Abstract
Objective
This research was aimed to preliminarily explore the clinical roles and potential molecular mechanisms of MIR99AHG and its significant transcripts in breast cancer (BRCA).
Methods
Public databases were utilized to analyze the expression and prognostic roles of MIR99AHG and its transcripts. Relationships between MIR99AHG expression and immune cells infiltration were analyzed in Xiantao platform. In addition, co-expressed genes and interacting proteins of MIR99AHG were predicted. CancerSEA analyzed its relationship with functional states. Next, CNV status, DNA methylation, interacting transcription factors (TFs) and ceRNA network were analyzed to explore its possible mechanisms. Then, RNA ISH and FISH assays were used to detect its expression and location in BRCA tissues and cell lines, respectively. Finally, qRT-PCR was utilized to investigate MIR99AHG expression in cell lines.
Results
Compared with the corresponding normal tissues, MIR99AHG expression levels were lower in all BRCA subtypes, and luminal B's was the lowest one. And MIR99AHG expression was negatively related to the tumor stage. In addition, 4 transcripts (ENST00000619222.4, ENST00000418813.6, ENST00000602901.5 and ENST00000453910.5) of MIR99AHG showed significant differences in the expression. Databases also suggested that the high MIR99AHG expression levels indicated good prognosis, especially in patients without lymph node metastasis. Xiantao found that MIR99AHG was positively related to 17 immune cells and negatively linked with 2 immune cells. CancerSEA analysis showed no relationships between MIR99AHG and functional states. From GEPIA and BCIP databases, 19 co-expressed genes were highly related to these four significant transcripts of MIR99AHG. StarBase, RNAct and HDOCK showed that several tumor-associated proteins, including U2AF65, hnRNPC, AEBP2, CHIC1 and so on, might interact with MIR99AHG. Genetically, BRCA had a higher proportion of MIR99AHG CNV loss than CNV gain, and the high level of DNA methylation indicated a good prognosis. Furthermore, 19 TFs were predicted to combine with the promoter of MIR99AHG. Then, we screened out 10 miRNAs potentially interacting with the significant transcripts of MIR99AHG, and five were significantly increased in breast tumors compared to normal tissues, including miR-194–5p, miR-320 b and so on, which could combine 14 mRNAs. Through ISH and FISH assays, we verified that MIR99AHG was down-regulated in BRCA samples and cell lines in comparison to non-tumor tissues and mammary epithelial cell line (MCF10A), and MIR99AHG was located both in cytoplasm and nucleus. qRT-PCR assay also showed the lower expression of MIR99AHG in breast cancer cells than that in MCF10A.
Conclusion
These results indicate that MIR99AHG can be a favorable prognostic indicator for BRCA. ENST00000619222.4, ENST00000418813.6, ENST00000602901.5 and ENST00000453910.5 are significant transcripts and their down-regulation may play crucial roles in the progression of BRCA.
Keywords: MIR99AHG, BRCA, Prognosis, Molecular mechanism, ceRNA, Transcription factor
1. Introduction
Breast cancer (BRCA) has become the most common cancer and a major public health problem worldwide [1]. Despite the adverse impact of the coronavirus disease 2019 (COVID-19) pandemic on cancer diagnosis, the number of new cases diagnosed with BRCA is still appallingly high and BRCA is one of the leading causes of cancer-associated deaths in women [2,3]. One of the main reasons is that the molecular pathogenesis of BRCA remains to be unravelled [4]. Therefore, it is critical to explore novel potential biomarkers for BRCA detection and management.
Due to the lack of open reading frames, non-coding RNAs (ncRNAs) are encoded but usually not translated into proteins [5]. Long non‐coding RNAs (lncRNAs), which are over 200 nucleotides in length, play critical functions in the modulation of gene expression, ubiquitination and cellular signal transduction [6,7]. Recently, more and more lncRNAs have been proved to be involved in the process of tumorigenesis, including proliferation, apoptosis, migration, invasion, epithelial-mesenchymal transition, stemness and so on [8,9]. Therefore, lncRNAs are considered as potent biomarkers and therapeutic targets for carcinomas. In breast carcinoma, the abnormal expression of lncRNAs not only promotes tumorigenesis, but also induces drug resistance [10,11].
MicroRNA-99a host gene (MIR99AHG), also known as LINC00478, has been proved to be a non-coding tumor suppressor gene in lung cancer [12]. It is located in chromosome 21q21.1 and encodes lncRNA MIR99AHG as well as the miR-99a/let-7c/miR-125b2 cluster [13]. Previous studies have reported that the expression levels of miR-99a, let-7c and miR-125b2 are down-regulated in breast tumors in comparison with normal tissues, and they may function as tumor suppressors and stand as diagnostic biomarkers in BRCA [13,14]. Another recent study has shown that MIR99AHG is negatively related to BRCA lymph node metastasis and may serve as a candidate biomarker for BRCA diagnosis [15]. Therefore, it is necessary to clarify the functions of lncRNA MIR99AHG and the mechanism of down-regulated expression of MIR99AHG gene in BRCA.
Interestingly, it is reported that the increased expression of MIR99AHG may also promote biological malignant behavior in several other tumor types [16,17]. Given that one lncRNA often has multiple transcripts, different transcripts may have diverse biological functions and play different roles in the diagnosis and treatment of diseases [18]. Therefore, based on bioinformatics, this study aims to initially identify the clinical roles of MIR99AHG in BRCA patients, and to explore potential significant transcripts of MIR99AHG and possible molecular mechanisms in BRCA through public databases.
2. Materials and methods
2.1. Database analysis of MIR99AHG expression in BRCA
Three public databases, Gene Expression Profiling Interactive Analysis 2 (GEPIA2, https://gepia2.cancer-pku.cn/), Breast Cancer Integrative Platform (BCIP, http://www.omicsnet.org/bcancer/database) and lncRNAs from cancer arrays (lnCAR, https://lncar.renlab.org/explorer) were utilized to analyze expression levels of MIR99AHG in breast normal tissues, breast tumors, and BRCA sub-types [[19], [20], [21]]. In addition, the levels of different transcripts of MIR99AHG were analyzed in GEPIA.
2.2. Prognostic and clinicopathological analyses in the databases
Prognostic roles of MIR99AHG were estimated via PrognoScan (http://dna00.bio.kyutech.ac.jp/PrognoScan/index.html), GEPIA, BCIP and Kaplan-Meier Plotter (https://kmplot.com/analysis/) databases [22,23]. The primary outcomes were overall survival (OS), recurrence/relapse free survival (RFS), distant metastasis free survival (DMFS), disease-specific survival (DS), post-progression survival (PPS) and disease-free survival (DFS). GEPIA was also utilized for survival and clinicopathological analyses of different MIR99AHG transcripts in BRCA.
2.3. CancerSEA analysis
CancerSEA (https://biocc.hrbmu.edu.cn/) database was used to analyze the relationships between MIR99AHG and 13 types of functional states, including angiogenesis, apoptosis, cell cycle, differentiation, DNA damage, DNA repair, EMT, hypoxia, inflammation, invasion, metastasis, proliferation, quiescence and stemness [24].
2.4. Xiantao analysis
Xiantao platform (https://www.xiantao.love/) was conducted to analyze possible clinicopathological roles of MIR99AHG in TNM (Tumor Node Metastasis) stages of 1109 BRCA samples. Besides, relationships between MIR99AHG expression and immune cells infiltration were analyzed by Spearman analysis to explore its potential roles in tumor immunity.
2.5. Co-expression analysis
We conducted GEPIA and BCIP databases, respectively, to explore top 100 co-expressed genes of MIR99AHG or its significant transcripts (ENST00000619222.4, ENST00000418813.6, ENST00000602901.5 and ENST00000453910.5). In GEPIA, we selected genes that were contained in at least three transcripts. According to these two databases, we screened out highly correlated genes.
2.6. Prediction of proteins interacting with MIR99AHG
StarBase (https://starbase.sysu.edu.cn/starbase2/index.php) was utilized to explore RNA-binding proteins (RBPs) which possibly interacted with MIR99AHG [25]. RNAct (https://rnact.crg.eu/) database could predict RBPs interacting with different MIR99AHG transcripts and we selected the top 100 ones of each transcript [26]. Then, we screened out candidate RBPs that were contained in all transcripts. According to the results of the databases above, we chose ENST00000418813 as a representative transcript, and HDOCK server was used to analyze RBP-ENST00000418813 docking [27].
