cell (4C), morula (MO), and blastocyst (BL) stages and observed that ATF6 was expressed in all stages. We used the mRNA level of ATF6 at 1C as a control and set it to 1. Results showed that ATF6 mRNA expression gradually decreased during embryonic development. Using ATF6 antibodies, we observed that ATF6 was accumulated around the nucleus in the form of dots at the 2C and 4C stages but was mainly localized in the nucleus at the BL stage.
Figure 1.
Effects of high-temperature (HT) on ATF6 expression pattern and localization
A: Immunofluorescence images for ATF6 expression at 2C, 4C, and BL stages. Blue, DAPI; Red, ATF6. Bar: 20 μm. B: RT-qPCR results of ATF6 mRNA expression levels during early porcine embryonic development. C: Blastocyst rate after HT exposure at 39.5 °C and 40.5 °C. **: P<0.01. D: Western blotting results of ATF6 protein expression after HT exposure at 4C stage. E: Band intensity analysis of ATF6 after HT exposure. **: P<0.01. F: Typical images of ATF6 intensity at 4C stage after HT exposure. Blue, DAPI; Red, ATF6. Bar: 20 μm. G: Fluorescence intensity of ATF6 at 4C stage after HT exposure. 1C: 1-cell; 2C: 2-cell; 4C: 4-cell; MO: Morula; BL: Blastocyst.
Based on the role of ATF6 in transducing stress-related signals to the ER, we studied the changes in ATF6 under HT conditions. As shown in Figure 1C, we tested the effects of different temperatures on early porcine embryonic development. Results revealed that the blastocyst rates in the 39.5 °C and 40.5 °C HT groups were significantly lower than those in the 38.5 °C control group (control group, 40.00%±4.45%, n=303; HT 39.5 °C group, 16.74%±4.60%, n=303, P<0.01; HT 40.5 °C group, 8.52%±2.46%, n=303, P<0.01). Therefore, 39.5 °C was selected as the HT in subsequent experiments. We also detected a significant increase in the protein expression level of ATF6 in the HT group compared with that in the control group using western blotting (control group, 1.00, n=280; HT group, 1.21±0.02, n=280, P<0.01; Figure 1D, E). In addition, immunofluorescence staining confirmed that fluorescence intensity of the ATF6 punctate signal around the nucleus increased significantly at the 4C stage (control group, 1.00, n=53; HT group, 1.26±0.05, n=60, P<0.05; Figure 1F, G). Furthermore, when the embryos were treated with intense HT (42.5 °C), most arrested at the 2C stage and ATF6 was mainly localized in the nucleus. Therefore, based on its expression and localization patterns in embryos, ATF6 appears to play an important role under HT conditions.
Effects of ATF6 knockdown on HT-embryonic development
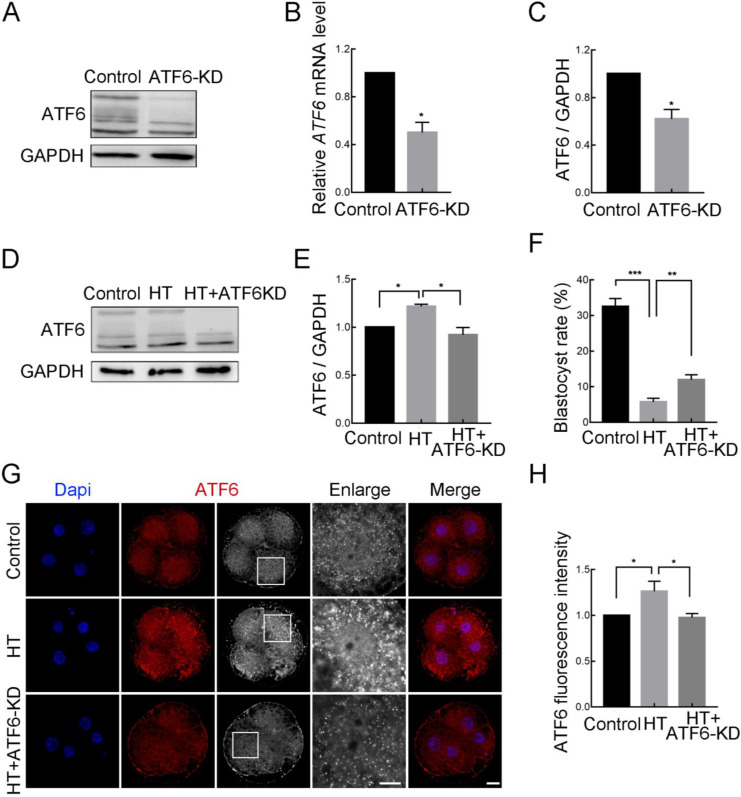
To further determine the potential role of ATF6 in embryos under HT conditions, we knocked down ATF6 expression and detected knockdown efficiency (Figure 2A, B). After microinjection of ATF6 dsRNA, the mRNA and protein expression levels at the 4C stage were significantly decreased in the ATF6-KD group compared with the control group, as detected by western blotting and RT-qPCR (control group, 1.00, n=120; ATF6-KD group, 0.50±0.08, n=120, P<0.05) and confirmed via densitometric analysis of the bands (control group, 1.00, n=280; ATF6-KD group, 0.62±0.08, n=280, P<0.05) (Figure 2C). In addition, western blotting revealed that the elevation in ATF6 expression at the 4C stage induced by HT could be reduced by microinjection of ATF6 dsRNA (control group, 1.00, n=210; HT group, 1.22±0.02, n=210, P<0.05; HT+ATF6-KD group, 0.92±0.08, n=210, P<0.05; Figure 2D, E). Moreover, compared with the HT group, ATF6-KD alleviated the decrease in the blastocyst rate caused by HT (control group, 32.54%±0.49%, n=246; HT group, 5.76%±1.04%, n=235, P<0.001; HT+ATF6-KD group, 11.98%±1.52%, n=239, P<0.01; Figure 2F). Immunofluorescence staining also revealed that compared to the increase in the ATF6 fluorescent punctate signal in the HT group, the signal in the HT+ATF6-KD group was significantly weakened at the 4C stage (control group, 1.00, n=65; HT group, 1.26±0.06, n=74, P<0.05; HT+ATF6-KD group, 0.98±0.02, n=64, P<0.05; Figure 2G, H). These results suggest that reducing ATF6 expression can attenuate HT damage in early porcine embryos.
