Abstract
Synaptic dysfunction is an important pathological hallmark and cause of Alzheimer’s disease (AD). High-frequency stimulation (HFS)-induced long-term potentiation (LTP) has been widely used to study synaptic plasticity, with impaired LTP found to be associated with AD. However, the exact molecular mechanism underlying synaptic plasticity has yet to be completely elucidated. Whether genes regulating synaptic plasticity are altered in AD and contribute to disease onset also remains unclear. Herein, we induced LTP in the hippocampal CA1 region of wild-type (WT) and AD model mice by administering HFS to the CA3 region and then studied transcriptome changes in the CA1 region. We identified 89 genes that may participate in normal synaptic plasticity by screening HFS-induced differentially expressed genes (DEGs) in mice with normal LTP, and 43 genes that may contribute to synaptic dysfunction in AD by comparing HFS-induced DEGs in mice with normal LTP and AD mice with impaired LTP. We further refined the 43 genes down to 14 by screening for genes with altered expression in pathological-stage AD mice without HFS induction. Among them, we found that the expression of Pygm, which catabolizes glycogen, was also decreased in AD patients. We further demonstrated that down-regulation of PYGM in neurons impaired synaptic plasticity and cognition in WT mice, while its overexpression attenuated synaptic dysfunction and cognitive deficits in AD mice. Moreover, we showed that PYGM directly regulated energy generation in neurons. Our study not only indicates that PYGM-mediated energy production in neurons plays an important role in synaptic function, but also provides a novel LTP-based strategy to systematically identify genes regulating synaptic plasticity under physiological and pathological conditions.
Keywords: Alzheimer’s disease, High-frequency stimulation, Long-term potentiation, PYGM, Synaptic plasticity, Transcriptome
INTRODUCTION
Alzheimer’s disease (AD) is the most common neurodegenerative disease and is characterized by progressive memory loss and cognitive function deterioration ( Guo et al., 2020a; Knopman et al., 2021). During learning and memory, neurons communicate with each other through synapses, enabling fast and point-to-point information transfer from presynaptic to postsynaptic neurons ( Sudhof, 2018). Synapse impairment is an early pathological hallmark of AD and is closely related to the decline of cognitive function ( Cuestas Torres & Cardenas, 2020; Koffie et al., 2011; Tzioras et al., 2023). Alterations in the expression and cellular distribution of proteins mediating synaptic transmission, including N-methyl-D-aspartate (NMDA) and α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid (AMPA) receptors, have been implicated in the occurrence and progression of AD ( Leuba et al., 2008; Liu et al., 2019; Proctor et al., 2010; Wang & Reddy, 2017).
Long-term potentiation (LTP) is a phenomenon in which brief high-frequency stimulation (HFS) of excitatory synapses on presynaptic neurons can induce a rapid and long-lasting increase in synaptic strength ( Nicoll, 2017; Wu et al., 2017). Given its involvement in the cellular processes underlying learning and memory, LTP, particularly in the hippocampus, has emerged as a valuable cellular model for studying synaptic plasticity associated with learning and memory ( Nicoll, 2017; Wu et al., 2017). In addition to learning and memory impairment, LTP is also reported to be impaired in AD animal models ( Viana Da Silva et al., 2016).
Synaptic plasticity is a highly regulated process involving the contribution of multiple genes. A thorough identification of such genes will strengthen our understanding of the molecular mechanisms underlying not only synaptic plasticity, but also the pathogenesis and progression of AD. In the present study, we hypothesize that gene expression in postsynaptic neurons will change in response to HFS in LTP induction. Although changes in the expression of most genes may only be an accompanying phenomenon, some expression changes may help stabilize and maintain LTP, thereby playing important roles in synaptic plasticity. Moreover, if the changes in gene expression differ between normal and impaired LTP, they may contribute to the dysfunction of synapses. Based on this hypothesis, we induced LTP in the hippocampal CA3-CA1 Schaffer collateral pathway in an AD mouse model and wild-type (WT) controls, and then compared transcriptome changes in the CA1 region. We identified a series of genes possibly involved in normal synaptic plasticity and synaptic dysfunction in AD. Specifically, we found that the expression of PYGM, which regulates glycogen catabolism and thus energy production, was decreased in AD. We further demonstrated that down-regulation of PYGM in neurons impaired synaptic plasticity and learning and memory in WT mice, while its overexpression attenuated synaptic impairment and cognitive deficits in AD mice.
MATERIALS AND METHODS
Animals
Male APP/PS1 mice carrying human APP Swedish and PSEN1 ΔE9 mutations on a C57BL/6 background, along with their WT control mice, were purchased from Cavins Laboratory Animal Co. Ltd. (Changzhou, Jiangsu, China) for behavioral and electrophysiological experiments, and from Wukong Biotechnology Co. Ltd. (Nanjing, Jiangsu, China) for confirmatory experiments. Some mice were crossed with female WT C57BL/6 mice obtained from the Xiamen University Laboratory Animal Center (Xiamen, Fujian, China) to generate offspring for harvesting primary neurons. Mice were housed under specific-pathogen-free (SPF) conditions and a 12:12 h light/dark cycle with free access to food and water. All animal procedures were approved by the Animal Ethics Committee of Xiamen University (#XMULAC20190077) and conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Electrophysiological recordings
Mice were anesthetized with isoflurane and rapidly decapitated for brain collection. Coronal brain slices (400 μm thick) were sectioned using a VT 1200S vibratory slicer (Leica, Germany) in ice-cold sectioning liquid constantly bubbled with a mixture of 95% O 2 and 5% CO 2 (vol/vol). The sectioning liquid contained 64 mmol/L NaCl, 2.5 mmol/L KCl, 1.25 mmol/L NaH 2PO 4, 10 mmol/L MgSO 4, 0.5 mmol/L CaCl 2, 26 mmol/L NaHCO 3, 10 mmol/L glucose, and 120 mmol/L sucrose. Acute brain slices were rapidly transferred to an incubator with artificial cerebrospinal fluid (ACSF) at 32 °C and saturated with a mixture of 95% O 2 and 5% CO 2 (vol/vol) for recovery. For field potential recordings, low-Mg 2+ ACSF containing 120 mmol/L NaCl, 3.5 mmol/L KCl, 1.25 mmol/L NaH 2PO 4, 1.3 mmol/L MgSO 4, 2.5 mmol/L CaCl 2, 26 mmol/L NaHCO 3, and 10 mmol/L glucose was used. For whole-cell recordings, normal ACSF containing 126 mmol/L NaCl, 2.5 mmol/L KCl, 1.2 mmol/L NaH 2PO 4, 2.4 mmol/L MgCl 2•6H 2O, 1.2 mmol/L CaCl 2, 18 mmol/L NaHCO 3, and 11 mmol/L glucose was used.
For LTP recordings, after 1 h of recovery at 32 °C and 1 h of culturing at room temperature, brain slices were positioned in a recording chamber filled with low-Mg 2+ ACSF and continuously perfused with 95% O 2/5% CO 2-saturated low-Mg 2+ ACSF at a rate of 4 mL/min. A concentric bipolar electrode was used to stimulate the CA3 region and field excitatory postsynaptic potentials (fEPSPs) were recorded in the CA1 stratum radiatum under current-clamp with glass pipettes (700 KΩ–1 MΩ) filled with the low-Mg 2+ ACSF. Baseline responses were acquired at a stimulation intensity that elicited a 30% maximum intensity reaction. After stable baseline recording for 10–20 min, two trains of HFS (100 Hz, 1 s) divided by a 30 s interval were used to establish LTP. The amplitudes of fEPSPs were recorded and analyzed using Clampfit v10.4 software (Molecular Device, USA), with three continuous responses averaged into one for later analysis. The amplitudes of the evoked fEPSPs were measured and normalized (as a percentage) to the mean amplitude of the baseline.
For whole-cell recordings, brain slices were recovered for 30 min at 32 °C, then held at room temperature. Brain slices, as well as coverslips containing treated primary neurons at 15–16 days in vitro (DIV), were mounted in a recording chamber filled with normal ACSF and continuously perfused with 95% O 2/5% CO 2-saturated normal ACSF at a rate of 2–3 mL/min. CA1 pyramidal neurons in hippocampal slices, as well as cultured primary neurons, both infected with green fluorescent protein (GFP)-expressing viruses, were identified using a 488 nm excitation/emission filter set on an upright microscope (BX51WI, Olympus, Japan). Patch pipettes (5–8 MΩ) were pulled from thick-walled borosilicate glass capillaries (Sutter Instrument, USA) on a P-97 Flaming Micropipette Puller (Sutter Instrument, USA). To record spontaneous excitatory postsynaptic currents (sEPSCs) and spontaneous inhibitory postsynaptic currents (sIPSCs), the caesium-based pipette internal solution contained 140 mmol/L CH 3O 3SCs, 2 mmol/L MgCl 2•6H 2O, 5 mmol/L TEA-Cl, 10 mmol/L HEPES, 1 mmol/L EDTA, 2.5 mmol/L Mg-ATP, and 0.3 mmol/L Na 2-GTP, adjusted to a pH of 7.35 using CsOH and osmolality set to 310–320 mOsm/kg H 2O. To record action potentials, the pipette internal solution included 140 mmol/L K-gluconate, 2 mmol/L MgCl 2•6H 2O, 0.1 mmol/L CaCl 2, 10 mmol/L HEPES, 1 mmol/L EDTA, 2.5 mmol/L Mg-ATP, and 0.3 mmol/L Na 2-GTP, calibrated to a pH of 7.35 with KOH and osmolality set to 305–315 mOsm/kg H 2O. All internal solutions were filtered using 0.22 μm filters before use. All whole-cell patch signals were obtained with an Axon MultiClamp 700B amplifier (Molecular Device, USA) and low-pass filtered at 2 kHz and digitally sampled at 10 kHz with a 1440A digitizer (Molecular Device, USA). Access resistance (Ra), membrane resistance (Rm), and membrane capacitance (Cm) were monitored using the membrane test protocol in Clampex v10.4 (Molecular Device, USA). Data were only included if they met the following conditions: preliminary Ra<20 MΩ, Rm>100 MΩ, Cm>100 pF, and <20% change in Ra at the later stage of recording. The sEPSCs and sIPSCs were recorded at holding potentials of −70 mV and 0 mV, respectively. The frequency and amplitude of sEPSC and sIPSC events were analyzed using Clampfit v10.4 (Molecular Device, USA) and Mini Analysis Program (Synaptosoft, USA) in cooperation. Action potentials were recorded using a current-clamp after neurons were injected directly with different intracellular depolarizing currents from −20 pA to 120 pA for 500 ms at an increasing magnitude of 10 pA. Action potential numbers were calculated using Clampfit v10.4 (Molecular Device, USA).
