Abstract
Background
Triple-negative breast cancer (TNBC), although the most intractable subtype, is characterized by abundant immunogenicity, which enhances responsiveness to immunotherapeutic measures.
Methods
First, we identified CD8+ T cell core genes (TRCG) based on single-cell sequence and traditional transcriptome sequencing and then used this data to develop a first-of-its-kind classification system based on CD8+ T cells in patients with TNBC. Next, TRCG-related patterns were systematically analyzed, and their correlation with genomic features, immune activity (microenvironment associated with immune infiltration), and clinicopathological characteristics were assessed in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), the Cancer Genome Atlas (TCGA), GSE103091, GSE96058 databases. Additionally, a CD8+ T cell-related prognostic signature (TRPS) was developed to quantify a patient-specific TRCG pattern. What's more, the genes-related TRPS was validated by polymerase chain reaction (PCR) experiment.
Results
This study, for the first time, distinguished two subsets in patients with TNBC based on the TRCG. The immune microenvironment and prognostic stratification between these have distinct heterogeneity. Furthermore, this study constructed a novel scoring system named TRPS, which we show to be a robust prognostic marker for TNBC that is related to the intensity of immune infiltration and immunotherapy. Moreover, the levels of genes related the TRPS were validated by quantitative Real-Time PCR.
Conclusions
Consequently, this study unraveled an association between the TRCG and the tumor microenvironment in TNBC. TRPS model represents an effective tool for survival prediction and treatment guidance in TNBC that can also help identify individual variations in TME and stratify patients who are sensitive to anticancer immunotherapy.
Keywords: Triple-negative breast cancer, CD8+ T cell, single cell sequencing, Heterogeneity, Tumor immune microenvironment
1. Introduction
Triple-negative breast cancer (TNBC), defined as the absence of expression of the estrogen receptor, the progesterone receptor, and ERBB2 (commonly referred to as human epidermal growth factor receptor 2, HER2), accounts for 12%–%17% of all breast cancer cases and is the most intractable subtype [1,2]. The major problems in TNBC treatment are early relapse and the absence of effective therapeutic targets [1,3]. Recent research has revealed that TNBC is characterized by greater immunogenicity than other subtypes, indicating the feasibility of using immunotherapeutic strategies [[4], [5], [6], [7], [8]]. Cancer cells can stimulate the expression of programmed death 1 (PD-1) on T cells and thereby exhaust activated T cells by upregulating PD-L1 [9,10]. Thus, clinical trials have tested antibodies that block PD-1/PD-L1 binding to inhibit immune evasion in various types of cancer [11]. Given the higher expression of PD-L1 in TNBC compared to other BC subtypes [12], clinical trials utilizing PD-L1 antibodies or combined therapies have been conducted [13]. The US FDA has approved the use of the PD-1 inhibitor, pembrolizumab, in combination with chemotherapy, as first-line treatment for metastatic PD-L1 + TNBC. Interestingly, atezolizumab, another PD-L1 inhibitor, has also demonstrated clinical activity; nonetheless, this clinical benefit is moderate because TNBCs are PD-L1+ only in about 40% of cases, and not all patients with advanced TNBC respond to this treatment. These unsatisfactory results may have contributed to the absence of patient screening based on tumor microenvironment landscapes. Additionally, as the entire landscape of TNBC microenvironment phenotypes remains unclear, it is essential to expand our comprehension of genomic heterogeneity. Thus, developing a new molecular classification is essential for improving prognosis because it can help appropriately categorize patients and provide the most effective treatment strategies.
Recently, single-cell RNAseq, an emerging technology, has been utilized to explore tumor heterogeneity and delineate genomic characteristics [14]. Several earlier studies have highlighted remarkable heterogeneity in the immune microenvironment, exosome, and cuproptosis in TNBC [2,15,16]. The tumor microenvironment (TME) includes abundant stromal cells, immune cells, and tumor cells, and greatly affects the malignant progression and tumor heterogeneity [17]. On the other hand, the dynamic interactions among immune cells are vital for the heterogeneity of the TME but eventually result in differential responses to therapies [17,18]. Notably, in TNBC, the contents of tumor-infiltrating lymphocytes (TILs) are associated with a favorable prognosis and an enhanced response to neoadjuvant chemotherapy [19,20]. Specifically, T cells play a crucial role in maintaining cellular integrity against intracellular tumors and pathogens [[21], [22], [23]], and immunotherapy that is based on blocking T cell inhibitory factors has become the paradigm [[24], [25], [26], [27], [28]]. These observations indicate that the development of better immunotherapies may be facilitated through a more wholesome understanding of the genetic landscape that governs T cell function. T cell receptors recognize cognate antigens on the surface of major histocompatibility complex (MHC) class I (MHC-I) and activate antigen-specific CD8 T cells [[29], [30], [31]], which then trigger T cell proliferation, cytokine production, and killing of the target cells [32]. Infections [[33], [34], [35], [36]] or cancer [[37], [38], [39]] can be attributed to defects in T cells, while immunopathology and autoimmunity can arise from dysregulated CD8+ T cell activation [[40], [41], [42], [43]]. Thus, CD8 T cells have become the central focus of emerging and novel cancer therapies [[44], [45], [46]].
Additionally, inhibitors of immune checkpoints that neutralize CTLA-4 and PD-1/PD-L1 also amplify CD8 T cell responses against tumors [28,30,35,[47], [48], [49]] and the discovery of CTLA-4 and PD-1 pathways has demonstrated that the mechanisms underlying immune evasion involve inhibiting CD8 T cell signaling [50,51]. Hence, it is possible that the level of CD8+ T cell infiltration can contribute to the prognostic and even predictive stratification of patients with TNBC. However, this necessitates a greater understanding of CD8+ T cell characteristics and their classification based on single-cell RNA-seq. Interestingly, two heterogeneous clusters have arisen in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort and these have been validated by three independent external cohorts. The differences in genomic variations, prognosis, biological characteristics, and the microenvironment were evident between the two clusters. Our study reveals novel aspects of the infiltration level of CD8+ T cells with underlying association of immunotherapy response and biological regulation stratification. Additionally, for the first time, we have developed a CD8+ T cell-related prognostic signature (TRPS). This study tested the prognostic value, clinical characteristics and tumor microenvironment (TME) of TRPS in TNBC. Ultimately, we examined the efficacy of TRPS in TNBC immunotherapy and chemotherapy.
2. Methods
2.1. Data collection
Data was mined from the METABRIC, the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and the Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) databases. Single-cell RNA-sequencing data of 6 TNBC samples in GSE118389 and 10 TNBC samples in GSE176078 were retrieved from the GEO database and were utilized to determine the CD8+ T cell marker genes in TNBC. METABRIC data of 310 patients with TNBC were downloaded from cBioPortal [52,53] and the Meta-TNBC cohort (TCGA-TNBC, GSE96058, GSE103091) [54] was utilized to validate the robustness of the results. The batch effects of non-biological technical biases from each dataset were mitigated by applying the ComBat algorithm in the package sva [55]. After excluding samples with the survival of fewer than 30 days, 315 samples were eventually selected for this study.
2.2. Identification of CD8+ T cell marker genes by scRNA-seq analysis
First, we implemented quality control in the data retrieved from GSE118389 and GSE176078. Specifically, three screening criteria were implemented at the raw matrix level for each cell, viz., (i) only genes that were expressed in at least five single cells were selected, (ii) cells that expressed less than 100 genes were removed, and (iii) cells with more than 15% mitochondrial genes were eliminated. Subsequently, the Seurat package was utilized to standardize the expression matrix of the GSE118389 and the GSE176078 datasets and “SeuratObject” was constructed. The two datasets were integrated to acquire a total of 5786 cells by sequentially performing “FindIntegrationAnchors” and “IntegrateData”. Next, using the “FindClusters” and “FindNeighbors” functions in the Seurat package, cell cluster analysis was implemented. Dimensionality reduction (Uniform Manifold Approximation and Projection, UMAP) analyses were run using the runUMAP function in the Seurat package. Subsequently, using the FindAllMarkers functionality in the Seurat package, marker genes for each cell cluster were identified based on the following screening criteria: log fold change (FC) ≥ 0.25 and p value < 0.05. For cluster annotation, we identified six cell subtypes, namely, B cells, endothelial cells, epithelial cells, fibroblasts, monocytes, and T cells. The Monocle 2 package was performed to reveal the pseudotime trajectory of single cells [56].