Figure 2.
Effects of ATF6 knockdown on high-temperature (HT)-embryonic development
A: Western blotting results of ATF6 protein expression after microinjection of ATF6 dsRNA. B: RT-qPCR results of ATF6 mRNA expression levels in control and ATF6‐KD groups. *: P<0.05. C: Band intensity analysis of ATF6 after ATF6 dsRNA microinjection. *: P<0.05. D: Western blotting results of ATF6 protein expression after HT exposure and ATF6 dsRNA microinjection at 4C stage. E: Band intensity analysis of ATF6 after HT exposure and ATF6 dsRNA microinjection at 4C stage. *: P<0.05. F: Blastocyst rate after HT exposure and ATF6 dsRNA microinjection. **: P<0.01; ***: P<0.001. G: Typical images of ATF6 intensity at 4C stage after HT exposure and ATF6 dsRNA microinjection. Blue, DAPI; Red, ATF6. Bar: 20 μm. H: Fluorescence intensity of ATF6 at 4C stage after HT exposure and ATF6 dsRNA microinjection. *: P<0.05.
Effects of ATF6 knockdown on ER function in HT-embryos
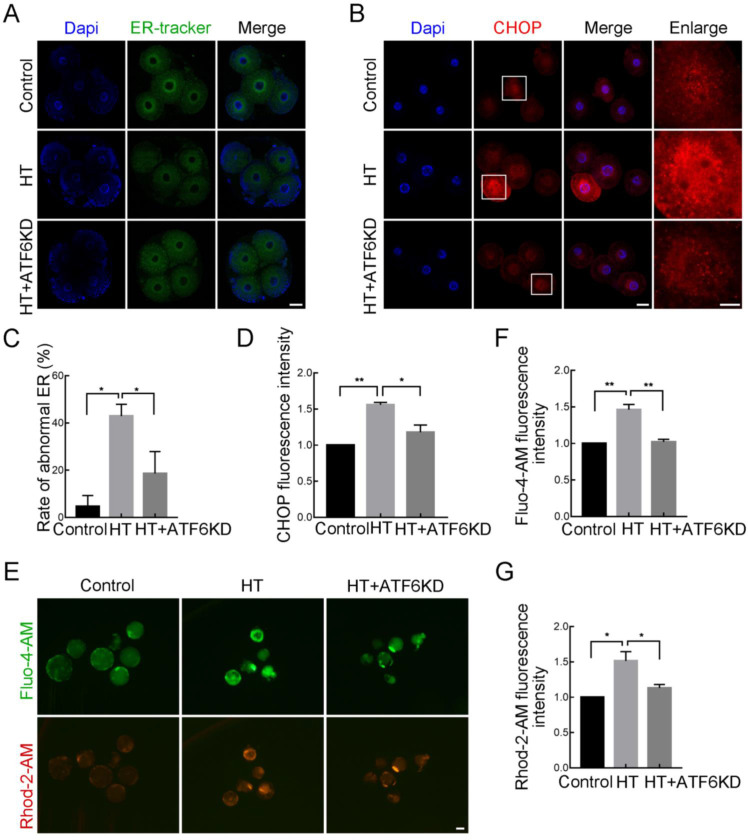
As ATF6 acts as an ER-localized protein, we next examined the effects of ATF6 on ER function under an HT environment. First, using an ER tracker to observe ER distribution, we found that in the control group embryos, the ER was mainly distributed around the nucleus at the 4C stage (Figure 3A, C); however, in the HT group, ER accumulation was lower in the peripheral region of the nucleus. We quantified the frequency of this abnormal distribution and discovered a significantly higher proportion in the HT group, but a decreased proportion in the HT+ATF6-KD group (control group, 4.46%±4.46%, n=25; HT group, 42.93%±4.97%, n=34, P<0.05; HT+ATF6-KD group, 18.61%±9.31%, n=27, P<0.05). In addition, CHOP, an important target gene of ATF6 in ER stress, showed a significant increase in fluorescence intensity in the perinuclear region at the 4C stage in the HT group compared with the control group, but a significant decrease in the HT+ATF6-KD group compared with the HT group (control group, 1.00, n=42; HT group, 1.56±0.03, n=43, P<0.01; HT+ATF6-KD group, 1.18±0.10, n=46, P<0.05; Figure 3B, D). ER stress results in the release of Ca2+ from the ER, emphasizing the important role of the ER in Ca2+ homeostasis. Therefore, we examined Ca2+ content in the cytosol and mitochondria using Fluo-4-AM and Rhod-2-AM, respectively. Results showed that the Fluo-4-AM and Rhod-2-AM fluorescence signals at the BL stage in the HT group were significantly increased compared with those in the control group, while signals in the HT+ATF6-KD group were significantly decreased compared with those in the HT group (Fluo-4-AM: control group, 1.00, n=60; HT group, 1.44±0.07, n=43, P<0.01; HT+ATF6-KD group, 1.02±0.04, n=62, P<0.01. Rhod-2-AM: control group, 1.00, n=60; HT group, 1.47±0.13, n=43, P<0.05; HT+ATF6-KD group, 1.13±0.05, n=62, P<0.05; Figure 3E–G). Thus, these findings suggest that ATF6 knockdown can alleviate ER stress and Ca2+ homeostatic imbalance induced by HT.
Figure 3.
