Abstract
Kinesin-5 crosslinks and slides apart microtubules to assemble, elongate, and maintain the mitotic spindle. Kinesin-5 is a tetramer, where two N-terminal motor domains are positioned at each end of the motor, and the coiled-coil stalk domains are organized into a tetrameric bundle through the bipolar assembly (BASS) domain. To dissect the function of the individual structural elements of the motor, we constructed a minimal kinesin-5 tetramer (mini-tetramer). We determined the x-ray structure of the extended, 34-nm BASS domain. Guided by these structural studies, we generated active bipolar kinesin-5 mini-tetramer motors from Drosophila melanogastor and human orthologues which are half the length of native kinesin-5. We then used these kinesin-5 mini-tetramers to examine the role of two unique structural adaptations of kinesin-5: 1) the length and flexibility of the tetramer, and 2) the C-terminal tails which interact with the motor domains to coordinate their ATPase activity. The C-terminal domain causes frequent pausing and clustering of kinesin-5. By comparing microtubule crosslinking and sliding by mini-tetramer and full-length kinesin-5, we find that both the length and flexibility of kinesin-5 and the C-terminal tails govern its ability to crosslink microtubules. Once crosslinked, stiffer mini-tetramers slide antiparallel microtubules more efficiently than full-length motors.
INTRODUCTION
Kinesin-5 motors are highly conserved across eukaryotes. These motors perform a series of steps to assemble and elongate the mitotic spindles. First, kinesin-5 moves along microtubules (MTs) and localizes to plus-ends. Second, Kinesin-5 crosslinks MTs and aligns them into antiparallel bundles. Finally, kinesin-5 slides antiparallel MTs apart to elongate the mitotic spindle. Each of these roles relies on unique structural adaptations, including a bipolar, tetrameric organization and mechanochemical gating through interactions between the C-terminal tail domains and N-terminal motor domains [1, 2]. Although kinesin-5 motors are conserved across eukaryotes, there is a remarkable diversity in their mechanisms across species. In various yeast species, kinesin-5 motors uniquely undergo minus-end directed motility as single motors and switch direction upon clustering or during MT sliding toward plus-end directed motility (Shapira et al., 2017; Singh et al., 2018; Pandey et al., 2021a; Pandey et al., 2021b). In contrast, metazoan Kinesin-5 motors undergo motility towards plus-ends both along single MTs and while sliding antiparallel MTs (Kapitein et al., 2005; Weinger et al., 2011; Bodrug et al., 2020).
The tetrameric-bipolar organization of kinesin-5 originates from the antiparallel folding of the four α-helices from the four subunits within its conserved bipolar assembly (BASS) domain (Scholey et al., 2014). The BASS domain lies at the center of the kinesin-5 minifilament and consists of the antiparallel four-helical bundle that uniquely swaps to form N-terminal dimeric coiled-coils that connect to motor domains on both ends. The BASS domain is the central force-bearing structure that coordinates between the two motile-motor ends as they simultaneously undergo hand-over-hand motility along two MTs. While the kinesin-5 MT sliding motility is linked to its bipolar-tetrameric organization, the functional relationship between the conserved kinesin-5 motor, tail, or BASS domains with the conserved and unique MT crosslinking and sliding mechanisms remains unknown (Kapitein et al., 2005).
The kinesin-5 C-terminal tail domains (termed tail from herein) emerge from bipolar-tetrameric minifilament near the motor domains of the antiparallel-dimeric folded subunits (Acar et al., 2013). The tail is essential for MT sliding activity across species (Hildebrandt et al., 2006; Weinger et al., 2011; Duselder et al., 2015). We recently discovered that the kinesin-5 tail regulates the MT-activated ATP hydrolysis within motor domains and this regulation is essential for kinesin-5 motors to transition from crosslinking to MT sliding motility (Bodrug et al., 2020). This allosteric tail-to-motor domain interaction may allow for multiple kinesin-5 motors to form multi-motor clusters through cross-interactions between kinesin-5 motors, leading to a clustering (Bodrug et al., 2020). Tail-mediated kinesin-5 clustering is critical in organizing the forces generated by multiple kinesin-5 motors, resulting MT crosslinking, alignment, and then sliding (Bodrug et al., 2020). However, it remains unknown how this tail-to-motor clustering regulates MT crosslinking, alignment, and sliding.
Here, we describe how the kinesin-5 tail-motor domain interaction and the length of the force-bearing bipolar tetrameric minifilament modulate kinesin-5 clustering, MT crosslinking, and MT sliding. We present a 34-nm X-ray structure of an extended BASS α-helical tetrameric-minifilament that reveals rigid N-terminal coiled-coil junctions that emerge from its central-tetrameric core with a clear pattern of heptad repeats. Using this pattern of coiled-coil heptads, we next engineered short and active human and Drosophila melanogaster bipolar 38-nm kinesin-5 mini-tetramers, which are roughly half the length of native kinesin-5 motors. In single-molecule motility assays, mini-tetramers without tails undergo processive motility with infrequent pauses. The tails promote kinesin-5 mini-tetramer frequent, static pausing. Multimotor clusters assemble when motile motors encounter paused motors along MTs. MT sliding assays reveal that unlike the native kinesin-5, mini-tetramers require the tail domain to crosslink, align, and slide two MTs. Kinesin-5 mini-tetramers are restricted in their ability to pair and align MTs. However, once MTs are paired, kinesin-5 mini-tetramers slide two MTs more efficiently than native kinesin-5. Our data demonstrate that the kinesin-5 tail and length of the tetrameric-bipolar minifilament are two critical structural adaptations for kinesin-5 motors to effectively crosslink and slide apart MTs. The tail-to-motor domain interactions promote clustering such that multiple motors can coordinate within MT overlaps, while the long-tetrameric minifilament provides flexibility and force transmission capacity required for efficient MT crosslinking leading to MT alignment and sliding.