Hippocampal CA1 tissue acquisition
After electrophysiological recordings, the hippocampal CA1 region in brain slices was collected using the UnipicK TM capillary-based vacuum-assisted cell and tissue acquisition system (NeuroInDx, USA) ( Kudo et al., 2012). Briefly, the CA1 region was recognized under an ECLIPSE Ts2 inverse microscope (Nikon, Japan) and captured by a disposable capillary unit (DCU) with a 40 μm inner diameter tip upon optimization of lighting, vacuum strength, and vacuum duration time. After capture, the DCUs containing the collected CA1 samples were removed from the UnipicK TM system and carefully placed into standard 200 μL test tubes, with the contents released using screw-in port syringes with filters.
RNA sequencing, data processing, and bioinformatics analysis
Hippocampal CA1 tissues collected from three brain slices (each from a different animal but with the same genotype and age) under the same HFS treatment were mixed. Control brain slices not induced by HFS but otherwise treated in a same manner as those subjected to HFS were used. Samples of different treatment groups were subjected to RNA extraction, mRNA library construction, RNA sequencing, and data processing, conducted by the Beijing Genomics Institute (BGI, Shenzhen, China). Briefly, total RNA was extracted from the tissues using an RNeasy Micro Kit (Qiagen, Germany) following the manufacturer’s instructions. Total RNA was qualified and quantified using a NanoDrop Spectrophotometer (Thermo Fisher Scientific, USA) and an Agilent 2100 Bioanalyzer (Thermo Fisher Scientific, USA), respectively. High-quality RNA with an RNA integrity number >7 was used for mRNA library construction.
High-quality RNA (more than 200 pg) was reverse transcribed into cDNA using oligo-dT and dNTP amplification by targeting the polyA tail. The template was switched to the 5’ end of the RNA and full-length cDNA was generated by polymerase chain reaction (PCR). The average length of the PCR products was determined using an Agilent 2100 Bioanalyzer. Subsequently, the purified cDNA from previous steps was fragmented into small pieces, then purified and selected using an Agencourt AMPure XP-Medium Kit (Thermo Fisher Scientific, USA). The Agilent 2100 Bioanalyzer was used to quantify cDNA. Following quality control (QC), the double-stranded PCR products were heat denatured and circularized using the splint oligo sequence. The single-strand circle DNA was formatted as the final library. The final library was quantitated in two ways to ensure the high quality of the sequencing data: determining average molecule length using the Agilent 2100 Bioanalyzer and quantifying the library by quantitative real-time PCR (qRT-PCR). The final library was amplified with phi29 (Thermo Fisher Scientific, USA) to form DNA nanoballs (DNBs), each containing more than 300 copies of a single molecule. The DNBs were loaded into the patterned nanoarray, and single-end reads of 50 bases were generated on the BGISEQ500 platform (BGI, China).
Raw sequencing data were filtered using SOAPnuke (v1.4.0) (https://github.com/BGI-flexlab/SOAPnuke) to remove reads with adaptors, reads with a low-quality base ratio of more than 20%, and reads with more than 5% of unknown bases. Clean reads were mapped to the Mus musculus reference genome (GCF_000001635.25_GRCm38.p5) using HISAT2 (v2.1.0) ( Kim et al., 2015) and aligned to the reference coding gene set using Bowtie2 (v2.2.5) ( Langmead & Salzberg, 2012). Gene expression levels were calculated and normalized to fragments per kilobase of transcript per million mapped reads (FPKM) using RSEM (v1.2.8) ( Li & Dewey, 2011). Differentially expressed genes (DEGs) were determined using DEGseq ( Wang et al., 2010), with a fold-change≥2 and false discovery rate (FDR)-adjusted P-value≤0.001. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using the Dr. Tom platform (https://report.bgi.com) of BGI (China).
Antibodies
Primary antibodies used for immunoblotting included: anti-APP (Thermo Fisher Scientific, 14-9749-82, 1:2 000, USA), anti-PYGM (4A-Biotech, 4ab095124, 1:1 000, China), anti-NeuN (Cell Signaling Technology, 94403S, 1:1 000, USA), anti-GFAP (Cell Signaling Technology, 3670S, 1:1 000, USA), anti-Iba1 (Cell Signaling Technology, 17198S, 1:1 000, USA), anti-NR1 (Cell Signaling Technology, 5704S, 1:1 000, USA), anti-NR2A (Cell Signaling Technology, 4205S, 1:1 000, USA), anti-NR2B (Cell Signaling Technology, 14544S, 1:1 000, USA), anti-GluA1 (Cell Signaling Technology, 13185S, 1:1 000, USA), anti-GluA2 (Cell Signaling Technology, 13607S, 1:1 000, USA), anti-HA (Abmart, M20003L, 1:1 000, USA), and anti-β-actin (Cell Signaling Technology, 8457S, 1:5 000, USA). Secondary antibodies used for immunoblotting included: goat anti-rabbit IgG (H+L)-HRP (Thermo Fisher Scientific, 31460, 1:4 000, USA) and goat anti-mouse IgG (H+L)-HRP (Thermo Fisher Scientific, 31430, 1:4 000, USA).
Primary antibodies used for immunofluorescence included: anti-NeuN (Cell Signaling Technology, 94403S, 1:200, USA), anti-Iba1 (Wako, 016-20001, 1:200, Japan), anti-GFAP (Proteintech, 16825-1-AP, 1:200, USA), and anti-HA (Proteintech, 51064-2-AP, 1:200, USA). Secondary antibodies used for immunofluorescence included: Alexa Fluor® 594 goat anti-mouse IgG (H+L) (Thermo Fisher Scientific, A-11005, 1:500, USA) and Alexa Fluor® 594 goat anti-rabbit IgG (H+L) (Thermo Fisher Scientific, A-11012, 1:500, USA).
Cultures of mouse primary neurons, astrocytes, and microglia
For mouse primary neurons, pregnant mice were anesthetized with isoflurane and embryos at embryonic day 15–16 (E15–E16) were rapidly removed and decapitated. The brains were dissected and placed in ice-cold Hanks’ balanced salt solution (HBSS, Gibco, USA) lacking Ca 2+ and Mg 2+. Brain tissues, including the cortex and hippocampus, were isolated and digested in fresh HBSS containing 0.025% trypsin (Gibco, USA) and 1 mg/mL DNase Ⅰ (Worthington, USA) for 15 min at 37 °C. After digestion, the tissue pellets were blown and triturated using glass pipettes, then centrifuged at 1 000 r/min for 5 min at room temperature. After removing the supernatant, cell pellets were re-suspended in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA). Cells were counted and plated onto Poly-L-Lysine (PLL, Sigma-Aldrich, USA)-coated plates or coverslips. After allowing neurons to adhere for 4 h, the culture medium was replaced with N+3 medium containing neurobasal medium (Gibco, USA) supplemented with 2% B-27 (Gibco, USA), 2 mmol/L L-glutamine (Gibco, USA), and 100 U/ mL penicillin and 100 µg/ mL streptomycin (Gibco, USA). Every 3 days, half of the cultured medium was replaced with fresh N+3 medium.
Mouse primary astrocytes and microglia were cultured as described previously ( Zhang et al., 2021). Briefly, the postnatal day 1 (P1) pup brains were dissected, sliced into small pieces, and digested to dissociate cells. Mix cells were cultured in flasks with medium containing DMEM (Gibco, USA), 10% heat-inactivated FBS (Gibco, USA), and 100×penicillin/streptomycin. After 10 days, the culture flasks were mechanically shaken at 200 r/min for 20 min at room temperature. Microglia floating in the medium were collected by centrifugation at 500 × g/min for 5 min at room temperature and cultured separately. Astrocytes attached to the flask bottom were isolated with trypsin- ethylenediaminetetraacetic acid (EDTA) and plated at a density of 2×10 6 cells per well in 6-well plates.
Viruses and viral infection in vitro and in vivo
Adeno-associated virus serotype 2/9 (AAV2/9) (viruses carrying mouse Pygm shRNA and a scrambled negative control shRNA were packaged by OBIO Technology (China). Their sequences are: Pygm shRNA (Y14949): 5’-CCAGGATAAGAGAAAGCAA-3’; Pygm shRNA (Y14950): 5’-GCTGAAGCAGGAGTACTTT-3’; Pygm shRNA (Y14951): 5’-CCTCGAGAGGGAATACAAA-3’, and scrambled negative control shRNA: 5’-CCTAAGGTTAAGTCGCCCTCG-3’. Because the mouse Pygm gene was 2 529 bp long and to ensure its efficient viral packaging and expression, we used lentivirus to express Pygm tagged with HA and driven by a human synapsin 1 promoter (hSyn-Pygm). Lentiviruses, including controls, were packaged by OBIO Technology. These viruses were used to infect primary neurons cultured for 3–4 DIV.
For in vivo infection, WT and APP/PS1 male mice (4–5 months of age) were anesthetized and immobilized in a stereotactic frame. The AAV2/9 viruses (1 µL, 1×10 12 VG/mL) carrying the Pygm shRNA (Y14949) or control shRNA were bilaterally delivered into the hippocampal CA1 coordinates (−2.0 anterior-posterior; ±1.6 media-lateral; −1.6 dorsal-ventral) of WT mice. Alternatively, lentiviruses (2 µL, 3×10 8 TU/mL) expressing Pygm or control were bilaterally delivered into the hippocampal CA1 coordinates of APP/PS1 mice. After surgery, the mice were positioned on a heating pad for recovery.
Immunoblotting
Cell and tissue samples were lysed in ice-cold 1% TNEN buffer (25 mmol/L Tris-HCl at pH 7.6, 150 mmol/L NaCl, 1% sodium deoxycholate, 1% Nonidet P-40, and 0.1% sodium dodecyl sulfate (SDS)), supplemented with protease and phosphatase inhibitors. Protein concentrations were determined using the bicinchoninic acid protein assay (BCA). Equal amounts of protein lysates were separated by acrylamide denaturing gels (SDS-polyacrylamide gel electrophoresis (SDS-PAGE)), then transferred onto polyvinylidene fluoride membranes (PVDF). After blocking with 5% nonfat milk in PBST (PBS buffer containing 0.05% Tween-20) at room temperature for 1 h, the membranes were incubated with primary antibodies overnight at 4 °C. On the second day, the membranes were incubated with appropriate horseradish peroxidase (HRP)-linked secondary antibodies at room temperature for 1 h. Enhanced chemiluminescence (ECL) kits were applied to detect blots on membranes through film development. Protein band intensity was quantified using ImageJ v1.8.0 (National Institutes of Health, USA).