Effects of ATF6 knockdown on ER function in high-temperature (HT)-embryos
A: Typical images of ER-tracker distribution at 4C stage after HT exposure and ATF6 dsRNA microinjection. Blue, DAPI; Green, ER-tracker. Bar: 20μm. B: Typical images of CHOP intensity at 4C stage after HT exposure and ATF6 dsRNA microinjection. Blue, DAPI; Red, CHOP. Bar: 20 μm. C: Abnormal ER distribution rate at 4C stage after HT exposure and ATF6 dsRNA microinjection. *: P<0.05. D: Fluorescence intensity of CHOP at 4C stage after HT exposure and ATF6 dsRNA microinjection. *: P<0.05; **: P<0.01. E: Typical images of Fluo-4-AM and Rhod-2-AM intensity at BL stage after HT exposure and ATF6 dsRNA microinjection. Red, Rhod-2-AM; Green, Fluo-4-AM. Bar: 100 μm. F: Fluorescence intensity of Fluo-4-AM at BL stage after HT exposure and ATF6 dsRNA microinjection. **: P<0.01. G: Fluorescence intensity of Rhod-2-AM at BL stage after HT exposure and ATF6 dsRNA microinjection. *: P<0.05.
Effects of ATF6 knockdown on Golgi function in HT-embryos
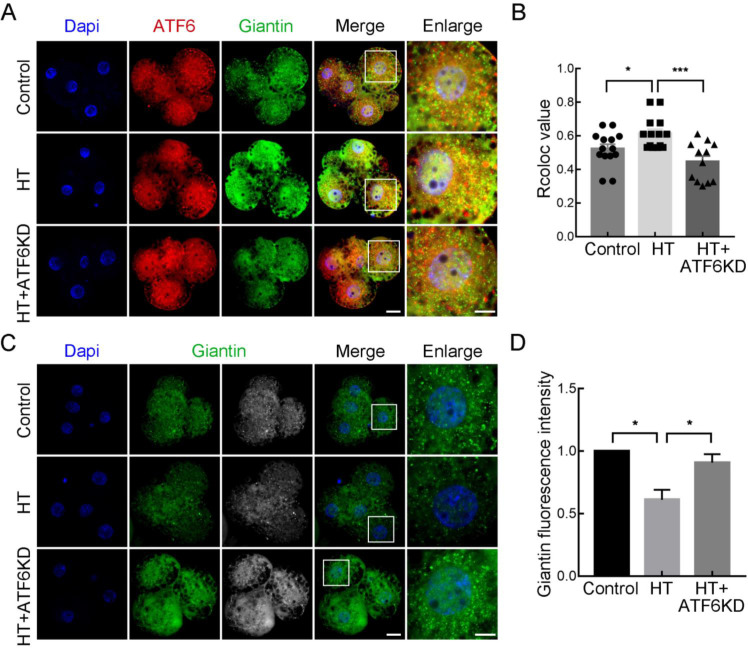
As ER stress can induce the dissociation of ATF6 from BiP, resulting in its translocation to the Golgi apparatus for further cleavage, we examined the localization of ATF6 and the Golgi apparatus at the 4C stage in different groups. As shown in Figure 4A, B, giantin was used as a Golgi marker to indicate its localization and correlation value. Results showed that the Rcoloc value (the Pearson's correlation coefficient) which measured by Image J software using “Coloc2” plugin to reflect the degree of fluorescence signal colocalization was significantly increased in the HT group compared with the control and HT+ATF6-KD groups (control group, 0.52±0.03, n=14; HT group, 0.61±0.03, n=14, P<0.05; HT+ATF6-KD group, 0.45±0.03, n=12, P<0.001), representing increased colocalization of ATF6 and giantin. In addition, giantin fluorescence intensity was significantly lower in the HT group than in the control group, but significantly higher in the HT+ATF6-KD group than in the HT group (control group, 1.00, n=60; HT group, 0.61±0.08, n=43, P<0.05; HT+ATF6-KD group, 0.91±0.07, n=12, P<0.05; Figure 4C, D). These findings suggest that HT induces the translocation of ATF6 to the Golgi apparatus and affects its function, which can be improved by ATF6 knockdown.
Figure 4.
Effects of ATF6 knockdown on Golgi function in high-temperature (HT)-embryos
A: Typical images of ATF6 and giantin location at 4C stage after HT exposure and ATF6 dsRNA microinjection. Blue, DAPI; Red, ATF6; Green, giantin. Bar: 20 μm. B: Rcoloc value of ATF6 and giantin at 4C stage after HT exposure and ATF6 dsRNA microinjection. *: P<0.05; ***: P<0.001. C: Typical images of giantin intensity at 4C stage after HT exposure and ATF6 dsRNA microinjection. Blue, DAPI; Green, giantin. Bar: 20 μm. D: Fluorescence intensity of giantin at 4C stage after HT exposure and ATF6 dsRNA microinjection. *: P<0.05.