RESULTS
An extended BASS X-ray structure reveals rigid and dimeric coiled-coils emerging from bipolar-tetramer junctions
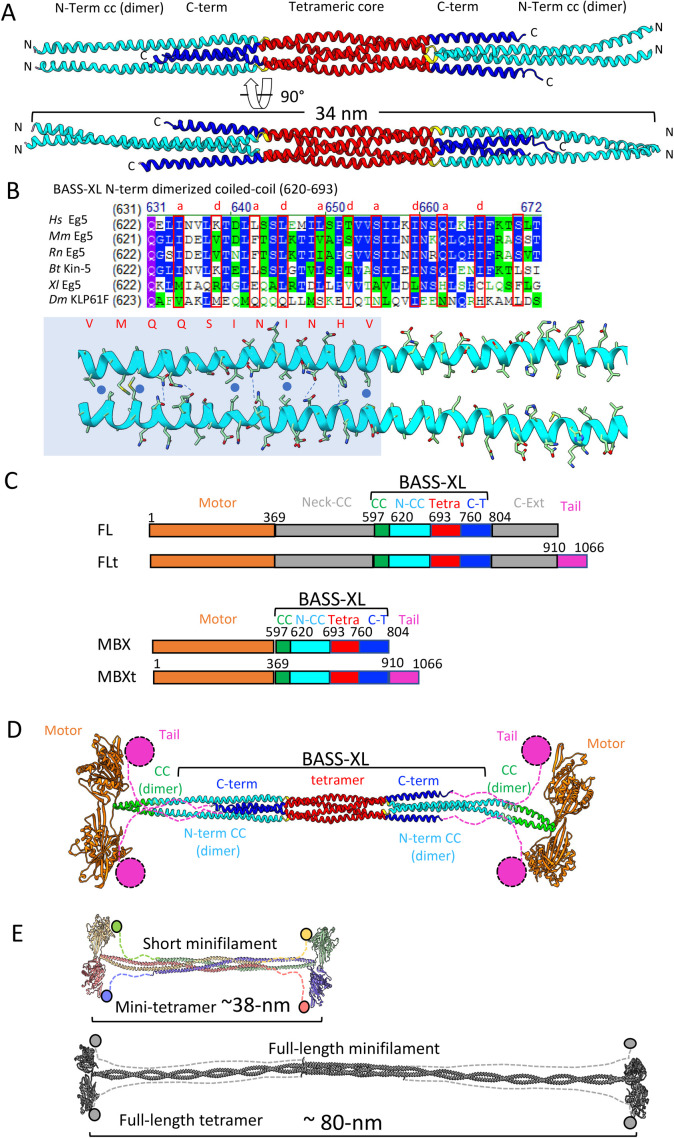
We first considered the overall domain architecture of the kinesin-5 motor protein, which consists of the motor domain, the tail domain, and the central-minifilament domain (Figure 1C). Examining the contribution of the tail domain to kinesin-5 motility can be performed by comparing constructs which have the full tail domain deleted to full-length wild-type constructs which contain the tail. To analyze the function of the tetramerization domain, we altered the length of the minifilament. We used X-ray crystallography to guide our design of a shortened-central minifilament containing the conserved BASS structural region. Although our previously solved BASS X-ray structure revealed parallel coiled-coil at its N-terminal ends, neither the orientation of N-terminal ends of the α-helices nor their supertwists match those observed in canonical coiled-coils (Kashina et al., 1996; Scholey et al., 2014). Thus, to understand how dimeric-parallel coiled-coils extend from N-terminal ends of the BASS junctions, we required a structure that includes a longer coiled-coil N-terminal region.
FIGURE 1:
The X-ray structure of the kinesin-5 BASS-XL minifilament reveals rigid dimeric coiled-coils emerge from a tetramer core and allow the designing of kinesin-5 minitetramers. A) Top, a side view of a 34-nm long KLP61F BASS-XL minifilament crystal structure reveals the formation of dimeric coiled-coils (cyan: 40-nm long) that are rigidly attached to the BASS-tetrameric core (red) and stabilized by the C-terminal extension (dark blue). Bottom panel, has a 90° rotated view compared with the top panel. B) Top, sequence alignment reveals the conservation, and the heptad repeat pattern of the dimeric coiled-coil (marked in “a” and “d” residues), mapped based on the X-ray BASS-XL structure. Bottom, view of the dimeric coiled-coil heptad interactions marked in B on the structural interface shown in A. C) Domain organization of the full-length and minitetramer kinesin-5 motors used in this study. Top, Full-length kinesin-5 with the tail domain (FLt), second full-length kinesin-5 without the tail domain (FL). Third, kinesin-5 minitetramer without tail (MBX). Fourth, kinesin-5 minitetramer with tail (MBXt). In FL and FLt, the Motor (residues 1–369; brown) is connected by a ∼230 residue neck coiled-coil (369–597 grey, Neck CC), then connected to the BASS-XL elements (described below) and connected to C-term extension (804–910). The kinesin-5 tail domain (tail, pink, 910–1066) is deleted in the FL construct. The minitetramer constructs connect the motor to the BASS-XL elements via a 20 residue N-terminal coiled-coils (cc, green, 597–620) and then fused to the N-terminal dimeric-coiled coil junctions (N-CC, cyan,620–693) of the BASS-XL and the tetramer core (tetra, red,693–760) followed by the C-terminal extension (C-t, blue, 760–804). The minitetramers may either include or lack the Kinesin-5 C-terminal tail domain (orange). D) The structural model for a 38-nm long kinesin-5 minitetrameric motor based on the fusion of domains based on matching the heptad repeats with neck helical coiled-coil. The tail domains extend near the motor domains of the subunits folded in antiparallel orientations. E) Scaled comparison of the minitetramer kinesin-5 motors (38 nm), shown on the top, reveals that they are half the length of the native kinesin-5 tetrameric motors (80 nm), shown on the bottom.
We, therefore, solved the X-ray structure of an extended D. melanogaster KLP61F segment (residues 610-804, termed BASS-XL) which is extended by 30 residues N-terminally compared with the previous BASS X-ray structure in the kinesin-5 sequence (Kashina et al., 1996; Scholey et al., 2014; Figure 1; Supplemental Figure S1). The BASS-XL X-ray structure was determined by molecular replacement, using the solved BASS structure as a starting model, and was refined to 4.4-Å resolution leading to a Rwork/Rfree (0.277/0.309; Figure 1; Supplemental Figure S1; Table 1). The BASS-XL structure reveals a 34-nm α-helical minifilament, compared with a 27-nm BASS minifilament (Figure 1; Supplemental Figure S1). In the BASS-XL structure, the additional N-terminal 30-residues form parallel-canonical coiled-coils that emerge from both ends of the BASS tetrameric core (Figure 1A; Scholey et al., 2014). In contrast to the BASS structure, the N-terminal parallel coiled-coils in BASS-XL form multiple heptad repeat interactions with clear a and d homotypic contact interfaces (residues 620–670; Figure 1B) and extend for 10 nm before they twist slightly out of register into a swap junction (residues 693–697) to form a four α-helical antiparallel bundle within the BASS core tetramer (residues 697–760; Figure 1A; Scholey et al., 2014). The BASS-XL C-termini form α-helices (residues 760–806) that stabilize the junctions of the N-terminal coiled-coil dimer on the opposite BASS-XL filament end (Figure 1A). Superimposing BASS-XL and BASS structures reveals a 5-Å outward supertwist of BASS-XL N-term coiled-coil compared with BASS, leading to a change in α-helical heptad pattern registry in BASS-XL compared with BASS (Supplemental Figure S1B). The extended N-terminal dimeric coiled coils in BASS-XL rigidly extend out of the tetrameric BASS core and lead their N-terminal ends to 180° rotated with respect to their orientation emerging from the opposite end (Figure 1A).
TABLE 1:
X-ray crystallographic data collection and refinement statistics.
| BASS-XL | |
|---|---|
| Data collection | |
| Resolution range | 126.55– 4.40 (4.64 – 4.40) |
| Wavelength (Å) | 0.979 |
| Space group | C 2 |
| Unit cell (Å): a, b, c (˚): β |
253.18, 84.89, 96.77, 91.44 |
| Total number of observed reflections | 55597 (7964) |
| Unique reflections | 13060 {10508}† |
| Average mosaicity | 0.33 |
| Multiplicity | 4.3 (4.3) |
| Completeness (%) | 98.7 (98.8) {79.76}† |
| Mean I/sigma(I) | 4.6 (1.8) |
| Wilson B-factor | 38.14 |
| Rmergea | 0.10 (0.47) |
| Structure refinement | |
| Rwork | 0.274 (0.251) |
| Rfree | 0.310 (0.368) |
| Molecules per asymmetric unit | 4 |
| Number of non-hydrogen atoms | 5037 |
| macromolecules | 5037 |
| Solvent | 0 |
| Protein residues | 702 |
| RMS bond lengths (Å) | 0.002 |
| RMS bond angles (°) | 0.56 |
| Ramachandran favored (%) | 96.2 |
| Ramachandran allowed (%) | 3.6 |
| Ramachandran outliers (%) | 0.43 |
| Rotamer outliers (%) | 0.41 |
| Clashscore | 11.42 |
| Mean B values (Å2) | |
| Overall | 113.93 |
| macromolecules | 113.93 |
| Number of TLS groups | 11 |
Parentheses numbers represent the highest-resolution shell.