2.3. Annotation and further segmentation of T cells
Data on T cells were extracted and clustered by implementing the “FindNeighbors” and the “FindClusters” functions (setting Resolution = 0.8). T cells were further divided into four subgroups and re-TSNE dimensionality was reduced by running the RunTSNE function.
2.4. Obtaining marker genes
The “FindMarkers” function in the “Seurat” package was utilized to identify differentially expressed genes (DEGs) between CD8+ and non-CD8 + T cells using the Wilcoxson test. The cutoff threshold values applied were log2 (fold change) > 0.25 and adjusted p value < 0.05.
Construction and Validation of CD8+ T cell linked Molecular Classification Based on Marker Genes.
The MCPcounter [57] and the CIBERSORT [58] algorithms were applied to evaluate differences in the infiltration level of CD8+ T cell in each sample in the METABRIC cohort. Given that the weighted gene coexpression network analysis (WGCNA) has the ability to identify associations between gene modules and characteristics [59], the gene co-expression network was shaped according to CD8+ T cell marker genes and then transformed as a scale-free network by identifying a soft threshold β (power of β = 3) and a scale-free R2 of 0.9. Based on the association between the module eigenvalue and characteristics, we curated an notable module by immune trait and module connectivity and further a coexpression network was shaped via the exportNetworkToCytoscape function in the WGCNA R package. Hub nodes were visualized using Cytoscape software [60] and CD8+ T cell related core genes (TRCG) were selected by implementing the cytoHubba plugin according to the maximal clique centrality (MCC) algorithm [61].
2.5. Identifying TRCG-related patterns
TNBC samples were classified into two subtypes based on TRCG using a consensus clustering algorithm in the METABRIC-TNBC cohort and further verified in the META-TNBC cohort. To ensure classification stability, this procedure was performed and repeated 1000 times using the package ConsensusClusterPlus.
2.6. Tumor microenvironment infiltration assessment
Based on immune cell type marker gene sets derived from Charoentong et al. [62], the relative amounts of the 28 immune cell types were quantified through single sample gene set enrichment analysis (ssGSEA) and the GSVA R package [63]. The gene set for each immune cell subset was acquired from Bindea et al. [64]. Additionally, we utilized the R package to assess stromal and immune infiltration levels between the two TRCG-related patterns [65]. The MCPcounter and the CIBERSORT algorithms were utilized to compare differences in immune cell infiltration levels between the two clusters in the METABRIC-TNBC cohort and the META-TNBC cohort. Immunotherapeutic efficacy among distinct clusters was assessed by two prevalent approaches that contained unsupervised subclass mapping (Submap) and T cell inflammatory signatures (TIS). Submap was used to analyze similarity in expression profiles between TNBC and immunotherapy patients [66], which corresponded to the similarity in clinical efficiency. TIS was calculated using the ssGSEA algorithm and contained 18 inflammatory genes with higher scores typically suggesting a more sensitive response to PD-1 inhibitor therapy [67]. Previous studies have identified characteristic signatures that indicate differential infiltration of immune cells based on TNBC subtypes.
2.7. Annotation and functional enrichment analyses
For the purpose of evaluating the correlation between immune markers and molecular subtypes [68], data for immunomodulators and inhibitory immune checkpoints, which included chemokines, MHC molecules, receptor molecules, immunostimulant molecules, and inhibitory immune checkpoint molecules, were retrieved from previous studies [62,69,70]. The Wilcoxson rank sum test was applied to determine whether these genes are differentially expressed among the TNBC subtypes. The gene sets obtained from h. all.v7.5.1. symbols, ontology gene sets in the Molecular Signatures Database v5.1 (MSigDB) (http://www.broad.mit.edu/gsea/msigdb/) were used to assess the association between molecular subtypes and specific biological processes.
2.8. Assessment of the Cancer–Immunity cycle
The cancer–immunity cycle, also called the anticancer immune response, has seven steps related to the TME. The Tracking Tumor Immunophenotype (TIP; http://biocc.hrbmu.edu.cn/TIP/index.jsp) program was used to assess the activity score of each step [71].
2.9. Construction of CD8+ T cell-related prognostic signature (TRPS)
To construct a quantification system based on TRCG, DEGs were extracted from two clusters in the METABRIC-TNBC cohort and intersected with genes involved in the merge-TNBC cohort. Subsequently, univariate Cox regression, a least absolute shrinkage and selection operator (LASSO), and multivariate Cox analysis were performed to build the TRPS:
TRPS = esum (each gene's expression × corresponding regression coefficient).
Cox regression models are usually tested for accuracy using the receiver operating characteristic (ROC). Further, based on the training and the test datasets, we performed Kaplan–Meier survival analyses of TRPS and calculated ROC curves using the R package (timeROC) for 1-, 3-, and 5-year survival. We also checked if TRPS is an independent prognostic factor using univariate and multivariate Cox regression analyses.
2.10. Establishment of a nomogram
Based on the results of multivariate analysis, the nomogram function in the “rms” package was used to shape the nomogram. Additionally, calibration plots were drawn with the rms package to estimate consistency among predicted 1-, 3-, and 5-year endpoint events and practical outcomes.
2.11. Cell cultures
Human breast cancer cell lines (MDA-MB-231, BT-549) and non-tumorigence epithelial cells (MCF-10 A) were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). MDA-MB-231 cells were maintained in DMEM medium (Gibco, Eggenstein, Germany), BT-549 cells were maintained in RPMI-1640 medium (Gibco). MCF-10 A were maintained in DMED-F12 with 500 μg/ml hydrocortisone, 20 ng/ml EGF, 10 μg/ml insulin, 0.3 g/L l-glutamine, 40 mg/L gentamicin. All media were supplemented with 10% fetal bovine serum (Gibco, USA).
2.12. RNA isolation and quantitative Real-Time PCR
qRT-PCR was used to estimate relative expression of N4BP2L1, IL18RAP, and KIR3DL3 in TNBC cells. Trizol reagent was used to extract RNA, which was then reverse transcribed using a kit (Toyobo, Osaka, Japan). Ultimately, qRT-PCR was performed using the THUN DERBIRD SYBR qPCR Mix (Toyobo, Osaka, Japan) on an Applied Biosystems 7500 cycler. Relative expression of N4BP2L1, IL18RAP, and KIR3DL3 was normalized to that of GAPDH, and each sample was analyzed at least in triplicate. The list of the primer sequences used for cDNA amplification in Supplement table 3.
2.13. Evaluation of clinical treatment
The pRRophetic package was used to forecast drug response according to gene expression data [72] and to estimate the half-maximal inhibitory concentration (IC50) of common chemotherapy drugs. Immunophenoscore (IPS) of samples in the TCGA was retrieved from The Cancer Immunome Atlas (https://tcia.at/).
2.14. Statistical analyses
All data analysis were performed using the R software (v4.1.3). Kaplan–Meier and Cox regression analyses were performed with the survival package. Log-rank test was utilized to compare survival statistics of categorical variables. Multivariate Cox regression analysis was employed to evaluate hazard ratio and validate the independent significance of multiple traits. The relationship between two continuous variables was assessed using Pearson's correlation analysis. ROC plots were generated using the pROC package to predict binary categorical variables. P < 0.05 was defined as statistically significant.
3. Results
3.1. Data processing
The flow chart in Fig. 1 illustrates how data was processed and analyzed for this study.
Fig. 1.
Schematic summary of the workflow.