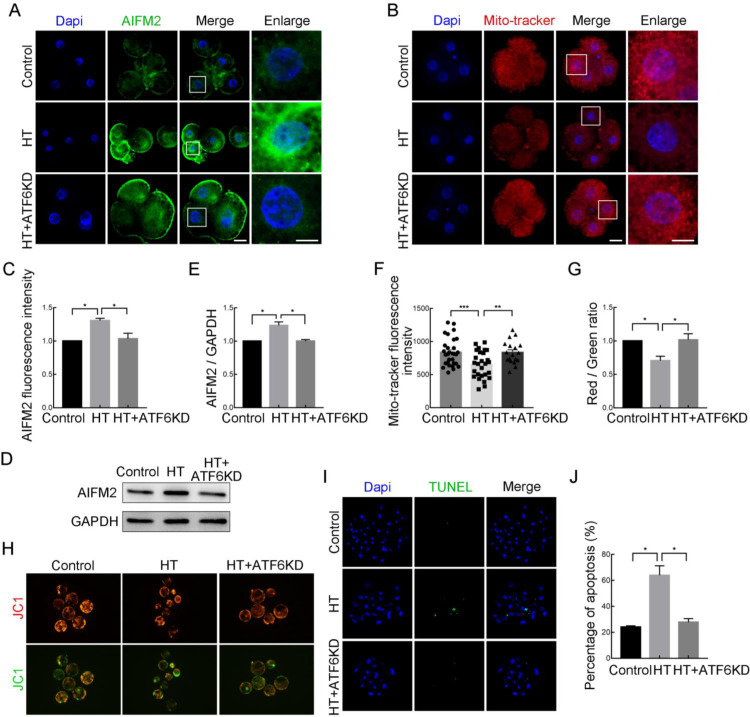
Effects of ATF6 on mitochondrial function and apoptosis in HT-embryos
To further explore the potential mechanism by which ATF6 regulates embryonic development under HT, we examined mitochondria-related functions. Apoptosis-inducing factor mitochondrial-associated 2 (AIFM2), localized in the outer mitochondrial membrane or freely distributed in the cytoplasm (Ohiro et al., 2002; Wu et al., 2004), is an important downstream regulatory partner of ATF6. As shown in Figure 5A, C, the fluorescence signal of AIFM2 at the 4C stage was significantly increased in the HT group compared with the control and HT+ATF6-KD group (control group, 1.00, n=30; HT group, 1.31±0.03, n=31, P<0.05; HT+ATF6-KD group, 1.03±0.08, n=29, P<0.05), as confirmed by western blot analysis (control group, 1.00, n=280; HT group, 1.24±0.05, n=280, P<0.05; HT+ATF6-KD group, 1.00±0.02, n=280, P<0.05; Figure 5D, E). The mitochondria and ER are dynamic organelles that interact physiologically and functionally. Based on the effects of ATF6 on ER function in early porcine embryos under HT conditions, we next examined mitochondrial function. Results showed that MitoTracker signal intensity in the 4C embryos was significantly decreased in the HT group compared with the control group but was significantly increased in the HT+ATF6-KD group compared with the HT group (control group, 857.35±49.29, n=30; HT group, 572.22±50.09, n=27, P<0.001; HT+ATF6-KD group, 802.90±58.70, n=11, P<0.01; Figure 5B, F). In addition, MMP (Δψ) was detected using JC1 staining. The J-aggregate (red) to J-monomer (green) ratio at the BL stage was significantly decreased in the HT-treated group compared with the control group, indicating a decrease in the MMP of embryos, whereas the MMP in the HT+ATF6-KD group was significantly increased compared with that in the HT group (control group, 1.00, n=30; HT group, 0.71±0.07, n=31, P<0.05; HT+ATF6-KD group, 1.02±0.09, n=29, P<0.05). Based on changes in AIFM2 expression and mitochondrial dysfunction caused by HT, we next detected embryonic apoptosis at the BL stage using TUNEL staining. As shown in Figure 5I, J, compared with the control and HT+ATF6-KD groups, the proportion of apoptotic cells in the HT group was significantly increased (control group, 24.07%±0.93%; HT group, 63.89%±7.35%, P<0.05; HT+ATF6-KD group, 27.78%±2.788%, P<0.05). These results suggest that ATF6 regulates embryonic apoptosis under HT conditions via AIFM2.
Figure 5.
Effects of ATF6 on mitochondrial function and apoptosis in high-temperature (HT)-embryos
A: Typical images of AIFM2 intensity at 4C stage after HT exposure and ATF6 dsRNA microinjection. Blue, DAPI; Green, AIFM2. Bar: 20 μm. B: Typical images of MitoTracker intensity at 4C stage after HT exposure and ATF6 dsRNA microinjection. Blue, DAPI; Red, MitoTracker. Bar: 20 μm. C: Fluorescence intensity of AIFM2 at 4C stage after HT exposure and ATF6 dsRNA microinjection. *: P<0.05. D: Western blotting results of AIFM2 protein expression after HT exposure and ATF6 dsRNA microinjection. E: Band intensity analysis of AIFM2 after HT exposure and ATF6 dsRNA microinjection at 4C stage. *: P<0.05. F: Fluorescence intensity of MitoTracker at 4C stage after HT exposure and ATF6 dsRNA microinjection. **: P<0.01; ***: P<0.001. G: JC-1 signal (red/green ratio) after HT exposure and ATF6 dsRNA microinjection. *: P<0.05. H: Typical images of JC-1 green and red channels at BL stage after HT exposure and ATF6 dsRNA microinjection. Bar: 100 μm. I: Typical images of TUNEL staining at BL stage after HT exposure and ATF6 dsRNA microinjection. Blue, DAPI; Green, TUNEL. Bar: 20 μm. J: Percentage of apoptosis at BL stage after HT exposure and ATF6 dsRNA microinjection. *: P<0.05.
