Abstract
Rationale
Fungal exposure has been associated with predisposing and protective effects on the development of childhood asthma.
Objectives
To study whether early-life house dust mycobiota composition is associated with the development of asthma.
Methods
Mycobiota were determined by amplicon sequencing from 382 dust samples collected from living room floors 2 months after birth in homes of the LUKAS cohort. Asthma status by 10.5 years of age was defined from questionnaires and assigned as ever asthma (n = 68) or current asthma (n = 27). Inhalant atopy was clinically determined at the same age. β-composition was analyzed using PERMANOVA-S, and asthma and atopy analyses were performed using discrete time hazard models and logistic regression, respectively.
Results
The house dust mycobiota composition based on Bray-Curtis distance was different in the homes of children who later did or did not develop asthma. The first and the fourth axes scores of principal coordinates analysis based on Bray-Curtis were associated with ever asthma. Of the genera with the strongest correlation with these axes, the relative abundance of Boeremia, Cladosporium, Microdochium, Mycosphaerella, and Pyrenochaetopsis showed protective associations with asthma. None of these associations remained significant after mutual adjustment among the five genera or when mutually adjusted for other microbial cell wall markers and previously identified asthma-protective bacterial indices. Neither fungal α-diversity nor load was associated with asthma in the whole population, but higher fungal richness was a risk factor among children on farms. Higher fungal loads (measured via quantitative polymerase chain reaction) in house dust were associated with the risk of inhalant atopy.
Conclusions
The results of our analyses from this well-characterized birth cohort suggest that the early-life house dust mycobiota in Finnish homes, characterized via DNA amplicon sequencing, do not have strong predisposing or protective effects on asthma development.
Keywords: atopy, environmental exposure, epidemiology, follow-up studies
Diverse microbial exposure in early life may instruct immune responses so that it reduces the risk of allergic diseases later in life (1). However, we are lacking knowledge on specific characteristics of beneficial environmental microbial communities, which may prevent the development of allergic diseases, impeding progress in the development of preventive therapeutic approaches.
Farming environments, rich in microbes, have been shown to decrease the risk of development of asthma and other allergic diseases (2). A vast majority of the previous epidemiologic studies on this topic have found protective associations mainly for bacteria and their cell wall components (2–6). Detailed characterization of bacterial exposures using DNA-based methods has shown a protective association of bacterial diversity with asthma in several studies (7–10), whereas other analyses did not reveal such associations (10, 11). In addition, patterns of farm-like bacterial microbiota have shown asthma-protective qualities in the present cohort, even when disconnected from the farming environments (5, 6). The effects of fungal species in this context have been much less explored. A study using cultivation of viable fungi has shown protective associations between early-life house dust mycobiota and asthma in the farming context (7), whereas other studies have mostly targeted fungal cell wall markers as surrogates for total fungal exposure (12).
Indoor fungal exposure and its effects on health have been closely intertwined with moisture damage and dampness in buildings, for which associations with adverse respiratory health symptoms, asthma exacerbation, and development of childhood asthma are well established (13–15). The concept of “indoor mold” is the key reason it has been hypothesized that increased exposure to fungal species could be harmful rather than beneficial for health (13, 16). However, previous studies relying on cultivation or cell wall markers have shown a tendency that fungal exposure during early life might have a protective effect on asthma development, but the absence of detailed taxonomical information has hindered our understanding of the specific exposures and mechanisms behind these associations (17). Associations between indoor mycobiota and inhalant atopy have rarely been explored, and the results are inconsistent (10, 11, 18). Thus, studies detailing the indoor mycobiota using current sequencing methods are needed. Currently, there are only a few studies that have attempted such mycobiota health profiling, limited by their cross-sectional design, short follow-up (11, 19), or low number of observations (8). The present work is the first large-scale prospective study determining associations between indoor mycobiota in early-life house dust—determined with next-generation sequencing—and its association with the development of asthma.
Our objective was to determine whether individual early-life fungal genera or other characteristics of mycobiota in house dust, such as diversity or community structure, are associated with the development of current and ever asthma until the age of 10.5 years and inhalant atopy at that time point. We hypothesized that applying in-depth profiling of mycobiota from early-life house dust samples of a well-defined cohort with a long follow-up time would result in identification of fungal taxa and/or community characteristics with both predisposing as well as protective associations with asthma. Based on earlier work (7), we expected to replicate the association of exposure to higher fungal richness with lower asthma incidence.
Methods
Study Cohort
The participants are from a Finnish birth cohort, LUKAS (20). Written informed consent was obtained from all parents. A detailed description of the cohort is provided in the supplementary methods. Ethical permission was granted by the hospital district of Northern Savo ethics committee (IORG00005196, LUKAS1:299/2017[33/2002], LUKAS2:300/2017[48/2004]).
Definition of Asthma and Other Outcomes
The development of childhood asthma was followed with questionnaires eight times during the follow-up of 10.5 years (6). Ever asthma (n = 68) and its timing were defined either as time of parent-reported doctor-diagnosed asthma or second diagnosed attack of asthmatic (or obstructive) bronchitis, whichever occurred first. If a child with ever asthma had used asthma medication and/or had wheezing in the past 12 months at the age of 10.5 years, the child was defined as having current asthma (n = 27). The statistical analyses included 361 children with ever asthma and 300 with current asthma definitions. In additional analyses, we used respiratory symptoms until the age of 10.5 years, and five different asthma phenotypes until the age of 6 years were defined by using latent class analyses (see details in the supplementary methods).
Allergen-specific immunoglobulin E (IgE) to 13 common inhalant allergens (two house dust mites [Dermatophagoides pteronyssinus and Dermatophagoides farinae]; seven pollens [alder, birch, European hazel, grass pollen mixture, rye, mugwort, and plantain]; cat, horse, and dog dander; and the mold Alternaria alternata) (Medwiss Analytic) (21) was measured from serum samples collected at the age of 10.5 years in 259 children with house dust mycobiota data. Inhalant atopy was defined as any of the tested inhalant allergen ⩾0.70 kU/L. This cutoff was also used for stratified analyses for atopic status in asthma models.
Sample Collection and Processing
A detailed description of the sample collection and processing, polymerase chain reaction (PCR) and MiSeq sequencing protocol and data processing, quantitative PCR (qPCR), and the molecular analyses of microbial cell wall markers is provided in the supplement. House dust samples were vacuumed from living room floors when the children were 2 months old.