3.2. Identification of CD8+ T cell marker genes
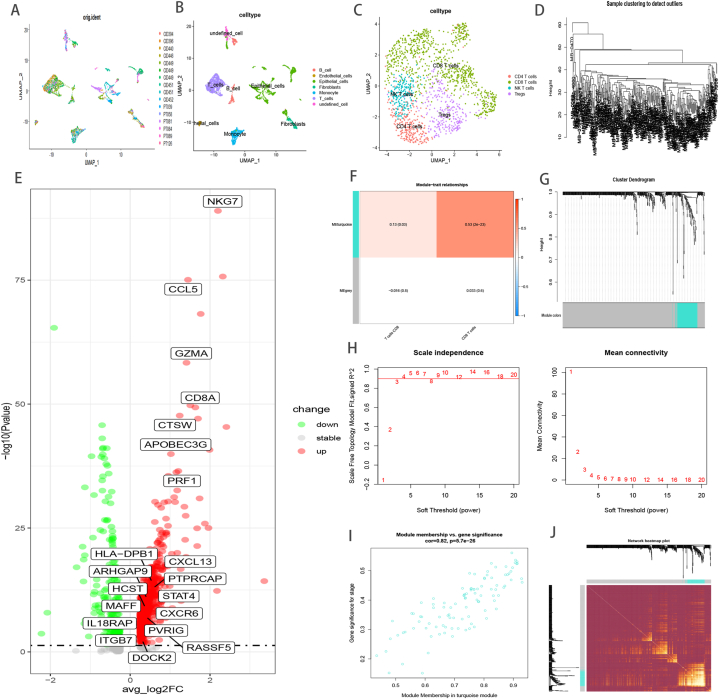
The scRNA-seq data available at GSE118386 and GSE176078 provided gene expression profiles of 5786 cells from 16 TNBC samples for follow up analysis (Fig. 2A). Dimensionality of all genes was reduced using the PCA algorithm and 27 cell clusters were identified. Next, the cell identity of each cluster was annotated using immune markers described previously [[73], [74], [75]] (Fig. 2B), and T cell subgroups were extracted and divided to four subgroups based on immune markers described elsewhere [74] (Fig. 2C). Finally, 1078 DEGs were curated between CD8+ T cell and non-CD8+ T cells, which were defined as TNBC-related CD8+ T cell marker genes (Fig. 2E, Supplement Table 1).
Fig. 2.
Single cell RNA data analysis; Umap diagram of 16 samples(A); umap distribution diagram of annotation cell subgroups(B); umap of four T cell subpopulations after cluster analysis (C); Analysis of the weighted co-expression network in metabric-TNBC cohort. Sample clustering of dataset metabric-TNBC(D). Volcano Plot illustrated the DEGs between CD8+ T cells and no CD8+ T cells based on single cell datasets, the genes of CD8+ T cell related classifier are exhibited(E). Analysis of correlations between modules and CD8+ T cell characteristic(F). Different modules are produced and shown in different colors by aggregating genes with strong correlations into a same module (G). Identification of optimal thresholds, which is 3(H). Scatter plot of module eigengenes in the turquoise module (I). Heatmap describing the topological overlap matrix among genes based on co-expression modules (J).
3.3. Shaping Co-expression network
Samples from the METABRIC-TNBC cohort were clustered using average linkage and Pearson's correlation (Fig. 2D). A scale-free network was ensured by utilizing the soft-thresholding parameter, which included a power of β = 3 and scale-free R2 of 0.9 (Fig. 2H). Subsequently, two modules were identified, namely, the turquoise module had a stronger correlation with levels of CD8+ T cell infiltration (Fig. 2F, G, 2I), and hence, this module was used for further analysis as a vital immune-related module. Eventually, an eigengene adjacency heatmap was drawn to illustrate the relationship between different modules (Fig. 2J). A total of 20 hub genes, which were screened using the cytoHubba plugin according to the MCC algorithm [61], were customized as CD8+ T cell related classifiers (Fig. 2E, Supplement Table 2).
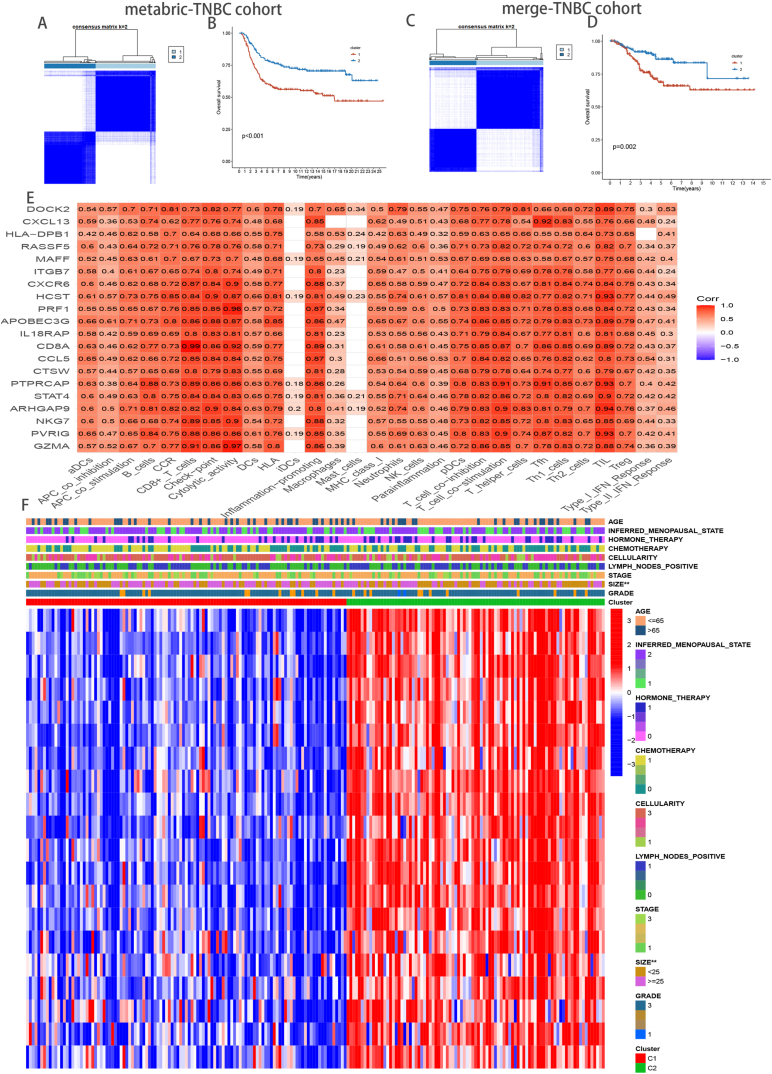
3.4. Unsupervised learning yielded two different TRCG-related patterns
Based on TRCG-related classifiers, two unique TRCG-related patterns were distinguished in the METABRIC-TNBC cohort upon application of an unsupervised clustering algorithm (Fig. 3A). Moreover, Kaplan–Meier analysis revealed that cases in Cluster 1 were associated with worse prognoses (Fig. 3B), and importantly, exploration of the merge-TNBC cohort attested to the robustness of the TRCG-related classifier (Fig. 3C and D). Intriguingly, there was a positive correlation between TRCG-related genes and the level of immune cell infiltration (Fig. 3E), and heatmap analysis revealed that TRCG-related genes were indeed abundant in cluster2, indicating an activated immune response (Fig. 3F).
Fig. 3.
Patients stratified into two clusters based on the CD8+ T cells hub genes. Consensus matrices of patients in the metabric-TNBC(A) and merge-TNBC(C) cohort via the unsupervised consensus clustering method (K-means). Survival analysis of the heterogeneous clusters in the metabric-TNBC (B) and merge-TNBC (D) cohort. Analysis of the hub genes–immune response relationships of TNBC in METABRIC data(E). Heatmap of the clinicopathological manifestations among the TRCG-related patterns (F).
3.5. The immune landscape of TRCG-related patterns
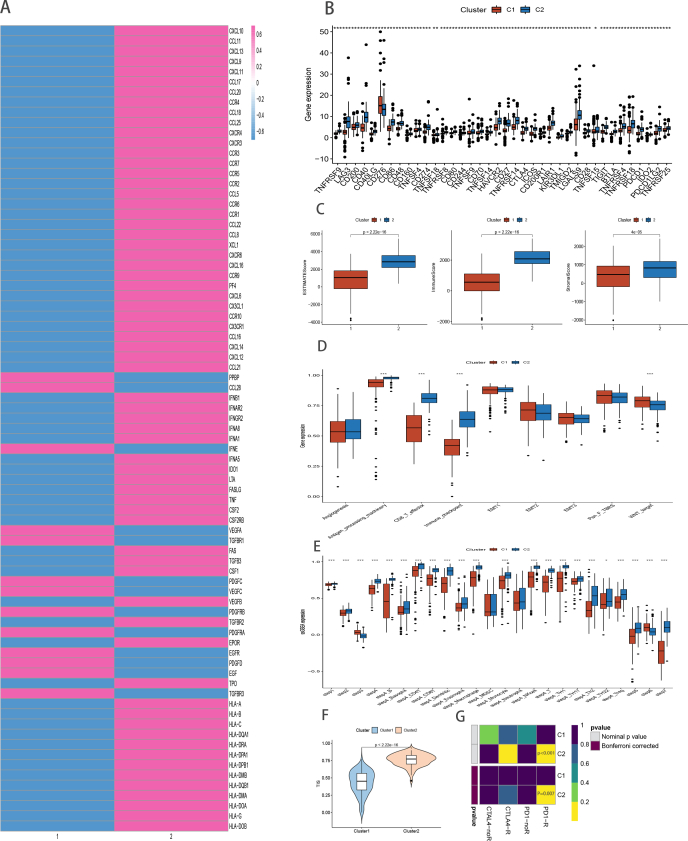
Firstly, a comparison of the proportion of immune cell infiltration of the TME between the two TRCG-related subtypes (supplement figure 1A) yielded the following crucial results. 1) Samples in Cluster 2, i.e., with a more favorable prognosis, mostly exhibited higher immune cell infiltration than samples in Cluster 1; and 2) immune cells, e.g., activated CD8+ T cells, dendritic cells, and type 1 T helper (Th1) cells, which have ability to mediate antitumor immune response, and multiple immunosuppressive cells, such as regulatory T cells (Treg), bone marrow derived suppressor cells, neutrophils, and immature dendritic cells, are abundant in the heatmap's immune infiltration zone, indicating that there may be a feedback mechanism, i.e., that the TME may stimulate the differentiation or recruitment of immunosuppressive cells.
