Abstract
Conjugative transfer of antibiotic resistance genes (ARGs) by plasmids is an important route for ARG dissemination. An increasing number of antibiotic and nonantibiotic compounds have been reported to aid the spread of ARGs, highlighting potential challenges for controlling this type of horizontal transfer. Development of conjugation inhibitors that block or delay the transfer of ARG-bearing plasmids is a promising strategy to control the propagation of antibiotic resistance. Although such inhibitors are rare, they typically exhibit relatively high toxicity and low efficacy in vivo and their mechanisms of action are inadequately understood. Here, we studied the effects of dihydroartemisinin (DHA), an artemisinin derivative used to treat malaria, on conjugation. DHA inhibited the conjugation of the IncI2 and IncX4 plasmids carrying the mobile colistin resistance gene (mcr-1) by more than 160-fold in vitro in Escherichia coli, and more than two-fold (IncI2 plasmid) in vivo in a mouse model. It also suppressed the transfer of the IncX3 plasmid carrying the carbapenem resistance gene blaNDM-5 by more than two-fold in vitro. Detection of intracellular adenosine triphosphate (ATP) and proton motive force (PMF), in combination with transcriptomic and metabolomic analyses, revealed that DHA impaired the function of the electron transport chain (ETC) by inhibiting the tricarboxylic acid (TCA) cycle pathway, thereby disrupting PMF and limiting the availability of intracellular ATP for plasmid conjugative transfer. Furthermore, expression levels of genes related to conjugation and pilus generation were significantly down-regulated during DHA exposure, indicating that the transfer apparatus for conjugation may be inhibited. Our findings provide new insights into the control of antibiotic resistance and the potential use of DHA.
Keywords: Dihydroartemisinin, Plasmid, mcr-1, bla NDM-5 , Conjugation inhibitors, TCA cycle
INTRODUCTION
Antimicrobial resistance (AMR) is a global health threat (Centers for Disease Control and Prevention (US) et al., 2013; O’Neill, 2016; WHO, 2017). Horizontal gene transfer mediated by plasmids, also known as conjugation, plays a critical role in the broad spread of AMR genes (ARGs) among animals, humans, and the environment (Lerminiaux & Cameron, 2019; Stevenson et al., 2017). For instance, naturally occurring plasmids, including IncI2, IncX4, and IncX3, increasingly contain the colistin resistance gene mcr, carbapenem resistance gene blaNDM, and tigecycline resistance gene tet(X4). Critically important antimicrobial agents used against severe infections in humans are destroyed in pathogens carrying these ARGs (He et al., 2019; Liu et al., 2016; Sun et al., 2016, 2019). During conjugation, DNA is directly transferred from the donor to recipient bacteria through a pilin bridge formed by the donor cell (Chee-Sanford et al., 2009; Ochman et al., 2000). The transfer of ARGs among pathogenic bacteria is a serious factor in both human and animal health, undermining the effectiveness of antimicrobial treatments and posing a significant challenge in controlling infections (Partridge et al., 2018). Therefore, efforts to prevent the horizontal transfer of plasmid-mediated ARGs, especially via conjugation, are essential to reduce their spread.
Notably, many antimicrobial agents (e.g., colistin), in combination with preservatives, disinfectants, nonantibiotic pharmaceuticals, and ionic liquids, have been shown to facilitate the conjugation of ARG-carrying plasmids (Cen et al., 2020; Wang et al., 2015, 2021, 2022; Xiao et al., 2022). This alarming trend has accelerated the dissemination of AMR, posing critical ecological and clinical challenges that necessitate the development of innovative strategies to counter the transmission of ARG-carrying plasmids. In this context, the development of novel compounds that can destabilize specific plasmids, ultimately leading to their eradication from bacterial populations, holds considerable promise (Buckner et al., 2018; Getino & de la Cruz, 2018). However, many of these compounds, with the exception of linoleic acid, ascorbic acid, chlorpromazine, and sodium dodecyl sulphate, exhibit toxicity to both humans and animals (Buckner et al., 2018). Thus, alternative strategies to address the challenge of ARG-carrying plasmids include the targeting of nonessential processes such as conjugation. Conjugation inhibitors may help reduce ARG propagation and revitalize antibiotic efficacy. For instance, CeO2 nanoparticles and MoS2-decorated nanocomposite particles can inhibit the conjugative transfer of the RP4 plasmid by decreasing both cell membrane permeability and transcription of transfer-related genes (Wang et al., 2020; Yu et al., 2020). While it is increasingly evident that adenosine triphosphate (ATP) is essential for driving DNA transfer and inducing conjugation, few reports have focused on the precise mechanism underlying energy supply for conjugation or conjugation inhibitors that target clinically important resistance plasmids (Jia et al., 2022; Li et al., 2021). Given their toxicity and poor efficacy in vivo, no previously identified conjugation inhibitor compound has been approved for clinical practice. Consequently, there remains an urgent need for the development of safer and more effective conjugation inhibitors that target energy metabolism to reduce the health impacts of ARG-carrying plasmids.
Herein, we identified and evaluated the effects of dihydroartemisinin (DHA), a derivative of artemisinin commonly used to treat malaria and reported to exhibit synergistic antibacterial effects with colistin against mcr-1-carrying pathogens (Tu, 2016; Zhou et al., 2022). Notably, our results showed that DHA significantly inhibited the conjugative transfer of ARG-carrying plasmids. Furthermore, DHA impaired the activity of the electron transport chain (ETC) by inhibiting the tricarboxylic acid (TCA) cycle pathway, thereby disrupting proton motive force (PMF) and reducing ATP content. These novel findings extend the current scope of DHA, a natural compound used in Traditional Chinese Medicine, to an antimicrobial adjuvant for the prevention of ARG dissemination through bacterial conjugation.
MATERIALS AND METHODS
Bacterial strains and culture media
Donor bacteria included two mcr-1-positive E. coli strains, ZJ807 (BioSample Accession Number: SAMN05437814) harboring the IncI2 plasmid pIncI2-mcr-1 and ZJ28 (SAMN05437813) carrying the IncX4 plasmid pIncX4-mcr-1, as well as E. coli harboring other ARG plasmids, including E. coli BW25113_IncX3 carrying the blaNDM-5-IncX3 plasmid pIncX3-blaNDM-5 and E. coli BW25113_IncX1 carrying the tet(X4)-IncX1 plasmid pIncX1-tet(X4). The recipient bacterium was E. coli J53 with sodium azide resistance.
Culture conditions for donor and recipient bacteria
Both the donor and recipient bacteria were cultured separately in brain heart infusion (BHI) broth at 37 °C overnight, supplemented with appropriate antibiotics. For donor bacteria, 2 μg/mL colistin for ZJ807 and ZJ28, 1 μg/mL meropenem for BW25113_IncX3, and 2 μg/mL tigecycline for BW25113_IncX1 were added to the BHI broth, respectively. For recipient bacteria, 200 μg/mL sodium azide was added to the BHI broth. The overnight cultures were diluted 10 times in 2 mL of BHI drug-free broth at 37 °C for 4 h to reach the logarithmic growth phase. The OD600 of the donor and recipient bacteria was adjusted to a consistent level (108–109 colony-forming units (CFUs)/mL). Afterwards, the donor and recipient bacteria were mixed at a ratio of 1:1 and centrifuged at 5 000 r/min for 5 min at room temperature, with the precipitate then collected and re-suspended in 2 mL of BHI broth for immediate use in the conjugation experiments.