Immunostaining
After deep anesthesia, the mice were transcardially perfused with 20 mL of 1×PBS. The brain was dissected, fixed with 0.1 mol/L sodium phosphate buffer containing 4% paraformaldehyde (PFA) overnight at 4 °C, sequentially dehydrated in 20%, 25%, and 30% sucrose, and embedded in optimal cutting temperature (OCT) compound for storage at −80 °C. Coronal brain sections (15 μm thick) were cut using a cryostat vibratome. Brain sections were washed three times in 1×PBS and blocked with 1×PBS containing 5% bovine serum albumin (BSA) and 0.2% Triton X-100 for 1 h at room temperature. Brain sections were then incubated overnight at 4 °C with primary antibodies in blocking solution. The following day, the sections were treated with appropriate fluorescence-labeled secondary antibodies in a dim environment for 1 h at room temperature. Subsequently, the cell nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI, 1:1 000) for 10 min. After mounting with anti-fade mounting agent, the sections were observed under a fluorescence confocal microscope (FV1000, Olympus, Japan) at 20× objective and XY scanning.
qRT-PCR
Total RNA from homogenized whole hippocampal tissues or cells was extracted with Trizol reagent (Sigma-Aldrich, USA) and reverse-transcribed into cDNA using a ReverTra Ace qPCR RT Kit (TOYOBO, Japan) following the manufacturers’ protocols. qRT-PCR was performed using FastStart Universal SYBR ® Green Master Mix (Rox) (Roche, Switzerland) and the LightCycler 480 ® Real-Time PCR system (Roche, Switzerland). Relative Pygm expression levels were analyzed using the 2 -ΔΔCт method and normalized to those of β-actin. The primers used were as follows: Pygm forward: 5’-TGCACTTCTATGGCCGAGTG-3’; Pygm reverse: 5’-GAGATGTTCTCGGCCAGGTT-3’; β-actin forward: 5’-AGCCATGTACGTAGCCATCCA-3’; β-actin reverse: 5’-TCTCCGGAGTCCATCACAATG-3’.
Oxygen consumption rate (OCR) measurement
Primary neurons derived from WT and APP/PS1 mice were plated on a Poly-L-lysine (PLL)-pre-coated Seahorse XF96 Cell Culture Microplate (Agilent, USA) at a density of 5×10 4 cells per well. WT neurons were infected with AAV2/9 containing Pygm shRNA or controls. APP/PS1 neurons were infected with lentiviruses expressing Pygm or controls. The OCR of 15–16 DIV neurons was analyzed using a Seahorse XFe96 Cellular Energy Metabolism Dynamic Analyzer (Agilent, USA) with the Seahorse XF Cell Mito Stress Test Kit (Agilent, USA) following the manufacturer’s instructions. Briefly, neuronal culture medium was changed to Seahorse XF Base Medium (Agilent, USA) supplemented with 1 mmol/L pyruvate, 2 mmol/L L-glutamine, 10 mmol/L D-glucose, and 5 mmol/L HEPES. Neurons were then maintained in a non-CO 2 incubator for 1 h at 37 °C before being placed into Seahorse XFe96 Analyzer Sensor cartridges. After baseline measurement of OCR, 20 μL of oligomycin, 22 μL of FCCP, and 25 μL of rotenone/antimycin A were sequentially injected to final concentrations of 1.5 μmol/L oligomycin, 1.5 μmol/L FCCP, and 0.5 μmol/L rotenone/antimycin A. The OCR was measured during the entire procedure. After assay, the neurons were immediately stained with DAPI (Invitrogen, P36935, USA). The OCR was normalized to total neuron number in each well (pmol/min/10 4 cells) using a Live Cell High Throughput Assay Platform Cytation-5&BioSpa8 (Biotek, USA). Post-normalization, data were exported from Wave v2.6.3 software (Agilent, USA) using the Seahorse XF Cell Mito Stress Test Report Generator for further analysis.
Behavioral tests
Y-maze test: The Y-maze test was employed to determine cognitive deficits of short-term working memory in mice. The Y-maze device consisted of three identical arms (30 cm×5 cm×15 cm each) with symmetrical angles of 120°. Each mouse was positioned in the center of the apparatus and given 5 min to freely explore the three arms. An arm entry was defined when the mouse body was completely within the arm. The number and sequence of arms entered were recorded using TopScanLite v2.0 video tracking software (CleverSys, USA). A correct alternation sequence was considered when a mouse entered the three different arms in succession. Alternation rate (% Alternation)=((total number of correct alternation sequence)/(total number of arm entry−2))×100%. A decrease in the rate of alternation is suggestive of a problem with short-term working memory.
Novel object recognition test: The novel object recognition test was used to evaluate the capacity of mice to discern a new object in a familiar setting. Each mouse was first acclimated to an empty acrylic box (40 cm×40 cm×40 cm) for 10 min on day 1. During the training session on day 2, two identical objects (A and A’) were placed in opposite corners of the box and each mouse was allowed to freely explore them for 10 min. In the probe test on day 3, one of the two objects (A’) was changed with one (B) that was noticeably different in color, shape, and texture, and each mouse was allowed to explore the two objects for 10 min. Exploration time of each mouse for the two old and one novel object was recorded and analyzed using TopScanLite v2.0 (CleverSys, USA). The recognition index was calculated using the formula: (time of novel object exploration/time of both novel and old object exploration)×100%.
Fear conditioning test: The fear conditioning test was used to assess mouse memory in response to conditioned environment and sound cues. On day 1, each mouse was placed in a transparent plexiglass cage (25 cm×25 cm×25 cm) with visual access to an environmental context. After 2 min of free exploration, a 30 s conditioning stimulus (CS) consisting of a 5 kHz, 80 dB tone was presented, accompanied by a co-terminated 2 s foot shock unconditioned stimulus (US) of 0.5 mA. Three CS-US pairs, separated by 60 s intervals, were given to each mouse. After the last CS-US pair, the mouse was left in the cage for another 90 s, then gently returned to its home cage. On day 2, each mouse was returned to the cage for 5 min to test their contextual fear response, during which neither CS nor US was delivered. On day 3, tiny white enclosed chambers with smooth walls and base were assembled inside the cage. Each mouse was placed into the chamber for 6 min, consisting of 3 min of free exploration followed by 3 min of the identical CS tone cue. Mouse behaviors, including freezing, on each day were recorded and analyzed using the FreezeScan v2.0 video tracking software (CleverSys, USA).
Morris water maze test: The Morris water maze test was applied to study spatial learning memory in mice. For this test, a circular water maze pool (150 cm in diameter and 60 cm in height) was filled with 20 °C tap water to a depth of 40 cm. The pool was divided equally into four quadrants. Different symbols were posted on the center of the quadrant walls, 10 cm above the water surface. An escape platform (10 cm in diameter) was placed in the target quadrant at 1 cm below the water surface. Titanium dioxide was added to the water and stirred well so that the platform was not visible to the mice. The water maze test was divided into two phases: spatial navigation training (first 6 days) and spatial exploration task (day 7). During the training phase, each mouse was released into the pool at a random site and allowed to find and climb onto the hidden platform within 60 s. If a mouse failed to reach the hidden platform within the timeline, it was guided to the platform. Two training trials per day were performed for each mouse, with a 15–20 min interval between each training session. Mouse behaviors were video recorded and analyzed with TopScanLite v2.0 (CleverSys, USA). Average escape latency for each training session was calculated. On day 7, the hidden platform was removed, and each mouse was placed into the water at the quadrant farthest from the platform. The time spent in each quadrant, number of crossings over the platform location, and swim speed within 60 s were recorded for comparison.
Statistical analysis
Results from three or more independent experiments were plotted as mean±standard error of the mean ( SEM) and analyzed using Prism v8.3.0 (GraphPad Software, USA). No outliers were excluded. Statistical differences between two groups were assessed using the unpaired t-test, and among more than two groups were assessed using one-way analysis of variance (ANOVA) with two-sided Holm correction or one-way ANOVA with Dunnett’s multiple comparisons. For statistical results, P<0.05 was considered significant.
RESULTS
APP/PS1 mice are characterized with normal LTP in the hippocampal CA3-CA1 Schaffer collateral pathway at 2–3 months of age, LTP defects at 5 months of age, and obvious Aβ plaques at 6–7 months of age ( Chen & Zhang, 2022; Gelman et al., 2018; Kim et al., 2022; Reiserer et al., 2007; Xu et al., 2018). Therefore, we studied LTP in the hippocampal CA3-CA1 Schaffer collateral pathway of APP/PS1 mice and WT controls at 2 and 6 months of age. Following HFS in the hippocampal CA3 region and monitoring fEPSPs at the CA1 region for 60 min, we found that LTP was comparable in 2-month-old APP/PS1 mice, 2-month-old WT mice, and 6-month-old WT mice, but significantly impaired in 6-month-old APP/PS1 mice (Supplementary Figure S1A–I). We then collected the CA1 regions from the treated mice and mice without HFS treatment for RNA sequencing (RNA-seq) (Supplementary Figure S1J). A total of 18 467 genes were identified in these samples (Supplementary Table S1).