2.7. Copy number variant (CNV) analysis
CNV gain and loss were two different statuses. We performed BCIP database to analyze CNV status of MIR99AHG in BRCA and compare the survival rates of different statuses, including OS and DS.
2.8. DNA methylation analysis
Epigenome-Wide Association Study (EWAS) Data Hub database (https://ngdc.cncb.ac.cn/ewas/datahub/index) was utilized to analyze the levels of MIR99AHG body DNA methylation and its prognostic role in BRCA and gynecological neoplasms, including endometrial cancer and cervical cancer [28].
2.9. Prediction of transcription factors (TFs) interacting with MIR99AHG promoter
According to MIR99AHG promoter sequence, we conducted PROMO database (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) to predict TFs and TF binding sites of MIR99AHG promoter [29]. Then, we compare the relationship between MIR99AHG and TFs through GEPIA.
2.10. Construction of MIR99AHG-miRNA-mRNA network
LncACTdb3 (http://www.bio-bigdata.net/LncACTdb/index.html) was used to predict the location of lncRNA MIR99AHG in cells [30]. Then, we used two databases, DIANA-LncBase v3 (https://diana.e-ce.uth.gr/lncbasev3) and lncRNASNP2 (http://bioinfo.life.hust.edu.cn/lncRNASNP#!/) to predict candidate miRNAs that could potentially combine with lncRNA MIR99AHG and to analyze possible binding sites of these miRNAs and significant MIR99AHG transcripts [31,32]. Among them, CancerMIRNome (http://bioinfo.jialab-ucr.org/CancerMIRNome/#tab-1929-4) was applied to identify differential miRNAs between normal tissues and breast tumors [33]. According to LncACTdb3 and miRDB (http://mirdb.org/), we screened out candidate mRNAs that could combine with lncRNA MIR99AHG-miRNAs in the competing endogenous RNA (ceRNA) network [34].
2.11. Clinical specimens and RNA in situ hybridization (ISH) assay
10 cases of breast tumors and non-tumor breast tissues were obtained from Kunshan First People's Hospital. This research was in accordance with the Kunshan First People's Hospital Ethics Committee and every patient signed the informed consent form. The ISH probe of MIR99AHG (QD646) was synthesized by BersinBio Technologies Co., Ltd. Briefly, after the course of dewaxing, rehydration, inactivation of endogenous enzymes and digestion, the sections were dealt with the prehybridization solution (AR0152, Boster). Mixed liquor of the probe and hybridization solution was then dropped onto the sections at 75 °C for 10min, and these sections were immediately transferred to 42 °C overnight. Following washing and sealing, sections were treated with biotinylated digoxin (AR0147, Boster), SABC (SA1020, Boster) and biotinylated peroxidase (AR0149, Boster) in turn. Finally, slides were stained with AEC (A2010, Solarbio) and counterstained with hematoxylin (G1004, Servicebio) for 10s. We evaluated MIR99AHG expression in these samples by using a semi-quantitative assessment system. This system was combined by a score of the percentage of tumor cells (P; 0, no positive tumor cells; 1, 1%–25% positive tumor cells; 2, 26%–50% positive tumor cells; 3, 51%–75% positive tumor cells; and 4, 76%–100% positive tumor cells), and a score of the staining intensity (SI; 0, no staining of the tumor cells; 1, weak staining; 2, moderate staining; and 3, strong staining). The formula of the staining score (SS) was “SS P*SI”, and the total score ranged from 0 to 12.
2.12. Cell lines and cell culture
The human mammary epithelial cell line (MCF10A) and the human breast cancer cell lines (MCF-7, ER+; SK-BR3, HER2+; MDA-MB-231 and SUM1315, ER-PR-HER2-) were stored in Central Laboratory, Kunshan First People's Hospital. MCF10A, MCF-7, SK-BR3 and MDA-MB-231 were derived from American Type Culture Collection, and SUM1315 was kindly provided by Xiaoan Liu (The First Affiliated Hospital of Nanjing Medical University, China). MCF10A were cultured in MCF10A cell specific medium (Procell, China). MDA-MB-231, SUM1315 and SK-BR3 were cultured in Dulbecco's modified Eagle's medium (DMEM; GIBCO Life Technologies, USA). And MCF-7 was cultured in minimum Eagle's medium (MEM; GIBCO Life Technologies, USA). All cell lines were cultured at 37 °C in a humidified atmosphere containing 5% CO2.
2.13. RNA fluorescence in situ hybridization (FISH) assay
0.5 × 105 cells per plate were seeded in 12-well plates with cell climbing sheets and cultured overnight at 37 °C in a 5% CO2 incubator. After washing, cells were fixed in 4% paraformaldehyde. RNA FISH Kit (GenePharma, China) was then used with the specific probe mixed liquor. The sequence of the probe was listed in Supplement file 1, which was designed and produced by GenePharma. Then, DAPI working solution was used for staining in the dark. After washing, a clean slide was treated with the anti-quench agent and covered the cell side of the sheet, and then the slice was observed under the fluorescence microscope.
2.14. Quantitative real-time PCR polymerase chain reaction (qRT-PCR) assay
Cells were collected to isolate total RNA through Ezol reagent (GenePharma, China). Then, RNA was reverse transcribed using the reverse transcription reaction system (GenePharma, China). Amplified DNA was measured by the universal reagent for real-time fluorescence quantification (GenePharma, China) and qPCR was performed using the ABI stepone plus real-time fluorescence quantitative PCR instrument (Life, USA). Then, we used 2−ΔΔCt value to calculate the relative RNA expression. cDNA was subjected to quantitative PCR using primers specific for MIR99AHG and GAPDH. Notably, the primer of MIR99AHG contained 16 main transcript variants in the NCBI database. The PCR primers were listed as follows:
MIR99AHG, Forward primer, 5′-ACAAGAGCACCTCAAAGGCA-3′, Reverse primer, 5′-AAGTTTTCTGCATCCGTGCG-3’; GAPDH, Forward primer, 5′-CATGAGAAGTATGACAACAGCCT-3′, Reverse primer, 5′-AGTCCTTCCACGATACCAAAGT-3’.
2.15. Statistical analysis
Difference of expression levels of RNAs between breast tumors or cancer cells and normal tissues or MCF10A was analyzed by t-test. Differences of MIR99AHG and its transcript expression among subtypes and stages were analyzed by ANOVA with LSD-t test. The database lnCAR used the aggregation rank score (AR score) to represent the integrated rank from the meta-analysis of fold change from different microarray studies, in which a larger AR score indicated an up-regulated lncRNA in most studies, whereas a down-regulated lncRNA. The HR and P value were calculated by univariate Cox regression model in PrognoScan, and by log-rank test in GEPIA and Kaplan-Meier Plotter. Similarity between genes is represented by Pearson correlation coefficient and BCIP used the adjusted P-value. Relationships between MIR99AHG expression and functional states were also analyzed by the Pearson correlation method. The lncRNA-miRNA-mRNA ceRNA network was drawn through Gephi. GraphPad Prism 6.0 was used to analyze the data in this study and P < 0.05 was set as a statistically significant threshold.
3. Results
3.1. Down-regulation of MIR99AHG in BRCA
In public databases, MIR99AHG was down-regulated in various carcinomas, including BRCA, CESC (cervical squamous cell carcinoma and endocervical adenocarcinoma), LUAD (lung adenocarcinoma), OV (ovarian serous cystadenocarcinoma), and so on (Fig. 1A). Compared to normal breast tissues, its expression was lower not only in breast tumors but also in all BRCA subtypes (Fig. 1B–H). In addition, luminal B BRCA had the lowest expression level of MIR99AHG (Fig. 1E, G and 1H).
Fig. 1.
Expression of MIR99AHG in BRCA and its sub-types in databases. (A) Expression of MIR99AHG in multiple cancers in GEPIA database; (B) Comparison of MIR99AHG expression between BRCA and normal breast tissues in GEPIA; (C) Comparison of MIR99AHG expression between BRCA and normal breast tissues in BCIP database; (D) Comparison of MIR99AHG expression between BRCA subtypes and normal breast tissues in GEPIA; (E) Comparison of MIR99AHG expression among BRCA subtypes in GEPIA; (F) Comparison of MIR99AHG expression between BRCA subtypes and normal breast tissues in BCIP; (G) Comparison of MIR99AHG expression among BRCA subtypes in BCIP; (H) Comparison of MIR99AHG expression between BRCA subtypes and normal breast tissues in lnCAR database. *P < 0.05, **P < 0.01, ***P < 0.001.