Growth curves
The concentration range at which DHA did not affect the growth of donor and recipient bacteria was determined based on growth curve analysis. Briefly, overnight cultures of donor (ZJ807 and ZJ28) and recipient (J53) bacteria were diluted 1:100 into fresh Luria-Bertani (LB) (Beijing Land Bridge Technology, China) broth and incubated at 37 °C for 4 h. The cultures were then diluted to a 0.5 McFarland turbidity standard and added to a 96-well plate. DHA was added to the cultures at 0, 10, 25, 50, 100, and 200 μg/mL. The bacteria were then cultured for up to 24 h, and their growth curves were recorded at 37 °C under a 600 nm wavelength and 30 min interval using an Infinite M200 microplate reader (Tecan, Switzerland). All experiments were performed with at least three biological replicates.
Conjugation experiments in the presence of DHA
Conjugation experiments between the donor and recipient bacteria were performed in the presence of DHA. Different doses of DHA were added to the donor and recipient mixtures (total volume of 1 mL) containing 108–109 CFUs/mL of bacteria to a final DHA concentration of 0, 10, 25, 50, 100, or 200 μg/mL. These concentrations were all sub-minimum inhibitory concentrations (MICs), which had no significant growth effects on the donor and recipient bacteria. The conjugation mating systems were incubated at 37 °C for 16 h without shaking, after which 100 μL of each mixture was spread onto BHI agar plates containing sodium azide (200 μg/mL) and colistin (2 μg/mL), sodium azide (200 μg/mL) and meropenem (1 μg/mL), or sodium azide (200 μg/mL) and tigecycline (2 μg/mL) at suitable concentrations for 24 h. The numbers of transconjugants and recipients were then counted, respectively. The conjugative transfer frequency was calculated by dividing the number of transconjugants detected by the total number of recipients. The donor and recipient bacteria were plated on double resistant plates to exclude spontaneous mutation. All experiments were performed with at least three biological replicates.
MIC determination and polymerase chain reaction (PCR) verification
The suspected transconjugant colonies on the double antibiotic plates were randomly selected and cultured in 1 mL of BHI broth at 37 °C for 4–6 h for MIC and PCR determination. The MIC experiment was performed with reference to EUCAST (v11.0), with E. coli ATCC 25922 used for quality control (EUCAST, 2020). For analysis of mcr-1, PCR was conducted using the upstream primer GCAGATGGCGTTGTTGGT (5'-3') and downstream primer GGTGGCGTTCAGCAGTCA (5'-3'). The PCR product length was 500 bp and the annealing temperature was 55 °C. The amplified products were then subjected to agarose gel electrophoresis for confirmation.
RNA extraction and genome-wide RNA sequencing
The conjugation mating system was established according to the abovementioned procedures. DHA concentrations of 0, 50, and 100 μg/mL were assigned to the control, low concentration, and high concentration groups, respectively. Total RNA was extracted from the conjugation mating system using a Bacterial RNA Rapid Extraction Kit (Beijing Adlai Biotechnology, China), and applied for cDNA library construction and Illumina double-terminal sequencing. The specific steps involved were as follows: after 2 h of conjugation, total RNA from each group was extracted using the RNA rapid extraction kit following the manufacturer’s recommendations. The RNA samples were subsequently submitted to Beijing Saimo Lily Biotechnology (China) for library preparation and sequenced with the Illumina Hiseq X Ten platform (150 bp paired-end module). The raw sequencing data were quality filtered using fastp software (v0.23.2) (Chen et al., 2018). The filtered fastq files were mapped to the reference genome using Rockhopper (v2.0) with default parameters (Tjaden, 2020). Novel transcripts and operon loci were then extracted using in-house python scripts. Ribosomal RNA (rRNA) reads were eliminated using the SortMeRNA (v2.1) program and Silva database (Kopylova et al., 2012). The reads were then mapped to rRNA genes by HISAT2 (v2.1.0) (Kim et al., 2015). Subsequently, FeatureCounts (v1.6.4) was employed for expression calibration (Liao et al., 2014). The DESeq2 suite of programs (v1.2.4) was used to analyze differential transcripts in a pairwise fashion (Love et al., 2014). The Benjamini-Hochberg false discovery rate was used to determine adjusted P-values for each pairwise comparison. Gene expression was calculated as fragments per kilobase of a gene per million mapped reads (FPKM). Fold-change differences between the control and DHA-treated samples were calculated using log2fold-change (LFC). Subsequently, genes exhibiting differential expression of greater than two-fold and statistical significance of P<0.05 were selected. The differentially expressed genes were then inputted into ClusterProfiler for enrichment analysis using hypergeometric distribution. Corrections for multiple comparisons were carried out using the Benjamini-Hochberg method to control the FDR.
Intracellular ATP determination
The levels of intracellular ATP in the donor and recipient bacteria treated with different concentrations of DHA (0, 25, 50, 100, and 200 μg/mL) were determined using an Enhanced ATP Assay Kit (Beyotime Biotechnology, China). Briefly, bacterial cultures grown overnight were diluted 1:100 in fresh LB and incubated at 37 °C for 4 h. The bacteria were collected by centrifugation (8000 r/min, 5 min, room temperature) and resuspended in 0.1 mol/L phosphate-buffered saline (PBS, pH 7.4) to achieve an OD600 of 0.5. Following treatment with different concentrations of DHA for 60 min, the bacterial cultures were collected by centrifugation (12000 r/min, 5 min, 4 °C). The precipitates were lysed by lysozyme dissolved in ddH2O (1 mg/mL) and recentrifuged (12000 r/min, 5 min, 4 °C). The supernatants were subsequently used to determine intracellular ATP levels. The detecting solution was added to a 96-well plate and incubated at 37 °C for 5 min. The supernatants were quickly mixed into the wells, and luminescence was measured using an Infinite M200 microplate reader (Tecan, Switzerland). All experiments were performed with at least three biological replicates.
PMF measurement assay
To assess the effects of DHA on PMF, comprising both transmembrane proton gradient (ΔpH) and membrane potential (ΔΨ), donor and recipient bacteria were first treated with different concentrations of DHA (0, 25, 50, 100, and 200 μg/mL), with ΔpH and ΔΨ then determined using the pH-sensitive fluorescence probe BCECF-AM (Beyotime Biotechnology, China) and membrane potential-sensitive cyanine dye DiSC3(5) (Aladdin, China), respectively. Briefly, bacterial cultures grown overnight were diluted 1:100 in fresh LB and incubated at 37 °C for 4 h. The bacteria were then collected by centrifugation (8 000 r/min, 5 min, room temperature) and resuspended in 5 mmol/L HEPES (pH 7.0, plus 5 mmol/L glucose) to achieve an OD600 of 0.5. The bacteria were loaded with BCECF-AM (final concentration of 20 μmol/L) and DiSC3(5) (final concentration of 1 μmol/L) and added to a 96-well plate to stabilize. The DHA was dispensed into the plate containing dye-loaded cells. To evaluate ΔpH, fluorescence intensity was immediately measured using an excitation wavelength of 488 nm and emission wavelength of 535 nm. To evaluate ΔΨ, fluorescence was monitored using an excitation wavelength of 622 nm and emission wavelength of 670 nm. All experiments were performed with at least three biological replicates.