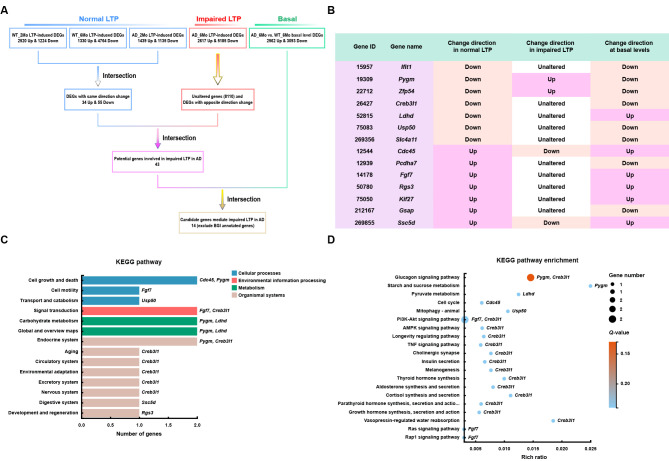
We identified 1 224 down-regulated and 2 520 up-regulated DEGs in 2-month-old WT mice subjected to HFS and 60-min recording (WT_2Mo_S60) compared to 2-month-old WT mice without HFS (WT_2Mo_NC), 4 764 down-regulated and 1 330 up-regulated DEGs in 6-month-old WT mice subjected to HFS and 60-min recording (WT_6Mo_S60) compared to 6-month-old WT mice without HFS (WT_6Mo_NC), and 1 135 down-regulated and 1 439 up-regulated DEGs in 2-month-old APP/PS1 mice subjected to HFS and 60-min recording (AD_2Mo_S60) compared to 2-month-old APP/PS1 mice without HFS (AD_2Mo_NC) ( Figure 1A; Supplementary Table S2A–C). Because LTP was comparable among the mice, genes consistently altered upon HFS may represent genes involved in synaptic plasticity. We identified a total of 89 such genes (55 down-regulated and 34 up-regulated) ( Figure 1A; Supplementary Table S3).
Figure 1.
LTP-based screening to identify genes involved in synaptic function and AD
A: Schematic of gene screening process. DEGs in response to HFS in mice with normal LTP (2-month-old WT and AD mice and 6-month-old WT mice) and impaired LTP (6-month-old AD mice), as well as DEGs in 6-month-old AD mice compared to WT control mice at basal levels (without HFS), were determined by RNA-seq. DEGs displaying consistent changes in the same direction in mice with normal LTP were screened as potential genes involved in normal synaptic plasticity. These genes were then intersected with those unchanged or changed in the opposite direction in mice with impaired LTP to screen for genes potentially involved in impaired LTP in AD, and further intersected with DEGs in 6-month-old AD mice at basal levels to identify final candidate genes. B: Expression alteration patterns of 14 identified final candidate genes under different conditions. C, D: KEGG (C) and KEGG enrichment (D) analyses of 14 candidate genes.
We also identified 5 185 down-regulated and 2 617 up-regulated DEGs and 8 110 unaltered genes in 6-month-old AD mice subjected to HFS and 60-min recording (AD_6Mo_S60) compared to 6-month-old AD mice without HFS (AD_6Mo_NC) ( Figure 1A; Supplementary Table S2D). By screening DEGs that showed consistent changes in samples with comparable LTP (WT_2Mo_S60 vs. WT_2Mo_NC, WT_6Mo_S60 vs. WT_6Mo_NC, and AD_2Mo_S60 vs. AD_2Mo_NC), but either no change or changes in the opposite direction in samples with impaired LTP (AD_6Mo_S60 vs. AD_6Mo_NC), we successfully identified 43 genes ( Figure 1A; Supplementary Table S4). The distinct alteration patterns observed in these genes between normal LTP and impaired LTP suggest potential involvement in synaptic dysfunction associated with AD.
Moreover, we identified 3 093 down-regulated and 2 562 up-regulated DEGs in AD_6Mo_NC vs. WT_6Mo_NC ( Figure 1A; Supplementary Table S2E). Genes already exhibiting altered expression at the basal level (without HFS) during early AD onset may also influence neuronal responses to LTP. Therefore, to select genes strongly correlated with AD, we further screened the 43 DEGs identified above, with 22 found to be altered in AD_6Mo_NC vs. WT_6Mo_NC (Supplementary Table S5). We excluded genes annotated by BGI without reported information in GenBank or assigned functions. Consequently, we identified 14 candidate genes that may mediate impaired LTP and thus synaptic plasticity in AD ( Figure 1A, B).
Based on KEGG pathway analysis of the 14 genes, we found that among the five pathways with more than one annotated gene, i.e., “cell growth and death”, “signal transduction”, “carbohydrate metabolism”, “global and overview maps”, and “endocrine system”, the Pygm gene was annotated in four of them ( Figure 1C). Although further KEGG pathway enrichment analysis of the 14 genes did not reveal any significant pathways, Pygm was annotated in the enriched “glucagon signaling pathway” with the lowest Q-value ( Q-value: 0.138; rich ratio: 0.015) ( Figure 1D). Therefore, we focused on Pygm in additional functional analyses.
PYGM expression is reduced in the brain of AD patients and model mice
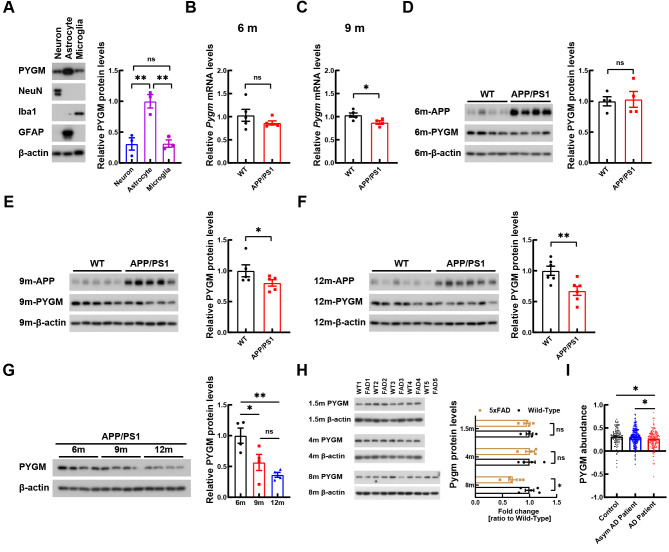
PYGM is a major enzyme involved in regulating glycogen catabolism in muscles. Previous studies have suggested that PYGM is primarily expressed in astrocytes in the central nervous system ( Migocka-Patrzalek & Elias, 2021; Uhlén et al., 2015). Our findings confirmed this observation and further revealed the expression of PYGM in neurons and microglia ( Figure 2A). The RNA-seq results showed that Pygm expression was decreased under normal LTP but increased under impaired LTP. Furthermore, in the basal state, Pygm expression was decreased in the hippocampal CA1 region of 6-month-old APP/PS1 mice ( Figure 1B). Pygm expression in the whole hippocampal tissue of 6-month-old APP/PS1 mice was not yet reduced ( Figure 2B) but was significantly decreased in 9-month-old APP/PS1 mice compared to controls ( Figure 2C). Expression of the PYGM protein also decreased in whole hippocampal tissues of AD mouse models with aging, including in the APP/PS1 ( Figure 2D–G) and 5xFAD ( Figure 2H) strains. Moreover, based on a large proteomic dataset ( Johnson et al., 2022), PYGM levels were decreased in the postmortem dorsolateral prefrontal cortex of AD patients compared to those of the controls and asymptomatic AD patients ( Figure 2I).
Figure 2.
PYGM expression is decreased in the brains of AD patients and APP/PS1 mice
A: Levels of PYGM in various primary neural cells were determined by immunoblotting and densitometry analyses. NeuN, Iba1, and GFAP were used as markers of neurons, microglia, and astrocytes, respectively. n=3 independent experiments. B, C: Pygm expression in whole hippocampal tissue of 6- (B, n=5) and 9-month-old (C, n=5) APP/PS1 and WT control mice were determined by qRT-PCR and normalized to β-actin levels for comparison. D–G: PYGM protein in whole hippocampal tissue of 6- (D, n=4), 9- (E, n=5), and 12-month-old (F, n=6) APP/PS1 and WT control mice, and of APP/PS1 mice at different ages (G, n=4), was immunoblotted, with PYGM levels normalized to β-actin levels for comparison. H: PYGM protein in whole hippocampal tissue of 1.5- ( n=4), 4- ( n=4), and 8-month-old ( n=5) 5xFAD and WT control mice was immunoblotted, with their levels normalized to β-actin levels for comparison. I: Values of PYGM abundance in dorsolateral prefrontal cortex of AD patients ( n=182), asymptomatic (Asym) AD patients ( n=200), and controls ( n=106) from a published proteomic dataset ( Johnson et al., 2022) were compared. Data represent mean± SEM and were analyzed by unpaired t-test (for B–F, and H) and one-way ANOVA with two-sided Holm correction (for A, G, and I). *: P<0.05; **: P<0.01; ns: Not significant.
Down-regulation of PYGM impairs neuronal synaptic transmission
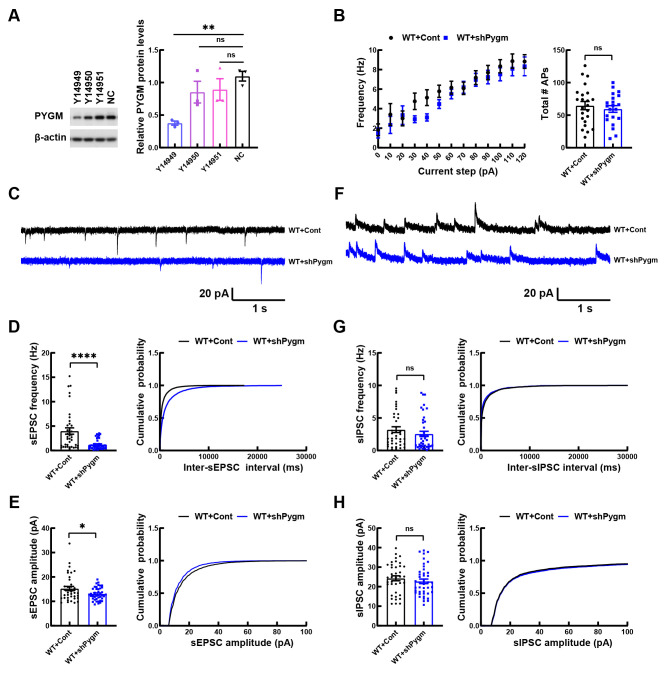
To study the function of PYGM, we first used AAV2/9 to deliver Pygm shRNAs into cultured primary neurons of C57BL/6 WT mice. Among the three tested AAV2/9 viruses (Y14949, Y14950, and Y14951) containing different Pygm shRNAs, Y14949 infection significantly down-regulated PYGM protein levels ( Figure 3A) and was thus used in the following studies. We found that down-regulation of PYGM had no effect on spontaneous firing activity in WT mouse primary neurons ( Figure 3B), but significantly decreased the frequency and amplitude of sEPSCs ( Figure 3C–E), without affecting the frequency and amplitude of sIPSCs ( Figure 3F–H). These findings suggest that the down-regulation of PYGM impairs neuronal excitatory synaptic transmission.