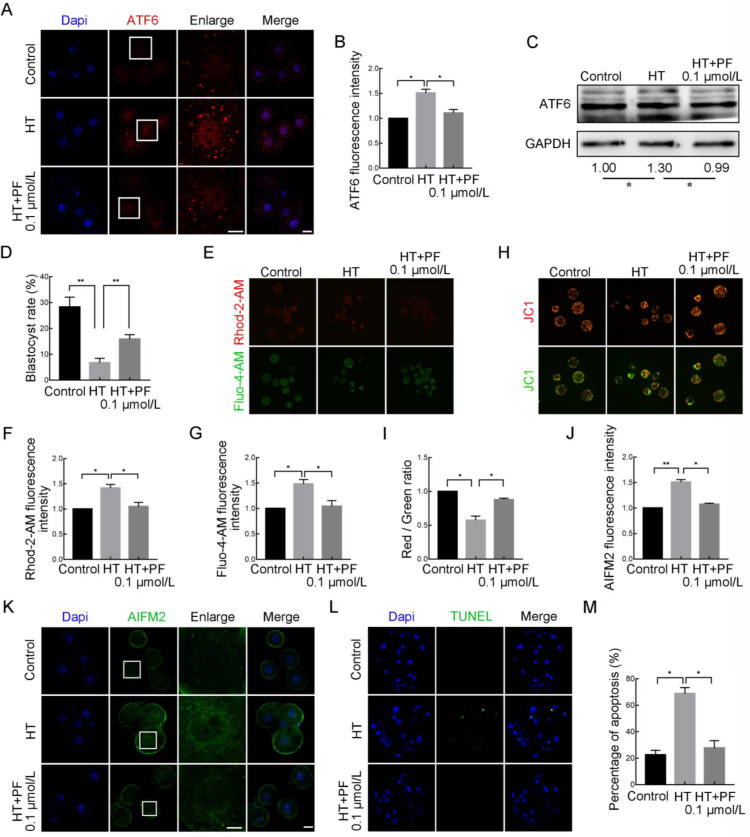
Effects of site-1 protease (S1P) inhibition on HT-embryonic development
After translocation to the Golgi apparatus, ATF6 is cleaved by two proteases, S1P and S2P. Here, we used a PF inhibitor (PF 429242 dihydrochloride) to inhibit S1P and verify the findings of the previous knockdown experiments. The HT embryos were treated with 0.1 μmol/L PF for 48 h until the 4C stage, then washed three times for subsequent experiments or placed in fresh IVC medium to continue culturing to the desired stage. As shown in Figure 6A, B, compared to the increase in the ATF6 fluorescent punctate signal around the nucleus at the 4C stage in the HT group, the signal in the HT+PF group was significantly attenuated (control group, 1.00, n=48; HT group, 1.51±0.07, n=45, P<0.05; HT+PF group, 1.11±0.07, n=43, P<0.05). Western blotting also revealed that the elevation in ATF6 expression at the 4C stage induced by HT could be reduced by PF treatment (control group, 1.00, n=280; HT group, 1.30±0.09, n=280, P<0.05; HT+PF group, 0.99±0.10, n=280, P<0.05; Figure 6C). In addition, PF treatment rescued the decrease in the blastocyst rate caused by HT (control group, 28.32%±3.81%, n=248; HT group, 6.75%±1.70%, n=254, P<0.01; HT+PF group, 15.87%±1.76%, n=252, P<0.01; Figure 6D). Regarding ER function, the Fluo-4-AM and Rhod-2-AM fluorescence signals at the BL stage were significantly decreased in the HT+PF group compared to the HT group (Fluo-4-AM: control group, 1.00, n=47; HT group, 1.56±0.09, n=56, P<0.05; HT+PF group, 1.04±0.11, n=51, P<0.05. Rhod-2-AM: control group, 1.00, n=70; HT group, 1.47±0.07, n=58, P<0.05; HT+PF group, 1.05±0.08, n=62, P<0.05; Figure 6E–G). In addition, the reduction in mitochondrial MMP at the BL stage in the HT group was alleviated in the HT+PF group (control group, 1.00, n=43; HT group, 0.57±0.06, n=34, P<0.05; HT+PF group, 0.88±0.02, n=35, P<0.05; Figure 6H, I). Furthermore, AIFM2 expression at the 4C stage was decreased in the HT+PF group compared to the HT group (control group, 1.00, n=45; HT group, 1.51±0.05, n=46, P<0.01; HT+PF group, 1.08±0.02, n=45, P<0.05), as was the proportion of AIFM2-mediated apoptosis at the BL stage (control group, 22.49%±3.44%; HT group, 68.81%±4.52%, P<0.05; HT+PF group, 27.78%±5.55%, P<0.05; Figure 6J–M). These results further confirm that ATF6 plays an important role in early embryonic development in pigs under HT conditions.
Figure 6.
Effects of S1P inhibition (PF) on high-temperature (HT)-embryonic development
A: Typical images of ATF6 intensity at 4C stage after HT exposure and treatment with PF at 0.1 μmol/L. Blue, DAPI; Red, ATF6. Bar: 20 μm. B: Fluorescence intensity of ATF6 at 4C stage after HT exposure and treatment with PF at 0.1 μmol/L. *: P<0.05. C: Western blotting results of ATF6 protein expression after HT exposure and treatment with PF at 0.1 μmol/L at 4C stage. *: P<0.05. D: Blastocyst rate after HT exposure and ATF6 treatment with PF at 0.1 μmol/L. **: P<0.01. E: Typical images of Fluo-4-AM and Rhod-2-AM intensity at BL stage after HT exposure and treatment with PF at 0.1 μmol/L. Red, Rhod-2-AM; Green, Fluo-4-AM. Bar: 100 μm. F: Fluorescence intensity of Rhod-2-AM at BL stage after HT exposure and treatment with PF at 0.1 μmol/L. *: P<0.05. G: Fluorescence intensity of Fluo-4-AM at BL stage after HT exposure and treatment with PF at 0.1 μmol/L. *: P<0.05. H: Typical images of JC-1 green and red channels at BL stage after HT exposure and treatment with PF at 0.1 μmol/L. Bar: 100 μm. I: JC-1 signal (red/green ratio) after HT exposure and treatment with PF at 0.1 μmol/L. *: P<0.05. J: Fluorescence intensity of AIFM2 at 4C stage after HT exposure and treatment with PF at 0.1 μmol/L. *: P<0.05; **: P<0.01. K: Typical images of AIFM2 intensity at 4C stage after HT exposure and treatment with PF at 0.1 μmol/L. Blue, DAPI; Green, AIFM2. Bar: 20 μm. L: Typical images of TUNEL staining at BL stage after HT exposure and treatment with PF at 0.1 μmol/L. Blue, DAPI; Green, TUNEL. Bar: 20 μm. M: Percentage of apoptosis at BL stage after HT exposure and treatment with PF at 0.1 μmol/L. *: P<0.05.