Iodonitrotetrazolium chloride (INT) reduction assay
As INT can be reduced by bacterial respiratory chain components to insoluble formazan (INF), it can be used as an indicator to assess bacterial respiratory capacity. Here, bacterial cultures grown overnight were diluted 1:100 in fresh LB and incubated at 37 °C for 4 h. The bacteria were then collected by centrifugation (8 000 r/min, 5 min, room temperature) and resuspended in 0.1 mol/L PBS (pH 7.4) to achieve an OD600 of 0.3 and maintained on ice. DHA (0, 25, 50, 100, and 200 μg/mL) was added to the centrifuge tube, followed by the addition of 1 mmol/L INT and 1 mL of bacteria to the buffer solution to a total volume of 3 mL. Subsequently, 200 µL of the solution was taken and added to a 96-well plate, with absorbance determined using an Infinite M200 microplate reader (Tecan, Switzerland) at 37 °C at a wavelength of 490 nm and an interval of 10 min. All experiments were performed with at least three biological replicates.
Targeted energy metabolomics
After treatment with different concentrations of DHA, the bacterial precipitate was sent to Shanghai Applied Protein Technology (China) for targeted energy metabolism measurement using liquid chromatography-tandem mass spectrometry (LC-MS/MS). In brief, the samples were thawed at 4 °C and 100 μL aliquots were mixed with 400 μL of cold methanol/acetonitrile (1:1, v/v) to remove the protein. The mixture was then centrifuged for 20 min (14 000 ×g, 4 °C) and the resulting supernatant was dried in a vacuum centrifuge. For LC-MS analysis, samples were re-dissolved in 100 μL of acetonitrile/water (1:1, v/v) and adequately vortexed and centrifuged (14 000 ×g, 4 °C, 15 min). The supernatants were then collected for LC-MS/MS analysis.
Reactive oxygen species (ROS) measurement
The ROS levels in the donor and recipient bacteria treated with different concentrations of DHA (0, 25, 50, 100, and 200 μg/mL) were detected with 10 μmol/L 2’,7’-dichlorofluorescein diacetate (DCFH-DA) following the manufacturer’s instructions (Beyotime Biotechnology, China). Bacterial cultures grown overnight were diluted 1:100 in fresh LB and incubated at 37 °C for 4 h. The bacteria were collected by centrifugation (8 000 r/min, 5 min, room temperature) and resuspended in 0.1 mol/L PBS (pH 7.4) to achieve an OD600 of 0.5. DCFH-DA was added to a final concentration of 10 μmol/L and the mixture was incubated at 37 °C for 30 min. After washing with 0.1 mol/L PBS three times, 190 μL of the probe-labeled bacterial cells and 10 μL of DHA at different concentrations were added to a 96-well plate. After incubation for 30 min at 37 °C, fluorescence intensity was immediately measured with an excitation wavelength of 488 nm and emission wavelength of 525 nm using an Infinite M200 microplate reader (Tecan, Switzerland). All experiments were performed with at least three biological replicates.
Membrane permeability assay
Cell membrane permeability was tested using propidium iodide (PI) fluorescent dye (Sigma-Aldrich, Cat No. P4170, USA) at a concentration of 10 μmol/L. Overnight grown donor and recipient bacteria were diluted 1:100 in fresh LB and incubated at 37 °C for 4 h. The bacteria were then collected by centrifugation (8 000 r/min, 5 min, room temperature) and resuspended in 0.1 mol/L PBS (pH 7.4) to achieve an OD600 of 0.5. PI was added to a final concentration of 10 μmol/L and the mixture was incubated at 37 °C for 30 min with shaking. Subsequently, 190 μL of the probe-labeled bacterial cells and 10 μL of DHA at different concentrations were added to a 96-well plate. After incubation for 30 min at 37 °C, fluorescence intensity was immediately measured with the excitation wavelength of 535 nm and emission wavelength of 615 nm using an Infinite M200 microplate reader (Tecan, Switzerland). All experiments were performed with at least three biological replicates.
Mice
Six–eight-week-old inbred female BALB/c mice (18–20 g) were purchased from Beijing Vital River Laboratory Animal Technology (China). The mice were allowed to acclimate for 5–7 days prior to experimentation at the Animal Laboratory at the China Agricultural University (CAU), Beijing. All animal procedures were approved by the Beijing Association for Science and Technology (approval ID SYXK (Beijing) 2016-0008) and conducted in strict accordance with the CAU Laboratory Animal Welfare and Animal Experimental Ethical Inspection issued by the CAU Committee on Animal Welfare and Experimental Ethics (approval no. AW01502202-2-1).
In vivo conjugation assay in mice
All mice were divided into two groups with each eight. All mice were pretreated with all four antibiotics, including ampicillin (1 mg/mL), vancomycin (0.5 mg/mL), clindamycin (0.5 mg/mL), and metronidazole (1 mg/mL), for 3 days to remove colonization resistance. Subsequently, eight mice were treated orogastrically with ~108 CFUs of E. coli J53 and ~108 CFUs of ZJ807, with a 24 h interval between the two treatments. After intragastric administration of donor and recipient bacteria, the mice were further treated via daily gavage of 10 mg/kg DHA into the intestinal tract, with the control group receiving PBS. Fecal samples were collected each day in 1 mL of PBS with glass beads for vortex mixing. Bacterial populations were then assessed by selective plating on China Blue media, facilitating the selection of donor bacteria and transconjugants with colistin (2 μg/mL), recipient bacteria and transconjugants with NaN3 (150 μg/mL), and transconjugants with colistin and NaN3. The final conjugative transfer frequency was calculated by dividing the number of transconjugants detected by the total number of recipients.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (v9.1.1). All data are expressed as mean±standard deviation (SD). For the in vitro studies, P-values were obtained by one-way analysis of variance (ANOVA) and Dunnett’s multiple comparisons. For the in vivo studies, statistical significance was determined using the Mann-Whitney U test. Significance levels were determined at P<0.05 (*), P<0.01 (**), P<0.001 (***), and P<0.0001 (****).
RESULTS
DHA inhibited conjugation of different Inc-type plasmids carrying mcr-1 and blaNDM-5
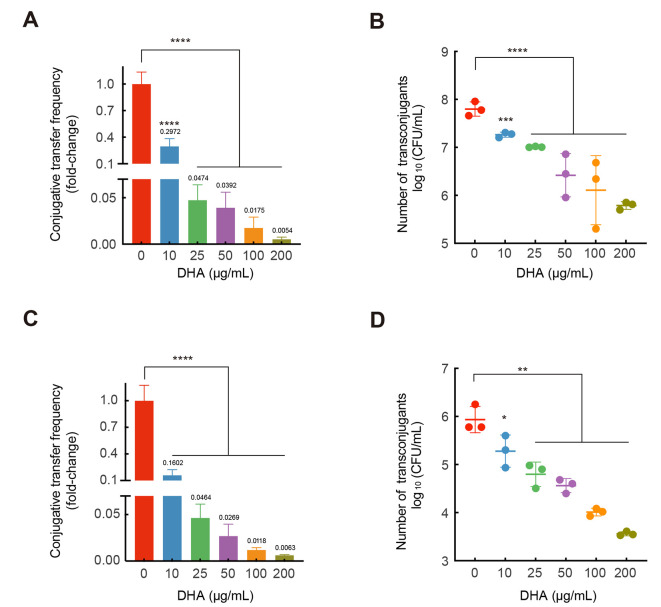
To explore the effects of DHA on conjugative transfer, we first evaluated its ability to inhibit horizontal gene transfer of a critically important ARG, mcr-1, using conjugation assays. At concentrations ranging from 10 to 200 μg/mL, DHA showed no significant bactericidal effects on the growth of either the donor or recipient strains (Supplementary Figures S1, S2). Next, we monitored the conjugative transfer frequency of the mcr-1-bearing plasmids under treatment with 0, 10, 25, 50, 100, and 200 μg/mL DHA. The MIC was determined, and PCR assays were performed to confirm the identity of the transconjugants (Supplementary Figure S3A, B). The spontaneous conjugative transfer frequency of pIncI2-mcr-1 in the absence of DHA was 2.01×10-1, similar to the previously reported transfer frequency (Liu et al., 2016). Notably, the conjugation frequencies and numbers of transconjugants of pIncI2-mcr-1 decreased significantly in a DHA concentration-dependent manner (P<0.01; Figure 1A, B). Specifically, at 10 and 200 μg/mL DHA, conjugation frequency and transconjugant number were approximately four-fold and 180-fold lower, respectively, than those at 0 μg/mL (control group). DHA also inhibited the conjugation frequency of pInX4-mcr-1 in a concentration-dependent manner (P<0.01; Figure 1C). Specifically, low (10 μg/mL) and high (200 μg/mL) concentrations of DHA resulted in a six-fold and 160-fold decrease in conjugation frequency, respectively. Furthermore, the numbers of transconjugants decreased significantly from 5.9 log10 (CFUs/mL) to 4.0 log10 (CFUs/mL) and 3.6 log10 (CFUs/mL) at DHA concentrations of 100 and 200 μg/mL, respectively (Figure 1D). We also explored the potential activity of DHA against the conjugative transfer of clinical resistance plasmids pIncX3-blaNDM-5 and pIncX1-tet(X4). High (200 μg/mL) concentration of DHA significantly inhibited the conjugative transfer frequency of pIncX3-blaNDM-5 by approximately 2.7-fold (P<0.05; Supplementary Figure S4A) but had no significant inhibitory effect on pIncX1-tet(X4) (Supplementary Figure S4B). Thus, these results showed that DHA significantly decreased the conjugative transfer of different Inc-type plasmids carrying mcr-1 and blaNDM-5.