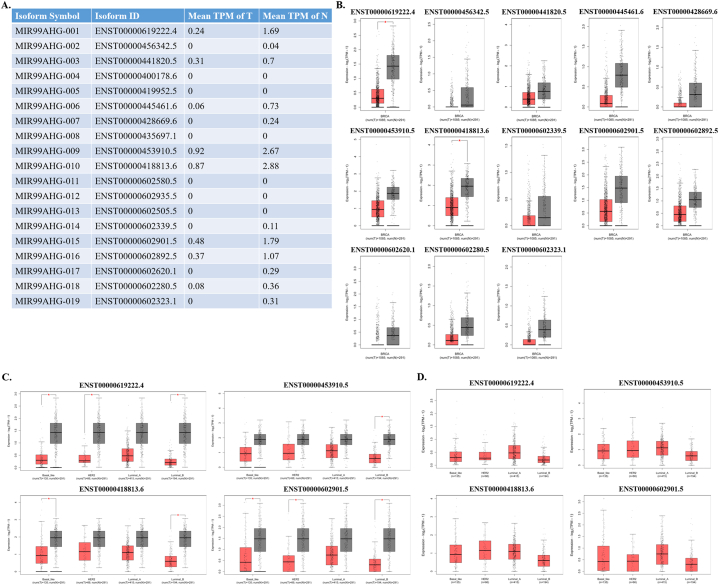
3.2. Differences of MIR99AHG transcripts between BRCA and normal tissues
Since one lncRNA usually has different transcripts, we searched GEPIA database for significant transcripts in BRCA. In GEPIA, 13 transcripts were detected in tissues, and only two transcripts were differentially expressed between tumors and normal tissues, including ENST00000619222.4 and ENST00000418813.6 (Fig. 2A and B). In the analyses of BRCA subtypes, basal like, HER2 positive and luminal B had significantly lower levels of ENST00000619222.4 and ENST00000602901.5 in tumors than those in normal tissues; the levels of ENST00000418813.6 in basal like and luminal B BRCA were clearly lower than normal tissues; and the level of ENST00000453910.5 was only lower in luminal B tumors in comparison of normal tissues (Fig. 2C). Although it seemed that the levels of these four transcripts in luminal B were the lowest, there was no statistic difference among BRCA subtypes (Fig. 2D).
Fig. 2.
Expression of different MIR99AHG transcripts in BRCA in GEPIA. (A) Expression of 19 MIR99AHG transcripts in BRCA and normal tissues; (B) Box plots of 13 transcripts; (C) Comparison of 4 significant transcripts expression between BRCA subtypes and normal breast tissues; (D) Comparison of 4 significant transcripts expression among BRCA subtypes. *P < 0.05.
Furthermore, we utilized Xiantao platform and GEPIA database to analyze possible clinicopathological roles of MIR99AHG and its transcripts in BRCA. In the Xiantao analysis, MIR99AHG expression in T2 (tumor stage 2) BRCA was obviously lower than that in T1 (tumor stage 1) and T3 (tumor stage 3) BRCA (P < 0.05, Fig. 3A). Though without statistical significance, T4 (tumor stage 4) BRCA had the least average expression level of MIR99AHG (Fig. 3A). However, there were no significant differences among N (node) stages or between M (metastasis) stages (Fig. 3A). In GEPIA database, during the increasing of stages, there was a downtrend of MIR99AHG expression (P < 0.05), and ENST00000619222.4 also had a downtrend but without significance (Fig. 3B). Although expression of ENST00000418813.6 varied between each stage, there was no significant downward trend with increasing BRCA stage (P < 0.05, Fig. 3B).
Fig. 3.
Relationships between MIR99AHG expression and stages. (A) Xiantao platform; (B) GEPIA.*P < 0.05; ***P < 0.001.
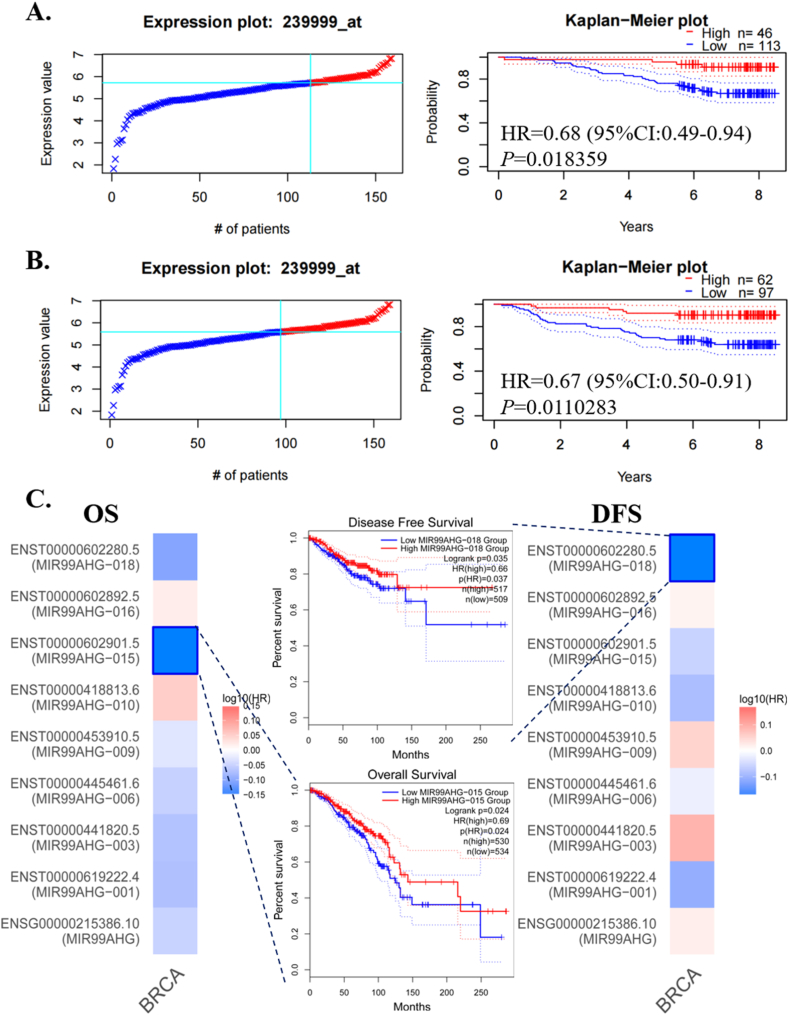
3.3. Prognostic role of MIR99AHG in BRCA
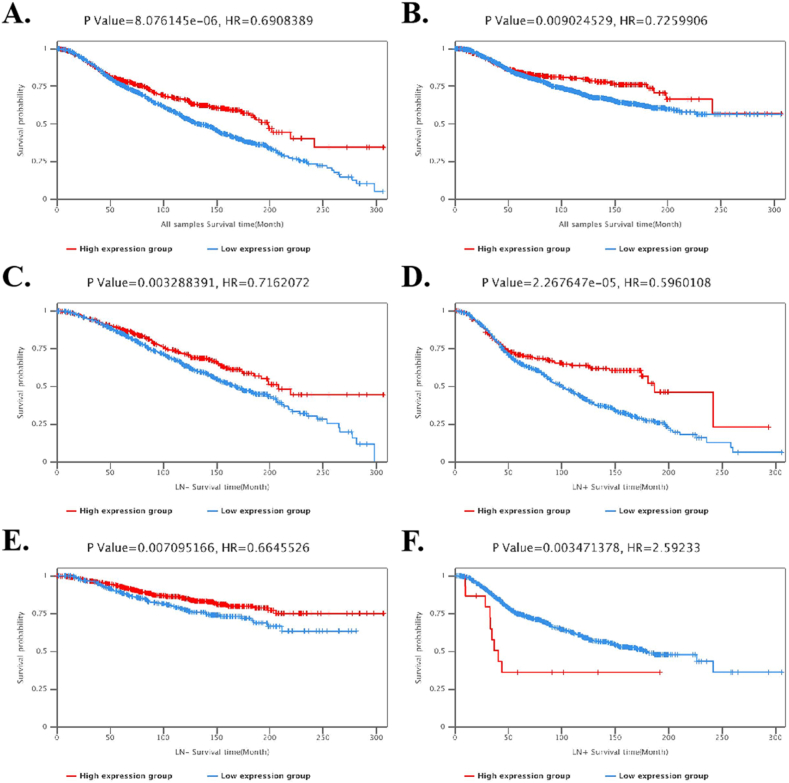
PrognoScan database showed that high level of MIR99AHG indicated good prognosis, including OS (HR = 0.68, 95%CI: 0.49–0.94, P = 0.018, Fig. 4A) and RFS (HR = 0.67, 95%CI: 0.50–0.91, P = 0.011, Fig. 4B). In addition, prognosis analyses of multiple cancers were listed in Supplement file 2. GEPIA also showed that high levels of ENST00000602901.5 and ENST00000602280.5 predicted good prognosis in BRCA (Fig. 4C). In Kaplan-Meier Plotter, levels of MIR99AHG also positively correlated with RFS, but without any significant relationship with OS, DMFS or PPS (Fig. 5A). Notably, we found that compared with the low level group, the high level of MIR99AHG predicted better outcomes, including OS, RFS and PPS, in BRCA patients without lymph node metastasis (Fig. 5B). However, there were no significant relationships between MIR99AHG expression and prognosis in BRCA patients with lymph node metastasis (Fig. 5C). In BCIP database, the high expression group of MIR99AHG also indicated better outcomes, including OS and DS, than the low expression group (Fig. 6A–B), especially patients without lymph node metastasis (Fig. 6C–E). High MIR99AHG expression in BRCA patients with lymph node metastasis predicted better OS but worse DS than the low expression group (Fig. 6D–F).