Sequencing and Sequence Data Processing
The fungal internal transcribed spacer (ITS) region was amplified using ITS1F/ITS2 primers (22) and sequenced with Illumina’s MiSeq PA300. The reads were preprocessed using Dada2 pipeline (23) and mapped against Unite database, version May 2021 (24). Fungal diversity in house dust was estimated with Chao1 richness and Shannon diversity indices. Fungal load in house dust was determined with qPCR (25).
Statistical Analyses
The statistical analysis is detailed in the supplementary methods. In short, principal coordinate analysis (PCoA) was used to visualize fungal β diversity based on a Bray-Curtis distance matrix, and permutational multivariate analysis of variance -S (PERMANOVA-S) (26) was used to assess differences between outcome groups from this matrix. PCoA axes (PC1–5) and their scores were used to find associations between mycobiota and the development of asthma. In most of the statistical testing, we used three levels of exposure classes.
The associations of dust mycobiota with both asthma diagnoses were determined with survival analysis discrete-time hazard models. Generalized estimating equations were used for respiratory symptoms, multinomial logistic regression for wheezing phenotypes, and logistic regression for inhalant atopy. The results are presented as adjusted odds ratios (aORs) and their 95% confidence intervals (95% CI). All models with health outcomes were adjusted for study cohort, farm living, maternal history of allergic diseases, sex, number of older siblings, and maternal smoking during pregnancy. The most relevant findings were tested for possible confounding factors (27) and a set of previously measured asthma-associated microbes (5, 6) or microbial markers in the house dust samples (28, 29).
Results
Altogether, 382 of the LUKAS cohort participants had house dust mycobiota assessed, and, of those, 361 children had data on ever asthma diagnosis and the information on the desired covariates. The prevalence of inhalant atopy assessed from 259 children was 45.9%. For detailed demographics of the study population, see Table E1 in the data supplement.
Fungal Diversity and Phylum-Level Composition in House Dust and Asthma
The house dust mycobiota was dominated by two phyla, Ascomycota (mean, 65.50%; standard deviation [SD], 18.77%) and Basidiomycota (mean, 33.22%; SD, 18.53%; Figure E1A). The relative abundance of these phyla showed high variability between samples, but there was no significant difference in the phylum-level taxa between the homes of subjects with asthma and those of subjects without asthma (Mann-Whitney, P > 0.15). Similarly, neither fungal richness and diversity nor the total fungal load in house dust differed significantly between the asthma groups (Figure E1B and Table E2).
Associations between Fungal Composition of House Dust and Asthma
To assess differences in the overall mycobiota composition in the house dust of subjects with and without asthma, we conducted PERMANOVA-S analysis on the compositional difference using Bray-Curtis dissimilarity, as well as Jaccard similarity coefficients. The first uses the relative abundance of taxa, whereas the latter uses the presence/absence of taxa in computing differences between samples. We found that the Jaccard similarity coefficient was significantly different in the early-life house dust of children who at 10.5 years had current asthma compared with children without asthma (P = 0.01). Bray-Curtis dissimilarity between samples also indicated a difference, although nonsignificant, between these two groups (P = 0.10). There was no statistical difference in the overall house dust mycobiota composition between the ever asthma and the non-asthma groups (Jaccard P = 0.21, Bray-Curtis P = 0.28).
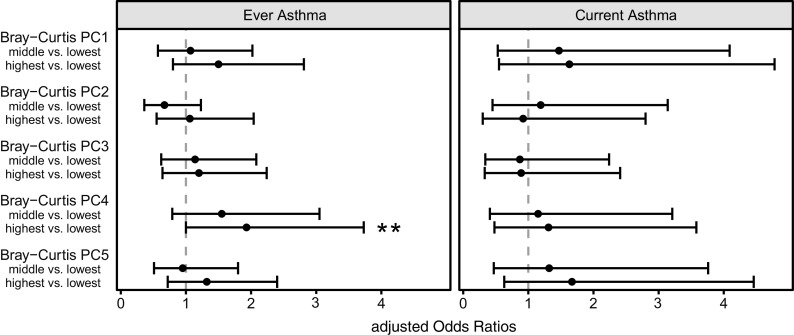
To better understand the drivers of the observed association with respect to specific fungal taxa and to guard against multiple testing, we first conducted PCoA using the Bray-Curtis dissimilarity matrix. We used the Bray-Curtis dissimilarity index rather than the binary Jaccard, as the latter does not facilitate downstream analyses of correlating taxa because of the presence/absence nature of this index. We computed survival analysis to test the associations between axes PC1 to PC5 on asthma development (Figure 1 and Table E3) and found axes PC4 to be significantly (P = 0.05) and PC1 to be tendentially (P = 0.20) associated with ever asthma diagnosis. The other axes showed neither a significant nor a dose–response type of association with asthma development. No significant associations were observed between PCoA axes and the current asthma definition.
Figure 1.
Associations between five fungal axis scores of Bray-Curtis dissimilarity index–based PC analysis and the development of ever asthma and current asthma until the age of 10.5 years. We compared the lowest tertile to the middle and highest tertile. Whiskers indicate the 95% confidence intervals. Models are adjusted for time of follow-up, farm living, cohort, sex, maternal history of allergic diseases, maternal smoking during pregnancy, and the number of older siblings. **Significance < 0.05. PC = principal coordinate.
We then aimed to determine which fungal taxa were associated with these two PC axes and explored correlations (Spearman) of abundant genera in house dust (86 genera with a mean relative abundance >0.1%) with the two axes. Eleven genera correlated (r >|0.4|) with PC1 and three genera with PC4 (Table 1). These taxa belonged to phyla Ascomycota or Basidiomycota, and their sum contributed on average 36% of the fungal sequences detected in the individual house dust samples.
Table 1.