DISCUSSION
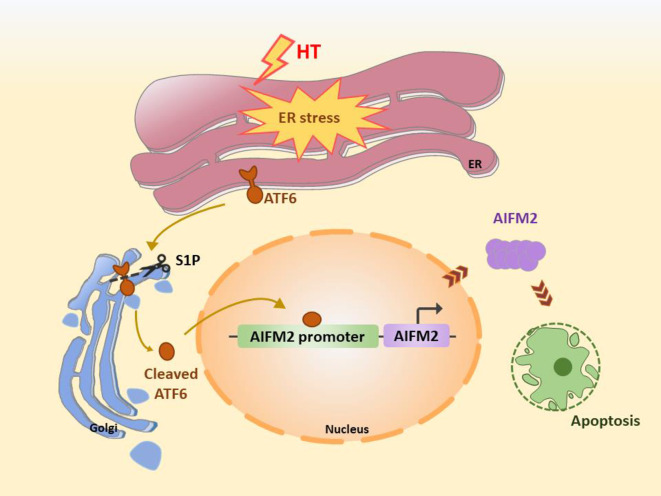
We investigated the role of ATF6 in early porcine embryonic development under HT conditions (Figure 7). Our results showed that HT induced ER stress in early porcine embryos, leading to an increase in ATF6 and its translocation to the Golgi apparatus, followed by organelle homeostasis disruption and Golgi and mitochondrial dysfunction. Additionally, AIFM2, which is involved in the induced apoptosis pathway, was up-regulated as an important downstream factor of ATF6. Knockdown of ATF6 expression promoted ER, Golgi, and mitochondrial homeostasis and down-regulated AIFM2 to reduce the occurrence of apoptosis and ultimately regulate embryo quality.
Figure 7.
Mechanistic representation of ATF6-mediated exacerbation of apoptosis in early porcine embryonic development through regulation of organelle homeostasis under HT conditions
HT-induced ER stress in early porcine embryos leads to increased expression of ATF6, followed by its translocation to the Golgi apparatus for cleavage by S1P, thereby inducing AIFM2-mediated apoptosis.
As an ER type II transmembrane protein, ATF6 contains a C-terminal region exposed to the ER lumen and an N-terminal cytoplasmic region containing DNA transcription activation and bZip domains (Shen et al., 2004). In the absence of cellular stress, ATF6 interacts with BiP in the ER and remains in an inactive state. However, when unfolded or misfolded proteins accumulate in the ER lumen due to stress, ATF6 dissociates from BiP and translocates from the ER to Golgi apparatus, where it undergoes cleavage by S1P and S2P (Read & Schroder, 2021; Shen et al., 2002). Following cleavage, active ATF6 translocates to the nucleus, triggering the transcriptional activation of ER stress response genes and ER stress response elements 1 and 2 (Shen et al., 2004). Our data showed that ATF6 was expressed at all stages of early porcine embryonic development, exhibiting a distribution pattern of dot-like formations around the nucleus during the 2C and 4C stages, and prominent localization within the nucleus during the BL stage. Under HT conditions, the blastocyst rate exhibited a decrease during embryonic development. In addition, HT stimulation led to an increase in ATF6 expression in the embryos, with stronger HT conditions leading to the arrest of embryos at the 2C stage, with localization of ATF6 in the nucleus. Previous studies have shown that heat stress can induce damage and activate the ATF6 signaling pathway in mouse testes (Qin et al., 2021). Similarly, investigations on zebrafish have revealed a significant increase in ATF6 expression in the spinal cord and head after 8 h of heat stress (Clark et al., 2020). Additionally, several studies have explored the translocation of ATF6 after stress. For example, in HuH7 cells, ER stress triggered by thapsigargin (TG) causes a shift in ATF6 localization from the perinuclear region to the nucleus and perinuclear region of the cell (Lebeau et al., 2018). Consistent with these results, our study further demonstrated the important role of ATF6 in early porcine embryonic development under stress.
To explore the potential function of ATF6 during early porcine embryo development under HT conditions, we microinjected embryos with ATF6 dsRNA to knockdown ATF6 expression. Results showed that HT-induced ATF6 knockdown improved the blastocyst rate and mitigated the increase in CHOP expression and Ca2+ release. Moreover, inhibition of S1P, a key enzyme involved in ATF6 cleavage, resulted in a decrease in ATF6 expression and an increase in the blastocyst rate and Ca2+ release in HT embryos. The ER serves as a major internal storage site for calcium ions and plays a critical role in calcium homeostasis (Mao et al., 2014). Similar to other signaling pathways, the UPR can play opposing roles in protection and destruction. Initially, the UPR functions to protect against and resolve ER stress. However, when ER stress remains unresolved, the activated UPR pathway can promote apoptosis (Xu et al., 2005). Key target genes of ATF6 involved in the UPR process include GRP78/BiP, X-box binding protein-1 (XBP1), and CHOP. Under normal circumstances, CHOP expression remains low but undergoes significant up-regulation in response to ER stress (Tabas & Ron, 2011). Previous studies have shown that inhibition of CHOP expression can prevent ER stress-induced overactivation and ultimately promote neuroprotection in a rat model of ischemic stroke (Gotoh et al., 2002). Other studies have also reported that the application of ATF6 siRNA or melatonin can significantly reduce CHOP expression, playing a protective role in a cerebral hemorrhage rat model (Xu et al., 2018). Consistent with these findings, our results showed the importance of ATF6 in response to ER stress under HT conditions.
As the largest multifunctional organelle within cells, the ER possesses unique structural elements consisting of both smooth and rough ER, which are structurally and functionally related to several other cellular organelles. The ER also contains domains that contact the plasma membrane and other organelles, including the Golgi apparatus, endosomes, mitochondria, lipid droplets, and peroxisomes (English & Voeltz, 2013). The ER serves as the primary site for the synthesis of newly formed molecules, which are subsequently transported via the secretory pathway to the Golgi apparatus (Passemard et al., 2019). The Golgi apparatus plays a central role in intracellular trafficking, including protein transportation and modification, and is often affected by ER stress. Our results indicated that HT induced the translocation of ATF6 from the ER to the Golgi apparatus and nucleus, as well as Golgi dysfunction, while knockdown of ATF6 expression ameliorated these effects. Similarly, previous studies have also found that HT can induce ER stress and lead to Golgi dysfunction in porcine oocytes (Lee et al., 2022). Moreover, it has been reported that ATF6 exhibits colocalization with the ER marker HSPA5 under normal conditions but redistributes to the Galnt2-marked Golgi compartment after TG treatment (Xu et al., 2019).