Fig. 4.
Prognosis analysis of MIR99AHG in PrognoScan and GEPIA. (A) OS in PrognoScan; (B) RFS in PrognoScan; (C) OS and DFS of MIR99AHG and its transcripts in GEPIA.
Fig. 5.
Prognosis analysis of MIR99AHG in Kaplan-Meier Plotter. (A) All BRCA patients; (B) BRCA patients without lymph node metastasis; (C) BRCA patients with lymph node metastasis.
Fig. 6.
Prognosis analysis of MIR99AHG in BCIP. (A) OS of all BRCA patients; (B) DS of all BRCA patients; (C) OS of BRCA patients without lymph node metastasis; (D) OS of BRCA patients with lymph node metastasis; (E) DS of BRCA patients without lymph node metastasis; (F) DS of BRCA patients with lymph node metastasis.
3.4. Relationship between MIR99AHG expression and different functional states
CancerSEA database showed the relationships of MIR99AHG expression with 14 functional states of eight cancer types. It seemed that the three types had significant relationships (Fig. 7), including HGG (high-grade glioma), LUAD and UM (uveal melanoma). In BRCA cells, only one single-cell dataset (EXP0054) showed a negative relationship of MIR99AHG expression with apoptosis (P < 0.05), but there was no relationship between MIR99AHG and apoptosis in the comprehensive analysis of the three datasets (Fig. 7). Besides, MIR99AHG expression wasn't correlated with any other functional state of BRCA cells.
Fig. 7.
CancerSEA analysis.
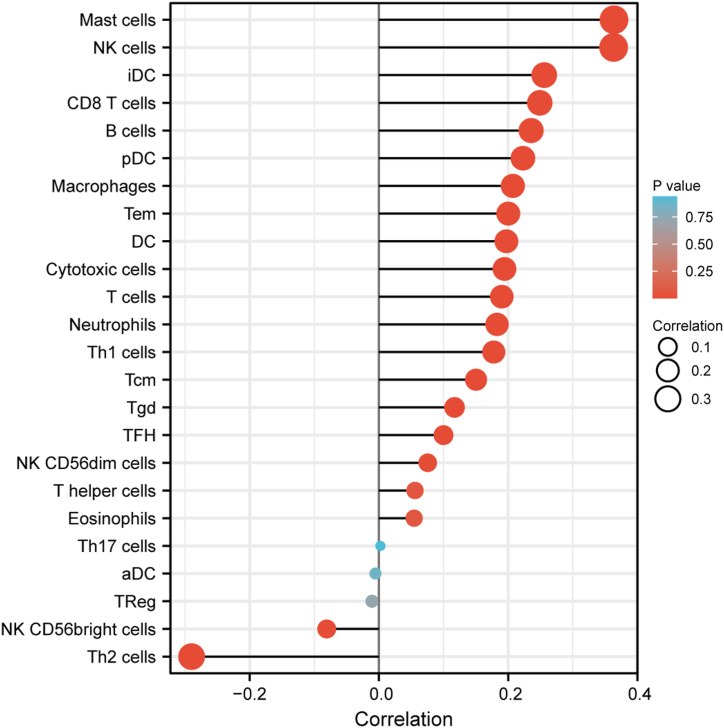
3.5. Correlation between MIR99AHG expression and immune infiltration levels
In the Xiantao platform, we analyzed the relationship between MIR99AHG expression and enrichment of immune cells. And we found 17 kinds of immune cells positively related to MIR99AHG expression (top 17 in Fig. 8, from Mast cells to NK CD56dim cells), especially Mast cells (R = 0.364, P < 0.001) and NK cells (R = 0.363, P < 0.001, Fig. 8). But levels of another 2 immune cells, Th2 cells and NK CD56bright cells, were negatively associated with MIR99AHG expression in BRCA (Fig. 8).
Fig. 8.
Relationships between MIR99AHG expression and enrichment of immune cells.
3.6. Analyses of co-expressed genes of MIR99AHG and its transcripts in databases
According to the top 100 similar genes of MIR99AHG and four significant transcripts in GEPIA, we screened out 33 highly co-expressed genes, including AP000473.5, COL14A1, SPARCL1 and so on (Fig. 9A). In addition, the top 100 co-expressed genes were elected from BCIP database, and 39 genes were both included in these two databases (Fig. 9B). Among them, 19 genes were also highly related to these four significant transcripts of MIR99AHG (Fig. 9B–C).
Fig. 9.
Co-expression analysis. (A) Heat map of relationships of MIR99AHG and its transcripts with their co-expressed genes expression; (B) Intersection of BCIP and GEPIA; (C) Correlation of MIR99AHG expression with 19 highly co-expressed genes in BCIP. “a” represented that 19 co-expressed genes were highly correlated with MIR99AHG expression both in BCIP and GEPIA; “a+b” represented 33 highly co-expressed genes of MIR99AHG transcripts in GEPIA.
3.7. Interactions of MIR99AHG with proteins in databases
StarBase showed 10 RBPs possibly interacting with MIR99AHG. Among them, U2AF65, hnRNPC, FUS and eIF4AIII could combine with significant transcripts, especially ENST00000418813 and ENST00000453910 (Table 1). According to the top 100 interacting proteins of each significant transcript in RNAct database, we screened out the five most interacting proteins, including AEBP2, CHIC1, DNAJC5B, OSCAR and PABPN1 (Table 2). These proteins were proved to be linked to tumorigenesis and chemoresistance, or to be necessary for the regulation of lncRNAs [[35], [36], [37], [38], [39]]. According to the results of docking between ENST00000418813 and RBPs, we found that PABPN1 had the most number of pairs of interface residues within 5.0 Å, and OSCAR had the highest docking score (Fig. 10). Besides, hnRNPC, AEBP2, U2AF65, eIF4AIII and DNAJC5B also had high docking scores, while CHIC1 and FUS had relatively low scores (Fig. 10).
Table 1.
The prediction of interactions between MIR99AHG transcripts and RBPs in StarBase. Red represented significant transcripts.
TIA1: T-cell-restricted intracellular antigen-1; UPF1: UP Frameshift 1; U2AF65: U2 (RNU2) small nuclear RNA auxiliary factor 2; hnRNPC: heterogeneous nuclear ribonucleoprotein C; DGCR8: DiGeorge syndrome critical region gene 8; TIAL1: TIA1 cytotoxic granule-associated RNA-binding protein-like 1; TDP43: TAR DNA-binding protein 43; LIN28B: lin-28 homolog B; FUS: Fused in sarcoma; eIF4AIII: eukaryotic translation initiation factor 4A3.
Table 2.