Spearman correlations between abundant (>0.1% relative abundance across house dust samples) fungal genera and the PC axes 1 and 4
| Phylum | Family | Genus | PC-1 Correlation | PC-4 Correlation | Mean Relative Abundance across Samples (%) |
|---|---|---|---|---|---|
| Ascomycota | Cladosporiaceae | Cladosporium | −0.41 | 0.26 | 9.80 |
| Mycosphaerellaceae | Mycosphaerella | −0.72 | 0.27 | 5.13 | |
| Cucurbitariaceae | Pyrenochaetopsis | −0.50 | 0.05 | 0.52 | |
| Didymellaceae | Boeremia | −0.52 | 0.05 | 0.72 | |
| Epicoccum | −0.44 | 0.26 | 1.45 | ||
| Neoascochyta | −0.63 | −0.04 | 1.31 | ||
| Helotiaceae | Articulospora | −0.51 | 0.09 | 0.52 | |
| Phaffomycetaceae | Cyberlindnera | 0.10 | 0.53 | 0.28 | |
| Saccharomycetales Incertae sedis | Candida | 0.12 | 0.50 | 3.82 | |
| Microdochiaceae | Microdochium | −0.45 | 0.09 | 0.64 | |
| Basidiomycota | Holtermanniales Incertae sedis | Holtermanniella | −0.69 | 0.10 | 1.44 |
| Bulleribasidiaceae | Vishniacozyma | −0.64 | 0.42 | 9.71 | |
| Trimorphomycetaceae | Saitozyma | −0.41 | 0.01 | 0.17 |
Definition of abbreviation: PC = principal coordinate.
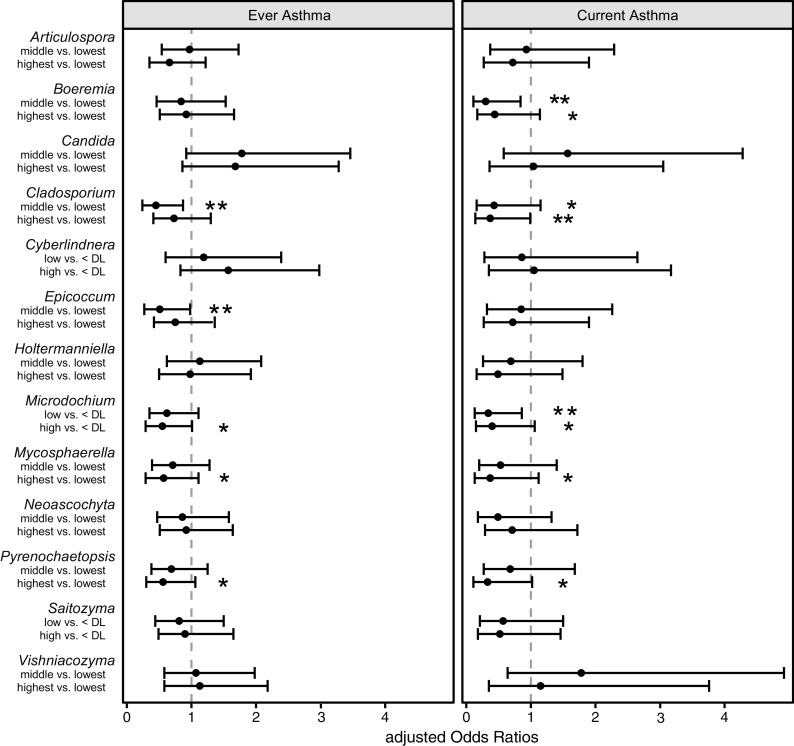
In the next step, we tested the individual associations of these 13 genera with ever asthma (Figure 2 and Table E4). A significant (linear trend test, P < 0.05) or borderline nonsignificant (P < 0.1) protective association was seen between the relative abundances of genera Mycosphaerella, Microdochium, and Pyrenochaetopsis and ever asthma. The associations tended to be dose responsive. Similar findings were detected with asthma type symptoms, including nocturnal cough and persistent wheeze phenotype (Tables E5 and E6). The additionally tested confounding factors did not affect the results among these three fungal taxa and ever asthma (changes in the estimates < 10%, data not shown). We then further adjusted the models individually with different microbial cell wall markers, the amount of dust, and asthma-protective bacterial indices. After restricting the data to those individuals who had all markers available (n = 301, including 51 children with asthma), none of the associations remained significant, and the individual microbial markers explained up to 62% of the associations between fungal taxa and ever asthma (Figure 3). The strongest effects were seen when the models were adjusted for an index representing the sum of 12 protective bacterial genera (6), bacterial richness (Chao1), and FaRMI (the farm-home resembling microbiota index) (5).
Figure 2.
Adjusted associations between relative abundances of 13 fungal genera and the development of asthma using ever asthma and current asthma definitions at the age of 10.5 years. Middle and highest tertiles were compared with lowest tertile, except in those taxa in which more than 1/3 of samples were below the <DL. Models are adjusted for time of follow-up, farm living, cohort, sex, maternal history of allergic diseases, maternal smoking during pregnancy, and the number of older siblings. Whiskers indicate the 95% confidence intervals. *Significance ⩽ 0.10. **Significance < 0.05. DL = detection limit.
Figure 3.