ER-mitochondrial connections form specific microdomains that play important roles in mitochondrial dynamics, inflammation, autophagy, and apoptosis. Consequently, perturbations of ER-mitochondrial connections contribute to disease progression (Veeresh et al., 2019). Considering the relationship between these two organelles, we assessed mitochondrial function. Our results indicated that HT-induced mitochondrial dysfunction can be ameliorated by ATF6 knockdown and S1P inhibition. In addition, as an important downstream protein factor of ATF6, AIFM2, also known as apoptosis-inducing factor-homologous mitochondrion-associated inducer of death (AMID), plays a vital role in mediating apoptotic responses and is considered an apoptosis-inducing factor-homologous mitochondrion-associated protein (Bilyy et al., 2008). Our results showed that AIFM2 mediated apoptosis under HT conditions and was regulated by ATF6. Previous studies have shown that ATF6 is an essential mediator of heat-induced apoptosis in mouse testes. Notably, decreased expression of ATF6 can lead to reduced levels of apoptosis-associated genes, including caspase12, caspase3, and CHOP, effectively mitigating heat stress-induced apoptosis in spermatocytes (Qin et al., 2021). In mice with severe acute pancreatitis (SAP), increased ATF6 expression is associated with pancreatic tissue apoptosis, ER disturbances, and mitochondrial dysfunction, while ATF6 knockdown can attenuate acinar injury, apoptosis, and ER disorder, suggesting that AIFM2 regulates SAP multi-organ injury via ATF6/p53-mediated apoptosis (Tan et al., 2020). Thus, our findings provide further confirmation that ATF6 regulates apoptosis mediated by AIFM2 under HT conditions.
Taken together, our research showed that AFT6 regulates homeostasis in the ER, Golgi apparatus, and mitochondria, as well as injury of early porcine embryos through AIFM2-mediated apoptosis under HT conditions. These findings provide new insights into ATF6-triggered signaling for targeted therapy of reproductive diseases under HT environmental stress.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
M.H.S. and X.S.C. designed the experiments; M.H.S. performed most of the experiments; D.Z., W.J.J., X.H.L., S.H.L., G.H., and Z.C. contributed to the materials; M.H.S. wrote the manuscript; and X.S.C. revised the manuscript. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the National Research Foundation (NRF) of Korea Grant funded by the Korean Government (MSIT) (2020R1A4A1017552, 2022R1A2C300769), Republic of Korea
References
- Al-Katanani YM, Paula-Lopes FF, Hansen PJ Effect of season and exposure to heat stress on oocyte competence in Holstein cows. Journal of Dairy Science. 2002;85(2):390–396. doi: 10.3168/jds.S0022-0302(02)74086-1. [DOI] [PubMed] [Google Scholar]
- Bilyy R, Kit Y, Hellman U, et al AMID: new insights on its intracellular localization and expression at apoptosis. Apoptosis. 2008;13(5):729–732. doi: 10.1007/s10495-008-0198-5. [DOI] [PubMed] [Google Scholar]
- Bischof LJ, Kao CY, Los FCO, et al. 2008. Activation of the unfolded protein response is required for defenses against bacterial pore-forming toxin in vivo. PLoS Pathogens, 4(10): e1000176.
- Clark EM, Nonarath HJT, Bostrom JR, et al Establishment and validation of an endoplasmic reticulum stress reporter to monitor zebrafish ATF6 activity in development and disease. Disease Models & Mechanisms. 2020;13(1):dmm041426. doi: 10.1242/dmm.041426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AR, Voeltz GK Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harbor Perspectives in Biology. 2013;5(4):a013227. doi: 10.1101/cshperspect.a013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frakes AE, Dillin A The UPRER: sensor and coordinator of organismal homeostasis. Molecular Cell. 2017;66(6):761–771. doi: 10.1016/j.molcel.2017.05.031. [DOI] [PubMed] [Google Scholar]
- Ghemrawi R, Battaglia-Hsu SF, Arnold C Endoplasmic reticulum stress in metabolic disorders. Cells. 2018;7(6):63. doi: 10.3390/cells7060063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh T, Oyadomari S, Mori K, et al Nitric oxide-induced apoptosis in RAW 264.7 macrophages is mediated by endoplasmic reticulum stress pathway involving ATF6 and CHOP. Journal of Biological Chemistry. 2002;277(14):12343–12350. doi: 10.1074/jbc.M107988200. [DOI] [PubMed] [Google Scholar]
- Guzel E, Arlier S, Guzeloglu-Kayisli O, et al Endoplasmic reticulum stress and homeostasis in reproductive physiology and pathology. International Journal of Molecular Sciences. 2017;18(4):792. doi: 10.3390/ijms18040792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Papa FR The unfolded protein response and cell fate control. Molecular Cell. 2018;69(2):169–181. doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- Lebeau P, Byun JH, Yousof T, et al Pharmacologic inhibition of S1P attenuates ATF6 expression, causes ER stress and contributes to apoptotic cell death. Toxicology and Applied Pharmacology. 2018;349:1–7. doi: 10.1016/j.taap.2018.04.020. [DOI] [PubMed] [Google Scholar]
- Lee SH, Sun MH, Zhou DJ, et al High temperature disrupts organelle distribution and functions affecting meiotic maturation in porcine oocytes. Frontiers in Cell and Developmental Biology. 2022;10:826801. doi: 10.3389/fcell.2022.826801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Lee JE, Kang JW, et al Endoplasmic reticulum (ER) stress and unfolded protein response (UPR) in mammalian oocyte maturation and preimplantation embryo development. International Journal of Molecular Sciences. 2019;20(2):409. doi: 10.3390/ijms20020409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao LN, Lou HY, Lou YY, et al Behaviour of cytoplasmic organelles and cytoskeleton during oocyte maturation. Reproductive Biomedicine Online. 2014;28(3):284–299. doi: 10.1016/j.rbmo.2013.10.016. [DOI] [PubMed] [Google Scholar]
- McAlpine CS, Werstuck GH The development and progression of atherosclerosis: evidence supporting a role for endoplasmic reticulum (ER) stress signaling. Cardiovascular & Hematological Disorders-Drug Targets. 2013;13(2):158–164. doi: 10.2174/1871529x11313020009. [DOI] [PubMed] [Google Scholar]
- Michalak M, Gye MC Endoplasmic reticulum stress in periimplantation embryos. Clinical and Experimental Reproductive Medicine. 2015;42(1):1–7. doi: 10.5653/cerm.2015.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu YJ, Zhou DJ, Cui XS S-nitrosoglutathione reductase maintains mitochondrial homeostasis by promoting clearance of damaged mitochondria in porcine preimplantation embryos. Cell Proliferation. 2021;54(3):e12990. doi: 10.1111/cpr.12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohiro Y, Garkavtsev I, Kobayashi S, et al. 2002. A novel p53-inducible apoptogenic gene, PRG3, encodes a homologue of the apoptosis-inducing factor (AIF). FEBS Letters, 524(1–3): 163–171.