Figure 3.
Knockdown of PYGM impairs synaptic transmission in mouse primary neurons
A: WT mouse primary neurons were infected with AAV2/9 containing three different Pygm shRNAs (Y14949, Y14950, and Y14951) or a scrambled negative control shRNA (NC) for 12 days. Equal amounts of protein lysates were immunoblotted to compare PYGM levels after β-actin normalization. n=3 independent experiments. B: WT mouse primary neurons were infected with AAV2/9 expressing control (Cont) or Pygm (shPygm) shRNA Y14949. Action potential firing frequency in response to increased input currents was recorded in infected neurons for comparison. WT+Cont, n=23 neurons from three independent experiments; WT+shPygm, n=21 neurons from three independent experiments. C–H: Representative sEPSC (C) and sIPSC (G) traces in GFP-positive neurons from WT+Cont and WT+shPygm cultures. Histogram plots and cumulative plots of frequencies (D, G) and amplitudes (E, H) of sEPSCs (D, E) and sIPSCs (G, H). WT+Cont, n=38 neurons from three independent experiments; WT+shPygm, n=42 neurons from three independent experiments. Data represent mean± SEM and were analyzed by one-way ANOVA with Dunnett’s multiple comparisons (for A) and unpaired t-test (for B, D, E, G, and H). *: P<0.05; **: P<0.01; ****: P<0.0001; ns: Not significant.
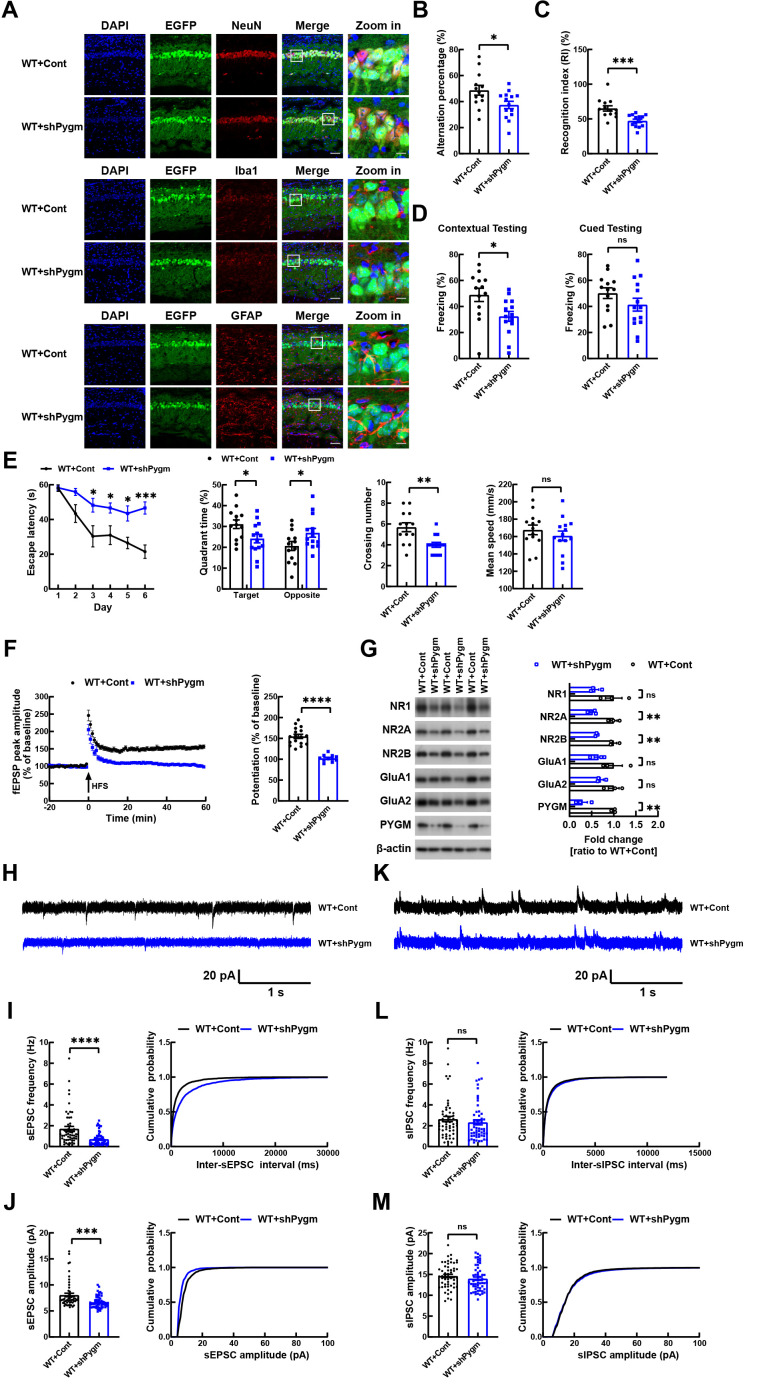
Down-regulation of PYGM in hippocampal CA1 region impairs learning and memory and synaptic function in WT mice
To study the function of neuronal PYGM in vivo, we injected AAV2/9 containing control or Pygm shRNAs, both expressing GFP, into the hippocampal CA1 region of C57BL/6 WT mice at 4–5 months of age (Supplementary Figure S2). The GFP signal was predominantly colocalized with the neuronal marker NeuN, but not with the microglial marker Iba1 or astrocytic marker GFAP ( Figure 4A), indicating that AAV2/9 primarily infected neurons rather than microglia or astrocytes. Consequently, PYGM expression was primarily down-regulated in neurons. The down-regulation of PYGM was also confirmed by immunoblotting ( Figure 4G). Treated mice were subjected to various behavioral tests after 4 months. In the Y-maze test, WT mice injected with AAV2/9 containing Pygm shRNA (WT+shPygm) showed a significantly reduced spontaneous alternation percentage compared to WT mice injected with control AAV2/9 (WT+Cont) ( Figure 4B). In the novel object recognition test, WT+shPygm mice exhibited significantly reduced recognition between a novel and familiar object compared to WT+Cont mice ( Figure 4C). In the fear conditioning test, WT+shPygm mice showed a decreased freezing percentage during contextual testing, but not during cued testing, compared to WT+Cont mice ( Figure 4D). In the Morris water maze test, WT+shPygm mice also demonstrated decreased escape latency during the 6 day training phase, and reduced time spent in the target quadrant and fewer platform crossings during the testing phase on day 7 compared to WT+Cont mice, although their mean swim speeds did no differ ( Figure 4E). Overall, these results indicate that PYGM down-regulation in the CA1 region impairs mouse learning and memory.
Figure 4.
Knockdown of PYGM in hippocampal CA1 neurons impairs synaptic function and learning and memory in WT mice
A: Representative images of hippocampal CA1 region in WT mice infected with control (Cont) or shPygm AAV2/9 expressing GFP (green). Samples were immunostained with NeuN (red), Iba1 (red), or GFAP (red), and stained with DAPI (blue). Scale bars: 50 µm for regular images; Scale bars: 10 µm for zoomed-in images. B: In Y-maze test, alternation percentage of treated mice was studied for comparison. C: In novel object recognition test, discrimination index of mice was determined for comparison. D: In fear conditioning test, freezing responses to context and cue were recorded for comparison. E: In Morris water maze test, escape latency during the 6 day training phase was recorded for comparison. On day 7, time spent in target and opposite quadrants and number of platform crossings were recorded for comparison. Mean swim speed was also compared. WT+Cont, n=13 mice; WT+shPygm, n=14 mice (for B–E). F: LTP of the Schaffer collateral circuit was studied by administering HFS in the CA3 region and recording fEPSPs in the CA1 region for 60 min. Average fEPSP peak amplitudes in the last 10 min of the recording were calculated for comparison. WT+Cont, n=17 slices from five mice; WT+shPygm, n=11 slices from four mice. G: NMDA and AMPA receptor subunits and PYGM in whole hippocampal regions of treated mice were immunoblotted and levels were quantified for comparison after β-actin normalization. n=3 per group. H–M: Representative sEPSC (H) and sIPSC (K) traces from whole-cell voltage-clamp recordings of GFP-positive hippocampal CA1 pyramidal neurons in brain slices from WT+Cont and WT+shPygm mice. Histogram plots and cumulative plots of frequencies (I, L) and amplitudes (J, M) of sEPSCs (I, J) and sIPSCs (L, M). WT+Cont, n=54 neurons from five mice; WT+shPygm, n=56 neurons from four mice. Data represent mean± SEM and were analyzed by unpaired t-test. *: P<0.05; **: P<0.01; ***: P<0.001; ****: P<0.0001; ns: Not significant.
Figure 4.
We also studied whether PYGM down-regulation in CA1 affects synaptic function in the CA3-CA1 Schaffer collateral pathway. When HFS was administered to the CA3 region, the fEPSPs in the CA1 region were much weaker in WT+shPygm mice than in WT+Cont mice ( Figure 4F), indicating LTP impairment upon PYGM down-regulation. Moreover, analysis of whole-cell sEPSCs and sIPSCs in infected hippocampal CA1 neurons confirmed that neurons with PYGM down-regulation in vivo exhibited impaired sEPSC frequency and amplitude ( Figure 4H–J), but unaffected sIPSC frequency and amplitude ( Figure 4K–M).
Synaptic plasticity is primarily regulated by AMPA and NMDA receptors. In our study, we found that PYGM down-regulation reduced protein levels of NMDA receptor subunits, including NR2A and NR2B ( Figure 4G).
Overexpression of PYGM attenuates learning and memory deficits and synaptic impairment in APP/PS1 mice
Given that PYGM expression is decreased in the brains of AD patients and model mice, we investigated whether PYGM overexpression exerts protection in APP/PS1 mice. We injected lentiviruses containing mouse Pygm driven by a human synapsin 1 promoter (hSyn-Pygm) or control lentiviruses bilaterally into the hippocampal CA1 region of APP/PS1 mice at 4–5 months of age and confirmed exogenous PYGM expression ( Figure 5A, G). As APP/PS1 mice are reported to display behavioral abnormalities as early as 7 months of age and as late as 9 months of age ( Hao et al., 2015; Hu et al., 2017; Yang et al., 2021), we carried out behavioral tests 4 months after lentivirus injection (at 8–9 months of age). In the Y-maze test, APP/PS1 mice injected with lentiviruses expressing hSyn-Pygm (APP/PS1+hSyn-Pygm) showed a significantly increased alternation percentage compared to APP/PS1 mice injected with control lentiviruses (APP/PS1+Cont) ( Figure 5B). In the novel object recognition test, APP/PS1+hSyn-Pygm and APP/PS1+Cont mice showed no significant differences ( Figure 5C). In the fear conditioning test, APP/PS1+hSyn-Pygm mice showed a significantly increased freezing percentage during contextual testing but not during cued testing compared to APP/PS1+Cont mice ( Figure 5D). In the Morris water maze test, APP/PS1+hSyn-Pygm mice demonstrated an increased escape latency during the 6 day training phase, as well as increased time spent in the target quadrant and greater number of platform crossings during the testing phase on day 7, but no differences in mean swim speed, compared to APP/PS1+Cont mice ( Figure 5E).