Figure 1.
Effects of DHA on conjugative transfer of pIncI2-mcr-1 and pIncX4-mcr-1
A: Fold-change of conjugative transfer frequency of pIncI2-mcr-1. B: Log10 values of transconjugants of pIncI2-mcr-1. C: Fold-change of conjugative transfer frequency of pIncX4-mcr-1. D: Log10 values of transconjugants of pIncI2-mcr-1. P-value was detected using one-way ANOVA and corrected using Dunnett’s method. *: P<0.05; **: P<0.01; ***: P<0.001; ****: P<0.0001.
DHA inhibited conjugation by interfering with energy metabolism
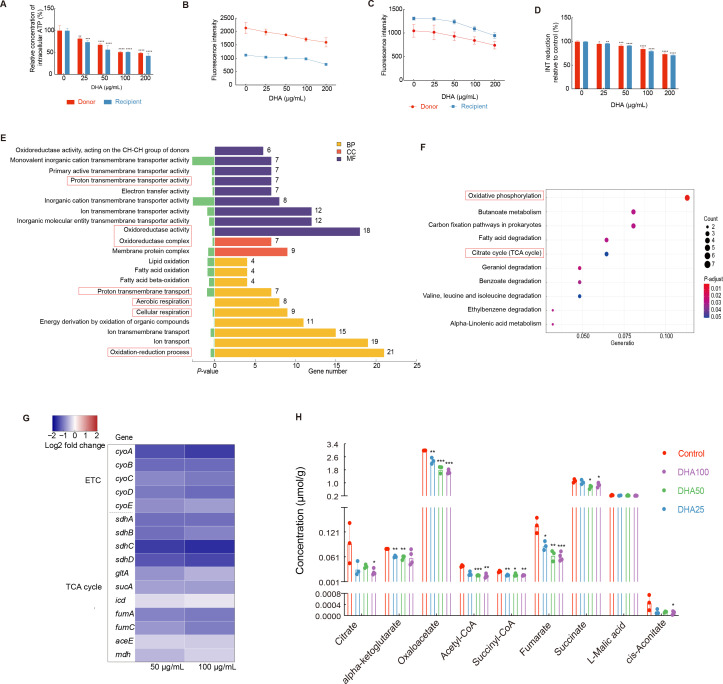
The strongest inhibitory effect of DHA on conjugative transfer was observed for pIncI2-mcr-1. Therefore, we conducted further investigations into the mechanism underlying this inhibition using pIncI2-mcr-1. Bacterial conjugative transfer is an energy-consuming process involving the replication of DNA and assembly of type IV secretion systems (Chen et al., 2005; Lu et al., 2018). Here, ATP determination analysis revealed that DHA treatment led to a dose-dependent decrease in intracellular ATP content in both donor (81% to 48%) and recipient (74% to 42%) bacteria (P<0.05; Figure 2A). To explore how DHA diminishes intracellular ATP levels, we evaluated PMF (ΔpH and ΔΨ), which plays a critical role in ATP generation (Domenech et al., 2020). Results demonstrated a dose-dependent reduction in ΔpH in the DHA-treated donor and recipient bacteria, implying an increase in H+ concentration (Figure 2B). Fluorescence intensity also showed a dose-dependent decrease in the DHA-treated donor and recipient bacteria, indicating an increase in ΔΨ and disruption of PMF (Figure 2C).
Figure 2.
Effects of DHA on bacterial energy metabolism
A: Relative intracellular ATP levels in donor and recipient bacteria. B: Relative intracellular ΔpH in donor and recipient bacteria. C: Relative intracellular ΔΨ in donor and recipient bacteria. D: Cellular respiration level as a measure of INT reduction by donor and recipient bacteria. E: GO enrichment analyses of down-regulated genes in donor bacteria treated with 50 μg/mL DHA. F: KEGG pathway enrichment analyses of down-regulated genes in donor bacteria treated with 50 μg/mL DHA. G: Fold-change in expression of core cellular respiratory-related and TCA cycle-related genes in donor bacteria. H: Abundance of metabolites of TCA cycle in bacteria treated with different DHA concentrations. *: P<0.05; **: P<0.01; ***: P<0.001; ****: P<0.0001.
To assess the activity of the ETC, a key component of PMF generation, we measured its ability to reduce INT (Altman, 1976). Compared to the untreated group, DHA treatment (25–200 μg/mL) led to a reduction in INT to red INF of 95% to 73% in donor bacteria and 96% to 71% in recipient bacteria (P<0.05, Figure 2D), indicating an impaired electron transport process. Based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses, the down-regulated genes were associated with electron transfer activity, oxidoreductase activity, oxidative phosphorylation, and the TCA cycle. These findings suggest disruption of the ETC, leading to inhibited cellular respiration in both donor (Figure 2E, F) and recipient bacteria (Supplementary Figure S5A, B). Moreover, the expression levels of cyoABCDE operon genes, responsible for encoding cytochrome bo3—an essential oxidoreductase involved in transferring electrons from quinol to O2 and a component of ETC complex IV in E. coli (Borisov & Verkhovsky, 2015; Kaila & Wikström, 2021)—were significantly down-regulated in the donor and recipient bacteria (1.7-fold and 2.8-fold, respectively). The expression levels of the succinate dehydrogenase-relevant gene sdhABCD, which encodes ETC complex II (Kaila & Wikström, 2021), were also significantly down-regulated in the donor and recipient E. coli (two-fold and three-fold, respectively) (Figure 2G; Supplementary Figure S5C and Tables S1, S2).