Additionally, we used the ssGSEA algorithm on 50 Hallmark gene sets to assess the underlying carcinogenic features of the two clusters [76] (Supplement figure 2A). In accordance with the above-mentioned findings, cluster 1(C1) was characterized by genes related to glycolysis, metabolic, and carcinogenic pathways, while cluster 2(C2) was abundant in immune inflammatory pathway genes via gene set enrichment analysis(GSEA) (Supplement figure 2B). These results show that cases in C2 might display greater responsiveness to immunotherapeutics. Consequently, we attempted to delineate immune checkpoint expression and the landscape of immune cell infiltration to understand the potential mechanism. C2, which was the “immune-hot” subtype, showed greater infiltration of immune cells, including CD8+ T cells, CD4+ T cells, B cells, nature killer cells, monocytes, macrophage M1 cells, and activated dendritic cells (P < 0.05) (Fig. 4B). Additionally, C2 also exhibited abundant expression of immune checkpoint genes, such as CTLA-4, CD274 (PD-L1) and LAG3, implying that C2 might be more responsive to immune checkpoint inhibitor (ICI) therapy (Supplement figure 2C). Our results also indicate enriched expression of chemokines in Cluster2, such as CXCL6, CXCL9, CXCL10, CXCL12, CXCL13, CXCL14, CXCR4, CXCR3, and CCL5 (Fig. 4A, top panel), which can attract DCs and CD8 + T cells. Concurrently, the expression of HLA molecules was also abundant in C2, suggesting that these patients may exhibit stronger antigen presentation (Fig. 4A, bottom panel). Kim et al. have revealed that MHC downregulation contributes to immune escape in cancer cells [77] and we found higher immune score and stromal score in C2 (Fig. 4C). The cancer immune cycle can be visualized as a series of step-by-step events during which the antitumor immune response effectively kills cancer cells. These pathways are evidently active in C2 (Fig. 4E), underscoring the significant role of TRCG in cancer infiltration. As patients with abundant expression of TRCG were indicated to have better tumor immunity, we subsequently compared heterogeneity in immune-associated gene set scores between the two subtypes. Intriguingly, gene sets scores for CD8 T effector cells, angiogenesis, antigen processing machinery, immune checkpoint, Epithelial–mesenchymal transition (EMT) 3, pan_F_TBRS, and WNT targets were higher in C2 (Fig. 4D).
Fig. 4.
Comparison of the two subtypes in TNBC patients. Expression of chemokines, receptors and MHC molecules(A) and Immune cell infiltration levels Immune cell infiltration levels(B). Evaluation of the TME in the two TTK related patterns(C). Boxplots depicting the difference of the TME related signatures(D), the cancer–immunity cycle(E) and TIS scores(F) amongs the two TRCG related patterns via ssGSEA. Response prediction to immunotherapy (anti-PD-1 and anti-CTLA4) amongs the two TRCG patterns based on SubMap algorithms(G).
Next, immune marker gene profiles were further explored in the two ways. Cluster 1 patients had high amounts of SPP1 and VEGFA expression, which can promote M2 macrophage polarization and give rise to vascular abnormalities, respectively, and facilitate immune escape [78,79] (Supplementary Figure 3). Conversely, patients in Cluster 2 had favorable survival outcomes that can be partly attributed to richer expression of cytolytic activity markers, such as PRF1 and GZMA, activated T cell markers, such as ZAP70, CD3E, IL2RB, and ITK, and exhaustion markers in T cells, such as PDCD1 (Supplementary Figure 3).
Additionally, sensitivity of the response to anti-PD1 and anti-CTLA4 immunotherapy was evaluated using TIS and the submap algorithm. As expected, C2 had higher TIS scores, indicating stronger response to ICIs and immune activation (P < 0.0001) (Fig. 4F). Furthermore, expression patterns of samples in C2 were strikingly similar to those that had responded to PD-L1 inhibitors (Bonferroni corrected P < 0.01), implying that C2 may also be responsive to anti-PD-L1 treatment. Taken together, these results indicate that precision immunotherapy may be an efficient treatment option in patients who are classified as C2 (Fig. 4G). These results was repeatable in merge-TNBC cohort(Supplement Figure 1B,4,5).
3.6. Construction of CD8+ T cell-related prognostic signature (TRPS)
To develop a more convenient scoring model for clinical applications, 1286 DEGs were screened in both TRCG-related subtypes after intersecting with genes involved in the merge-TNBC cohort (Supplement Table 4). Additionally, for exploring biological pathways related to DEGs, GO and KEGG analyses were performed, and we show that DEGs were enriched in immune-related biological pathways (Supplement Figure 6). Next, univariate Cox regression (Supplement Table 5), LASSO, and multivariate Cox regression analysis identified three genes, viz, N4BP2L1, IL18RAP, and KIR3DL3, which were used to construct the TRPS using the following formula:
TRPS = (−0.19699 * expression of N4BP2L1) + (−0.22011 * expression of IL18RAP) + (−0.23479 * expression of KIR3DL3)
Patients with TNBC from the training (METABRIC) and the validating (merge - TNBC, GSE103091, GSE96058) cohorts were separated into high- and low-TRPS groups based on median TRPS value. Compared to patients with high TRPS in the training (Fig. 5A) and validating (Supplement Fig. 7A-C) sets, Kaplan–Meier survival curves revealed favorable survival outcomes in patients with low-TRPS. Area under the ROC curve values for predicting 1-, 3-, 5-year survival times were 0.695, 0.661, and 0.666 in METABRIC-TNBC cohort (Fig. 5B and C), 0.672, 0.656, and 0.665 in merge-TNBC cohort, 0.602, 0.703, 0.726 in GSE103091, and 0.728, 0.644, 0.634 in GSE96058, respectively. These values attest to the outstanding ability of the model to forecast overall survival (OS) in patients with TNBC (supplement Fig. 7D-F). The hazard ratio and 95% CI of TRSP in the univariate (P = 0.002) and multivariate Cox regression analyses (P = 0.011), indicate that it is indeed an independent prognostic index of OS in patients with TNBC (Supplement Fig. 7G and H). Likewise, the heatmap delineates the association between TRSP and clinicopathological traits (Fig. 5D). Surprisedly, Kaplan–Meier survival curves also revealed beneficial survival outcomes in patients with high-IL18RAP(Fig. 5E), high-N4BP2L1(Fig. 5F), high-KIR3DL(Fig. 5G) groups in multiple cohorts, respectively. A hybrid nomogram containing TRSP and clinicopathological manifestations is delineated in Supplementary Figure 7I. Both practical and predicted 1-, 3-, and 5-year survival rates, generated using calibration curve analysis, are delineated in Supplementary Figure 7J. These results indicate that the nomogram is both robust and precise, and can, consequently, be applied in clinical settings, i.e., in patients with TNBC.
Fig. 5.
TRSP in metabric-TNBC cohort analysis. Kaplan–Meier curves (A), time-dependent ROC analysis (B) and risk score(C). The association between TRSP and special clinicopathological traits(D). Kaplan–Meier curves of TRSP related genes in METABRIC-TNBC and merge - TNBC cohort (E, F, G).