The prediction of RBPs interacting with 4 candidate transcripts of MIR99AHG in RNAct database.
| Proteins | UniProt Accession | Length | Prediction Score | Functions in cancers or other diseases | |
|---|---|---|---|---|---|
| AEBP2 | Q6ZN18 | 517 aa | ENST00000418813 | 35.56 | As a zinc finger protein, AEBP2 was proved to be an oncogene which promoted ovarian cancer cell proliferation and increased cisplatin resistance [35]. |
| ENST00000619222 | 8.00 | ||||
| ENST00000602901 | 22.82 | ||||
| ENST00000453910 | 18.18 | ||||
| CHIC1 | Q5VXU3 | 224 aa | ENST00000418813 | 34.21 | CHIC1 was a cis regulator of XIST and involve in the Sjögren's syndrome X-chromosome dose effect [36]. |
| ENST00000619222 | 8.65 | ||||
| ENST00000602901 | 22.32 | ||||
| ENST00000453910 | 18.08 | ||||
| DNAJC5B | Q9UF47 | 199 aa | ENST00000418813 | 22.60 | DNAJC5B was associated with microenvironment and its high expression was related to shorter overall survival of esophageal cancer [37]. |
| ENST00000619222 | 8.90 | ||||
| ENST00000602901 | 15.06 | ||||
| ENST00000453910 | 18.33 | ||||
| OSCAR | Q8IYS5 | 282 aa | ENST00000418813 | 19.93 | OSCAR was involved in osteoclast differentiation and maturation in multiple myeloma [38]. |
| ENST00000619222 | 7.91 | ||||
| ENST00000602901 | 12.96 | ||||
| ENST00000453910 | 16.02 | ||||
| PABPN1 | Q86U42 | 306 aa | ENST00000418813 | 19.20 | PABPN1 regulated alternative polyadenylation events and was associated with tumorigenesis, proliferation, metastasis and chemosensitivity in breast cancer [39]. |
| ENST00000619222 | 8.70 | ||||
| ENST00000602901 | 12.64 | ||||
| ENST00000453910 | 18.11 | ||||
AEBP2: AE binding protein 2; CHIC1: cysteine rich hydrophobic domain 1; DNAJC5B: DnaJ heat shock protein family (Hsp40) member C5 beta; OSCAR: osteoclast associated Ig-like receptor; PABPN1: poly(A) binding protein nuclear 1.
Fig. 10.
The whole view of ENST00000418813 interaction with RBPs. Yellow: ENST00000418813; Brown: RBPs; Number of pairs: the number of receptor-ligand pairs with interface residues within 5.0 Å.
3.8. Prediction of molecular genetic and epigenetic mechanisms of MIR99AHG
BCIP database also showed a higher proportion of MIR99AHG CNV loss than CNV gain in BRCA (Fig. 11A). However, there were no significant relationships between MIR99AHG CNV status and prognosis of BRCA patients (Fig. 11B–C). EWAS Data Hub database investigated MIR99AHG gene body methylation levels through various probes, 16 of which could detect its gene body DNA methylation (Fig. 11D, Supplement file 3). In this database, the high level of methylation indicated longer survival time of BRCA patients in comparison to low level group (Fig. 11E). However, there wasn't any significant relationship between gene body DNA methylation and prognosis of other female reproductive system cancers. In addition, 19 TFs were predicted to combine with the promoter of MIR99AHG (Fig. 11F). According to their relationships with MIR99AHG expression in GEPIA, we divided these TFs into three groups, namely no significance, positive relationship and negative relationship.
Fig. 11.
Prediction of MIR99AHG gene regulation. (A) Comparison of proportions between CNV loss and gain in BRCA in BCIP; (B) OS of BRCA patients stratified according to CNV statuses; (C) DS of BRCA patients stratified according to CNV statuses; (D) The diagram of MIR99AHG gene and probes in EWAS Data Hub database; (E) Survival curves of BRCA and other female reproductive system cancers stratified according to methylation levels; (F) The diagram of interactions between MIR99AHG promoter and TFs.
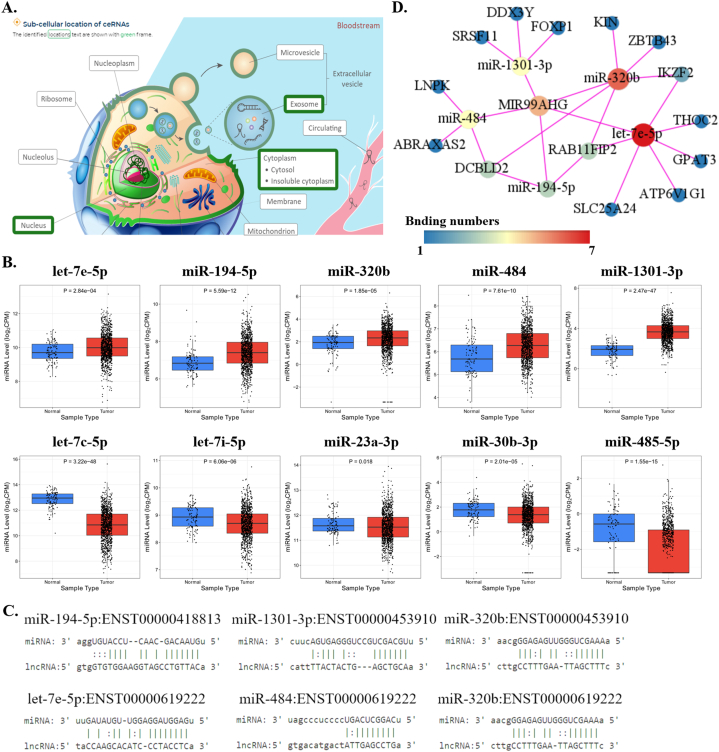
Fig. 12A showed that MIR99AHG was located in the cytoplasm, nucleus and exosomes of cells, which meant that it might act as a miRNA sponge. According to miRNA-lncRNA interactions of DIANA-LncBase v3, we found a total of 83 miRNAs possibly combined with MIR99AHG (Supplement file 4). Then, we screened out 10 miRNAs potentially interacting with significant transcripts of MIR99AHG by comparing DIANA-LncBase v3 and lncRNASNP databases, including miR-194–5p, miR-320 b, miR-23a-3p and so on (Supplement file 5). CancerMIRNome showed that the levels of five miRNAs (let-7e-5p, miR-194–5p, miR-320 b, miR-484 and miR-1301–3p) were visibly higher in breast tumors in comparison of normal tissues, while levels of the other five were obviously lower in BRCA (Fig. 12B). Thus, under-expression of MIR99AHG might cause the up-regulation of these five miRNAs (let-7e-5p, miR-194–5p and so on) in BRCA and their predicted binding sites were listed in Fig. 12C. Then, we found 14 mRNAs possibly interacting with these miRNAs through miRDB and lncACTdb databases and built the lncRNA-miRNA-mRNA ceRNA network (Supplement file 6, Supplement file 7 and Fig. 12D).
Fig. 12.
Prediction of interactions between lncRNA MIR99AHG and miRNAs. (A) The diagram of possible locations of MIR99AHG in cancer cells; (B) Comparison of expression of 10 potentially interacting miRNAs between BRCA and normal tissues in CancerMIRNome; (C) Predicted binding sites of MIR99AHG transcripts and candidate miRNAs; (D) The lncRNA-miRNA-mRNA ceRNA network.
3.9. Validation of MIR99AHG expression and location in BRCA
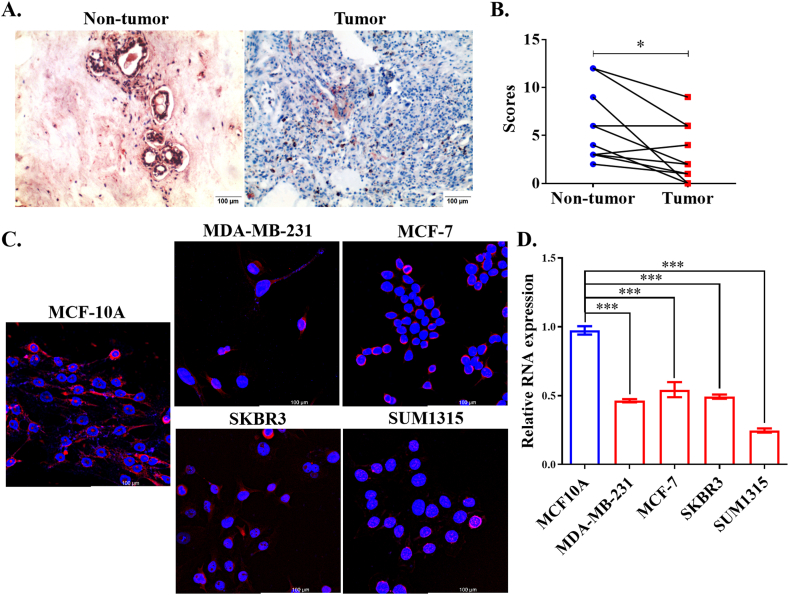
Due to the functions of significant transcripts of MIR99AHG, we designed a relatively specific ISH probe which could detect several MIR99AHG transcripts containing ENST00000418813, ENST00000453910 and so on. Through ISH assay, we found that lncRNA MIR99AHG was located in the cytoplasm and nucleus, and its expression was down-regulated in breast tumors in comparison of non-tumor tissues (Fig. 13A–B). Then, we used a set of FISH probes containing most transcripts of MIR99AHG to investigate its expression and locations in cell lines. Fig. 13C proved that MIR99AHG was located in the cytoplasm and nucleus of cell lines as well. In addition, its expression in MCF10A seemed to be stronger than breast cancer cell lines (MDA-MB-231, MCF-7, SKBR3 and SUM1315). Through qRT-PCR, we identified that MIR99AHG expression in cancer cell lines was obviously lower than that in MCF10A (Fig. 13D).