†Numbers represent the truncated data after being treated with ellipsoidal truncation and anisotropic scaling.
aRmerge = ΣhklΣi|Ii(hkl)-Iav(hkl)|/ΣhklΣiIi(hkl).
Guided by the pattern of N-terminal heptad repeats in the BASS-XL structure, we next engineered shortened bipolar-tetrameric kinesin-5 motors by fusing active-dimeric motors onto an extended BASS-XL minifilament, and henceforth refer to these as “mini-tetramers” (Figure 1C). The BASS-XL minifilament was extended by three heptad repeats to a total length of 38 nm to ensure coil–coil stability, and then fused to the C-terminal end of a motor-neck linker region (Figure 1C green). On its C-terminus, the BASS-XL mini-tetramer sequence was either terminated (termed MBX) or was C-terminally fused to the N-terminal end of the kinesin-5 tail domain (termed MBXt; Figure 1C; Supplemental Figures S1 and S2). We present a structural model for the 38-nm kinesin-5 mini-tetramer revealing that is roughly half the length of native kinesin-5 (38 nm vs. 80 nm; Acar et al., 2013; Scholey et al., 2014; Figure 1D; Supplemental Figure S2, C and D). With this structural information in hand and with the goal of dissecting the function of both the minifilament and tail domains across species, we generated D. melanogaster KLP61F mini-tetramers fused to Neon Green (-tail: kMBX, or +tail: kMBXt) and human Eg5 kinesin-5 mini-tetramers labeled with Tetra-methyl Rhodamine (TMR) fluorophores at reactive Cysteine residues (-tail: hMBX, or +tail: hMBXt) (Supplemental Figure S1, A–C; Figure 1C) for further study in fluorescence-based in vitro motility assays. D. melanogaster KLP61F and human Eg5 minitetramers were purified using bacteria (Supplemental Figure S2B, top panel). Mass-photometry measurements of the D. melanogaster KLP61F minitetramers revealed masses supporting these motors are tetrameric assemblies (Figure S2B, lower panel).
Kinesin-5 mini-tetramers undergo processive plus-end-directed motility along with pausing
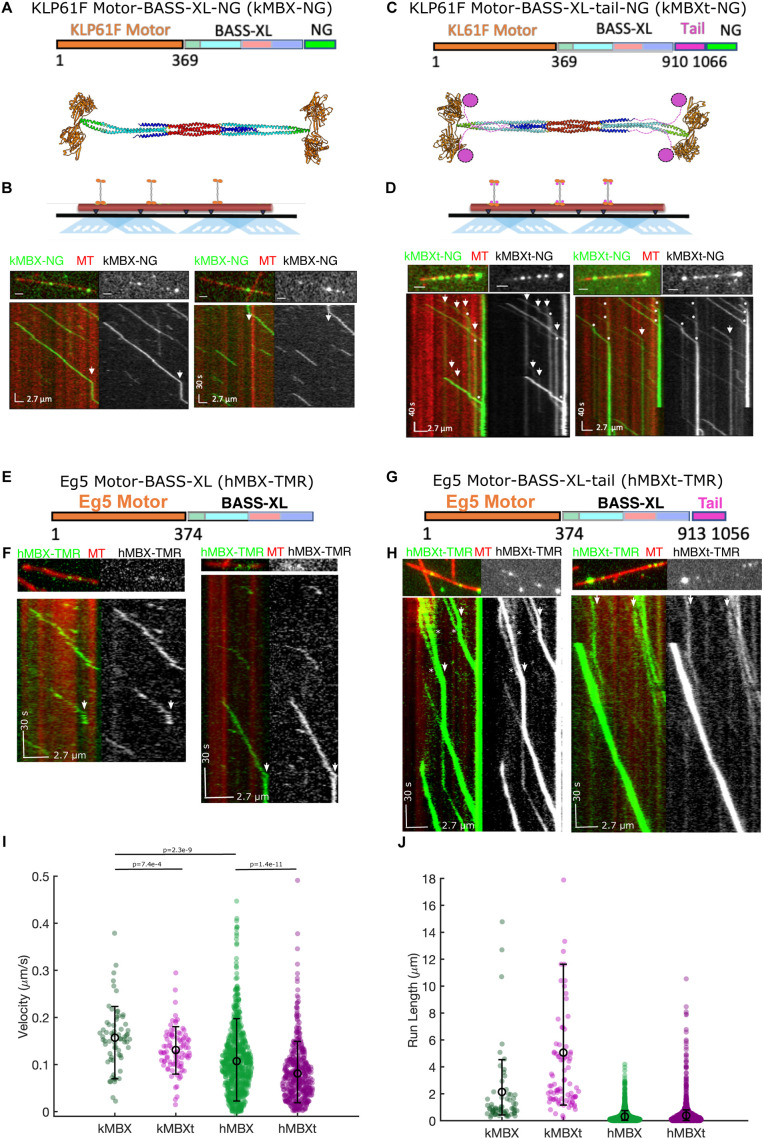
We next set out to determine the motility of our mini-tetramer constructs and examine the functional role of the C-terminal tail domain. We reconstituted the motility of kMBX and kMBXt mini-tetramer motors along single MTs using total internal reflection fluorescence (TIRF) microscopy, as previously described (Bodrug et al., 2020). Single kMBX motors undergo slow-processive motility along MTs interspersed with rare, brief pauses (Figure 2, A and B). In contrast, kMBXt motors undergo processive motility towards MT plus-ends and pause for extended periods (Figure 2, C and D). While paused along MTs, kMBXt motors often encounter other motile motors and can occasionally merge with paused ones to form brighter multi-motor clusters. Once clusters of kMBXt motors do form, they can undergo motility as clusters until reaching MT plus-ends, where they bind for extensive time periods. These data suggest that the kinesin-5 tail-motor domain interaction promotes pausing and multimotor clustering, recapitulating activities observed for full-length Eg5 (Bodrug et al., 2020).