3.7. Estimation of TME and immunotherapy efficacy in high- and low- TRSP
We attempted to understand the differences in immune characteristics between high- and low- TRSP subgroups and found that both immune and ESTIMATE scores were conspicuously upregulated in the low-TRSP subgroup, suggesting greater proportion of infiltrating immune cells (Fig. 6A). To further explore the correlation between TRSP and TME, we evaluated heterogeneity among immune infiltrating cells and immune function to more precisely assess the TME landscape. Our results demonstrate higher levels of immune cells equipped for antigen-presenting, antigen processing, and tumor-killing in low-TRSP group, such as CD8 T cells, T cells, B cells, and cytotoxic lymphocytes (Fig. 6B). As expected, we found that the signaling for antigen recognition, processing and presentation, and antitumor effects, containing APC co-stimulation, cytolytic activity, MHC class I, HLA, type I IFN response, type II IFN response, and T cell co-stimulation, were more active in the low-TRSP group (Fig. 6C). The TRSP is negative correlation with the expression of immune checkpoint genes, suggesting the low-TRSP set possible sensitivity to ICI therapy (Fig. 6D). Furthermore, the cancer immune cycle related pathways are evidently active in low-TRSP group (Fig. 6E). Additionally, compared to those with high TRPS scores, patients in the low-TRSP group had higher TIS and IPS, (Fig. 6F and G). Moreover, expression patterns of samples in TRSP-low subgroup were significantly similar to those that had responded to PD-L1 inhibitors (Fig. 6H) (Bonferroni corrected P < 0.01). Together, these results indicate that patients with a low-TRSP score can possibly benefit from immunotherapy.
Fig. 6.
The comparison of the TRSP -high and -low subgroups. Evaluation of the TME in the TRSP -high and -low subgroups(A). The heterogeneity of Immune cell infiltration levels amongs TRSP -high and -low subgroups in METABRIC(B) cohort by MCPcounter. Boxplots depicting the difference of the TME related signatures(C) amongs the two TRCG related patterns via ssGSEA. Correlation between TRSP and immune checkpoint-related genes(D). A comparison of the relative sensitivity of responding to anti-PD-1/PD-L1 as well as anti-CTLA-4 treatment in the TRSP high and low subgroups(E). The distinction of TIS scores amongs TRSP -high and -low subgroups(F). Response prediction to immunotherapy (anti-PD-1 and anti-CTLA4) amongs the TRSP – high and – low subgroups based on TIDE(G) and SubMap algorithms(H). The TRSP related genes are all low expressed in TNBC cells compared to normal breast cancer cell by Student's t-test. 2−ΔΔCt is used to present the fold change in qRT-PCR experiment(I). (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
3.8. Chemotherapy sensitivity related to the TRSP
A comparison was made of the IC50 values of chemotherapeutic drugs amongs patients in high- and low- TRSP subgroups, which can indicate the sensitivity to them. Patients in the low-TRSP group were more applicable to Cisplatin and Gefitinib, Methotrexate, Rapamycin, Sunitinib, Vinblastine, Vinorelbine. On the contrary, patients in the high-TRSP set were more responsive to Docetaxel, Doxorubicin and Sorafenib(Supplement Figure 8).
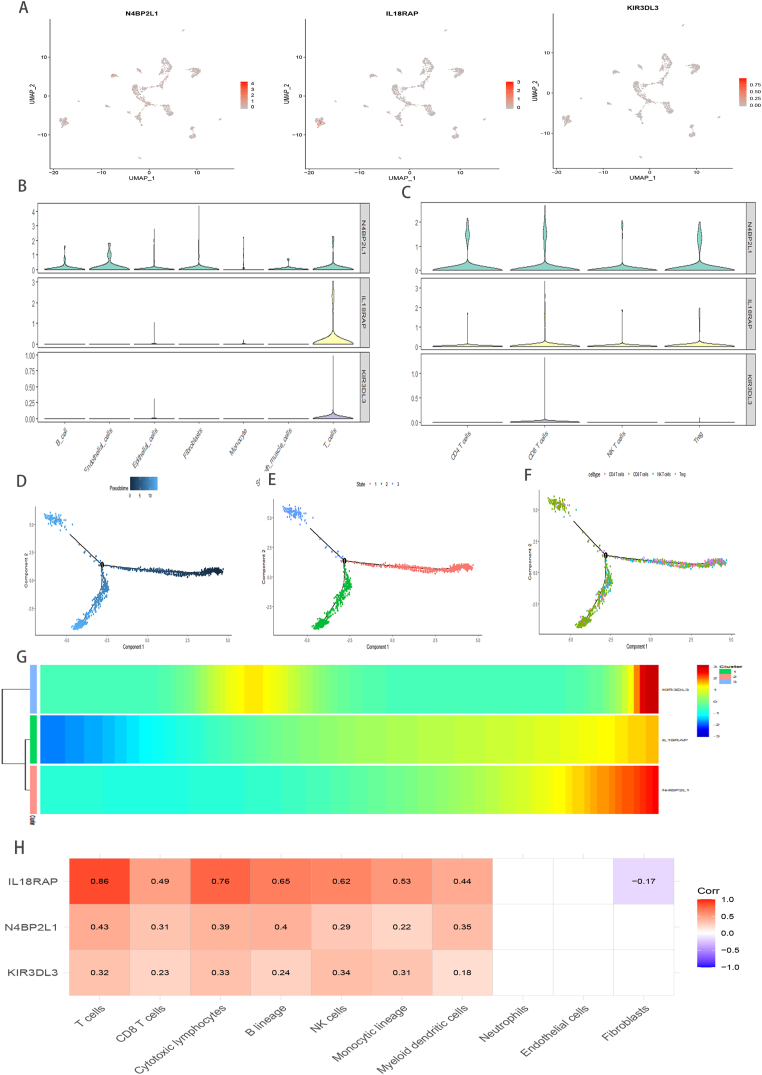
3.9. Validation of TRSP related genes expression via qRT-PCR, scRNA-seq analysis and bulk analysis
The qRT-PCR unraveled that these TRSP related genes all are poorer expression in TNBC cells than normal breast cell(Fig. 6I). To further confirm the type of cells expressing these TRSP related genes in the TME, we analyzed previously available TNBC scRNA-seq data and, as expected, found that N4BP2L1, IL18RAP, and KIR3DL3 were predominantly expressed in T cells(Fig. 7A–C). Moreover, to shed light on the underlying correlation between the TRPS related genes and the dynamic evolutionary trajectory of T cells in TNBC, we reordered single cells into a pseudo‐temporal timeline based on Monocle2 toolkit, and the result clearly unrevealed the uniform development route T cells(Fig. 7D–F). In the heatmap, the pseudo-time-dependent genes are displayed based on their pseudo-timeline trends(Fig. 7G). Notably, these genes are remarkably positive association with the infiltration levels of immunocompetent cells, specially CD8+ T cells, based on MCPcounter algorithm in metabric-TNBC cohort(Fig. 7H). These results again indicated that the TRSP related genes take crucial roles on TME in TNBC.
Fig. 7.
The exploration of TRSP relaed genes. UMPA (A) plots showing different TRSP related genes expression distribution based on scRNA-Seq and Violin plots revealing the difference of the expression of N4BP2L1, IL18RAP, KIR3DL3 in all cell subgroup (B) and T cell subgroup (C), respectively. Trajectory reconstruction of all T cells in TNBC, with a color code for pseudo-time (D), clusters (E), cell subtypes (F), respectively. The branched heatmap indicates the dynamics of the expression of N4BP2L1, IL18RAP, KIR3DL3 during T cells transdifferentiation(G), the redder the color, the higher the expression. The correlation between N4BP2L1, IL18RAP, KIR3DL3 and the level of immune cell infiltration (H).