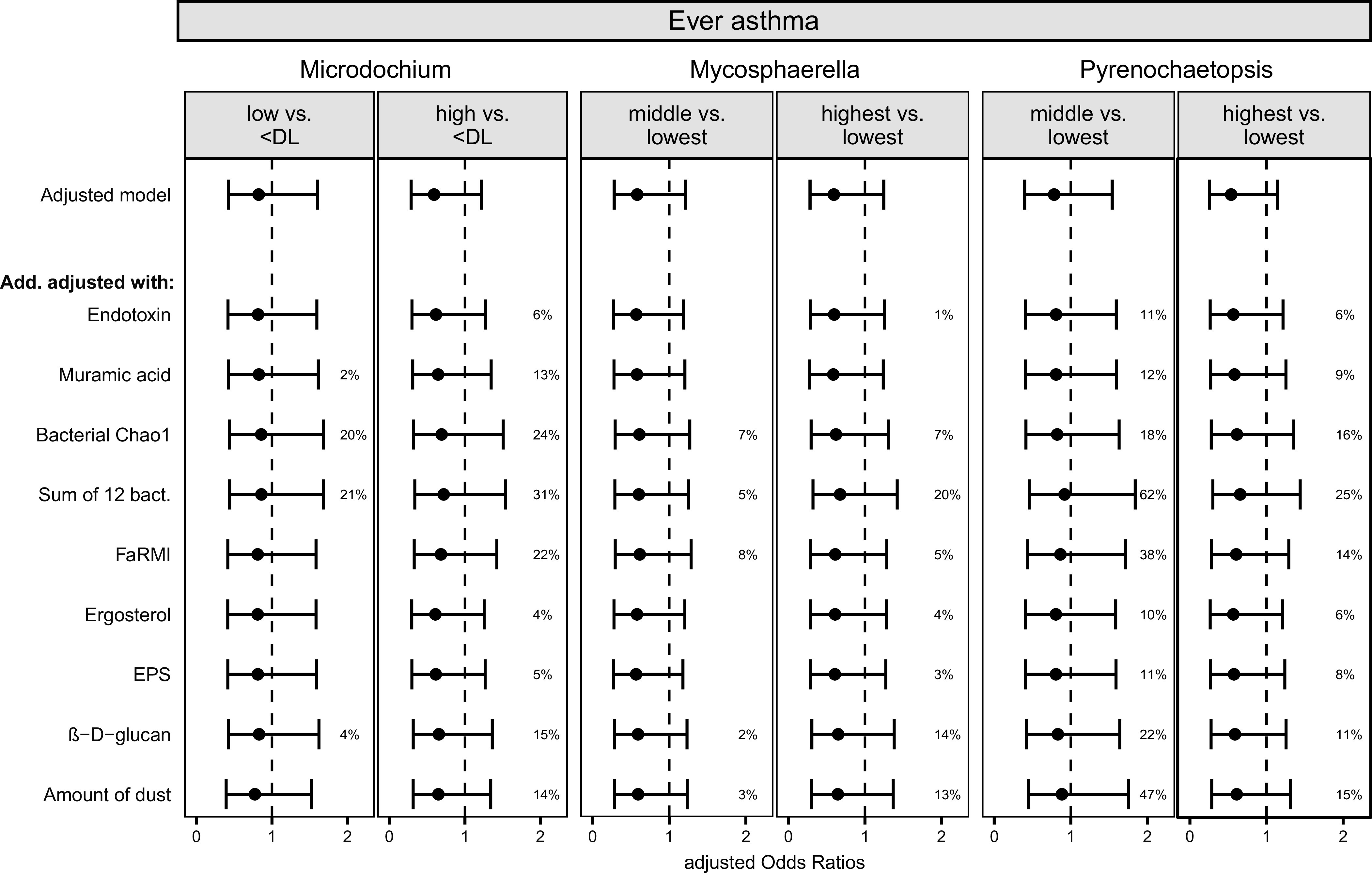
Additionally adjusted associations between relative abundances of three fungal genera and the development of asthma until the age of 10.5 years. The middle and highest tertiles are compared with the lowest tertile, except in Microdochium, in which more than 1/3 of samples were below the <DL. Whiskers indicate the 95% confidence intervals. Adjusted model = models are adjusted for time of follow-up, farm living, cohort, sex, maternal history of allergic diseases, maternal smoking during pregnancy, number of older siblings. Add. adjusted with = adjusted models were also adjusted for one bacterial or fungal microbial marker or amount of dust at a time. % indicates how much the given microbial marker explained the association between fungal taxon and asthma. If % is missing, the microbial marker did not explain the association. Data were restricted to those observations that had no missing information in the additionally adjusted markers (N = 301). Chao1 = richness; DL = detection limit; EPS = the extracellular polysaccharides of Penicillium and Aspergillus spp.; FaRMI = farm home resembling microbiota index.
For the ever asthma outcome, the middle tertile of the relative abundance of genera Epicoccum and Cladosporium showed significant protective associations. Moreover, for current asthma, Boeremia and Cladosporium showed significant protective associations, in addition to the three genera that were also associated with ever asthma (Figure 2). We summed up the relative abundances of three (Mycosphaerella, Pyrenochaetopsis, and Microdochium for ever asthma) or five (adding Boeremia and Cladosporium for current asthma) asthma-associated taxa (linear trend test P values < 0.1); neither of the two sum variables was associated with the given asthma outcome (Table E7).
The abundances of the five taxa associated with the development of asthma correlated significantly among each other (r > 0.3; P < 0.05; Figure E2). When performing mutual adjustments, we observed that the protective associations of these taxa on asthma outcome were not independent of each other (Figure E3).
We performed two additional analyses to 1) overcome the limitation of amplicon sequencing in expressing relative rather than the total abundance of taxa; and 2) improve taxonomic specificity in our results. Because the ITS amplicon sequencing can generate only information on the relative differences of taxa abundance, we additionally transformed the relative abundance of reads with the total fungal load of samples to calculate the absolute taxa abundances. This adjustment strengthened the associations between Mycosphaerella, Microdochium, and Pyrenochaetopsis and asthma (Table E8). However, also here, the associations were mostly weakened when adjusting with other microbial markers (data not shown).
To further increase taxonomic resolution within the identified genera, we analyzed amplicon sequencing variants (ASVs) within the selected genera. We tested a total of 18 individual ASVs within the five selected genera: Mycosphaerella, Pyrenochaetopsis, Microdochium, Boeremia, and Cladosporium. We found protective associations with ever asthma and current asthma (P < 0.10), and these ASVs were taxonomically assigned to Mycosphaerella tassiana, Pyrenochaetopsis leptospora, Pyrenochaetopsis pratorum, and Cladosporium delicatulum (Table E9).
As inhalant atopy is a known risk factor for asthma, we also tested the association between house dust mycobiota and ever asthma for interaction with the atopic status of the study subjects. We observed significant interaction (P < 0.1) between atopic status and three genera (Articulospora, Boeremia, and Microdochium) and fungal load in ever asthma, but not with richness or Shannon diversity (Tables E2 and E4) or with the sum of three fungal genera (Table E7). In atopic children, Boeremia and Microdochium genera were inversely associated with asthma (linear trend test, P < 0.1), and the shape of the association between Articulospora and asthma tended to be inverted-U (Table E10). No significant associations were found in children without atopy. With fungal load, the middle versus lowest tertile was inversely associated with asthma in atopic children.
Fungi and Inhalant Atopy
We found no association between fungal richness (Chao1) and Shannon diversity (Table E2) with inhalant atopy at the age of 10.5 years, similar to the results we reported earlier based on the approach using operational taxonomic units rather than ASVs that we used here (30). Fungal load (measured via qPCR) was positively associated with inhalant atopy (aOR [95% CI]: 1.45 [0.76–2.79] for middle vs. lowest and 2.13 [1.01–4.50] for highest vs. lowest tertile; linear trend test, P = 0.05; Table E2). β-diversity (Jaccard P = 0.54; Bray-Curtis P = 0.88) and five PCoA axes based on Bray-Curtis dissimilarity (P > 0.2; Table E3) were not associated with inhalant atopy, and therefore associations with individual taxa were not tested further.