FIGURE 2:
Kinesin-5 minitetramers undergo processive motility interrupted by static pauses along MTs in vitro. A) Top, kMBX domain organization. Motor- and neck-linker domains (1–365, blue) extended coiled-coil (green), BASS-XL minifilament with its dimerized zone (orange), tetrameric zone (red), and C-terminal zone (cyan). Middle, structural model for the kMBX minitetramer as shown in Figure 1D. B) Top, TIRF microscopy reconstitution setup to examine kMBX motors motility along MTs. Middle panel, image of individual MTs (red) with kMBX motors (green). Bottom panel, kymographs of the above image with kMBX motor motility along MTs revealing their processive motility with extended pauses (arrows) and do not accumulate at MT plus-ends. C) Top, kMBX domain organization. Motor- and neck-linker domains (1–365, orange) extended coiled-coil (green), BASS-XL minifilament with its dimerized zone (cyan), tetrameric zone(red), C-terminal zone (blue) and C-terminal tail domain (pink). Middle, structural model for the kMBX minitetramer as shown in Figure 1D. D) Top, TIRF microscopy setup for kMBXt MT-motility assays. Middle panel, images of individual MTs (red) with kMBXt motors (green). Bottom panel, kymograph of the above image with kMBXt motor motility along MTs. Motor assembly into clusters is marked by an asterisk, and motor pauses are marked by arrowheads. E) The hMBX minitetramer consists of the human Eg5 motor-neck linker domain (1–374, orange), the D. melanogaster BASS domain (597–799) with its extended coiled-coil (green), with its dimerized zone (cyan), tetrameric zone (red), C-terminal zone (blue). F) Top panel, image of individual MT (red) with hMBX motors (green). Bottom panel, kymographs of the above image with hMBX motor motility along MTs revealing their processive motility with short pauses (arrows) and do not accumulate at MT plus-ends. G) The hMBXt-TMR minitetramer consists of the human Eg5 motor–neck linker domain (1–374, orange), the D. melanogaster BASS-XL minifilament (597–799) with its extended coiled-coil (green), with its dimerized zone (cyan), tetrameric zone(red), C-terminal zone (blue) and the human Eg5 C-terminal tail domain (913–1056, pink). H) Top panel, images of individual MTs (red) with hMBXt-TMR motors (green). Bottom panel, kymographs of the above image with hMBXt motor motility along MTs. Motor assembly into clusters is marked by an asterisk, and motor pauses are marked by arrowheads. I) Histogram distributions for velocity (μm/s) of the kMBX (green), kMBXt (pink), hMBX (green), and hMBXt (pink) motors along MTs showing that the kMBXt or hMBXt undergo slower motility than the kMBX and hMBX motors. T tests show highly significant differences between kMBX and kMBXt as well as hMBX and hMBXt. There are no significant differences between hMBXt and kMBXt motor velocities. J) Histogram distributions for motility run lengths (μm) of the kMBX (green), kMBXt (pink), hMBX (green), and hMBXt (pink) motors along MTs revealing that the kMBXt or hMBXt motors are generally more processive than the hMBX and kMBX motors.
To further understand the nature of the D. melanogaster kinesin-5 tail domain in regulating minitetramer activity, we studied the impact of changing the ionic strength in the buffer on the motility of both kMBX and kMBXt. At the lowest concentration used (25 mM KCl), both motors underwent slow motility, while increasing the concentration up to 100 mM KCl tended to increase the speed at which both motors moved. Beyond 100 mM KCl, we observed a decrease in kMBXt velocities, while kMBX motors dissociated rapidly and did not processively move under these high-ionic strength conditions (Table 2). At the lowest ionic strengths, both motors exhibited extremely long-run lengths, while increasing the ionic strength reduced the run lengths and total run times observed for kMBX to a higher degree than for kMBXt motors (Table 2). These data suggest that the presence of the kinesin-5 tail domain leads to slower motility but an increase in run lengths and run times during each processive-motility event.
TABLE 2:
D. melanogaster Kinesin-5 minitetramer motor-motility properties
| kMBX | Velocity (nm/s) | Run length (nm) | Run time (s) | Merging freq (μm–1min–1) | MT plus-end dwell time(s) |
|---|---|---|---|---|---|
| 25 mM KCl | 115 ± 3n = 340 | N/A | N/A | N/A | N/A |
| 50 mM KCl | 144 ± 6n = 136 | 1014 ± 222 | 9 ± 1 | 0.006 ± 0.001 | 78 ± 19 |
| 100 mM KCl | 156 ± 8n = 66 | 2137 ± 428 | 16 ± 4 | 0.001 ± 0.0007 | 25 ± 18 |
| kMBXt | |||||
| 25 mM KCl | 78 ± 4n = 52 | 3832 ± 433 | 56 ± 8 | 0.007 ± 0.002 | 125 ± 51 |
| 50 mM KCl | 127 ± 4n = 136 | 3135 ± 222 | 29 ± 1 | 0.011 ± 0.002 | 117 ± 15 |
| 100 mM KCl | 114 ± 5n = 88 | 5060 ± 605 | 51 ± 7 | 0.050 ± 0.008 | 267 ± 45 |
| 125 mM KCl | 136 ± 4n = 149 | 5440 ± 362 | 45 ± 4 | 0.023 ± 0.006 | 147 ± 34 |
| 150 mM KCl | 119 ± 6n = 19 | 3347 ± 538 | 29 ± 5 | 0.001 ± 0.0008 | 25 ± 17 |
To explore whether tail-motor regulation is a conserved behavior across species, we next studied the activities of human Eg5 motor minitetramers hMBX (-tail) and hMBXt (+tail; Figure 2, E–H). The hMBX and hMBXt motors were engineered with reactive cysteines for labeling with TMR fluorophores to allow for robust, high-resolution, single-molecule tracking analyses (see Materials and Methods). The hMBX motors undergo processive motility towards MT plus-ends over long distances with infrequent pauses (Figure 2, E and F), while pauses are more frequent for the hMBXt motors, similar to our observation with the kMBXt motors (Figure 2, G and H). As with the kMBXt motors, motile hMBXt motors merge with paused motors forming processive clusters with higher intensities, leading to bright- and dim-motile motor complexes which accumulate at MT plus-ends (wide view shown in Supplemental Movie S1, and close up view shown in Supplemental Movie S2). The differences between the motility velocities of hMBX/hMBXt or kMBX/kMBXt motors are highly significant, but there are no significant differences between the velocities of hMBXt and kMBXt motors (Figure 2I). Thus, for both D. melanogaster and human minitetramers, the presence of the kinesin-5 tail results in slower, processive motility, frequent pauses, and assembly into multi-motor clusters upon encounters along MTs (Figure 2, I and J).
Movie S1.
Large‐scale view of hMBXt motors (green) undergoing motility along MTs (magenta) revealing kinesin‐5 mini‐tetramer motor pausing, clustering, and MT plus end association.
Movie S2.
Close‐view of hMBX motors (green) undergoing motility, forming clusters, with pausing, and while dwelling at MT (magenta) plus‐ends.