The TCA cycle is a crucial bioprocess for generating reducing power. To explore metabolic changes in donor bacteria under DHA treatment (25–100 μg/mL), we performed targeted energy metabolomic analysis. Results showed that DHA significantly decreased the levels of four TCA-related metabolites (citrate, alpha-ketoglutarate, oxaloacetate, and acetyl-CoA) by 1.35-fold to four-fold (Figure 2H). Furthermore, genes encoding enzymes involved in the TCA cycle were also down-regulated to varying degrees. For instance, the alpha-ketoglutarate dehydrogenase-encoding gene sucA and citrate synthase-encoding gene gltA were down-regulated by 1.6-fold and 1.7-fold, respectively, in the donor bacteria (Figure 2G). Given that low energy sources can inhibit conjugation frequency (Huang et al., 2019), we propose that DHA may inhibit ETC activity by down-regulating the TCA cycle pathway, which, in turn, disrupts the PMF and reduces intracellular ATP content, thereby contributing to a decline in the conjugative transfer frequency of mcr-1-carrying plasmids.
DHA suppressed expression of conjugation-related genes in pIncI2-mcr-1
The conjugative plasmid pIncI2-mcr-1 carries two sets of genetic components: i.e., DNA transfer and replication (Dtr) system for conjugative DNA processing and mating-pair formation (Mpf) system for DNA delivery via donor and recipient bacterial membranes (Getino & de la Cruz, 2018; Meinersmann, 2019). Compared with the control group, the expression levels of the Dtr genes nikB and traC were down-regulated by 1.3-fold and 1.6-fold under 50 and 100 µg/mL DHA treatment, respectively (Figure 3A; Supplementary Table S3). Plasmid transfer from the donor to recipient requires a pilin bridge, established by Mpf during the conjugation process (Schröder & Lanka, 2005). Pilin formation genes on pIncI2-mcr-1 include virB1, virB2, and virB5, which were down-regulated in the presence of 50 or 100 µg/mL DHA (1.4–2.0-fold). Additionally, the expression of virB7, which encodes a core protein of the Mpf system, was down-regulated by 2.3–2.4-fold. Only bacteria carrying the IncI-type plasmid display the unique type IV pilus, which participates in conjugative transfer (Sekizuka et al., 2017). Here, pilus-encoding genes, such as pilU, were also down-regulated (1.6–1.7-fold) following DHA exposure (Figure 3A). Mutual physical contact is essential for the conjugation process (Cabezón et al., 2015; Chee-Sanford et al., 2009). We found that the adhesion-relevant bacterial operons fim and yeh were significantly down-regulated in the donor bacteria following DHA exposure (Figure 3B; Supplementary Table S4), including the yehB gene (by 5.3-fold). The fim and yeh genes are involved in a critical step of the direct cell-to-cell contact required for plasmid DNA transfer (Rendón et al., 2007). Based on the abovementioned changes in gene transcription in donor bacteria, we propose that DHA inhibits conjugative transfer of plasmids by suppressing the expression of pilus-formation genes, conjugative transfer-related regulatory genes, and adhesion-related genes.
Figure 3.
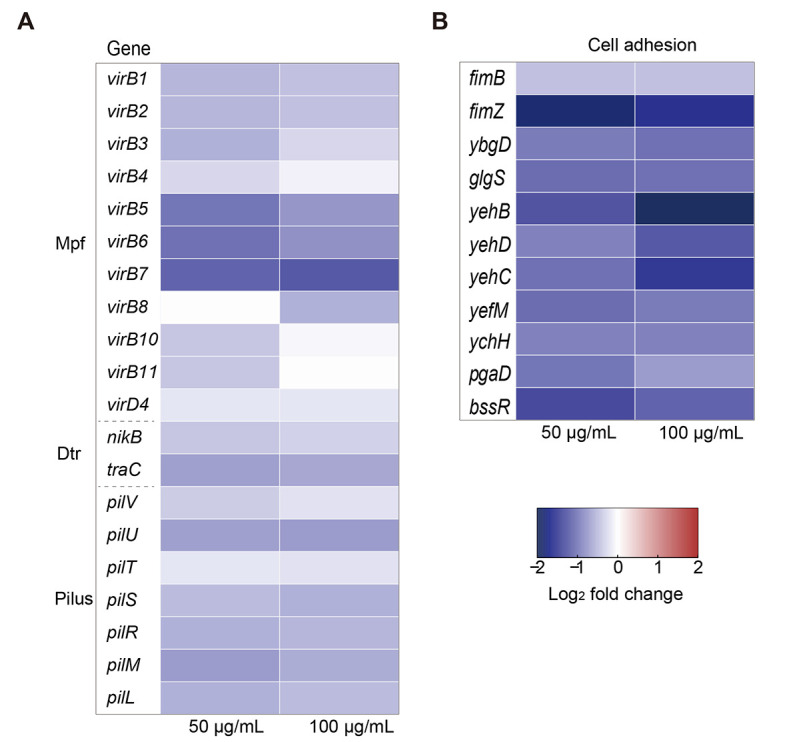
Effects of DHA on expression of pIncI2-mcr-1-encoded genes and pilus generation in donor bacteria
A: Fold-change in expression of core conjugation-associated genes in pIncI2-mcr-1 plasmid in donor bacteria treated with 50 and 100 µg/mL DHA. B: Fold-change in expression of core genes related to adhesive-pilus generation in donor bacteria treated with 50 and 100 µg/mL DHA.
DHA had no effect on SOS response or membrane permeability
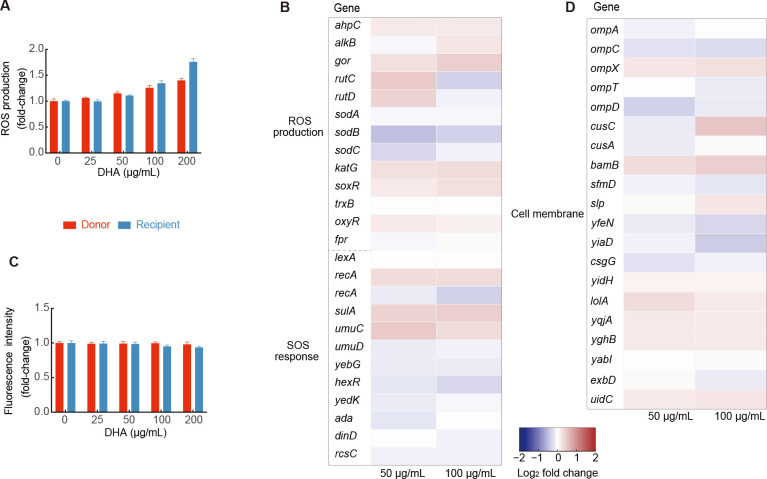
The SOS response triggered by oxidative damage and enhancement of membrane permeability are known to promote bacterial conjugation (Beaber et al., 2004; Yu et al., 2021). Furthermore, disruption of the ETC can lead to the production of ROS (Nolfi-Donegan et al., 2020). Thus, to investigate the impact of DHA on ROS production, we examined bacteria exposed to varying DHA concentrations. Results revealed that ROS production was enhanced with DHA concentration, increasing 1.1-fold to 1.8-fold in both donor and recipient strains (Figure 4A). We also found no significant changes in the expression levels of ROS-related and SOS response-related genes in either the donor or recipient strains (Figure 4B; Supplementary Figure S6A and Tables S5, S6). Certain exogenous compounds can promote the conjugative transfer of plasmids by damaging or increasing cell membrane permeability (Feng et al., 2021), which is an important factor affecting conjugative transfer frequency. Thus, we examined membrane permeability of the bacterial strains using PI dye, which only penetrates dead or damaged cells. However, no significant change in permeability was found in response to DHA treatment (Figure 4C). Furthermore, compared to the control group, transcriptomic analysis revealed that the expression levels of inner and outer cell membrane-related genes did not change significantly under 50 or 100 µg/mL DHA in either the donor or recipient bacteria (Figure 4D; Supplementary Figure S6B and Tables S7, S8). Therefore, we concluded that DHA limited the energy available for conjugation and suppressed the expression of conjugation- and pilus-related genes independently of the SOS response and cell membrane.