Fig. 13.
Expression of MIR99AHG in specimens and cell lines. (A) MIR99AHG expression was detected by ISH in breast non-tumor and tumor tissues ( × 100); (B) Diagram of the comparison of MIR99AHG expression between breast non-tumor and tumor tissues (n = 10). *P < 0.05; (C) MIR99AHG expression was detected by FISH in MCF10A, MDA-MB-231, MCF-7, SKBR3 and SUM1315 ( × 630); (D) MIR99AHG expression was investigated by qRT-PCR in cell lines. ***P < 0.001.
4. Discussion
Numerous ncRNAs have been proved to be critical for the occurrence and development of breast carcinoma [8]. As the host gene of the miR-99a/let-7c/miR-125b-2 cluster, MIR99AHG gene may play vital anti-cancer effects not only through the activation of these tumor suppressor miRNAs, but also via the transcription of lncRNA MIR99AHG. Han CC et al. showed that copy number deletion of the MIR99AHG gene could lead to the down-regulation of these ncRNAs in lung cancer [12]. In our study, the percentage of CNV loss was higher than CNV gain in BRCA. However, we failed to find any difference of survival rates between CNV loss and gain. Further researches need to be conducted to verify the relationships of expression and regulation between MIR99AHG and these three miRNAs in BRCA in order to elucidate novel mechanisms of BRCA carcinogenesis.
Although MIR99AHG was over-expressed in several gastrointestinal tumors in previous studies, our research showed the down-regulation of MIR99AHG in BRCA in comparison with normal breast tissues. Notably, luminal B BRCA had the lowest expression level of MIR99AHG and its level in luminal A was obviously higher than luminal B. Patients with luminal B BRCA often had a worse prognosis than those with luminal A BRCA [40]. Therefore, MIR99AHG could become a novel biomarker for BRCA, especially luminal breast cancer. However, whether there is a mutual regulation between MIR99AHG and Ki-67 (the biomarker for differentiation between luminal A and B) needs to be confirmed via further experiments. In addition, MIR99AHG was down-regulated in breast cancer cell lines in comparison to MCF10A both through FISH and qRT-PCR assays. Notably, triple-negative breast cancer (TNBC) cell lines, SUM1315 and MDA-MB-231, had the lowest expression of MIR99AHG. Therefore, abnormal down-regulation of MIR99AHG might be involved in BRCA tumorigenesis, especially TNBC. In further research, we will select these cell lines to conduct in vitro experiments to prove its tumor suppressor function.
Due to various transcripts of MIR99AHG, we screened out four significant transcripts (ENST00000418813, ENST00000453910, ENST00000619222 and ENST00000602901) from GEPIA database. Then, we designed the probe that could detect several MIR99AHG transcripts containing ENST00000418813, ENST00000453910 and so on, and detected its down-regulated expression and location in BRCA tissues through RNA ISH assay. Although there were statistically significant differences in expression levels of ENST00000418813.6 among different stages of BRCA, there was no obvious downward trend of ENST00000418813.6 expression with the increase of stages. Therefore, we need to enlarge BRCA samples and identify its clinicopathological and prognostic value.
Generally, our study showed that high expression level of MIR99AHG predicted good prognosis of BRCA patients. Particularly, MIR99AHG played a greater role in BRCA patients with no lymph node metastasis. Zhang X et al. demonstrated that MIR99AHG stood as a novel biomarker for BRCA diagnosis due to its negative relationship with lymph node metastasis [15]. These meant that MIR99AHG might become a significant biomarker for early-stage BRCA. However, there was no difference of MIR99AHG expression between BRCA cases with and without lymph node metastasis. Furthermore, high levels of ENST00000602901.5 and ENST00000602280.5 indicated longer OS and DFS, respectively. However, ENST00000418813.6 and the other two significant transcripts failed to become indicators of the prognosis of BRCA in the database GEPIA. Therefore, it is essential to perform further researches with larger samples to identify the prognostic roles of different MIR99AHG transcripts in BRCA and to verify their functions in early-stage and advanced BRCA.
In the single-cell analysis of CancerSEA database, MIR99AHG expression was negatively related to half of the functional states in lung cancer, especially angiogenesis, EMT and metastasis, which was in accordance with the previous study [12]. However, MIR99AHG was negatively linked with apoptosis only in one dataset of BRCA, which was contradictory to its anti-cancer function. Combining the three datasets, MIR99AHG had no relationship with apoptosis in BRCA. It is uncertain whether different transcripts of MIR99AHG possess diverse functions in cancer cells. Therefore, what we need to do is to precisely analyze the biological characteristics of BRCA cells through the regulation of MIR99AHG transcript expression.
LncRNAs are proved to be involved in regulating gene expression [41]. Our study explored out 19 highly co-expressed genes through two databases, most of which played critical roles in the regulation of the extracellular microenvironment [[42], [43], [44], [45]]. For example, follistatin-like 1 (FSTL1) is one of the members of the secreted extracellular glycoprotein rich in the cysteine (SPARC) family and involved in the development of the nervous system [42,46]. RECK was also a cysteine-rich extracellular protein and repressed the invasion of MCF-7 cells by modulating MMP9 expression [43]. Secreted protein acidic and rich in cysteine-like 1 (SPARCL1) also belongs to SPARC family, and it was associated with focal adhesion and tumor microenvironment [44]. Moreover, in the Xiantao analysis, we found that MIR99AHG was associated with tumor immunity which was involved in tumor microenvironment. Especially, MIR99AHG expression was positively related with the level of Th1 cells, but was negatively associated with Th2 cells. It was supposed that abnormally low expression of MIR99AHG might suppress the tumor immunity by reducing the Th1/Th2 value. Therefore, decreased MIR99AHG expression might induce the occurrence and development of BRCA through the alteration of extracellular and immune microenvironment. Thus, in further studies, we need to identify whether MIR99AHG can regulate these proteins in a common way, such as transcriptional control, ceRNA network and so on, and to illustrate its possible mechanisms of tumor immunity and microenvironment.
It is proved that RBPs play important roles in regulating the molecular biological functions of lncRNAs [47]. Through the prediction of databases, we failed to find any of these 19 co-expressed genes being combined with significant MIR99AHG transcripts. Besides, these two databases, StarBase and RNAct, showed two different groups of RBPs related to MIR99AHG. Therefore, HDOCK server was applied to the docking between ENST00000418813 and RBPs. Except for CHIC1 and FUS, the other seven RBPs had high docking scores, especially PABPN1 and OSCAR. PABPN1 was considered as an oncogenic RBP and its depletion could suppress cell proliferation and enhance apoptosis in BRCA cells [39,48]. Therefore, whether MIR99AHG transcript expression has a relationship with these possibly interacting RBPs and what is the mechanism of their interaction remains to be uncovered.
Though without any CpG island in the promoter of MIR99AHG, EWAS showed its gene body DNA methylation and a favorable prognostic role of its high methylation level in BRCA, which was the same to high level of MIR99AHG expression. Yang X et al. figured that gene body DNA methylation had the opposite effect on gene expression compared to promoter methylation and increased gene expression [49]. Therefore, MIR99AHG gene body DNA methylation may be one of the initial factors of the regulation of lncRNA MIR99AHG expression and may become a novel indicator of BRCA. However, whether BRCA has a low level of MIR99AHG gene body DNA methylation needs to be verified in further experiments.
In the sequence of MIR99AHG promoter, we found dozens of binding sites that TFs could interact with. According to the correlation analysis of GEPIA, we found that several TFs, including GR, C/EBPbeta, c-Jun and so on, had positive relationships with MIR99AHG, which implied that these TFs possibly up-regulated the level of MIR99AHG by interacting with its promoter. Nevertheless, dual-luciferase reporter assays, chromatin immunoprecipitation and other confirmatory experiments are needed to be conducted to verify their functions in directly controlling MIR99AHG expression. In addition, further researches should be performed to identify whether abnormal expression of these TFs influences MIR99AHG transcription.