4. Discussion
TNBC is the most invasive breast cancer subtype and is characterized by inferior prognosis and distinct heterogeneity [1,80]. Irrespective of cancer stage, chemotherapy remains the preferential treatment modality; however, TNBC displays resistance to chemotherapy and high rates of relapse. Further, compared to other breast cancer subtypes, TNBC has greater expression of PD-L1; this fact has contributed to the recent approval of immunotherapy with anti-PD-L1 monoclonal antibodies. Recent clinical trials with ICIs have revealed poor efficacy of immunotherapy in TNBC patients [[81], [82], [83]], which may be attributed to molecular heterogeneity in these patients. Thus, TNBC lesions with abundant TILs [84] may exhibit potentially greater benefits from chemotherapy and a lower possibility of disease relapse, indicating that it is imperative to investigate genomic characteristics and develop appropriate molecular classification. Notably, the infiltrating density of CD8+ T cells (preferred immune cells for targeting cancers), is a predictive marker of the efficacy of ICIs therapy [85]. Recent developments in single-cell RNA seq have helped explore tumor heterogeneity, and we have used this emerging technology to identify immune cell subpopulations and features of CD8+ T cells. Previous studies have proved that TME is intricately associated with and actively modulates antitumor immune responses and tumor heterogeneity [1,86], and as CD8+ T cells are known to possess cytotoxic properties that kill tumor cells [87], they are associated with the effectiveness of immunotherapy and prognosis [87,88].
Our results point toward a close relationship between the characteristics of CD8+ T cells, prognosis, and heterogeneity in clinical outcomes, which provide a rationale for constructing a molecular classification. Thence, the unsupervised clustering algorithm was implemented to obtain different molecular clusters based on CD8+ T cell marker genes. Ultimately, two robust clusters were distinguished in the METABRIC-TNBC cohort, and the results indicate that both clusters had heterogeneous survival and that prognosis was robust and reproducible in the merge-TNBC cohort. Delineating the intrinsic biological features of the two clusters according to GSVA and GSEA enrichment analyses revealed that while cluster 1 was significantly enriched in glycolysis metabolic pathways, cluster 2 was predominantly related to immune-inflammatory pathways. Additionally, the two clusters showed diverse genomic characteristics. Owing to stronger immune inflammatory activity, greater stable genomic characteristics, and more favorable clinical prognosis, patients in cluster2 may display better antitumor activity and profit from immunotherapy. It is known that molecular characteristics can predict clinical prognosis and facilitate personalized treatment approaches; nonetheless, to tailor a clinical plan for patients with TNBC, it is essential to identify individual variations and account for the same. The efficacy of distinct clinical treatments was assessed and compared among the two clusters. Cluster2 was defined as the “immune-hot” subtype with abundant immune cells, such as CD4+ T cells, CD8+ T cells, and activated dendritic cells, along with greater expression of immune checkpoint genes, such as CD274 (PD-L1), CTLA- 4, and LAG3. Activated CD8 T cells are capable of anti-tumor activity, and dendritic cells trigger immune responses after eliminating tumor cells [88,89]. Additionally, PD-L1 releases negative regulatory signals and promotes immune escape [88]. As multiple approaches used here have demonstrated that cluster 2 patients may be more responsive to immunotherapy, a more proactive strategy for immunotherapy should be recommended to patients classified as cluster 2.
Furthermore, DEGs between TRCG-related patterns were shown to be enriched in immune-related biological processes; this, when combined with individual heterogeneity, facilitated the construction of a scoring system, TRPS, to estimate and quantify TRCG-related patterns in TNBC patients. We reveal that, while those with low TRPS were enriched in immune activation pathways, indicating an immune-inflamed condition, those with high TRPS showed higher expression in carcinogenic pathways. Concurrently, cluster 2, defined as an immune-inflamed phenotype, is characterized by a favorable prognosis and a lower TRPS, whereas cluster 1 features an adverse prognosis and the higher TRPS. Together, these results indicate that TRPS is a robust and reliable device for the broad evaluation of personalized immune response patterns and could be utilized to further explore tumor immune phenotypes and immune cell infiltration in the TME. Moreover, patients with high TRPS were more prone to suffer malignancy and adverse clinicopathological manifestations and molecular subtypes; in contrast, opposite patterns were found in the low TRPS subgroup. Additionally, patients with lower TRPS, defined by immune activation, were strikingly associated with greater sensitivity to ICI immunotherapy.
Multiple trials have shown that immune checkpoint inhibitors and chemotherapy are synergistic because chemotherapy can not only damage the activity of immunosuppressive cells, such as Tregs and myeloid suppressor cells (MDSC), but also facilitate tumor cell apoptosis, enhance tumor antigen cross-presentation ability, and promote dendritic cell (DC) maturation and CD8+T cell infiltration to increase immune response. Currently, a series of clinical studies are exploring the potential benefits of immunotherapy, combined with chemotherapy, for advanced TNBC [90].
It is imperative to seek valid biomarkers that can predict if TNBC patients will benefit from combined chemotherapy and immunotherapy. We show that TRPS is negatively associated with immune checkpoint molecules, TMB, and TME scores, which indirectly suggests that the TRPS may play a crucial role in evaluating the efficiency of immunotherapy. The sensitivity of some chemotherapeutic and targeted drugs is linked to TRSP, implying that TNBC patients may be able to select more efficacious chemotherapeutics based on TRPS.
5. Conclusion
In conclusion, we describe the presence of two distinct heterogeneous clusters of TNBC patients that are characterized by differences in molecular characteristics and heterogeneity in clinical features, biological characteristics, genomic variations, and immune landscapes. Moreover, we identify TRPS as a useful prognostic marker for TNBC that is also linked to immune cell infiltration and immunotherapy. We hypothesize that this TRSP model is a valid tool for predicting survival and providing treatment guidance in TNBC and that it can help elucidate individual variations in TME and to identify patients who would benefit most from anticancer immunotherapy. Future research should focus on understanding the specific mechanisms involved in TRPS and immune cells. Despite the above, TRPS has some limitations. First, all the samples used in this study were derived from different public datasets and were retrospective. Proper validation with actual clinical data of eligible patients who had undergone immunotherapy was not possible. Second, the median value of TRPS was used as the cutoff to divide TNBC samples into high and low TRPS, but stratifying TNBC patients based on an optimal cutoff value may be more appropriate. Third, there is still a lack of clarity regarding the detailed signaling pathways of the target genes in the immune microenvironment. Our future research will focus on understanding the specific mechanisms involved in TRPS and immune cells.
Ethics approval and consent to participate
Not applicable.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author contribution statement
Yin-Wei Dai: Conceived and designed the experiments; Performed the experiments. Wei-Ming Wang: Analyzed and interpreted the data. Xiang Zhou: Wrote the paper.
Data availability statement
Data associated with this study has been deposited at The datasets used and/or analyzed during the current study are available in the cBio Cancer Genomics Protal(cBioportal, https://www.cbioportal.org/), the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA) network (https://cancergenome.nih.gov/). R and other custom scripts for analyzing data are available upon reasonable request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the funding of the Zhejiang provincial natural science foundation of china under grant (NO.LQ22H160022).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e19798.
Contributor Information
Yin-wei Dai, Email: 601716404@qq.com.
Wei-ming Wang, Email: wwm_boy2010@163.com.
Xiang Zhou, Email: zhouxiang36@outlook.com.
Abbreviations
- TNBC
Triple-negative breast cancer
- TRCG
CD8+ T cell core genes
- TRPS
CD8+ T cell-related prognostic signature
- ICI
immune checkpoint inhibitors
- PD-1
programmed cell death protein 1
- TME
tumor microenvironment
- PCR
polymerase chain reaction
- GSVA
gene set variation analysis
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OS
overall survival
- ssGSEA
single-sample gene set enrichment analysis
- scRNA-seq
Single-cell RNA sequencing
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
figs7.
figs8.