The kinesin-5 tail domain promotes static pausing
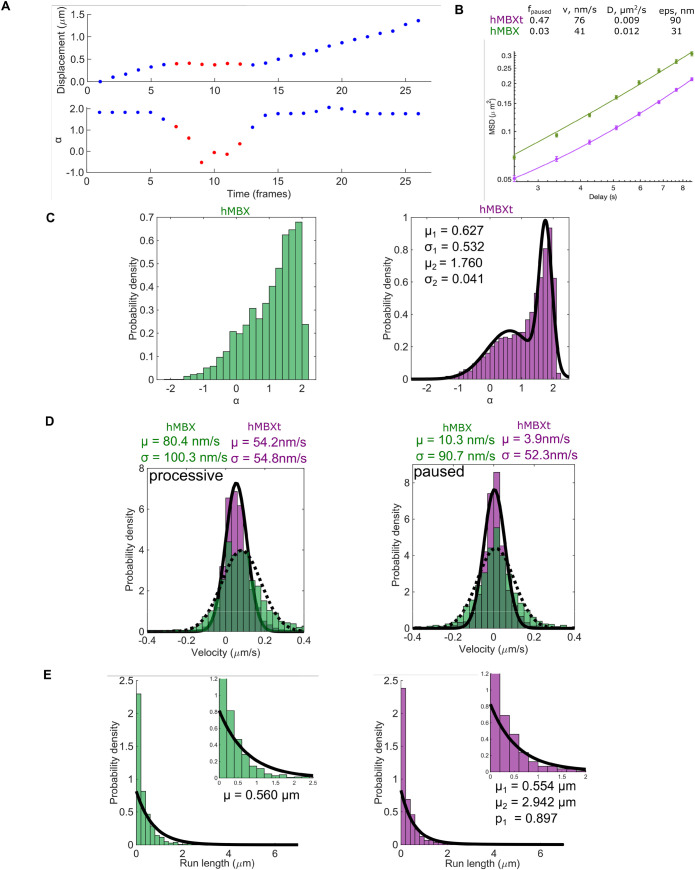
We next sought to further understand the nature of kinesin-5 minitetramer pausing at the single-molecule level. We, therefore, analyzed the hMBX and hMBXt motility tracks by sub-pixel localization enabled by the bright TMR labels and linked these into trajectories, as previously described (Arcizet et al., 2008; Zajac et al., 2013; Hafner et al., 2016; Tinevez et al., 2017). The mean-squared displacements were compared as a function of time on a log–log plot, where a slope of α = 1 indicates a purely diffusive process, α < 1 indicates confined diffusion, and α = 2 describes processive transport (Arcizet et al., 2008; Ruthardt et al., 2011; Zajac et al., 2013; Hafner et al., 2016). We calculated the MSD for all hMBX and hMBXt motility events (paused and processive segments) and fit it to the function 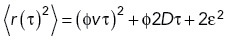 where φ is the fraction of time in the processive state (Chugh et al., 2018). From this global analysis, the fraction of time paused is 0.47 for hMBXt compared with 0.03 for hMBX motors (Figure 3B). We then used a sliding window to calculate a local α-value for each point in the trajectory (Figure 3A; Supplemental Figure S3). For hMBX, we observe a distribution centered around ∼1.8 indicating primarily processive motility. In contrast, hMBXt exhibits a bimodal α-value distribution with peaks at 0.63 ± 0.04 and 1.76 ± 0.04 corresponding to paused and processive segments (Figure 3C). We then applied change-point analysis to identify segments of processive and paused motility (Beausang et al., 2011). From these analyses, we determined that ∼41%, ∼27%, and ∼12% of long trajectories contained zero, one, or two pauses, respectively (Supplemental Figure S3), with a mean pause time of ∼13.5 s (Supplemental Figure S3). The velocity of the hMBXt motor during processive segments displayed a single distribution (Figure 3D; 54.2 ± 1.4 nm/s) while the mean velocity during paused segments is near zero (μ = 3.9 ± 1.8 nm/s and σ = 52.3 ± 1.3 nm/s) suggesting static motors that are strongly attached to MTs (Figure 3D). To confirm this interpretation, we performed motility assays on hMBXt with the slowly hydrolyzable ATP analog, AMPPNP, which is expected to trap motors in a strongly bound state (Chen et al., 2016). As expected, hMBXt exhibited only static binding to MTs in the presence of AMPPNP, with a normally distributed α-value (0.10 ± 0.32), indicative of static binding (Supplemental Figure S3, F and G). Together, these analyses indicate that pauses are due to the static binding of hMBXt motors to MTs while processive motility is dominated by active transport. We also propose that the paused states represent the nucleotide-free rather than the ATP-like bound state, in both of which the motor domain has a high affinity for MT lattice sites (Cross and McAinsh, 2014).
where φ is the fraction of time in the processive state (Chugh et al., 2018). From this global analysis, the fraction of time paused is 0.47 for hMBXt compared with 0.03 for hMBX motors (Figure 3B). We then used a sliding window to calculate a local α-value for each point in the trajectory (Figure 3A; Supplemental Figure S3). For hMBX, we observe a distribution centered around ∼1.8 indicating primarily processive motility. In contrast, hMBXt exhibits a bimodal α-value distribution with peaks at 0.63 ± 0.04 and 1.76 ± 0.04 corresponding to paused and processive segments (Figure 3C). We then applied change-point analysis to identify segments of processive and paused motility (Beausang et al., 2011). From these analyses, we determined that ∼41%, ∼27%, and ∼12% of long trajectories contained zero, one, or two pauses, respectively (Supplemental Figure S3), with a mean pause time of ∼13.5 s (Supplemental Figure S3). The velocity of the hMBXt motor during processive segments displayed a single distribution (Figure 3D; 54.2 ± 1.4 nm/s) while the mean velocity during paused segments is near zero (μ = 3.9 ± 1.8 nm/s and σ = 52.3 ± 1.3 nm/s) suggesting static motors that are strongly attached to MTs (Figure 3D). To confirm this interpretation, we performed motility assays on hMBXt with the slowly hydrolyzable ATP analog, AMPPNP, which is expected to trap motors in a strongly bound state (Chen et al., 2016). As expected, hMBXt exhibited only static binding to MTs in the presence of AMPPNP, with a normally distributed α-value (0.10 ± 0.32), indicative of static binding (Supplemental Figure S3, F and G). Together, these analyses indicate that pauses are due to the static binding of hMBXt motors to MTs while processive motility is dominated by active transport. We also propose that the paused states represent the nucleotide-free rather than the ATP-like bound state, in both of which the motor domain has a high affinity for MT lattice sites (Cross and McAinsh, 2014).
FIGURE 3:
The Kinesin-5 tail domain induces pauses, increases run lengths, and decreases motility velocities of the minitetramer kinesin-5. A) Pauses (red) and periods of processive motility for hMBX motor (blue) were identified in trajectories based on the slope of the mean-squared displacement (α) calculated within a sliding window along the trajectory. Alpha fluctuates between ∼2 and ∼0 indicating processive and stationary motility (bottom). B) The MSD for the entire trajectories of hMBX motors were fit to an expression that describes periods of stationary pauses and motile events consisting of processive and diffusive movements, MSD = (φvt)^2+φ2Dt+2ε^2 where φ is the fraction of time in a motile state. HMBX motors pause more frequently. C) Plotting all local α-values reveals primarily processive motility (green), while hMBXt exhibits a bimodal distribution with peaks consistent with static binding (μ1) and processive motility (μ2; purple). D) Histograms showing the frame-to-frame velocity of hMBXt motors (purple; solid line) and hMBX motors (green; dotted line). hMBX motors move faster than hMBXt motors during the processive sections of trajectories. Velocities are similar during paused sections. E) The total run length for hMBX motors was fit with a single exponential distribution with mean μ. All trajectories > 3 frames were included (n = 2056 trajectories). Unless otherwise specified, n = 111 trajectories from three independent experiments for hMBX constructs and n = 143 trajectories from nine independent experiments for hMBXt constructs.