Zhang L et al. discovered that MIR99AHG could competitively consume miR-29 b-3p and accelerate adipocyte differentiation [50]. In our research, we found that MIR99AHG was mainly located in the cytoplasm and nucleus of BRCA cells through ISH assay and databases, which was the basis of being a miRNA sponge. Thus, we speculated that MIR99AHG might suppress cancer cell proliferation and invasion by diminishing the levels of onco-miRNAs, such as miR-194–5p, miR-320 b and so on [51,52]. Notably, ENST00000619222 could act as a sponge of let-7c-5p, but the level of let-7c-5p was obviously lower in breast tumors than normal tissues, which was the same to MIR99AHG and ENST00000619222. Since MIR99AHG gene was the host gene of the miR-99a/let-7c/miR-125b-2 cluster, we speculated that when MIR99AHG gene was activated, lncRNA MIR99AHG and let-7c-5p simultaneously increased in spite of their interaction. This might explain why let-7c-5p was down-regulated in BRCA.
The ceRNA network was built according to the prediction of databases, and showed that the number of mRNAs that let-7e-5p might combine with was the largest. Let-7e-5p was proved to be an onco-miRNA in rectal carcinoma [53]. It was the most significant up-regulated miRNA in rectal tumors with heterochrony hepatic metastasis and led to the down-regulation of dozens of target genes which were related to tumor metastasis [53]. In addition, DCBLD2 and RAB11FIP2 could both combine with three miRNAs in the network. Previous studies reported that dysregulation of them was involved in the development of carcinomas [54,55]. Concretely, Rab11FIP2 suppressed tumor growth in lung cancer, while DCBLD2 expression was linked with prognosis of patients with glioblastoma [54,55]. Therefore, MIR99AHG might regulate complicated ceRNA networks through targeting miRNAs. In future research, we need to illustrate the relationships and interactions between MIR99AHG and miRNAs, miRNAs and target mRNAs, and MIR99AHG and these mRNAs in further in vitro and in vivo researches.
Admittedly, our study still had several limitations. Firstly, since our findings were mainly based on bioinformatic analyses, this theoretical work remains to be verified. Therefore, in vitro and in vivo experiments should be conducted in the further. Secondly, we only selected 10 pairs of BRCA samples for the preliminary investigation of MIR99AHG through ISH assay. And most of our bioinformatic results were derived from the TCGA data. Thus, other cohorts should be used to prove our findings. In the next study, we need to enlarge our samples and use clinical data to validate prognostic and clinicopathological roles of MIR99AHG in BRCA. Thirdly, the results might be varied and these databases lack concrete information of classical covariates, such as age, cancer stage, treatment and so on. Thus, we failed to use the multivariate Cox regression to assess the prognostic role of MIR99AHG in the databases. Therefore, it is essential to collect the clinicopathological data and follow-up information of our samples being enlarged and to enhance the reliability and repeatability of our results in further researches.
In conclusion, MIR99AHG was down-regulated in BRCA and its high level indicated good prognosis. ENST00000619222.4, ENST00000418813.6, ENST00000602901.5 and ENST00000453910.5 were potential significant transcripts and might play vital roles in molecular regulation and extracellular microenvironment of BRCA (Fig. 14). Further researches need to be carried out to identify the molecular mechanisms of MIR99AHG in BRCA.
Fig. 14.
Schematic model of the possible mechanism of MIR99AHG in BRCA. CNV: Copy number variant; CEP: Co-expressed proteins; RBP: RNA-binding protein.
Ethics statement
This study was approved by the Kunshan First People's Hospital Ethics Committee (2022-03-015-K01) and was given an exemption from informed consent since this research was a retrospective study. Although we obtained the informed consent from the initial collection but our ethics committee still thought that a retrospective study needed the waiver of informed consent.
Author contribution statement
Wei Han: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Chun-tao Shi: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Hua Chen: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data. Qin Zhou; Wei Ding; Fang Chen: Analyzed and interpreted the data. Zhi-wei Liang; Ya-jie Teng: Performed the experiments. Qi-xiang Shao: Conceived and designed the experiments; Performed the experiments. Xiao-qiang Dong: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Data availability statement
Data is available at GEPIA, BCIP, lnCAR, PrognoScan, Kaplan-Meier Plotter, CancerSEA, Xiantao platform, StarBase, EWAS, LncACTdb3, DIANA-LncBase v3, lncRNASNP2, CancerMIRNome and miRDB.
Funding
National Natural Science Foundation of China (No. 81901579), Kunshan Major Project of Social Research and Development (No. KS2308), Suzhou Scientific and Technological Development Plan (People's Livelihood Science and Technology - Basic Research Project of Medical Treatment and Health Application) (No. SYS2020060), Medical Education Collaborative Innovation Fund of Jiangsu University (No. JDY2023014), Suzhou Basic Research Plan (Basic Research in Medical Application) (No.SKY2023094) and High-Level Medical Talents in Kunshan (No. ksgccrc2007).
Additional information
Databases we utilized in this research were listed as follow: GEPIA, BCIP, lnCAR, PrognoScan, Kaplan-Meier Plotter, CancerSEA, StarBase, RNAct, EWAS Data Hub, PROMO, LncACTdb3, DIANA-LncBase v3, lncRNASNP2, CancerMIRNome and miRDB.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the staff at the Department of Pathology of Kunshan First People's Hospital, the staff of BersinBio Technologies Co., Ltd and the staff of GenePharma for providing technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e19805.
Contributor Information
Qi-xiang Shao, Email: shao_qx@ujs.edu.cn.
Xiao-qiang Dong, Email: dongxq@hotmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fuchs H.E., et al. Cancer statistics, 2022. CA A Cancer J. Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 3.Cairns A., Jones V.M., Cronin K., et al. Impact of the COVID-19 pandemic on breast cancer screening and operative treatment. Am. Surg. 2022;88(6):1051–1053. doi: 10.1177/00031348221087920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasha F., Sharma M., Pruitt K. Mechanisms of endocrine therapy resistance in breast cancer. Mol. Cell. Endocrinol. 2021;532 doi: 10.1016/j.mce.2021.111322. [DOI] [PubMed] [Google Scholar]
- 5.Khalil A.M., Guttman M., Huarte M., et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U. S. A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He R.Z., Luo D.X., Mo Y.Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019;6(1):6–15. doi: 10.1016/j.gendis.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hua Q., Wang D., Zhao L., et al. AL355338 acts as an oncogenic lncRNA by interacting with protein ENO1 to regulate EGFR/AKT pathway in NSCLC. Cancer Cell Int. 2021;21(1):525. doi: 10.1186/s12935-021-02232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian X., Jiang C., Zhu Z., et al. Long non-coding RNA LINC00511 facilitates colon cancer development through regulating microRNA-625-5p to target WEE1. Cell Death Dis. 2022;8(1):233. doi: 10.1038/s41420-021-00790-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavlič A., Hauptman N., Boštjančič E., et al. Long non-coding RNAs as potential regulators of EMT-related transcription factors in colorectal cancer-A systematic review and bioinformatics analysis. Cancers. 2022;14(9):2280. doi: 10.3390/cancers14092280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miraghel S.A., Ebrahimi N., Khani L., et al. Crosstalk between non-coding RNAs expression profile, drug resistance and immune response in breast cancer. Pharmacol. Res. 2022;176 doi: 10.1016/j.phrs.2021.106041. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y., Yuan C., Peng J., et al. LncRNA MIR205HG expression predicts efficacy of neoadjuvant chemotherapy for patients with locally advanced breast cancer. Genes Dis. 2021;9(4):837–840. doi: 10.1016/j.gendis.2021.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han C., Li H., Ma Z., et al. MIR99AHG is a noncoding tumor suppressor gene in lung adenocarcinoma. Cell Death Dis. 2021;12(5):424. doi: 10.1038/s41419-021-03715-7. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forés-Martos J., Cervera-Vidal R., Chirivella E., et al. A genomic approach to study down syndrome and cancer inverse comorbidity: untangling the chromosome 21. Front. Physiol. 2015;6:10. doi: 10.3389/fphys.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J. Identification of MicroRNAs as diagnostic biomarkers for breast cancer based on the cancer genome atlas. Diagnostics. 2021;11(1):107. doi: 10.3390/diagnostics11010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X., Zhuang J., Liu L., et al. Integrative transcriptome data mining for identification of core lncRNAs in breast cancer. PeerJ. 