Figure 4.
Effects of DHA on ROS production, SOS response, and cell membrane permeability
A: Fold-change in ROS production in donor and recipient bacteria. B: Expression levels of core genes related to ROS production and SOS response in donor bacteria. C: Fold-change in cell membrane permeability in donor and recipient bacteria. D: Expression levels of core genes related to inner and outer cell membranes in donor bacteria.
DHA inhibited conjugation of pIncI2-mcr-1 in vivo
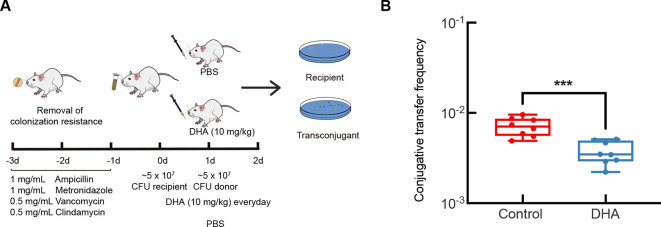
To verify the effects of DHA on the conjugation of pIncI2-mcr-1 in a complex environment, we targeted the mammalian gut for bacterial interaction (Benz et al., 2021). Conjugation experiments were performed using E. coli ZJ807 as the donor and E. coli J53 as the recipient in the intestines of mice in the presence of DHA (Figure 5A). After treatment with multiple antibiotics to remove endogenous intestinal bacteria, the mice were exposed to recipient and donor bacteria via oral gavage, followed by treatment with 10 mg/kg DHA or PBS (control). Results showed a downward trend in conjugative transfer, consistent with the in vitro experiment results. Average conjugative transfer frequency in the control group (7.12×10-3) was approximately twice that of the DHA group (3.73×10-3) (P=0.0006; Figure 5B), suggesting that DHA is effective in a murine intestinal infection model.
Figure 5.
Inhibitory effects of DHA on conjugative transfer frequency of pIncI2-mcr-1 in vivo in a murine intestinal infection model
A: Schematic of experimental protocol for the measurement of conjugative transfer in vivo. B: Conjugative transfer frequency in vivo (n=8). P-value was determined using a two-sided Mann-Whitney U test. ***: P<0.001.
DISCUSSION
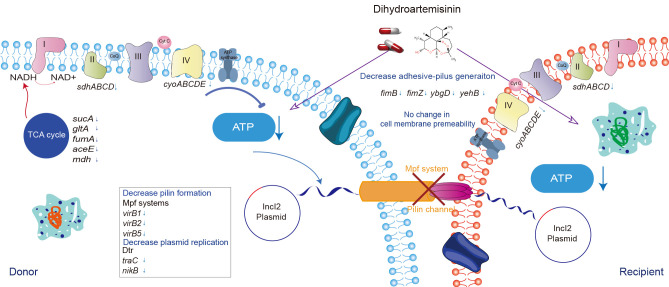
The widespread dissemination and colonization of ARGs in the human and animal gut present a global threat to public health and the environment (Ling et al., 2020). Various antimicrobial- and non-antimicrobial agents are known to promote the dissemination of ARGs. For example, we previously demonstrated that colistin can promote conjugative transfer of mcr-1-bearing IncI2 plasmids (Wang et al., 2022). Consequently, inhibiting the conjugative transfer of ARG-carrying plasmids has emerged as a crucial strategy to delay AMR dissemination. In the current study, we demonstrated that DHA, a derivative of artemisinin, effectively inhibited the dissemination of ARG-bearing plasmids. Notably, DHA impaired cellular respiration and decreased ATP levels by inhibiting TCA cycle pathways, and down-regulated genes involved in the conjugation process (Figure 6). The inhibitory effects of DHA on conjugative transfer, both in vitro and in vivo, suggest that reducing the spread of ARGs may be a valuable approach for extending the life of crucial antimicrobial agents, such as colistin, in both human and veterinary medicine.
Figure 6.
Proposed model of mechanism underlying DHA-mediated inhibition of conjugative transfer of pIncI2-mcr-1
DHA is well-known for its anti-malarial properties against Plasmodium, as well as its anti-tumor, anti-parasitic, and anti-autoimmune disease effects (Efferth, 2017; Kiani et al., 2020). DHA tablets exhibit remarkable clinical efficacy, with a wide range of dosing formulations and low toxicity and side effects (Ansari et al., 2013; Slezakova & Ruda-Kucerova, 2017). Moreover, artemisinin, the active compound in veterinary drugs derived from Artemisia annua, such as A. annua powder and A. annua Changshan granules, is widely used for the prevention and treatment of chicken coccidiosis in livestock and poultry breeding in China (Jiao et al., 2018; Niu et al., 2018). DHA is also known to produce an antibacterial synergistic effect with colistin against mcr-1-carrying gram-negative bacteria in vitro and in vivo (Zhou et al., 2022). Notably, DHA treatment is reported to significantly inhibit the conjugative transfer of mcr-1-carrying plasmids, making it a promising candidate not only for preventing bacterial infections during parasitic treatment but also for preventing the spread of colistin resistance and prolonging the lifespan of colistin in veterinary medicine. In clinical settings, children infected with Plasmodium falciparum are often co-infected with nontyphoidal Salmonella, which can lead to systemic intestinal infections with serious and sometimes fatal consequences (Mooney et al., 2019). Hence, DHA shows considerable potential not only for treating malaria infections but also for delaying the emergence of AMR bacteria, thereby enhancing the efficacy of antimicrobial agents against bacterial infections.
In this study, we found that DHA exhibited a strong inhibitory effect on the conjugative transfer of the pIncI2-mcr-1 and pIncX4-mcr-1 plasmids (180-fold and 160-fold, respectively) as well as the pIncX3-blaNDM-5 plasmid (2.7-fold), showing substantially higher efficacy compared to previously reported compounds (Jia et al., 2022; Li et al., 2021). However, it showed lower effects against pIncX3-blaNDM-5 and had no impact on pIncX1-tet(X4). These results suggest that DHA works more effectively against plasmids with higher conjugation efficiencies, although the exact mechanism remains to be explored. Similar to DHA, melatonin also displays significant inhibitory effects on clinical mcr-1-positive plasmids, with research suggesting that disruption of PMF contributes to the suppressive effects of melatonin on conjugation (Jia et al., 2022). Although our results showed that DHA perturbed PMF by enhancing ΔΨ and decreasing ΔpH, whereas melatonin decreased both ΔΨ and ΔpH (Jia et al., 2022), both treatments ultimately resulted in decreased intracellular ATP content. This may be because melatonin destroys cellular membranes, while DHA does not. Moreover, the perturbation of PMF by DHA in the current study was attributed to impaired ETC function resulting from the inhibition of the TCA cycle pathway, as shown by the down-regulation of genes involved in the respiratory chain and TCA cycle through transcriptomic analysis. Our metabolomic data provided further evidence that DHA significantly reduced TCA cycle intermediates. Additionally, DHA blocked the conjugation-related pathways involving the Dtr and Mpf systems in pIncI2-mcr-1. This repression of gene expression was likely a consequence of decreased ATP levels, as the expression and translation of plasmid-encoding genes consume additional ATP and metabolites (Chen et al., 2005; San Millan, 2018). Interestingly, although DHA contributed to the production of excess ROS, it did not induce the SOS response, possibly because the DHA-related ATP decrease (Figure 3A) reduced the energy available for transcriptional expression of genes involved in the pIncI2-mcr-1-encoded transfer apparatus, ROS detoxification, and SOS response (Huang et al., 2019; Wu et al., 2022). Thus, our results indicate that impairing the ETC by inhibiting the TCA cycle to limit ATP generation represents a promising strategy to combat the dissemination of ARGs.