In addition to promoting pausing, the tail domain also enhances the processive run lengths of hMBXt motors. The run length distribution of hMBX is described well by a single exponential with a characteristic run length of 0.56 ± µm, Figure 3E). In contrast, the run length distribution for hMBXt is described by a double exponential with run lengths of 0.55 ± 0.06 µm and 2.94 ± 1.11 µm, with 90 ± 5% of runs in the shorter population (Figure 3E). Plotting the interpause run lengths revealed a single-exponential distribution with a mean run length of 1.26 ± 0.17 µm (Figure 3E; Supplemental Figure S3). Thus, the shorter of the two estimates for total run lengths likely corresponds to the interpause run length. Together, these data suggest that hMBXt motors may string together multiple shorter runs interspersed with pauses, allowing for a longer total-run length. Additionally, both hMBXt and kMBXt motors exhibit very similar pausing behavior, suggesting conservation across motors from different species (Figure 2, D and H).
The kinesin-5 tail domain promotes motor clustering
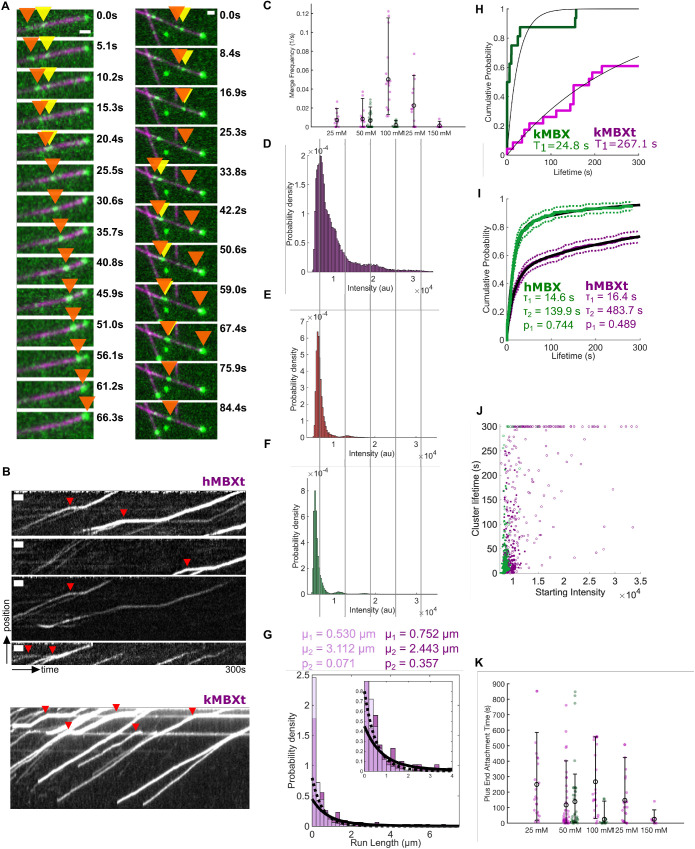
We next sought to determine the molecular mechanism behind multi-motor clustering for both hMBXt and kMBXt motors observed during their traversal along MTs (Figure 4, A and B). We compared the clustering properties of kMBXt and kMBX across different ionic-strength conditions (Figure 4C; Supplemental Movie S3). For each condition, we normalized the intensities of spots corresponding to motile motors along MTs visible in each field of view and analyzed their distribution to determine an average intensity of individual minitetramers and identify motors per cluster stoichiometries (see Materials and Methods; Pandey et al., 2021a; Supplemental Figure S4A). The analysis revealed a major peak of spot intensities likely representing individual kMBXt minitetramers, and a higher and broad-intensity peak representing a collection of brighter clusters composed of multiple motors. At the lowest-ionic strengths examined, most single kMBXt motors merged with either one or a cluster of two paused motors, which we denoted as (motile + static; Supplemental Figure S4F). However, at higher-ionic strengths, a wider range of clusters was observed, representing 5–10 motors per cluster. We next calculated the total frequency for kMBXt motor merging into multimotor clusters per unit MT length across the range of ionic strengths tested, which revealed a peak around 100 mM KCl, and low-clustering frequency at both 25 mM and 150 mM KCl (Figure 4C). In contrast, few whether any clustering events were observed for kMBX motors across all conditions examined. Likewise, we calculated a near-zero merging frequency for kMBX motors (Figure 4C). Finally, we examined the relationship between kMBXt motor-cluster size and average velocities, run lengths, and run times of the clusters. At low-ionic strength, we found that clusters contained fewer motors while run lengths and run times were shorter. In contrast, at intermediate ionic strength, we found brighter clusters containing more motors and observed an 80–100% increase in run lengths and run times (Supplemental Figure S4G). Together, these data suggest that the tail is involved in kinesin-5 assembly into multimotor clusters and that the formation of higher-order clusters can enhance both run length and run time in an ionic strength-dependent manner.
FIGURE 4:
Kinesin-5 minitetramers with the tail domain, but not those without the tail, form multimotor clusters while undergoing motility along an MT. A) hMBXt minitetramers form clusters while moving along the MT. Two example montages are shown. A subset of hMBXt motors is paused (yellow arrow), while others are processively moving (orange arrow). These two types of motors merge and then continue moving together as clusters (orange arrow). Six (left) or 10 (right) frames between timepoints. Scale bars 1 μm wide. B) Kymographs showing the assembly of hMBXt and KMBXt motor clusters. Note that many of these clusters form while one set of motors involved is paused (red triangles). Scale bars 10 s wide and 1 μm tall. C) The cumulative frequencies and their average for kMBX (green) and kMBXt (pink) motors to merge into clusters in relationship to solution-salt concentration (mM). The extended data for kMBX and kMBXt motor clustering are shown in Supplemental Figure S4., D–F) Intensity distribution for constructs D) with the tail domain (hMBXt) in the presence of ATP, E) with the tail domain (hMBXt) in the presence of AMP-PNP, and F) without the tail domain (hMBX) in the presence of ATP, suggesting that the minitetramers require the tail domain and motility to form clusters. Clusters likely correspond to two to three motors, and the blurring of the intensity distribution for motors in the presence of ATP is likely due to the quenching and unquenching of the TMR sensors used as labels during motor stepping. Lines are ∼6500 au apart. For D, n = 1406 trajectories from nine independent experiments. For E, n = 1298 trajectories from two independent experiments. For F, n = 2056 trajectories from three independent experiments. G) Total run-length distributions for single hMBXt motors (light purple; dotted line) and clusters (dark purple; solid line). Distributions were fit with a double-exponential distribution, and both groups have a short (μ1) and long (μ2) run length population. However, the fraction of motors in the long-run length population (p2) is larger for clusters. H, I) hMBXt and hMBXt Motors often reach the plus-ends of stabilized MTs and remain bound there for some time. Lifetimes were fit to a Weibull distribution. The hMBXt motors were best fit by a double Weibull distribution, suggesting the presence of populations with a short and long lifetime. The short lifetime (τ1) was similar for both hMBX and hMBXt, whereas the long lifetime (τ2) was longer for hMBXt than hMBX motors. J) Distribution of lifetimes of motors/clusters at the ends of MTs as a function of their starting intensity (average intensity of the first five frames they are detected). hMBX motors (green) generally arrive as single motors, whereas hMBXt motors (purple) often arrive as motor clusters. Open circles denote censored lifetimes (present in the first or last frame of the movie, indicating they are at least as long or longer than the point plotted). K) The cumulative MT plus-end association times for kMBX and kMBXt motors and their average values in relationship to the salt concentration (mM). The extended data for kMBX and kMBXt motor clustering are shown in Supplemental Figure S4, F and G).