References
- 1.Bianchini G., Balko J.M., Mayer I.A., Sanders M.E., Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016;13(11):674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao Y., Ma D., Zhao S., Suo C., Shi J., Xue M.Z., Ruan M., Wang H., Zhao J., Li Q., et al. Multi-omics profiling reveals distinct microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin. Cancer Res. : an official journal of the American Association for Cancer Research. 2019;25(16):5002–5014. doi: 10.1158/1078-0432.CCR-18-3524. [DOI] [PubMed] [Google Scholar]
- 3.Denkert C., Liedtke C., Tutt A., von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet (London, England) 2017;389(10087):2430–2442. doi: 10.1016/S0140-6736(16)32454-0. [DOI] [PubMed] [Google Scholar]
- 4.Denkert C., von Minckwitz G., Darb-Esfahani S., Lederer B., Heppner B.I., Weber K.E., Budczies J., Huober J., Klauschen F., Furlanetto J., et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19(1):40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 5.Loi S., Michiels S., Salgado R., Sirtaine N., Jose V., Fumagalli D., Kellokumpu-Lehtinen P.L., Bono P., Kataja V., Desmedt C., et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann. Oncol. : official journal of the European Society for Medical Oncology. 2014;25(8):1544–1550. doi: 10.1093/annonc/mdu112. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann B.D., Bauer J.A., Chen X., Sanders M.E., Chakravarthy A.B., Shyr Y., Pietenpol J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burstein M.D., Tsimelzon A., Poage G.M., Covington K.R., Contreras A., Fuqua S.A., Savage M.I., Osborne C.K., Hilsenbeck S.G., Chang J.C., et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. : an official journal of the American Association for Cancer Research. 2015;21(7):1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y.R., Jiang Y.Z., Xu X.E., Yu K.D., Jin X., Hu X., Zuo W.J., Hao S., Wu J., Liu G.Y., et al. Comprehensive transcriptome analysis identifies novel molecular subtypes and subtype-specific RNAs of triple-negative breast cancer. Breast Cancer Res. 2016;18(1):33. doi: 10.1186/s13058-016-0690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J., Jiang C.C., Jin L., Zhang X.D. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann. Oncol. : official journal of the European Society for Medical Oncology. 2016;27(3):409–416. doi: 10.1093/annonc/mdv615. [DOI] [PubMed] [Google Scholar]
- 10.Ritprajak P., Azuma M. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol. 2015;51(3):221–228. doi: 10.1016/j.oraloncology.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Gong J., Chehrazi-Raffle A., Reddi S., Salgia R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. Journal for immunotherapy of cancer. 2018;6(1):8. doi: 10.1186/s40425-018-0316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mittendorf E.A., Philips A.V., Meric-Bernstam F., Qiao N., Wu Y., Harrington S., Su X., Wang Y., Gonzalez-Angulo A.M., Akcakanat A., et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol. Res. 2014;2(4):361–370. doi: 10.1158/2326-6066.CIR-13-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteva F.J., Hubbard-Lucey V.M., Tang J., Pusztai L. Immunotherapy and targeted therapy combinations in metastatic breast cancer. Lancet Oncol. 2019;20(3):e175–e186. doi: 10.1016/S1470-2045(19)30026-9. [DOI] [PubMed] [Google Scholar]
- 14.Rich-Griffin C., Stechemesser A., Finch J., Lucas E., Ott S., Schäfer P. Single-cell transcriptomics: a high-resolution avenue for plant functional genomics. Trends Plant Sci. 2020;25(2):186–197. doi: 10.1016/j.tplants.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Qiu P., Guo Q., Yao Q., Chen J., Lin J. Characterization of exosome-related gene risk model to evaluate the tumor immune microenvironment and predict prognosis in triple-negative breast cancer. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.736030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sha S., Si L., Wu X., Chen Y., Xiong H., Xu Y., Liu W., Mei H., Wang T., Li M. Prognostic analysis of cuproptosis-related gene in triple-negative breast cancer. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.922780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., Porta-Pardo E., Gao G.F., Plaisier C.L., Eddy J.A., et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830.e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M., Coussens L.M., Gabrilovich D.I., Ostrand-Rosenberg S., Hedrick C.C., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salgado R., Denkert C., Demaria S., Sirtaine N., Klauschen F., Pruneri G., Wienert S., Van den Eynden G., Baehner F.L., Penault-Llorca F., et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. : official journal of the European Society for Medical Oncology. 2015;26(2):259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali H.R., Provenzano E., Dawson S.J., Blows F.M., Liu B., Shah M., Earl H.M., Poole C.J., Hiller L., Dunn J.A., et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann. Oncol. : official journal of the European Society for Medical Oncology. 2014;25(8):1536–1543. doi: 10.1093/annonc/mdu191. [DOI] [PubMed] [Google Scholar]
- 21.Kaech S.M., Wherry E.J., Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2002;2(4):251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 22.Karaca E., Harel T., Pehlivan D., Jhangiani S.N., Gambin T., Coban Akdemir Z., Gonzaga-Jauregui C., Erdin S., Bayram Y., Campbell I.M., et al. Genes that affect brain structure and function identified by rare variant analyses of mendelian neurologic disease. Neuron. 2015;88(3):499–513. doi: 10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang N., Bevan M.J. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35(2):161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fesnak A.D., June C.H., Levine B.L. Engineered T cells: the promise and challenges of cancer immunotherapy. Nat. Rev. Cancer. 2016;16(9):566–581. doi: 10.1038/nrc.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maude S.L., Frey N., Shaw P.A., Aplenc R., Barrett D.M., Bunin N.J., Chew A., Gonzalez V.E., Zheng Z., Lacey S.F., et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014;371(16):1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maus M.V., June C.H. Making better chimeric antigen receptors for adoptive T-cell therapy. Clin. Cancer Res. : an official journal of the American Association for Cancer Research. 2016;22(8):1875–1884. doi: 10.1158/1078-0432.CCR-15-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellman I., Coukos G., Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480(7378):480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribas A. Tumor immunotherapy directed at PD-1. N. Engl. J. Med. 2012;366(26):2517–2519. doi: 10.1056/NEJMe1205943. [DOI] [PubMed] [Google Scholar]
- 29.Blum J.S., Wearsch P.A., Cresswell P. Pathways of antigen processing. Annu. Rev. Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brahmer J.R., Drake C.G., Wollner I., Powderly J.D., Picus J., Sharfman W.H., Stankevich E., Pons A., Salay T.M., McMiller T.L., et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J. Clin. Oncol. : official journal of the American Society of Clinical Oncology. 2010;28(19):3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tscharke D.C., Croft N.P., Doherty P.C., La Gruta N.L. Sizing up the key determinants of the CD8(+) T cell response. Nat. Rev. Immunol. 2015;15(11):705–716. doi: 10.1038/nri3905. [DOI] [PubMed] [Google Scholar]
- 32.Wong P., Pamer E.G. CD8 T cell responses to infectious pathogens. Annu. Rev. Immunol. 2003;21:29–70. doi: 10.1146/annurev.immunol.21.120601.141114. [DOI] [PubMed] [Google Scholar]
- 33.Dumontet E., Osman J., Guillemont-Lambert N., Cros G., Moshous D., Picard C. Recurrent respiratory infections revealing CD8α deficiency. J. Clin. Immunol. 2015;35(8):692–695. doi: 10.1007/s10875-015-0213-x. [DOI] [PubMed] [Google Scholar]
- 34.Hersperger A.R., Martin J.N., Shin L.Y., Sheth P.M., Kovacs C.M., Cosma G.L., Makedonas G., Pereyra F., Walker B.D., Kaul R., et al. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood. 2011;117(14):3799–3808. doi: 10.1182/blood-2010-12-322727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurktschiev P.D., Raziorrouh B., Schraut W., Backmund M., Wächtler M., Wendtner C.M., Bengsch B., Thimme R., Denk G., Zachoval R., et al. Dysfunctional CD8+ T cells in hepatitis B and C are characterized by a lack of antigen-specific T-bet induction. J. Exp. Med. 2014;211(10):2047–2059. doi: 10.1084/jem.20131333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q., Davis J.C., Lamborn I.T., Freeman A.F., Jing H., Favreau A.J., Matthews H.F., Davis J., Turner M.L., Uzel G., et al. Combined immunodeficiency associated with DOCK8 mutations. N. Engl. J. Med. 2009;361(21):2046–2055. doi: 10.1056/NEJMoa0905506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chia J., Yeo K.P., Whisstock J.C., Dunstone M.A., Trapani J.A., Voskoboinik I. Temperature sensitivity of human perforin mutants unmasks subtotal loss of cytotoxicity, delayed FHL, and a predisposition to cancer. Proc. Natl. Acad. Sci. U.S.A. 2009;106(24):9809–9814. doi: 10.1073/pnas.0903815106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clementi R., Locatelli F., Dupré L., Garaventa A., Emmi L., Bregni M., Cefalo G., Moretta A., Danesino C., Comis M., et al. A proportion of patients with lymphoma may harbor mutations of the perforin gene. Blood. 2005;105(11):4424–4428. doi: 10.1182/blood-2004-04-1477. [DOI] [PubMed] [Google Scholar]