The Effect of Farming on Fungal House Dust Composition and Associations with Asthma
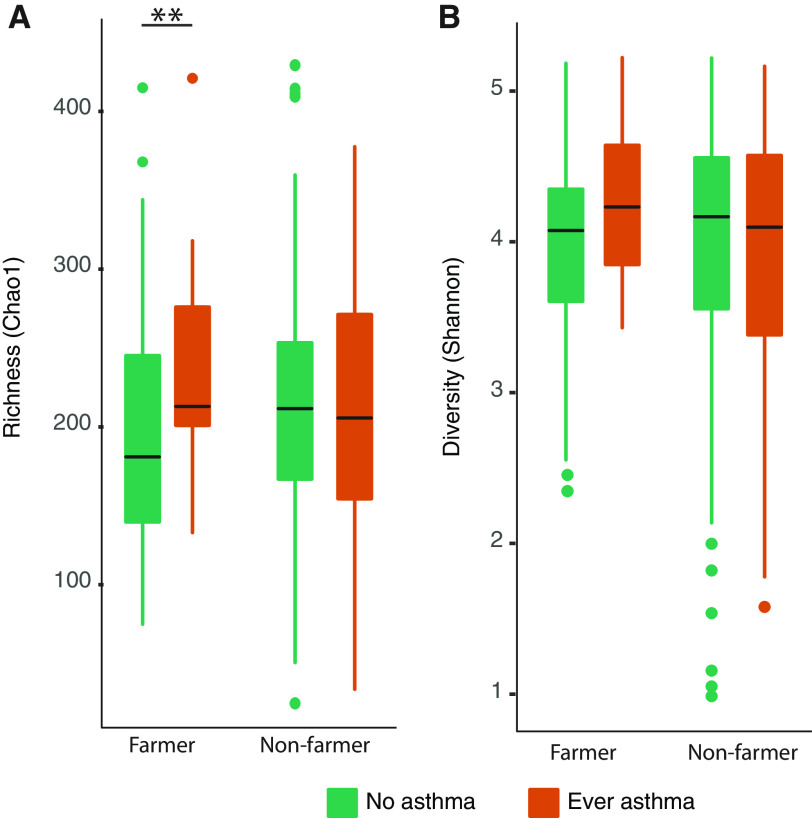
Because of the observed significant interaction between farming status and Chao1 richness on asthma (Table E2), we further explored the effect of farming on house dust mycobiota. The difference in the Bray-Curtis dissimilarity between farm and non-farm homes was significant and explained 3% of mycobiota variation (PERMANOVA-S, P < 0.001). We observed a significant difference in the fungal richness (Mann-Whitney, P = 0.03; Figure 4A), but there was no significant difference in the Shannon diversity (P = 0.2) between farmers and non-farmers (Figure 4B).
Figure 4.
The levels of (A) fungal richness (Chao1), and (B) Shannon diversity index in the house dust from homes of children with and without asthma, stratified by farm living. **Significance < 0.05.
Children living on a farm were less frequently diagnosed with asthma (12.4%) than those who were not living on a farm (21.5%; chi-square test, P = 0.04). Our data suggest that higher fungal richness is a potential risk factor for asthma development in farm but not in non-farm homes (aOR, 2.32 [95% CI, 1.05–5.09] per interquartile range change among farmers, and aOR, 0.96 [95% CI, 0.66–1.40] among non-farmers; P value for interaction term = 0.02). No such interaction was observed with Shannon diversity (P = 0.62; Table E2). The number of current subjects with asthma was too low to perform a similar stratified analysis.
We additionally adjusted the analysis of fungal richness against ever asthma among farmers for moisture damage, bacterial richness, diversity, the sum of 12 previously asthma-associated bacterial genera (6), FaRMI (5), and other microbial markers (26). Our results indicate that fungal cell wall markers, extracellular polysaccharides of Penicillium and Aspergillus spp. (EPS) and β-d-glucan were the only factors, which explained part of the associations (17% and 8%, respectively). Most of the adjusted factors strengthened the effect between fungal richness and asthma (Figure E4).
We also explored the possibility of a difference in house dust mycobiota composition between subjects with asthma and those without asthma within farmers but did not observe significant difference between the groups (PERMANOVA-S, P > 0.05). To understand the interrelations between richness and asthma risk among farmers, we stratified the compositional data according to high richness (>median 207.5) and farm/non-farm status. In this analysis among homes with elevated fungal richness, we observed a significant difference in the mycobiota composition between farmers and non-farmers (PERMANOVA-S, P < 0.001), indicating a general shift in the fungal composition (Figure E5). To further explain the observed difference, we explored differences between farmers with high versus low richness with 35 determinants and observed that newly constructed houses and larger farms tended to have higher richness (Table E11).
Discussion
In this study, we aimed to detail how early-life indoor mycobiota exposure affects the later development of childhood asthma. We used a well-defined Finnish birth cohort and analyzed the mycobiota from 382 floor dust samples. To date, this is the largest study detailing the associations between indoor fungal composition and asthma development using next-generation sequencing methods. Against our hypothesis, we did not identify fungal taxa in early-life house dust that would be predisposing for the development of asthma. On the contrary, all the associations between asthma development were protective, but the significance was lost when adjusting with other microbial markers, pointing toward little relevance of the fungal indoor mycobiota in asthma development.
Fungal species have allergenic and irritating properties, and their presence has been linked to adverse health effects when living in moisture-damaged buildings (13, 15). However, the contribution of fungi and microbes to the well-established association between moisture damage and dampness has not been sufficiently clarified in epidemiological studies (13, 19). It has been indicated that mycobiota exposure in dust does not predispose to asthma development (17). The majority of earlier studies measuring fungal cell wall markers have reported at least a tendency of inverse associations with asthma (16), as also seen in earlier analysis within the current cohort (28). These somewhat conflicting results may be due to insufficient resolution within the earlier measures of indoor fungal exposure, likely consisting of hundreds of different fungal species. This controversy was one of the motivations for our current study, as we hypothesized that different fungal taxa in early-life house dust may show both risk and protective associations with asthma development.
Previous studies using crude measurements of fungal diversity suggested a protective quality of higher fungal diversity in the development of childhood asthma and respiratory symptoms (7, 10), although several of the recent studies using sequencing technology for more refined mycobiota characterization have failed to replicate the initial finding (8, 11, 19). This was also the case in our study in which, against our initial hypothesis, we did not observe associations between fungal richness or diversity and the incidence of asthma. The contrast between the earlier findings and the data created with modern sequencing methods can be explained with differing study protocols and methods of determining diversity, where sequencing methods generate richer taxa profiles, arguably resulting in better estimation of true mycobiota diversity. Ours and others’ results suggest that fungal diversity and richness measures in early-life fungal exposure are not consistently associated with the development of asthma.