Movie S3.
motility of kMBXt motors at four different ionic strength conditions (25‐150 mM KCl) revealing the impact of ionic strength on motor clustering, processive motility, and plus‐end attachment. Clustering is improved at 50 mM KCl and decreases at 100‐150 mM KCl. MT plus end accumulation occurs at 25‐100 mM KCl and is diminished at 150 mM KCl.
We next examined the cluster and motility properties of hMBXt motors, which also formed clusters when encountering a subset of the paused hMBXt motor along MTs (Figure 4, A and B; Supplemental Figure S4, F–G). Because the hMBXt were chemically labeled with TMR, we were able to estimate the number of hMBXt motors in these clusters more precisely using multimodal distribution analyses. We observed broad peak at ∼6500au, representing single hMBXt minitetramers, and additional peaks at two and three higher intensity (∼13,000 au and ∼19,500 au) representing approximately clusters with two or three minitetramers (Figure 4D). In the presence of AMPPNP, however, intensity distribution was predominantly unimodal (major peak at ∼6100 au and a minor peak at ∼13,000 au), suggesting that motility is required for hMBXt clustering along MTs (Figure 4E). The multimodal distribution of hMBX intensities (Figure 4F), was similar to that of the hMBXt motors in the presence of AMPPNP, with a major peak at ∼5300 au and without higher-intensity clusters. The small shift in average intensity is due to a difference in TMR labeling ratios for the two constructs. We next measured the run length of hMBXt motor clusters moving along MTs. We quantified the total run length of clusters with intensity ≥1.5-fold higher than hMBXt minitetramer intensity, revealing run lengths of 0.75 ± 0.39 µm and 2.44 ± 1.65 µm, similar to the run lengths of 0.53 ± 0.06 µm and 3.11 ± 1.49 µm for single hMBXt motors. However, only 7 ± 4% of single hMBXt motors were in the longer lifetime population, whereas 36 ± 40% of hMBXt clusters were in this population (Figure 4G; Table 3). Together, these data are consistent with those from the kMBX/kMBXt motors, suggesting clustering and enhanced processivity are conserved properties of kinesin-5.
TABLE 3:
Weibull distribution fits of lifetimes for hMBX and hMBXt motors (Figure 3I). 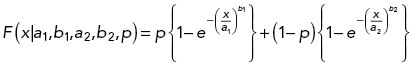
| a1 | b1 | a2 | b2 | p | |
|---|---|---|---|---|---|
| hMBX | 14.6 ± 3.2 | 1.07 ± 0.20 | 140 ± 138 | 0.76 ± 0.42 | 0.74 ± 0.18 |
| hMBXt | 16.4 ± 4.9 | 0.88 ± 0.12 | 484 ± 153 | 0.89 ± 0.64 | 0.49 ± 0.15 |
The kinesin-5 tail induced clusters to accumulate at MT plus-ends
We next analyzed the nature of the accumulation of minitetramers at MT plus-ends. Both hMBXt and kMBXt motors are highly processive, with run lengths that extend nearly the full lengths of MTs examined (McHugh et al., 2018). We observe an accumulation of both hMBXt and kMBXt at MT plus-ends, indicative of their stable association (Figure 4A). We quantified the dwell times of hMBXt at MT plus-ends by measuring the intensity centered at MT plus-ends and defining cluster lifetime as the period for which the intensity was > 3 SDs higher than the background. The hMBXt motors pause for 10 times as long as hMBX motors at MT plus-ends, suggesting the tail domain enables MT plus-end association (Figure 4, H and I). We used mixed Weibull distribution to fit hMBX MT plus-end dwell-times and revealed two populations of motors at MT plus-ends with different dwell times (16.4 ± 4.9 s and 483.7 ± 153.2 s; Figure 4I, Figure 4, H–K) with 49 ± 15% of them having shorter lifetimes. Next, we analyzed MT plus-end lifetimes as a function of the intensity of the first five frames of cluster arrival at MT ends, revealing that motors that arrived “preformed” clusters along the MT were likely to remain at MT plus-end (Figure 4J). We used the starting spot intensity rather than the mean spot intensity to distinguish between “preformed” clusters and motors arriving individually and gradually accumulating at MT plus-end. Intensity traces in the region near MT plus-ends increased above baseline or decreased to baseline in one or a few frames, regardless of the plateau intensity. This suggests “preformed” clusters arrive at MT plus-ends (Figure 4J; Supplemental Figure S4, J and K). The shorter lifetime likely represents single hMBXt motors residing at the ends of MTs, while the longer lifetime represents hMBXt clusters. The hMBXt clusters may drag MT plus end along aother MT acting to align the two MTs (Supplemental Movie S4).
Movie S4.
An example of a cluster of kinesin‐5 hMBXt motors dragging the plus‐end at which the motors accumulated along another MT, acting to align the two MTs. This demonstrates that the plus‐end clusters are still motile and can withstand load (the MTs are quite bent and still anchored to the coverslip at some sites by an avidin‐biotin interaction). Note that to observe this, a higher density of MTs was immobilized using a lower density of Neutravidin. 50 frames between time points. Scale bar 2μm wide.
The hMBX and kMBX motors did not accumulate as robustly at MT plus-ends, supporting the critical role of the tail regulation in this process (Figure 4I). The hMBX dwell times at MT plus-ends were 14.6 ± 3.2 s and 139.9 ± 137.5 s, with 74 ± 18% of motors in the shorter lifetime population (Figure 4I; Supplemental Figure S4K). Thus, the short dwell times for hMBX and hMBXt are similar, in agreement with the idea that this represents the lifetime of single motors at MT plus-ends. In contrast, the longer hMBX lifetimes remain short due to their inability to form clusters (Figure 4H, I and J). To determine the possibility of hMBXt clusters reaching MT plus-ends, we next compared the distribution of frame-to-frame velocities for single motors and clusters in the paused and processive states (Figure 4). Our analysis agrees with the visual observations, that hMBXt clusters move more slowly than single hMBXt motors: whereas the mean paused velocities of single motors (4.6 ± 3.1 nm/s and σ = 64.8 ± 2.2 nm/s) and clusters (3.6 ± 1.9 nm/s and σ = 37.6 ± 1.3 nm/s) were similar, the processive velocity of single motors (63.3 ± 2.3 nm/s) was higher than that for clusters (43.2 ± 1.6 nm/s). Additionally, the velocities of single motors were more variable than that of clusters. During the processive sections of trajectories, the SD of single motor velocities was 65.3 ± 1.6 nm/s, whereas that of clusters was 39.5 ± 1.2 nm/s. The effect on slow motility in clusters versus single motors might contribute to the observation that the velocity of hMBXt (54.2 ± 1.0 nm/s) is slower than that of hMBX (80.4 ± 4.4 nm/s).
We next analyzed the impact of ionic strength on kMBXt MT plus-end residence. The kMBX motors dwell at MT ends for 78 ± 19 s at 50 mM KCl and for 25 ± 18 s at 100 mM KCl, showing a three-fold decrease upon a two-fold increase in ionic strength (Figure 4K). In contrast, kMBXt showed consistently longer dwell time at MT plus-ends, which is two-three folds higher than kMBX (125–250 s) and is not influenced by changes in ionic strength (25–150 mM KCl; Figure 4K). The majority of kMBXt clustering events also correlated with the motors arriving at MT plus-ends, particularly at 100 mM KCl. This is likely due to the enhanced multimotor clustering for kMBXt, coupled with its enhanced dwell time at MT plus-ends at 100 mM KCl. Taken together, our data suggests that the MT plus-end dwell time is enhanced by tail-induced clustering and pausing.