- 39.Mortaz E., Tabarsi P., Mansouri D., Khosravi A., Garssen J., Velayati A., Adcock I.M. Cancers related to immunodeficiencies: update and perspectives. Front. Immunol. 2016;7:365. doi: 10.3389/fimmu.2016.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gravano D.M., Hoyer K.K. Promotion and prevention of autoimmune disease by CD8+ T cells. J. Autoimmun. 2013;45:68–79. doi: 10.1016/j.jaut.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 41.Holzelova E., Vonarbourg C., Stolzenberg M.C., Arkwright P.D., Selz F., Prieur A.M., Blanche S., Bartunkova J., Vilmer E., Fischer A., et al. Autoimmune lymphoproliferative syndrome with somatic Fas mutations. N. Engl. J. Med. 2004;351(14):1409–1418. doi: 10.1056/NEJMoa040036. [DOI] [PubMed] [Google Scholar]
- 42.Valori M., Jansson L., Kiviharju A., Ellonen P., Rajala H., Awad S.A., Mustjoki S., Tienari P.J. A novel class of somatic mutations in blood detected preferentially in CD8+ cells. Clin. Immunol. 2017;175:75–81. doi: 10.1016/j.clim.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walter U., Santamaria P. CD8+ T cells in autoimmunity. Curr. Opin. Immunol. 2005;17(6):624–631. doi: 10.1016/j.coi.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 44.Blankenstein T., Coulie P.G., Gilboa E., Jaffee E.M. The determinants of tumour immunogenicity. Nat. Rev. Cancer. 2012;12(4):307–313. doi: 10.1038/nrc3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen D.S., Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39(1):1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Fridman W.H., Pagès F., Sautès-Fridman C., Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer. 2012;12(4):298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 47.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swart M., Verbrugge I., Beltman J.B. Combination approaches with immune-checkpoint blockade in cancer therapy. Front. Oncol. 2016;6:233. doi: 10.3389/fonc.2016.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tumeh P.C., Harview C.L., Yearley J.H., Shintaku I.P., Taylor E.J., Robert L., Chmielowski B., Spasic M., Henry G., Ciobanu V., et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishida Y., Agata Y., Shibahara K., Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992;11(11):3887–3895. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leach D.R., Krummel M.F., Allison J.P. Enhancement of antitumor immunity by CTLA-4 blockade. Science (New York, N.Y.) 1996;271(5256):1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 52.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clough E., Barrett T. The gene expression Omnibus database. Methods Mol. Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14(10):979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Becht E., Giraldo N.A., Lacroix L., Buttard B., Elarouci N., Petitprez F., Selves J., Laurent-Puig P., Sautès-Fridman C., Fridman W.H., et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. Suppl 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 63.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C., Angell H., Fredriksen T., Lafontaine L., Berger A., et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 65.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoshida Y., Brunet J.P., Tamayo P., Golub T.R., Mesirov J.P. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS One. 2007;2(11):e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R., Albright A., Cheng J.D., Kang S.P., Shankaran V., et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong W.F., Liu M.Y., Liang L., Zhang Y., Li Z.J., Han K., Du S.S., Chen Y.J., Ma L.H. Molecular characteristics of T cell-mediated tumor killing in hepatocellular carcinoma. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.868480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Auslander N., Zhang G., Lee J.S., Frederick D.T., Miao B., Moll T., Tian T., Wei Z., Madan S., Sullivan R.J., et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med. 2018;24(10):1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen P.L., Roh W., Reuben A., Cooper Z.A., Spencer C.N., Prieto P.A., Miller J.P., Bassett R.L., Gopalakrishnan V., Wani K., et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6(8):827–837. doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu L., Deng C., Pang B., Zhang X., Liu W., Liao G., Yuan H., Cheng P., Li F., Long Z., et al. TIP: a web server for resolving tumor Immunophenotype profiling. Cancer Res. 2018;78(23):6575–6580. doi: 10.1158/0008-5472.CAN-18-0689. [DOI] [PubMed] [Google Scholar]
- 72.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pal B., Chen Y., Vaillant F., Capaldo B.D., Joyce R., Song X., Bryant V.L., Penington J.S., Di Stefano L., Tubau Ribera N., et al. A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. EMBO J. 2021;40(11) doi: 10.15252/embj.2020107333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jia L., Wang T., Zhao Y., Zhang S., Ba T., Kuai X., Wang B., Zhang N., Zhao W., Yang Z., et al. Single-cell profiling of infiltrating B cells and tertiary lymphoid structures in the TME of gastric adenocarcinomas. OncoImmunology. 2021;10(1) doi: 10.1080/2162402X.2021.1969767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karaayvaz M., Cristea S., Gillespie S.M., Patel A.P., Mylvaganam R., Luo C.C., Specht M.C., Bernstein B.E., Michor F., Ellisen L.W. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun. 2018;9(1):3588. doi: 10.1038/s41467-018-06052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen K., Wang Q., Li M., Guo H., Liu W., Wang F., Tian X., Yang Y. Single-cell RNA-seq reveals dynamic change in tumor microenvironment during pancreatic ductal adenocarcinoma malignant progression. EBioMedicine. 2021;66 doi: 10.1016/j.ebiom.2021.103315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim J.M., Chen D.S. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure) Ann. Oncol. : official journal of the European Society for Medical Oncology. 2016;27(8):1492–1504. doi: 10.1093/annonc/mdw217. [DOI] [PubMed] [Google Scholar]
- 78.Zhang Y., Du W., Chen Z., Xiang C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp. Cell Res. 2017;359(2):449–457. doi: 10.1016/j.yexcr.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 79.Fukumura D., Kloepper J., Amoozgar Z., Duda D.G., Jain R.K. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat. Rev. Clin. Oncol. 2018;15(5):325–340. doi: 10.1038/nrclinonc.2018.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou S., Huang Y.E., Liu H., Zhou X., Yuan M., Hou F., Wang L., Jiang W. Single-cell RNA-seq dissects the intratumoral heterogeneity of triple-negative breast cancer based on gene regulatory networks. Mol. Ther. Nucleic Acids. 2021;23:682–690. doi: 10.1016/j.omtn.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Emens L.A. Breast cancer immunotherapy: facts and hopes. Clin. Cancer Res. : an official journal of the American Association for Cancer Research. 2018;24(3):511–520. doi: 10.1158/1078-0432.CCR-16-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nanda R., Chow L.Q., Dees E.C., Berger R., Gupta S., Geva R., Pusztai L., Pathiraja K., Aktan G., Cheng J.D., et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase ib KEYNOTE-012 study. J. Clin. Oncol. official journal of the American Society of Clinical Oncology. 2016;34(21):2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schmid P., Rugo H.S., Adams S., Schneeweiss A., Barrios C.H., Iwata H., Diéras V., Henschel V., Molinero L., Chui S.Y., et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2020;21(1):44–59. doi: 10.1016/S1470-2045(19)30689-8. [DOI] [PubMed] [Google Scholar]
- 84.Chen C., Li S., Xue J., Qi M., Liu X., Huang Y., Hu J., Dong H., Ling K. PD-L1 tumor-intrinsic signaling and its therapeutic implication in triple-negative breast cancer. JCI insight. 2021;6(8) doi: 10.1172/jci.insight.131458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Farhood B., Najafi M., Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: a review. J. Cell. Physiol. 2019;234(6):8509–8521. doi: 10.1002/jcp.27782. [DOI] [PubMed] [Google Scholar]
- 86.Fu Y., Liu S., Zeng S., Shen H. From bench to bed: the tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J. Exp. Clin. Cancer Res. : CRN. 2019;38(1):396. doi: 10.1186/s13046-019-1396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumar B.V., Connors T.J., Farber D.L. Human T cell development, localization, and function throughout life. Immunity. 2018;48(2):202–213. doi: 10.1016/j.immuni.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mariathasan S., Turley S.J., Nickles D., Castiglioni A., Yuen K., Wang Y., Kadel E.E., III, Koeppen H., Astarita J.L., Cubas R., et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee Y.S., Radford K.J. The role of dendritic cells in cancer. International review of cell and molecular biology. 2019;348:123–178. doi: 10.1016/bs.ircmb.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 90.Zhu Y., Zhu X., Tang C., Guan X., Zhang W. Progress and challenges of immunotherapy in triple-negative breast cancer. Biochim. Biophys. Acta, Rev. Cancer. 2021;1876(2) doi: 10.1016/j.bbcan.2021.188593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at The datasets used and/or analyzed during the current study are available in the cBio Cancer Genomics Protal(cBioportal, https://www.cbioportal.org/), the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA) network (https://cancergenome.nih.gov/). R and other custom scripts for analyzing data are available upon reasonable request.