Currently, there are only a few known conjugation inhibitors, and their mechanisms of action are varied and specific. For instance, certain unsaturated fatty acids can inhibit conjugation by binding to the plasmid-encoded TrwD, a type IV secretion ATPase and a homologue of the Agrobacterium tumefaciens gene virB11 (Getino et al., 2015; Ripoll-Rozada et al., 2016). While the TCA cycle is essential for bacterial energy metabolism, it has rarely been identified as a potential target for inhibiting conjugation. Recent studies have suggested that melatonin disrupts the ΔΨ component of PMF, which may be the mechanism by which it inhibits conjugation (Jia et al., 2022). Additionally, certain competence (COM)-blockers can disrupt PMF to prevent bacterial transformation via horizontal DNA transfer (Getino et al., 2015). However, their effects on the TCA cycle pathway have not been fully explored. Here, we confirmed that DHA may impair ETC function by inhibiting the TCA cycle pathway, which could potentially serve as a universal target to inhibit the spread of ARG-carrying plasmids.
In conclusion, we identified strong activity of DHA, a nonantibiotic, plant-derived pharmaceutical, against the horizontal gene transfer of ARG-bearing plasmids. DHA impaired energy generation and the conjugative transfer apparatus, demonstrating potential as a conjugation inhibitor. This study provides guidance for the screening of effective conjugation inhibitors that target the TCA cycle and delay the spread of AMR.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
DATA AVAILABILITY
All raw RNA-seq data were deposited in the National Center for Biotechnology Information (NCBI) sequence read archive (SRA) under BioProjectID PRJNA971775, Genome Sequence Archive under Accession No. CRA011728, and Science Data Bank under DOI: 10.57760/sciencedb.j00139.00055. All data generated in this study are available within the article and the Supplementary Data files.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
The study was conceived and supervised by Y.W. X.Y.W., H.W.S. and T.Y. performed all the experiments. X.Y.W. analyzed the data under the guidance of Y.B.S., C.S.D., C.T.S., D.J.L., C.M.W. and J.Z.S.. Y.W. and X.Y.W. drafted most of the manuscript. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We thank Mu-Chen Zhang, Si-Yuan Yang, and Zhi-Yu Zou for their kind help in finishing the conjugation experiments. We also thank Shi-Zhen Ma and Teng-Fei Ma for their valuable contributions during manuscript writing.
Funding Statement
This work was supported in part by grants from the Laboratory of Lingnan Modern Agriculture Project (NT2021006) and National Key Research and Development Program of China (2022YFD1800400)
References
- Altman FP. 1976. Tetrazolium salts and formazans. Progress in Histochemistry and Cytochemistry, 9(3): III–VI, 1–51.
- Ansari MT, Saify ZS, Sultana N, et al Malaria and artemisinin derivatives: an updated review. Mini-Reviews in Medicinal Chemistry. 2013;13(13):1879–1902. doi: 10.2174/13895575113136660097. [DOI] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427(6969):72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- Benz F, Huisman JS, Bakkeren E, et al Plasmid- and strain-specific factors drive variation in ESBL-plasmid spread in vitro and in vivo. The ISME Journal. 2021;15(3):862–878. doi: 10.1038/s41396-020-00819-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov VB, Verkhovsky MI. 2015. Oxygen as acceptor. EcoSal Plus, 6(2),doi: 10.1128/ecosalplus.ESP-0012-2015.
- Buckner MMC, Ciusa ML, Piddock LJV Strategies to combat antimicrobial resistance: anti-plasmid and plasmid curing. Fems Microbiology Reviews. 2018;42(6):781–804. doi: 10.1093/femsre/fuy031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezón E, Ripoll-Rozada J, Peña A, et al Towards an integrated model of bacterial conjugation. Fems Microbiology Reviews. 2015;39(1):81–95. doi: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- Cen TY, Zhang XY, Xie SS, et al Preservatives accelerate the horizontal transfer of plasmid-mediated antimicrobial resistance genes via differential mechanisms. Environment International. 2020;138:105544. doi: 10.1016/j.envint.2020.105544. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (U. S. ), National Center for Emerging Zoonotic and Infectious Diseases (U. S. ), National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention (U. S. ), et al. 2013. Antibiotic Resistance Threats in the United States, 2013. Atlanta: CDC.
- Chee-Sanford JC, Mackie RI, Koike S, et al Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. Journal of Environmental Quality. 2009;38(3):1086–1108. doi: 10.2134/jeq2008.0128. [DOI] [PubMed] [Google Scholar]
- Chen I, Christie PJ, Dubnau D The ins and outs of DNA transfer in bacteria. Science. 2005;310(5753):1456–1460. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SF, Zhou YQ, Chen YR, et al fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech A, Brochado AR, Sender V, et al Proton motive force disruptors block bacterial competence and horizontal gene transfer. Cell Host & Microbe. 2020;27(4):544–555.e3. doi: 10.1016/j.chom.2020.02.002. [DOI] [PubMed] [Google Scholar]
- Efferth T From ancient herb to modern drug: Artemisia annua and artemisinin for cancer therapy. Seminars in Cancer Biology. 2017;46:65–83. doi: 10.1016/j.semcancer.2017.02.009. [DOI] [PubMed] [Google Scholar]
- EUCAST. 2020. Breakpoint tables for interpretation of MICs and zone diameters, version 11.0. The European Committee on Antimicrobial Susceptibility Testing.
- Feng GQ, Huang HN, Chen YG Effects of emerging pollutants on the occurrence and transfer of antibiotic resistance genes: a review. Journal of Hazardous Materials. 2021;420:126602. doi: 10.1016/j.jhazmat.2021.126602. [DOI] [PubMed] [Google Scholar]
- Getino M, de la Cruz F. 2018. Natural and artificial strategies to control the conjugative transmission of plasmids. Microbiology Spectrum, 6(1),doi: 10.1128/microbiolspec.MTBP-0015-2016.