In this cohort, the general mycobiota composition differed between those who later developed or did not develop asthma when considering the presence and absence, but not when using the relative abundance, of taxa. We have earlier investigated from the same cohort the bacterial composition in indoor dust using similar measurements. There we found a more striking difference in the overall bacterial composition between subjects with and without asthma when compared with what we observed here with the fungal data (6). In both the bacterial and fungal microbiota studies (6, 11), it was the individual taxa rather than overall compositional characteristics that showed stronger associations with asthma, but the associations were weak for fungi, and they lost significance when adjusted with other microbial markers or asthma-related bacterial taxa.
We identified five fungal genera (Cladosporium, Boeremia, Microdochium, Mycosphaerella, and Pyrenochaetopsis), which were inversely associated with asthma development. Those represent taxonomic groups generally observed in the outdoor environment containing multiple plant pathogenic species. The most abundant of these, Cladosporium, has not been associated with asthma in earlier studies (10, 11, 17, 19, 28, 31). In addition, a recent study is in line with our finding by showing that the homes of subjects with asthma had lower levels of two Mycosphaerella (teleomorphs of Cladosporium) species (32). However, the associations observed in the current study between individual taxa and asthma outcome were lost when adjusting with each other or with other microbial markers (5, 6, 28). This suggests that the protective association between the individual genera and asthma may reflect other protective microbial exposures.
Our present study suggests that associations between fungal taxa and total fungal load in house dust may be different depending on the atopic status of the children. We observed more pronounced protective associations of fungal genera and fungal load on asthma in atopic compared with nonatopic children. This is in contrast to previous cross-sectional studies among atopic children with asthma (33) and in adults (34). However, all three studies identified in principle different genera or species of fungi that were linked to asthma or asthma severity, making comparisons challenging. Our indicative findings should motivate targeted follow-up investigation.
Reflecting on our previous work (6), the compositional qualities of house dust mycobiota do not seem to match the bacterial microbiota with respect to their asthma-preventive potential. It is well understood that taxonomic profiling with amplicon sequencing cannot differentiate metabolically active from nonactive or dead fungal cells in house dust (35). Recent work on moisture damage has suggested considerable differences in the conclusions of health effects that would be drawn from compositional studies of dust mycobiota versus functional profiling from meta-transcriptome data (36). This study has pointed out that assessments not taking into account the elevated health impacts suggested by functional profiling might not accurately reflect exposure, which could lead to false conclusions. The nature of our current study does not allow us to further substantiate such a suggestion. However, our results first suggest that the early-life house dust mycobiota or even metabolites (37) in Finnish homes do not have a strong predisposing or protective quality in asthma development. Second, we hypothesize that approaches including functional rather than compositional profiling are more likely to identify novel and potentially stronger associations between the indoor mycobiome and asthma.
We observed a dose–response risk association between total fungal load in house dust and inhalant atopy at the age of 10.5 years. In contrast to our finding, other studies have reported mostly an inverse association (38, 39) or no association (12, 40, 41) when they used other fungal markers such as ergosterol, β-d-glucan, or extracellular polysaccharides to measure fungal exposure. In our study, we did not find association between the mycobiota composition or fungal richness or diversity in house dust and inhalant atopy. The diversity finding confirms our earlier analysis (30) and is also in line with observations from another birth cohort with results on atopic status at 7 and 12 years (11). Because of the absence of compositional differences in β-diversity metrics and our analysis strategy that focused on guarding against multiple testing, we did not explore associations between individual fungal taxa and inhalant atopy in the present study.
Interestingly, we observed a predisposing association between fungal richness and ever asthma in farming, but not in non-farming, homes. This is a novel finding and has not been observed in any previous study. The observed difference was not explained by other fungal or cell wall and bacterial exposures or by moisture damage in the child’s main living areas. We observed that the mycobiota composition contributing to high richness was different between farm and non-farm homes. The measured determinants did not explain the observed variation in richness. For example, moisture damage, a potential candidate to explain our observation, was not the cause for the increased richness. The numbers in this study were too low to conclude on the actual taxa causing the observed predisposing factor, but this warrants further study to understand the potential risk factors with high richness in farm environment and asthma development.
Strengths and Limitations
The main strengths of this study include the prospective birth cohort with an extensive set of microbial exposure measurements and deep sequencing of the fungal ITS region. Another strength was that we collected our study samples from living room floors when the participants were 2 months of age, which has been shown to be when intensive maturation of the adaptive immunity occurs. Although only some of the dust on the floors is contributing to inhalation exposure of the children (25), floor dust can be considered a surrogate of total infant exposure, including also oral and dermal exposure routes. The main limitation of the study is the small number of children with current asthma, which is why our results need to be confirmed in further studies.
Conclusions
We did not identify any early-life fungal taxa that would be predisposing to asthma. Our data suggest that the composition and the abundance of specific common environmental genera in the early-life indoor mycobiota may have a small protective effect on the development of asthma, but that effect was not independent from other asthma-protective microbial exposures identified from that same cohort. We conclude that the early-life house dust mycobiota in Finnish homes neither strongly nor independently contributes to the development of asthma, with the exception of the identified risk association of higher fungal richness in the farming subgroup. Our results, taken together with previous observations, point toward a limited potential for breakthrough findings on studies purely focusing on compositional aspects of fungal exposures early in life. Widening the scope of such studies toward including functional profiling of the indoor mycobiome might be recommendable.
Acknowledgments
Acknowledgment
The authors thank the families for their participation in the study; the study nurses Raija Juntunen, Riikka Juola, Anneli Paloranta, and Seija Antikainen for field work; Environmental microbiology lab personnel at Finnish Institite for health and welfare (THL); Jutta Kauttio for sample processing and DNA extraction; and Asko Vepsäläinen, Pekka Tiittanen, and Timo Kauppila for data management.