- Passemard S, Perez F, Gressens P, et al Endoplasmic reticulum and Golgi stress in microcephaly. Cell Stress. 2019;3(12):369–384. doi: 10.15698/cst2019.12.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin DZ, Cai H, He C, et al Melatonin relieves heat-induced spermatocyte apoptosis in mouse testes by inhibition of ATF6 and PERK signaling pathways. Zoological Research. 2021;42(4):514–524. doi: 10.24272/j.issn.2095-8137.2021.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read A, Schroder M The unfolded protein response: an overview. Biology. 2021;10(5):384. doi: 10.3390/biology10050384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T, Livak K Analyzing real-time PCR data by the comparative CT method. Nature Protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Senft D, Ronai ZA UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends in Biochemical Sciences. 2015;40(3):141–148. doi: 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JS, Chen X, Hendershot L, et al ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Developmental Cell. 2002;3(1):99–111. doi: 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Shen XH, Ellis RE, Sakaki K, et al. 2005. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genetics, 1(3): e37.
- Shen XH, Zhang KZ, Kaufman RJ. 2004. The unfolded protein response-a stress signaling pathway of the endoplasmic reticulum. Journal of Chemical Neuroanatomy, 28(1–2): 79–92.
- Song SL, Tan J, Miao YY, et al Crosstalk of ER stress-mediated autophagy and ER-phagy: involvement of UPR and the core autophagy machinery. Journal of Cellular Physiology. 2018;233(5):3867–3874. doi: 10.1002/jcp.26137. [DOI] [PubMed] [Google Scholar]
- Tabas I, Ron D Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nature Cell Biology. 2011;13(3):184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JH, Cao RC, Zhou L, et al ATF6 aggravates acinar cell apoptosis and injury by regulating p53/AIFM2 transcription in Severe Acute Pancreatitis. Theranostics. 2020;10(18):8298–8314. doi: 10.7150/thno.46934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeresh P, Kaur H, Sarmah D, et al Endoplasmic reticulum-mitochondria crosstalk: from junction to function across neurological disorders. Annals of the New York Academy of Sciences. 2019;1457(1):41–60. doi: 10.1111/nyas.14212. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, et al ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Developmental Cell. 2007;13(3):351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Wu M, Xu LG, Su T, et al AMID is a p53-inducible gene downregulated in tumors. Oncogene. 2004;23(40):6815–6819. doi: 10.1038/sj.onc.1207909. [DOI] [PubMed] [Google Scholar]
- Xu CY, Bailly-Maitre B, Reed JC Endoplasmic reticulum stress: cell life and death decisions. The Journal of Clinical Investigation. 2005;115(10):2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Meng XB, Wu F, et al. 2019. ER stress drives ER-to-Golgi trafficking of ATF6 by blocking its membrane insertion. bioRxiv,doi: 10.1101/822965.
- Xu WL, Lu XY, Zheng JW, et al Melatonin protects against neuronal apoptosis via suppression of the ATF6/CHOP pathway in a rat model of intracerebral hemorrhage. Frontiers in Neuroscience. 2018;12:638. doi: 10.3389/fnins.2018.00638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, et al Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Developmental Cell. 2007;13(3):365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Zeng FY, Schultz RM RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Developmental Biology. 2005;283(1):40–57. doi: 10.1016/j.ydbio.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Zhang KZ, Wong HN, Song BB, et al The unfolded protein response sensor IRE1α is required at 2 distinct steps in B cell lymphopoiesis. The Journal of Clinical Investigation. 2005;115(2):268–281. doi: 10.1172/JCI200521848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PC, McGrath B, Li SA, et al The PERK eukaryotic initiation factor 2α kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Molecular and Cellular Biology. 2002;22(11):3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou DJ, Li XH, Lee SH, et al Effects of alpha-linolenic acid and essential amino acids on the proliferation and differentiation of C2C12 myoblasts. Journal of Animal Reproduction and Biotechnology. 2022;37(1):17–26. doi: 10.12750/JARB.37.1.17. [DOI] [Google Scholar]
- Zhou DJ, Niu YJ, Cui XS M-RAS regulate CDH1 function in blastomere compaction during porcine embryonic development. Journal of Animal Reproduction and Biotechnology. 2020;35(1):12–20. doi: 10.12750/JARB.35.1.12. [DOI] [Google Scholar]
- Zhou DJ, Sun MH, Lee SH, et al ROMO1 is required for mitochondrial metabolism during preimplantation embryo development in pigs. Cell Division. 2021;16(1):7. doi: 10.1186/s13008-021-00076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]