Figure 5.
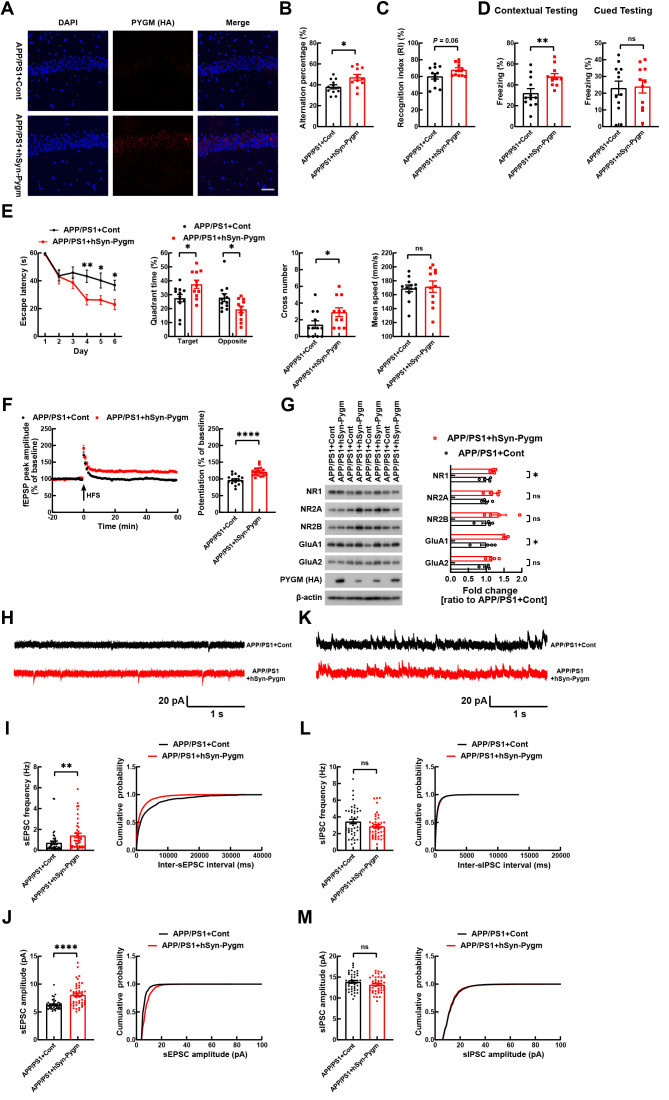
Overexpression of PYGM in hippocampal CA1 region ameliorates synaptic dysfunction and cognitive deficits in APP/PS1 mice
A: Representative images of hippocampal CA1 region in APP/PS1 mice receiving injection of LV-Cont or LV-hSyn-Pygm (HA). Samples were immunostained with anti-HA antibody (red) and nuclei were stained with DAPI (blue). Scale bar: 50 µm. B: In Y-maze test, alternation percentage of treated mice was studied for comparison. C: In novel object recognition test, discrimination index of mice was determined for comparison. D: In fear conditioning test, freezing responses to context and cue were recorded for comparison. E: In Morris water maze test, escape latency during the 6 day training phase was recorded for comparison. On day 7, time spent in target and opposite quadrants and number of platform crossings were recorded for comparison. Mean swim speed was also compared. APP/PS1+Cont, n=12 mice; APP/PS1+hSyn-Pygm, n=11 mice (for B–E). F: LTP of the Schaffer collateral circuit was studied by administering HFS in the CA3 region and then recording fEPSPs at the CA1 region for 60 min. Average fEPSP peak amplitudes in the last 10 min of the recording were calculated for comparison. APP/PS1+Cont, n=16 slices from five mice; APP/PS1+hSyn-PYGM, n=18 slices from six mice. G: NMDA and AMPA receptor subunits and PYGM in the whole hippocampal regions of treated mice were immunoblotted and levels were quantified for comparison after β-actin normalization. n=4 per group. H–M: Representative sEPSC (H) and sIPSC (K) traces from whole-cell voltage-clamp recordings of GFP-positive hippocampal CA1 pyramidal neurons in brain slices from APP/PS1+Cont and APP/PS1+hSyn-Pygm mice. Histogram plots and cumulative plots of frequencies (I, L) and amplitudes (J, M) of sEPSCs (I, J) and sIPSCs (L, M). APP/PS1+Cont, n=44 neurons from four mice; APP/PS1+hSyn-Pygm, n=44 neurons from four mice. Data represent mean± SEM and were analyzed by unpaired t test. *: P<0.05; **: P<0.01; ****: P<0.0001; ns: Not significant.
Figure 5.
Upon analyzing LTP in treated mice, we observed that HFS in the CA3 region resulted in much stronger fEPSPs, indicative of elevated LTP in the CA1 region, in APP/PS1+hSyn-Pygm mice compared to APP/PS1+Cont mice ( Figure 5F). In addition, based on whole-cell recordings in infected hippocampal CA1 neurons, we found that neurons with PYGM overexpression in vivo exhibited increased frequency and amplitude of sEPSCs ( Figure 5H–J), but no change in the frequency and amplitude of sIPSCs ( Figure 5K–M).
Moreover, we found that PYGM overexpression increased the protein levels of the NMDA receptor subunit NR1 and AMPA receptor subunit GluA1 ( Figure 5G). Together, these results demonstrate that PYGM overexpression can attenuate synaptic dysfunction and thus learning and memory deficits in AD mice.
PYGM regulates neuronal energy metabolism
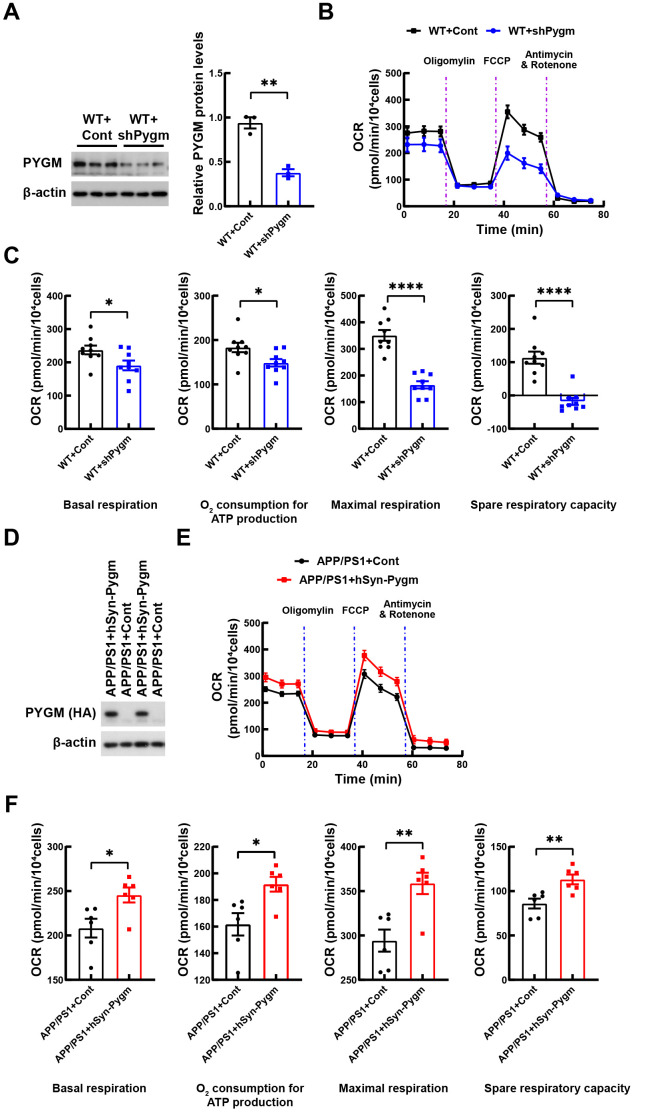
Neuronal synaptic transmission requires energy. As PYGM catabolizes glycogen for energy production, we explored whether PYGM regulates energy metabolism in neurons. Cultured WT mouse primary neurons were first infected with AAV2/9 to down-regulate PYGM expression ( Figure 6A), then subjected to OCR measurement over time, following the administration of four inhibitory compounds targeting key enzymes in the mitochondrial electron transport chain. Comparisons of key parameters related to mitochondrial respiratory capability, obtained from the OCR kinetics curve, showed that PYGM down-regulation significantly reduced basal respiration, adenosine triphosphate (ATP) production-associated respiration, maximal respiration, and spare respiratory capacity ( Figure 6B, C).
Figure 6.
PYGM regulates neuronal oxygen consumption rate
A: WT mouse primary neurons were infected with AAV2/9 expressing control (Cont) or Pygm (shPygm) shRNAs for 12 days. Equal amounts of protein lysates were immunoblotted and PYGM levels were quantified for comparison after β-actin normalization. B: Representative curves of changes in oxygen consumption rate (OCR) over time after injection of various compounds that inhibit key enzymes in the mitochondrial electron transport chain in WT+Cont and WT+shPygm neurons. C: OCR changes at basal respiration, ATP production, maximal respiration, and spare respiratory capacity stages in (B) were compared, n=9 (three mice per group with triplicates). D: APP/PS1 mouse primary neurons were infected with lentiviruses expressing control (Cont) or hSyn-Pygm tagged with HA for 12 days. Equal amounts of protein lysates were immunoblotted using anti-HA antibodies. E: Representative curves of changes in OCR at different stages in APP/PS1+Cont and APP/PS1+hSyn-Pygm neurons. F: OCR changes at basal respiration, ATP production, maximal respiration, and spare respiratory capacity stages in (E) were compared, n=6 (three mice per group with duplicates). Data represent mean± SEM and were analyzed by unpaired t-test. *: P<0.05; **: P<0.01; ****: P<0.0001.