Footnotes
Supported by Academy of Finland grants 139021, 287675, 296587, 296814, 296817, 308254, 339666, and 349427; the Juho Vainio Foundation; the Yrjö Jahnssonin Foundation; the Foundation for Pediatric Research; the Research Committee of the Kuopio University Hospital Catchment Area for the State Research Funding project 5041730, Kuopio, Finland; the Päivikki and Sakari Sohlberg Foundation; the Farmers’ Social Insurance Institution of Finland; The Finnish Cultural Foundation; Kuopio Area Respiratory Foundation; European Commission grant QLK4-CT-2001-00250; Commission of the European Communities under the Seventh Framework Programme grant KBBE-2007-2-2-06; and the Finnish Institute for Health and Welfare, Finland. C.S. is supported by the Universities Giessen and Marburg Lung Center, the German Center for Lung Research (DZL), University Hospital Giessen and Marburg (UKGM) research funding according to article 2, section 3 cooperation agreement, and Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)-SFB 1021 project number 197785619, KFO 309 project number 284237345, and SK 317/1-1 project number 428518790, as well as by the Foundation for Pathobiochemistry and Molecular Diagnostics.
Author Contributions: M.T.: sample processing and molecular analysis, sequencing assessment, and manuscript preparation and revisions. J.J.: data analysis, manuscript preparation, and drafting. P.V.K.: sequencing assessment and manuscript preparation. P.T.: data handling and preprocessing and manuscript preparation. A.H.: dust sampling assessment and manuscript preparation. C.S.: specific IgE measurements and manuscript preparation. E.P.-S.: authority on pediatric issues and manuscript preparation. J.P.: study concept and design and manuscript preparation. A.M.K.: data collecting, statistical data analyses, manuscript preparation, and drafting. All authors critically reviewed the final draft of the paper.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. von Mutius E. The microbial environment and its influence on asthma prevention in early life. J Allergy Clin Immunol . 2016;137:680–689. doi: 10.1016/j.jaci.2015.12.1301. [DOI] [PubMed] [Google Scholar]
- 2. von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol . 2010;10:861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 3. Deckers J, Marsland BJ, von Mutius E. Protection against allergies: microbes, immunity, and the farming effect. Eur J Immunol . 2021;51:2387–2398. doi: 10.1002/eji.202048938. [DOI] [PubMed] [Google Scholar]
- 4. Stein MM, Hrusch CL, Gozdz J, Igartua C, Pivniouk V, Murray SE, et al. Innate immunity and asthma risk in Amish and Hutterite farm children. N Engl J Med . 2016;375:411–421. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kirjavainen PV, Karvonen AM, Adams RI, Täubel M, Roponen M, Tuoresmäki P, et al. Farm-like indoor microbiota in non-farm homes protects children from asthma development. Nat Med . 2019;25:1089–1095. doi: 10.1038/s41591-019-0469-4. [DOI] [PubMed] [Google Scholar]
- 6. Karvonen AM, Kirjavainen PV, Täubel M, Jayaprakash B, Adams RI, Sordillo JE, et al. Indoor bacterial microbiota and development of asthma by 10.5 years of age. J Allergy Clin Immunol . 2019;144:1402–1410. doi: 10.1016/j.jaci.2019.07.035. [DOI] [PubMed] [Google Scholar]
- 7. Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrländer C, et al. GABRIELA Transregio 22 Study Group Exposure to environmental microorganisms and childhood asthma. N Engl J Med . 2011;364:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 8. Dannemiller KC, Mendell MJ, Macher JM, Kumagai K, Bradman A, Holland N, et al. Next-generation DNA sequencing reveals that low fungal diversity in house dust is associated with childhood asthma development. Indoor Air . 2014;24:236–247. doi: 10.1111/ina.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Birzele LT, Depner M, Ege MJ, Engel M, Kublik S, Bernau C, et al. Environmental and mucosal microbiota and their role in childhood asthma. Allergy . 2017;72:109–119. doi: 10.1111/all.13002. [DOI] [PubMed] [Google Scholar]
- 10. Tischer C, Weikl F, Probst AJ, Standl M, Heinrich J, Pritsch K. Urban dust microbiome: impact on later atopy and wheezing. Environ Health Perspect . 2016;124:1919–1923. doi: 10.1289/EHP158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cox J, Stone T, Ryan P, Burkle J, Jandarov R, Mendell MJ, et al. Residential bacteria and fungi identified by high-throughput sequencing and childhood respiratory health. Environ Res . 2022;204:112377. doi: 10.1016/j.envres.2021.112377. [DOI] [PubMed] [Google Scholar]
- 12. Ege MJ, Frei R, Bieli C, Schram-Bijkerk D, Waser M, Benz MR, et al. PARSIFAL Study team Not all farming environments protect against the development of asthma and wheeze in children. J Allergy Clin Immunol . 2007;119:1140–1147. doi: 10.1016/j.jaci.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 13. Mendell MJ, Mirer AG, Cheung K, Tong M, Douwes J. Respiratory and allergic health effects of dampness, mold, and dampness-related agents: a review of the epidemiologic evidence. Environ Health Perspect . 2011;119:748–756. doi: 10.1289/ehp.1002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caillaud D, Leynaert B, Keirsbulck M, Nadif R, Mould ANSES Working Group Indoor mould exposure, asthma and rhinitis: findings from systematic reviews and recent longitudinal studies. Eur Respir Rev . 2018;27:170137. doi: 10.1183/16000617.0137-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization. WHO guidelines for indoor air quality: dampness and mold. Copenhagen: World Health Organization; 2009. [PubMed] [Google Scholar]
- 16. Tischer C, Chen CM, Heinrich J. Association between domestic mould and mould components, and asthma and allergy in children: a systematic review. Eur Respir J . 2011;38:812–824. doi: 10.1183/09031936.00184010. [DOI] [PubMed] [Google Scholar]
- 17. Behbod B, Sordillo JE, Hoffman EB, Datta S, Webb TE, Kwan DL, et al. Asthma and allergy development: contrasting influences of yeasts and other fungal exposures. Clin Exp Allergy . 2015;45:154–163. doi: 10.1111/cea.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Valkonen M, Täubel M, Pekkanen J, Tischer C, Rintala H, Zock JP, et al. Microbial characteristics in homes of asthmatic and non-asthmatic adults in the ECRHS cohort. Indoor Air . 2018;28:16–27. doi: 10.1111/ina.12427. [DOI] [PubMed] [Google Scholar]