2019;7 doi: 10.7717/peerj.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng Q., Wang X., Xue T., et al. Long noncoding RNA MIR99AHG promotes gastric cancer progression by inducing EMT and inhibiting apoptosis via miR577/FOXP1 axis. Cancer Cell Int. 2020;20:414. doi: 10.1186/s12935-020-01510-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J., Xu W., Yang X., et al. LncRNA MIR99AHG mediated by FOXA1 modulates NOTCH2/Notch signaling pathway to accelerate pancreatic cancer through sponging miR-3129-5p and recruiting ELAVL1. Cancer Cell Int. 2021;21(1):674. doi: 10.1186/s12935-021-02189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Traversa D., Simonetti G., Tolomeo D., et al. Unravelling similarities and differences in the role of circular and linear PVT1 in cancer and human disease. Br. J. Cancer. 2022;126(6):835–850. doi: 10.1038/s41416-021-01584-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Z., Kang B., Li C., et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J., Hu S., Chen Y., et al. BCIP: a gene-centered platform for identifying potential regulatory genes in breast cancer. Sci. Rep. 2017;7 doi: 10.1038/srep45235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Y., Xu Q., Liu M., et al. lnCAR: a comprehensive resource for lncRNAs from cancer arrays. Cancer Res. 2019;79(8):2076–2083. doi: 10.1158/0008-5472.CAN-18-2169. [DOI] [PubMed] [Google Scholar]
- 22.Mizuno H., Kitada K., Nakai K., et al. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lánczky A., Győrffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J. Med. Internet Res. 2021;23(7) doi: 10.2196/27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan H., Yan M., Zhang G., et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47(D1):D900–D908. doi: 10.1093/nar/gky939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J.H., Liu S., Zhou H., et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang B., Armaos A., Tartaglia G.G. RNAct: protein-RNA interaction predictions for model organisms with supporting experimental data. Nucleic Acids Res. 2019;47(D1):D601–D606. doi: 10.1093/nar/gky967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan Y., Tao H., He J., et al. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020;15(5):1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 28.Xiong Z., Li M., Yang F., et al. EWAS Data Hub: a resource of DNA methylation array data and metadata. Nucleic Acids Res. 2020;48(D1):D890–D895. doi: 10.1093/nar/gkz840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messeguer X., Escudero R., Farré D., et al. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18(2):333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 30.Wang P., Guo Q., Qi Y., et al. LncACTdb 3.0: an updated database of experimentally supported ceRNA interactions and personalized networks contributing to precision medicine. Nucleic Acids Res. 2022;50(D1):D183–D189. doi: 10.1093/nar/gkab1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karagkouni D., Paraskevopoulou M.D., Tastsoglou S., et al. DIANA-LncBase v3: indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 2020;48(D1):D101–D110. doi: 10.1093/nar/gkz1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao Y.R., Liu W., Zhang Q., et al. lncRNASNP2: an updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Res. 2018;46(D1):D276–D280. doi: 10.1093/nar/gkx1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li R., Qu H., Wang S., et al. CancerMIRNome: an interactive analysis and visualization database for miRNome profiles of human cancer. Nucleic Acids Res. 2022;50(D1):D1139–D1146. doi: 10.1093/nar/gkab784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q., Wang W., Gao Q. beta-TRCP-mediated AEBP2 ubiquitination and destruction controls cisplatin resistance in ovarian cancer. Biochem. Biophys. Res. Commun. 2020;523(1):274–279. doi: 10.1016/j.bbrc.2019.12.050. [DOI] [PubMed] [Google Scholar]
- 36.Mougeot J.L., Noll B.D., Bahrani Mougeot F.K. Sjögren's syndrome X-chromosome dose effect: an epigenetic perspective. Oral Dis. 2019;25(2):372–384. doi: 10.1111/odi.12825. [DOI] [PubMed] [Google Scholar]
- 37.Zhou M.H., Wang X.K. Microenvironment-related prognostic genes in esophageal cancer. Transl. Cancer Res. 2020;9(12):7531–7539. doi: 10.21037/tcr-20-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z., Liu H., Li Y., et al. Adiponectin inhibits the differentiation and maturation of osteoclasts via the mTOR pathway in multiple myeloma. Int. J. Mol. Med. 2020;45(4):1112–1120. doi: 10.3892/ijmm.2020.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Lang G.T., Xue M.Z., et al. Dissecting the heterogeneity of the alternative polyadenylation profiles in triple-negative breast cancers. Theranostics. 2020;10(23):10531–10547. doi: 10.7150/thno.40944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennecke H., Yerushalmi R., Woods R., et al. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 2010;28(20):3271–3277. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Huang X., Liu J., et al. New insight into long non-coding RNAs associated with bone metastasis of breast cancer based on an integrated analysis. Cancer Cell Int. 2021;21(1):372. doi: 10.1186/s12935-021-02068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J., Liao M., Hatta T., et al. Identification of a follistatin-related protein from the tick Haemaphysalis longicornis and its effect on tick oviposition. Gene. 2006;372:191–198. doi: 10.1016/j.gene.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 43.Shen Z., Jiao K., Teng M., et al. Activation of STAT-3 signalling by RECK downregulation via ROS is involved in the 27-hydroxycholesterol-induced invasion in breast cancer cells. Free Radic. Res. 2020;54(2–3):126–136. doi: 10.1080/10715762.2020.1715965. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H.P., Wu J., Liu Z.F., et al. SPARCL1 is a novel prognostic biomarker and correlates with tumor microenvironment in colorectal cancer. BioMed Res. Int. 2022;2022 doi: 10.1155/2022/1398268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yue X., Lan F., Xia T. Hypoxic glioma cell-secreted exosomal miR-301a activates wnt/β-catenin signaling and promotes radiation resistance by targeting TCEAL7. Mol. Ther. 2019;27(11):1939–1949. doi: 10.1016/j.ymthe.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang S., Zhang Y., Jiang T., et al. Knockdown of Follistatin-like 1 disrupts synaptic transmission in hippocampus and leads to cognitive impairments. Exp. Neurol. 2020;333 doi: 10.1016/j.expneurol.2020.113412. [DOI] [PubMed] [Google Scholar]
- 47.Xu S., Xie J., Zhou Y., et al. Integrated analysis of RNA binding protein-related lncRNA prognostic signature for breast cancer patients. Genes. 2022;13(2):345. doi: 10.3390/genes13020345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wigington C.P., Williams K.R., Meers M.P., et al. Poly(A) RNA-binding proteins and polyadenosine RNA: new members and novel functions. Wiley Interdiscip Rev RNA. 2014;5(5):601–622. doi: 10.1002/wrna.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X., Han H., De Carvalho D.D., et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26(4):577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L., Ma J., Pan X., et al. LncRNA MIR99AHG enhances adipocyte differentiation by targeting miR-29b-3p to upregulate PPARγ. Mol. Cell. Endocrinol. 2022;550 doi: 10.1016/j.mce.2022.111648. [DOI] [PubMed] [Google Scholar]
- 51.Mao Q., Lv M., Li L., et al. Long intergenic noncoding RNA 00641 inhibits breast cancer cell proliferation, migration, and invasion by sponging miR-194-5p. J. Cell. Physiol. 2020;235(3):2668–2675. doi: 10.1002/jcp.29170. [DOI] [PubMed] [Google Scholar]
- 52.Luo M., Deng S., Han T., et al. LncRNA NR2F2-AS1 functions as a tumor suppressor in gastric cancer through targeting miR-320b/PDCD4 pathway. Histol. Histopathol. 2022;37(6):575–585. doi: 10.14670/HH-18-429. [DOI] [PubMed] [Google Scholar]
- 53.Chen W., Lin G., Yao Y., et al. MicroRNA hsa-let-7e-5p as a potential prognosis marker for rectal carcinoma with liver metastases. Oncol. Lett. 2018;15(5):6913–6924. doi: 10.3892/ol.2018.8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng S., Wang L.Y., Wang C.H., et al. Transmembrane protein DCBLD2 is correlated with poor prognosis and affects phenotype by regulating epithelial-mesenchymal transition in human glioblastoma cells. Neuroreport. 2021;32(6):507–517. doi: 10.1097/WNR.0000000000001611. [DOI] [PubMed] [Google Scholar]
- 55.Dong W., Li H., Wu X. Rab11-FIP2 suppressed tumor growth via regulation of PGK1 ubiquitination in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2019;508(1):60–65. doi: 10.1016/j.bbrc.2018.11.108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available at GEPIA, BCIP, lnCAR, PrognoScan, Kaplan-Meier Plotter, CancerSEA, Xiantao platform, StarBase, EWAS, LncACTdb3, DIANA-LncBase v3, lncRNASNP2, CancerMIRNome and miRDB.