We also infected cultured primary neurons from APP/PS1 mice with lentiviruses to overexpress PYGM ( Figure 6D). Measuring OCR over time, we found that PYGM overexpression significantly increased basal respiration, ATP production-associated respiration, maximal respiration, and spare respiratory capacity in APP/PS1 neurons ( Figure 6E, F). These results suggest that PYGM may regulate energy metabolism in neurons, which may be the mechanism underlying its regulation of synaptic function.
DISCUSSION
LTP is a widely used cellular model for studying synaptic plasticity in learning and memory. Impaired synaptic plasticity is not only implicated in cognitive impairment but is also associated with AD ( Cuestas Torres & Cardenas, 2020; Koffie et al., 2011; Tzioras et al., 2023). Herein, based on LTP induction, we developed a novel method to systematically identify genes responsible for mediating synaptic plasticity and whose abnormal alteration contributes to synaptic dysfunction in AD. To achieve this, we employed an LTP induction protocol by delivering HFS to the hippocampal CA3 region, recording fEPSPs in the CA1 region, and collecting CA1 region samples for RNA-seq. By comparing 2-month-old WT and APP/PS1 mice, as well as 6-month-old WT mice, all of which exhibited normal LTP, we identified 89 DEGs with consistent changes in expression among the mice. While many of the changes in gene expression may simply accompany the LTP process, some may help stabilize and maintain LTP, thereby playing important roles in synaptic plasticity.
Among the 89 DEGs, 43 exhibited either no change or a change in the opposite direction upon HFS treatment in 6-month-old APP/PS1 mice with impaired LTP. These genes may be associated with synaptic dysfunction in AD. To further refine the selection and identify genes strongly link to AD, we screened the 43 genes based on their altered expression in 6-month-old APP/PS1 mice without HFS compared to WT controls. After excluding genes annotated by BGI but lacking known features, we ultimately identified 14 candidate genes. These candidate genes may play a role in regulating synaptic plasticity, and their abnormality may contribute to synaptic dysfunction in AD.
Among the 14 candidate genes, Pygm encodes PYGM, a glycogen phosphorylase involved in the catabolism of glycogen. Glycogen, a vital energy storage molecule in mammals, is synthesized through glucose polymerization by glycogen synthase. When required, glycogen can be rapidly decomposed into glucose or lactate to provide energy for the body. Glycogen phosphorylases, the enzymes responsible for glycogen degradation, can be divided into muscle type (PYGM), liver type (PYGL), and brain type (PYGB) ( Benarroch, 2010; Brown, 2004; Öz et al., 2007). Although PYGM is predominantly expressed in the muscle, it can also be detected in the brain ( Migocka-Patrzalek & Elias, 2021; Uhlén et al., 2015), and was found to be expressed in various neural cells in our study.
Glycogen serves as a major energy reserve in the brain and is almost exclusively stored in astrocytes. When required, astrocytes can rapidly degrade glycogen into lactate and release it as an energy source to support neurons ( Belanger et al., 2011; Brooks, 2002; Duran et al., 2019a; Turner & Adamson, 2011). The impact of glycogen metabolism disruption on synaptic function appears to vary with age. Inhibition of glycogen breakdown can impair LTP in the Schaffer collateral pathway and inhibit memory consolidation in young animals, but favor LTP in the Schaffer collateral pathway in aged animals ( Drulis-Fajdasz et al., 2015; Gibbs et al., 2006). In our study, we observed a decrease in Pygm expression after HFS in mice with normal LTP. Similarly, Drulis-Fajdasz et al. ( 2015) compared the expression levels of Pygb and Gys1 in the hippocampal CA1 region of young and aged rats before and after LTP induction and found a decrease in the expression of both after HFS (LTP vs. CS in their Figure 3). Collectively, these findings and our results suggest that glycogen metabolism is important for physiological LTP and thus synaptic plasticity.
However, we also found that Pygm expression was increased after HFS in APP/PS1 mice with impaired LTP and that PYGM expression was decreased in the brain of AD patients and model mice. These results suggest that dysregulation of PYGM may play a significant role in AD. Mutations in PYGM can lead to the rare condition known as McArdle disease, in which patients exhibit exercise intolerance due to reduced glycogen catabolism and energy supply in the muscle ( Migocka-Patrzalek & Elias, 2021). Interestingly, a previous study identified cognitive impairment in a McArdle patient ( Mancuso et al., 2011). If further investigations reveal a high prevalence of cognitive deficits in individuals with McArdle disease, it may provide additional support for the notion that dysregulated glycogen catabolism caused by PYGM deficiency impairs synaptic function and cognition.
Glycogen, primarily stored in astrocytes, serves as a source of lactate that neurons preferentially utilize for energy supply ( Belanger et al., 2011; Brooks, 2002; Duran et al., 2019a; Turner & Adamson, 2011). As expected, we observed predominant expression of PYGM in astrocytes; however, we also detected PYGM expression in neurons and microglia. Previous studies have reported that GYS1 and PYGB are expressed in neurons, along with glycogen metabolism ( Drulis-Fajdasz et al., 2018; Saez et al., 2014). Knockout of Gys1 in neurons impairs synaptic plasticity and memory formation in mice ( Duran et al., 2019b), and renders neurons more susceptible to hypoxia ( Saez et al., 2014). Conversely, abnormally synthesized glycogen in neurons can induce neuronal apoptosis ( Magistretti & Allaman, 2007; Vilchez et al., 2007) and glycogen accumulation in neurons can occur in patients with Lafora disease ( Duran et al., 2019a). Therefore, neuronal glycogen metabolism holds considerable biological significance. To explore whether PYGM can directly catabolize glycogen to produce energy in neurons, we modulated PYGM levels and found that down-regulation of PYGM expression decreased OCRs in cultured WT mouse primary neurons, while PYGM overexpression promoted OCRs in cultured APP/PS1 mouse primary neurons. These findings provide evidence that PYGM directly regulates energy generation in neurons.
Energy supply is essential for synaptic activity. Consistent with compromised energy generation, we found that down-regulation of PYGM impaired synaptic transmission in cultured primary neurons. Importantly, we showed that down-regulation of PYGM in hippocampal CA1 neurons markedly impaired synaptic and cognitive functions in WT mice, and that overexpression of PYGM in hippocampal CA1 neurons attenuated synaptic dysfunction and cognitive deficits in APP/PS1 mice. These results demonstrate that PYGM-mediated energy production in neurons plays an important role in synaptic plasticity and learning and memory.
Among the other candidate genes identified, Ldhd encodes D-lactate-dehydrogenase (LDHD), the enzyme responsible for converting D-lactate to pyruvate for downstream metabolic pathways ( Drabkin et al., 2019; Flick & Konieczny, 2002). In addition, Slc4a11 encodes the NH 3:H + cotransporter SLC4A11, mutations of which can cause corneal endothelial dystrophy and sensorineural deafness ( Zhang et al., 2017). An earlier study found that glutamine catabolism is severely disrupted in SLC4A11-deficient cells, suggesting that SLC4A11 is important for energy production ( Zhang et al., 2017). The identification of these two energy production-related genes, in addition to Pygm, further supports the potential association between dysregulated energy metabolism and synaptic dysfunction in AD.
Our study has several limitations. Firstly, we focused solely on exploring the function of PYGM in neurons, despite its predominant expression in cultured primary astrocytes compared to cultured primary neurons and microglia. Future investigations should study whether PYGM exhibits a similar expression pattern in adult organisms, whether the decline in PYGM expression observed in AD is present in all cell types or restricted to specific ones, and whether PYGM in other cell types, particularly astrocytes, also plays a role in synaptic plasticity and cognition. Secondly, we focused solely on the effect of neuronal PYGM on LTP. Both NMDA receptor- and AMPA receptor-mediated synaptic transmissions are involved in LTP ( Cuestas Torres & Cardenas, 2020). While our results suggest that changes in PYGM affect NMDA and AMPA receptor subunits, whether PYGM regulates LTP by affecting either NMDA receptor- or AMPA receptor-mediated synaptic transmission, or both, deserves further scrutiny. Thirdly, our study indicated that neuronal PYGM overexpression attenuates deficits in APP/PS1 mice; however, whether this is a partial or complete rescue remains unclear. Future studies incorporating WT controls may help determine this. Lastly, we only employed male mice as a proof-of-concept study to validate the viability of our screening strategy. Given the significant roles of sex differences in AD, future studies using animals from both sexes may identify new genes that not only regulate synaptic plasticity in AD, but also exert their effects in sex-dependent or sex-independent manners.
In summary, our study presents a novel strategy for identifying genes involved in synaptic plasticity and their potential contribution to AD. By demonstrating the role of PYGM in the regulation of synaptic activity and learning and memory in both WT and AD model mice, we provide compelling evidence for the effectiveness of this strategy.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
DATA AVAILABILITY
RNA-seq data were deposited into the Sequence Archive of the China National GeneBank Database ( Chen et al., 2020; Guo et al., 2020b) with accession number CNP0003942, Sequence Read Archive of the National Center for Biotechnology Information with accession number PRJNA972124, and Genome Sequence Archive of the China National Center for Bioinformation with accession number CRA011030. These data were also submitted to the ZR Journal Community portal at https://www.scidb.cn/c/zoores with a Data DOI of 10.57760/sciencedb.08216.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
T.W. carried out most electrophysiological, biochemical, and animal behavioral studies. Y.Q.Z., Y.W., L.Z., Xiang Z., and X.Y.W. helped with animal work. J.H.W. helped with electrophysiological analysis. L.K.H. and J.M. helped with biochemical studies. Xian Z. and H.L. provided technical support. Q.L.M. and Z.X.W. provided intellectual contributions. Y.W.Z. conceived and supervised the project. T.W. and Y.W.Z. wrote the manuscript. All authors reviewed the manuscript. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the National Natural Science Foundation of China (U21A20361 and 82130039 to Y.W.Z.), Fundamental Research Funds for the Central Universities (20720220133 to Y.W.Z.), Natural Science Foundation of Fujian Province (2021J02057 to Q.L.M.), Science and Technology Plan Projects of Fujian Province (2020Y2015 to Z.X.W.), and 2020 Joint Support of Key Projects on Health Care (3502Z20209005 to Z.X.W.)