- 19. Adams RI, Leppänen H, Karvonen AM, Jacobs J, Borràs-Santos A, Valkonen M, et al. Microbial exposures in moisture-damaged schools and associations with respiratory symptoms in students: a multi-country environmental exposure study. Indoor Air . 2021;31:1952–1966. doi: 10.1111/ina.12865. [DOI] [PubMed] [Google Scholar]
- 20. Karvonen AM, Hyvärinen A, Roponen M, Hoffmann M, Korppi M, Remes S, et al. Confirmed moisture damage at home, respiratory symptoms and atopy in early life: a birth-cohort study. Pediatrics . 2009;124:e329–e338. doi: 10.1542/peds.2008-1590. [DOI] [PubMed] [Google Scholar]
- 21. Herzum I, Blümer N, Kersten W, Renz H. Diagnostic and analytical performance of a screening panel for allergy. Clin Chem Lab Med . 2005;43:963–966. doi: 10.1515/CCLM.2005.165. [DOI] [PubMed] [Google Scholar]
- 22. Wopereis H, Oozeer R, Knipping K, Belzer C, Knol J. The first thousand days. Intestinal microbiology of early life: establishing a symbiosis. Pediatr Allergy Immunol . 2014;25:428–438. doi: 10.1111/pai.12232. [DOI] [PubMed] [Google Scholar]
- 23. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods . 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nilsson RH, Larsson KH, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res . 2019;47:D259–D264. doi: 10.1093/nar/gky1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hyytiäinen HK, Jayaprakash B, Kirjavainen PV, Saari SE, Holopainen R, Keskinen J, et al. Crawling-induced floor dust resuspension affects the microbiota of the infant breathing zone. Microbiome . 2018;6:25–28. doi: 10.1186/s40168-018-0405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tang ZZ, Chen G, Alekseyenko AV. PERMANOVA-S: association test for microbial community composition that accommodates confounders and multiple distances. Bioinformatics . 2016;32:2618–2625. doi: 10.1093/bioinformatics/btw311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karvonen AM, Hyvärinen A, Korppi M, Haverinen-Shaughnessy U, Renz H, Pfefferle PI, et al. Moisture damage and asthma: a birth cohort study. Pediatrics . 2015;135:e598–e606. doi: 10.1542/peds.2014-1239. [DOI] [PubMed] [Google Scholar]
- 28. Karvonen AM, Hyvärinen A, Rintala H, Korppi M, Täubel M, Doekes G, et al. Quantity and diversity of environmental microbial exposure and development of asthma: a birth cohort study. Allergy . 2014;69:1092–1101. doi: 10.1111/all.12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karvonen AM, Hyvärinen A, Gehring U, Korppi M, Doekes G, Riedler J, et al. PASTURE Study Group Exposure to microbial agents in house dust and wheezing, atopic dermatitis and atopic sensitization in early childhood: a birth cohort study in rural areas. Clin Exp Allergy . 2012;42:1246–1256. doi: 10.1111/j.1365-2222.2012.04002.x. [DOI] [PubMed] [Google Scholar]
- 30. Hyytiäinen H, Kirjavainen PV, Täubel M, Tuoresmäki P, Casas L, Heinrich J, et al. Microbial diversity in homes and the risk of allergic rhinitis and inhalant atopy in two European birth cohorts. Environ Res . 2021;196:110835. doi: 10.1016/j.envres.2021.110835. [DOI] [PubMed] [Google Scholar]
- 31. Sharpe RA, Bearman N, Thornton CR, Husk K, Osborne NJ. Indoor fungal diversity and asthma: a meta-analysis and systematic review of risk factors. J Allergy Clin Immunol . 2015;135:110–122. doi: 10.1016/j.jaci.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 32. Wardlaw AJ, Rick EM, Pur Ozyigit L, Scadding A, Gaillard EA, Pashley CH. New perspectives in the diagnosis and management of allergic fungal airway disease. J Asthma Allergy . 2021;14:557–573. doi: 10.2147/JAA.S251709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dannemiller KC, Gent JF, Leaderer BP, Peccia J. Indoor microbial communities: influence on asthma severity in atopic and nonatopic children. J Allergy Clin Immunol . 2016;138:76–83.e1. doi: 10.1016/j.jaci.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Juel Holst G, Pørneki A, Lindgreen J, Thuesen B, Bønløkke J, Hyvärinen A, et al. Household dampness and microbial exposure related to allergy and respiratory health in Danish adults. Eur Clin Respir J . 2020;7:1706235. doi: 10.1080/20018525.2019.1706235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Emerson JB, Adams RI, Román CMB, Brooks B, Coil DA, Dahlhausen K, et al. Schrödinger’s microbes: tools for distinguishing the living from the dead in microbial ecosystems. Microbiome . 2017;5:86. doi: 10.1186/s40168-017-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hegarty B, Dannemiller KC, Peccia J. Gene expression of indoor fungal communities under damp building conditions: implications for human health. Indoor Air . 2018;28:548–558. doi: 10.1111/ina.12459. [DOI] [PubMed] [Google Scholar]
- 37. Kirjavainen PV, Täubel M, Karvonen AM, Sulyok M, Tiittanen P, Krska R, et al. Microbial secondary metabolites in homes in association with moisture damage and asthma. Indoor Air . 2016;26:448–456. doi: 10.1111/ina.12213. [DOI] [PubMed] [Google Scholar]
- 38. Douwes J, van Strien R, Doekes G, Smit J, Kerkhof M, Gerritsen J, et al. Does early indoor microbial exposure reduce the risk of asthma? The Prevention and Incidence of Asthma and Mite Allergy birth cohort study. J Allergy Clin Immunol . 2006;117:1067–1073. doi: 10.1016/j.jaci.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 39. Gehring U, Heinrich J, Hoek G, Giovannangelo M, Nordling E, Bellander T, et al. Bacteria and mould components in house dust and children’s allergic sensitisation. Eur Respir J . 2007;29:1144–1153. doi: 10.1183/09031936.00118806. [DOI] [PubMed] [Google Scholar]
- 40. Bertelsen RJ, Carlsen KCL, Carlsen KH, Granum B, Doekes G, Håland G, et al. Childhood asthma and early life exposure to indoor allergens, endotoxin and beta(1,3)-glucans. Clin Exp Allergy . 2010;40:307–316. doi: 10.1111/j.1365-2222.2009.03424.x. [DOI] [PubMed] [Google Scholar]
- 41. Hsu NY, Lee CC, Wang JY, Li YC, Chang HW, Chen CY, et al. Predicted risk of childhood allergy, asthma, and reported symptoms using measured phthalate exposure in dust and urine. Indoor Air . 2012;22:186–199. doi: 10.1111/j.1600-0668.2011.00753.x. [DOI] [PubMed] [Google Scholar]