- Getino M, Sanabria-Ríos DJ, Fernández-López R, et al Synthetic fatty acids prevent plasmid-mediated horizontal gene transfer. mBio. 2015;6(5):e01032–15. doi: 10.1128/mBio.01032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T, Wang R, Liu DJ, et al Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nature Microbiology. 2019;4(9):1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- Huang HN, Liao JQ, Zheng X, et al Low-level free nitrous acid efficiently inhibits the conjugative transfer of antibiotic resistance by altering intracellular ions and disabling transfer apparatus. Water Research. 2019;158:383–391. doi: 10.1016/j.watres.2019.04.046. [DOI] [PubMed] [Google Scholar]
- Jia YQ, Yang BQ, Shi JR, et al Melatonin prevents conjugative transfer of plasmid-mediated antibiotic resistance genes by disrupting proton motive force. Pharmacological Research. 2022;175:105978. doi: 10.1016/j.phrs.2021.105978. [DOI] [PubMed] [Google Scholar]
- Jiao JY, Yang YQ, Liu MJ, et al Artemisinin and Artemisia annua leaves alleviate Eimeria tenella infection by facilitating apoptosis of host cells and suppressing inflammatory response. Veterinary Parasitology. 2018;254:172–177. doi: 10.1016/j.vetpar.2018.03.017. [DOI] [PubMed] [Google Scholar]
- Kaila VRI, Wikström M Architecture of bacterial respiratory chains. Nature Reviews Microbiology. 2021;19(5):319–330. doi: 10.1038/s41579-020-00486-4. [DOI] [PubMed] [Google Scholar]
- Kiani BH, Kayani WK, Khayam AU, et al Artemisinin and its derivatives: a promising cancer therapy. Molecular Biology Reports. 2020;47(8):6321–6336. doi: 10.1007/s11033-020-05669-z. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28(24):3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- Lerminiaux NA, Cameron ADS Horizontal transfer of antibiotic resistance genes in clinical environments. Canadian Journal of Microbiology. 2019;65(1):34–44. doi: 10.1139/cjm-2018-0275. [DOI] [PubMed] [Google Scholar]
- Li G, Xia LJ, Zhou SY, et al. 2021. Linoleic acid and α-linolenic acid inhibit conjugative transfer of an IncX4 plasmid carrying mcr-1. Journal of Applied Microbiology, 130(6): 1893–1901.
- Liao Y, Smyth GK, Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Ling ZR, Yin WJ, Shen ZQ, et al. 2020. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. Journal of Antimicrobial Chemotherapy, 75(11): 3087–3095.
- Liu YY, Wang Y, Walsh TR, et al Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infectious Diseases. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Wang Y, Li J, et al Triclosan at environmentally relevant concentrations promotes horizontal transfer of multidrug resistance genes within and across bacterial genera. Environment International. 2018;121:1217–1226. doi: 10.1016/j.envint.2018.10.040. [DOI] [PubMed] [Google Scholar]
- Meinersmann RJ The biology of IncI2 plasmids shown by whole-plasmid multi-locus sequence typing. Plasmid. 2019;106:102444. doi: 10.1016/j.plasmid.2019.102444. [DOI] [PubMed] [Google Scholar]
- Mooney JP, Galloway LJ, Riley EM Malaria, anemia, and invasive bacterial disease: a neutrophil problem? Journal of Leukocyte Biology. 2019;105(4):645–655. doi: 10.1002/JLB.3RI1018-400R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B, Qin WW, Guo WZ, et al Study on anticoccidial efficacy of artemisia annua powder. China Animal Husbandry & Veterinary Medicine. 2018;45(6):1683–1691. [Google Scholar]
- Nolfi-Donegan D, Braganza A, Shiva S Mitochondrial electron transport chain: oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biology. 2020;37:101674. doi: 10.1016/j.redox.2020.101674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J. 2016. Tackling drug-resistant infections globally: final report and recommendations.
- Ochman H, Lawrence JG, Groisman EA Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Partridge SR, Kwong SM, Firth N, et al Mobile genetic elements associated with antimicrobial resistance. Clinical Microbiology Reviews. 2018;31(4):e00088–17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendón MA, Saldaña Z, Erdem AL, et al Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(25):10637–10642. doi: 10.1073/pnas.0704104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll-Rozada J, García-Cazorla Y, Getino M, et al Type IV traffic ATPase TrwD as molecular target to inhibit bacterial conjugation. Molecular Microbiology. 2016;100(5):912–921. doi: 10.1111/mmi.13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Millan A Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends in Microbiology. 2018;26(12):978–985. doi: 10.1016/j.tim.2018.06.007. [DOI] [PubMed] [Google Scholar]
- Schröder G, Lanka E The mating pair formation system of conjugative plasmids-a versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54(1):1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Sekizuka T, Kawanishi M, Ohnishi M, et al Elucidation of quantitative structural diversity of remarkable rearrangement regions, shufflons, in IncI2 plasmids. Scientific Reports. 2017;7(1):928. doi: 10.1038/s41598-017-01082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slezakova S, Ruda-Kucerova J Anticancer activity of artemisinin and its derivatives. Anticancer Research. 2017;37(11):5995–6003. doi: 10.21873/anticanres.12046. [DOI] [PubMed] [Google Scholar]
- Stevenson C, Hall JP, Harrison E, et al Gene mobility promotes the spread of resistance in bacterial populations. The ISME Journal. 2017;11(8):1930–1932. doi: 10.1038/ismej.2017.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Chen C, Cui CY, et al. 2019. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nature Microbiology, 4(9): 1457–1464.
- Sun J, Yang RS, Zhang QJ, et al. 2016. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nature Microbiology, 1(12): 16176.
- Tjaden B A computational system for identifying operons based on RNA-seq data. Methods. 2020;176:62–70. doi: 10.1016/j.ymeth.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu YY Artemisinin-a gift from traditional chinese medicine to the world (Nobel lecture) Angewandte Chemie International Edition. 2016;55(35):10210–10226. doi: 10.1002/anie.201601967. [DOI] [PubMed] [Google Scholar]
- Wang HG, Qi HC, Gong SJ, et al Fe3O4 composited with MoS2 blocks horizontal gene transfer. Colloids and Surfaces B:Biointerfaces. 2020;185:110569. doi: 10.1016/j.colsurfb.2019.110569. [DOI] [PubMed] [Google Scholar]
- Wang Q, Mao DQ, Luo Y Ionic liquid facilitates the conjugative transfer of antibiotic resistance genes mediated by plasmid RP4. Environmental Science & Technology. 2015;49(14):8731–8740. doi: 10.1021/acs.est.5b01129. [DOI] [PubMed] [Google Scholar]
- Wang XY, Jiang JY, Yang L, et al. 2022. Colistin promotes mcr-1-positive IncI2 plasmid conjugation between Escherichia coli. Scientia Agricultura Sinica, 55(14): 2862–2874. (in Chinese)
- Wang Y, Lu J, Zhang S, et al Non-antibiotic pharmaceuticals promote the transmission of multidrug resistance plasmids through intra- and intergenera conjugation. The ISME Journal. 2021;15(9):2493–2508. doi: 10.1038/s41396-021-00945-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Geneva: WHO.
- Wu YJ, Yan HC, Zhu XM, et al Biochar effectively inhibits the horizontal transfer of antibiotic resistance genes via restraining the energy supply for conjugative plasmid transfer. Environmental Science & Technology. 2022;56(17):12573–12583. doi: 10.1021/acs.est.2c02701. [DOI] [PubMed] [Google Scholar]
- Xiao X, Zeng FX, Li RC, et al Subinhibitory concentration of colistin promotes the conjugation frequencies of Mcr-1- and blaNDM-5-positive plasmids. Microbiology Spectrum. 2022;10(2):e0216021. doi: 10.1128/spectrum.02160-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu KQ, Chen FR, Yue L, et al CeO2 Nanoparticles regulate the propagation of antibiotic resistance genes by altering cellular contact and plasmid transfer. Environmental Science & Technology. 2020;54(16):10012–10021. doi: 10.1021/acs.est.0c01870. [DOI] [PubMed] [Google Scholar]
- Yu ZG, Wang Y, Lu J, et al Nonnutritive sweeteners can promote the dissemination of antibiotic resistance through conjugative gene transfer. The ISME Journal. 2021;15(7):2117–2130. doi: 10.1038/s41396-021-00909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YL, Liu BC, Chu XL, et al Commercialized artemisinin derivatives combined with colistin protect against critical Gram-negative bacterial infection. Communications Biology. 2022;5(1):931. doi: 10.1038/s42003-022-03898-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.