References
- Belanger M, Allaman I, Magistretti PJ Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metabolism. 2011;14(6):724–738. doi: 10.1016/j.cmet.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Benarroch EE Glycogen metabolism: metabolic coupling between astrocytes and neurons. Neurology. 2010;74(11):919–923. doi: 10.1212/WNL.0b013e3181d3e44b. [DOI] [PubMed] [Google Scholar]
- Brooks GA Lactate shuttles in nature. Biochemical Society Transactions. 2002;30(2):258–264. doi: 10.1042/bst0300258. [DOI] [PubMed] [Google Scholar]
- Brown AM Brain glycogen re-awakened. Journal of Neurochemistry. 2004;89(3):537–552. doi: 10.1111/j.1471-4159.2004.02421.x. [DOI] [PubMed] [Google Scholar]
- Chen FZ, You LJ, Yang F, et al. 2020. CNGBdb: China national GeneBank DataBase. Hereditas ( Beijing), 42(8): 799–809. (in Chinese)
- Chen ZY, Zhang Y Animal models of Alzheimer’s disease: Applications, evaluation, and perspectives. Zoological Research. 2022;43(6):1026–1040. doi: 10.24272/j.issn.2095-8137.2022.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuestas Torres DM, Cardenas FP Synaptic plasticity in Alzheimer's disease and healthy aging. Reviews in the Neurosciences. 2020;31(3):245–268. doi: 10.1515/revneuro-2019-0058. [DOI] [PubMed] [Google Scholar]
- Drabkin M, Yogev Y, Zeller L, et al Hyperuricemia and gout caused by missense mutation in D-lactate dehydrogenase . The Journal of Clinical Investigation. 2019;129(12):5163–5168. doi: 10.1172/JCI129057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drulis-Fajdasz D, Gizak A, Wójtowicz T, et al Aging-associated changes in hippocampal glycogen metabolism in mice. Evidence for and against astrocyte-to-neuron lactate shuttle. Glia. 2018;66(7):1481–1495. doi: 10.1002/glia.23319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drulis-Fajdasz D, Wójtowicz T, Wawrzyniak M, et al Involvement of cellular metabolism in age-related LTP modifications in rat hippocampal slices. Oncotarget. 2015;6(16):14065–14081. doi: 10.18632/oncotarget.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran J, Gruart A, López-Ramos JC, et al. 2019a. Glycogen in astrocytes and neurons: physiological and pathological aspects. In: Fitter AH. Brain Glycogen Metabolism. Cham: Springer, 311–329.
- Duran J, Gruart A, Varea O, et al Lack of neuronal glycogen impairs memory formation and learning-dependent synaptic plasticity in mice. Frontiers in Cellular Neuroscience. 2019b;13:374. doi: 10.3389/fncel.2019.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick MJ, Konieczny SF Identification of putative mammalian D-lactate dehydrogenase enzymes . Biochemical and Biophysical Research Communications. 2002;295(4):910–916. doi: 10.1016/S0006-291X(02)00768-4. [DOI] [PubMed] [Google Scholar]
- Gelman S, Palma J, Tombaugh G, et al Differences in synaptic dysfunction between rTg4510 and APP/PS1 mouse models of Alzheimer's disease. Journal of Alzheimer's Disease. 2018;61(1):195–208. doi: 10.3233/JAD-170457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs ME, Anderson DG, Hertz L Inhibition of glycogenolysis in astrocytes interrupts memory consolidation in young chickens. Glia. 2006;54(3):214–222. doi: 10.1002/glia.20377. [DOI] [PubMed] [Google Scholar]
- Guo TT, Zhang DH, Zeng YZ, et al Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer's disease. Molecular Neurodegeneration. 2020a;15(1):40. doi: 10.1186/s13024-020-00391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo XQ, Chen FZ, Gao F, et al. 2020b. CNSA: a data repository for archiving omics data. Database ( Oxford), 2020: baaa055.
- Hao JR, Sun N, Lei L, et al L-Stepholidine rescues memory deficit and synaptic plasticity in models of Alzheimer's disease via activating dopamine D1 receptor/PKA signaling pathway. Cell Death & Disease. 2015;6(11):e1965. doi: 10.1038/cddis.2015.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Wei P, Jin L, et al Overexpression of EphB2 in hippocampus rescues impaired NMDA receptors trafficking and cognitive dysfunction in Alzheimer model. Cell Death & Disease. 2017;8(3):e2717. doi: 10.1038/cddis.2017.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ECB, Carter EK, Dammer EB, et al Large-scale deep multi-layer analysis of Alzheimer's disease brain reveals strong proteomic disease-related changes not observed at the RNA level. Nature Neuroscience. 2022;25(2):213–225. doi: 10.1038/s41593-021-00999-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TA, Syty MD, Wu K, et al Adult hippocampal neurogenesis and its impairment in Alzheimer’s disease. Zoological Research. 2022;43(3):481–496. doi: 10.24272/j.issn.2095-8137.2021.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopman DS, Amieva H, Petersen RC, et al Alzheimer disease. Nature Reviews Disease Primers. 2021;7(1):33. doi: 10.1038/s41572-021-00269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffie RM, Hyman BT, Spires-Jones TL Alzheimer's disease: synapses gone cold. Molecular Neurodegeneration. 2011;6(1):63. doi: 10.1186/1750-1326-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo LC, Vi N, Ma ZC, et al Novel cell and tissue acquisition system (CTAS): microdissection of live and frozen brain tissues. PLoS One. 2012;7(7):e41564. doi: 10.1371/journal.pone.0041564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuba G, Savioz A, Vernay A, et al Differential changes in synaptic proteins in the Alzheimer frontal cortex with marked increase in PSD-95 postsynaptic protein. Journal of Alzheimer's Disease. 2008;15(1):139–151. doi: 10.3233/JAD-2008-15112. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Chang LR, Song YZ, et al The role of NMDA receptors in alzheimer's disease. Frontiers in Neuroscience. 2019;13:43. doi: 10.3389/fnins.2019.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistretti PJ, Allaman I Glycogen: a Trojan horse for neurons. Nature Neuroscience. 2007;10(11):1341–1342. doi: 10.1038/nn1107-1341. [DOI] [PubMed] [Google Scholar]
- Mancuso M, Orsucci D, Volterrani D, et al Cognitive impairment and McArdle disease: is there a link? Neuromuscular Disorders. 2011;21(5):356–358. doi: 10.1016/j.nmd.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Migocka-Patrzalek M, Elias M Muscle glycogen phosphorylase and its functional partners in health and disease. Cells. 2021;10(4):883. doi: 10.3390/cells10040883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll RA A brief history of long-term potentiation. Neuron. 2017;93(2):281–290. doi: 10.1016/j.neuron.2016.12.015. [DOI] [PubMed] [Google Scholar]
- Öz G, Seaquist ER, Kumar A, et al Human brain glycogen content and metabolism: implications on its role in brain energy metabolism. American Journal of Physiology-Endocrinology and Metabolism. 2007;292(3):E946–E951. doi: 10.1152/ajpendo.00424.2006. [DOI] [PubMed] [Google Scholar]
- Proctor DT, Coulson EJ, Dodd PR Reduction in post-synaptic scaffolding PSD-95 and SAP-102 protein levels in the Alzheimer inferior temporal cortex is correlated with disease pathology. Journal of Alzheimer's Disease. 2010;21(3):795–811. doi: 10.3233/JAD-2010-100090. [DOI] [PubMed] [Google Scholar]
- Reiserer RS, Harrison FE, Syverud DC, et al Impaired spatial learning in the APP Swe + PSEN1ΔE9 bigenic mouse model of Alzheimer's disease . Genes, Brain and Behavior. 2007;6(1):54–65. doi: 10.1111/j.1601-183X.2006.00221.x. [DOI] [PubMed] [Google Scholar]
- Saez I, Duran J, Sinadinos C, et al Neurons have an active glycogen metabolism that contributes to tolerance to hypoxia. Journal of Cerebral Blood Flow and Metabolism. 2014;34(6):945–955. doi: 10.1038/jcbfm.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC Towards an understanding of synapse formation. Neuron. 2018;100(2):276–293. doi: 10.1016/j.neuron.2018.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DA, Adamson DC Neuronal-astrocyte metabolic interactions: understanding the transition into abnormal astrocytoma metabolism. Journal of Neuropathology & Experimental Neurology. 2011;70(3):167–176. doi: 10.1097/NEN.0b013e31820e1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzioras M, McGeachan RI, Durrant CS, et al Synaptic degeneration in Alzheimer disease. Nature Reviews Neurology. 2023;19(1):19–38. doi: 10.1038/s41582-022-00749-z. [DOI] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, et al Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Viana Da Silva S, Haberl MG, Zhang P, et al Early synaptic deficits in the APP/PS1 mouse model of Alzheimer's disease involve neuronal adenosine A 2A receptors . Nature Communications. 2016;7:11915. doi: 10.1038/ncomms11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Ros S, Cifuentes D, et al Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nature Neuroscience. 2007;10(11):1407–1413. doi: 10.1038/nn1998. [DOI] [PubMed] [Google Scholar]
- Wang LK, Feng ZX, Wang X, et al DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Wang R, Reddy PH Role of glutamate and NMDA receptors in Alzheimer's disease. Journal of Alzheimer's Disease. 2017;57(4):1041–1048. doi: 10.3233/JAD-160763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Bacaj T, Morishita W, et al Postsynaptic synaptotagmins mediate AMPA receptor exocytosis during LTP. Nature. 2017;544(7650):316–321. doi: 10.1038/nature21720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Chen P, Wang XJ, et al miR-34a deficiency in APP/PS1 mice promotes cognitive function by increasing synaptic plasticity via AMPA and NMDA receptors. Neuroscience Letters. 2018;670:94–104. doi: 10.1016/j.neulet.2018.01.045. [DOI] [PubMed] [Google Scholar]
- Yang X, Li X, Liu L, et al Transferrin-Pep63-liposomes accelerate the clearance of Aβ and rescue impaired synaptic plasticity in early Alzheimer's disease models. Cell Death Discovery. 2021;7(1):256. doi: 10.1038/s41420-021-00639-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MX, Zhou YQ, Jiang YR, et al Profiling of sexually dimorphic genes in neural cells to identify Eif2s3y, whose overexpression causes autism-like behaviors in male mice . Frontiers in Cell and Developmental Biology. 2021;9:669798. doi: 10.3389/fcell.2021.669798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WL, Li HD, Ogando DG, et al Glutaminolysis is essential for energy production and ion transport in human corneal endothelium. EBioMedicine. 2017;16:292–301. doi: 10.1016/j.ebiom.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.