Short kinesin-5 minitetramers show unique MT crosslinking and MT sliding features
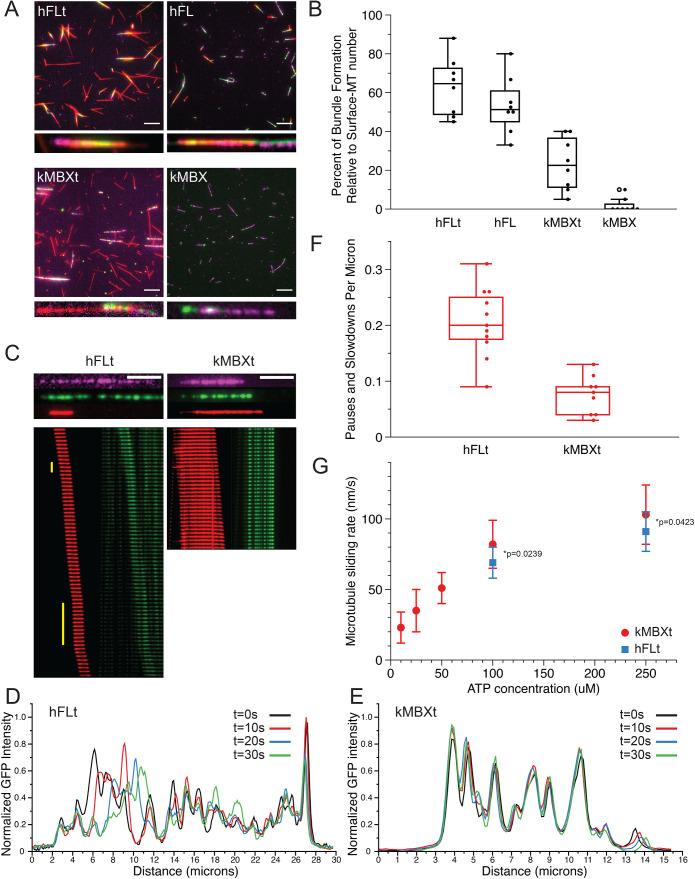
We next sought to determine whether kinesin-5 minitetramers are capable of crosslinking, aligning, and sliding pairs of MTs, and how their activities compare with native full-length kinesin-5 motors. For these assays, we immobilized taxol-stabilized HiLyte647- and biotin-labeled MTs on a coverslip via a streptavidin-biotin linkage. We found that to achieve efficient MT bundling, particularly for the minitetramer motors (see below), we required high-velocity flow of MTs within flow cells such that they were well nearly aligned with the direction of flow. Therefore, MT pairing under rapid flow appears to enhance parallel-crosslinking orientation. MT pairs were most likely to form at low relative angles and crosslink as nearly aligned pairs. Next, motors were introduced into the sample chamber at specified concentrations and allowed to decorate the MTs which are attached to coverslips. Finally, kinesin-5 motor, Rhodamine-labeled MTs and ATP were introduced with rapid-flow rates, and kinesin-5 mediated crosslinked MT bundles were allowed to form, and then motors initiated relative MT sliding. We then imaged both the coverslip attached MT (purple), paired MTs (red) and kinesin-5 motors (green) using the three different channels via TIRF microscopy at 2–5 s frame rates.
We first set out to determine how efficiently four different kinesin-5 motor constructs can crosslink and pair MTs by direct comparison: full-length human kinesin-5 (Eg5) with tail (hFLt), full-length kinesin-5 without the tail (hFL), as previously prepared and described (Bodrug et al., 2020), D. melanogaster kinesin-5 minitetramer with tail (kMBXt), and kinesin-5 minitetramer without a tail (kMBX). Due to being similarly fused to green fluorescent proteins (GFP or NG), the D. melanogaster minitetramer motors (kMBX and kMBXt) were much more suitable to compare to human full-length kinesin-5 motors (hFLt and hFL) in the MT-sliding assays. The hMBX and hMBXt were chemically labeled with TMR, and thus their localization could not be directly compared with the hFLt motors in our multicolor MT-sliding assay imaging setup. We then imaged multiple fields-of-view immediately after flowing in ATP and Rhodamine-MTs and measured how many MT bundles had formed relative to the population of surface-bound MTs (Figure 5, A and B). We found that both hFLt and hFL motors recruited free MTs from the solution, forming MT pairs that underwent sliding at relatively high rates, with 63 ± 5% and 54 ± 16% in the hFLt and hFL motor conditions, respectively. This is consistent with observations we made previously at similar concentrations (Bodrug et al., 2020). In contrast, the kMBXt motor formed MT bundles three-fold less frequently (24 ± 14%). Whereas the kMBX almost never recruited a free MT from solution and rarely formed any MT pairs, with only 2% of all surface MTs observed to bundle with free MTs (Figure 5, A and B). Together, these data suggest that the shorter and stiffer kinesin-5 minitetramers are less efficient than the full-length kinesin-5 motors at forming aligned antiparallel-MT bundles and that the tail domain increases the probability of establishing MT crosslinked geometries, especially for minitetramers. These data suggest that the presumed higher-torsional flexibility of the 60–80 nm kinesin-5 bipolar minifilament is critical for crosslinking and aligning pairs of MTs. Decreasing the length of the bipolar minifilament is believed to increase torsional rigidity, leading to a three-fold decrease in the efficiency for MT crosslinking and alignment even when incident angles of the two MTs are very low due to the rapid flow in our imaging system. Our data also show that unlike the hFLt motors which only requires the tail to transition from crosslinking to sliding states and for generating high sliding forces (Bodrug et al., 2020), kinesin-5 tail is essential for the kMBXt to form crosslinks and then further pair MTs.
FIGURE 5:
Kinesin-5 minitetramers show defects in crosslinking MTs but enhanced MT sliding once aligned, compared with native kinesin-5. A) Representative TIRF images of surface-immobilized MTs (magenta) crosslinked via kinesin-5 (green) constructs to free MTs (red). Four different kinesin-5 constructs were examined: hFLt, hFL, kMBXt, and kMBX. Scale bar = 10 microns. B) Percentage of surface-immobilized MTs that engaged in kinesin-5 mediated crosslinking with free MTs for each kinesin-5 construct. N = 8 fields of view analyzed per condition. C) Sample MT pairs (top) and kymographs (bottom) depicting MT sliding driven by either hFLt (left) or kMBXt (right). Free MT (red) and kinesin-5-GFP or nNG (green) are shown. Pauses are identified by the vertical yellow bar. Scale bar = 4 microns; frame rate for hFLT = 10 s per frame; for kMBXt = 5 s per frame. D) Linescan analysis of the GFP signal from hFLt data shown in C). Normalized GFP intensity is plotted against distance along the microtubule for four different timepoints. E) Linescan analysis of the GFP signal from kMBXt data shown in C). Normalized GFP intensity is plotted against distance along the microtubule for four different timepoints. F) Number of pauses or velocity reduction events observed per micron for hFLt and kMBXt driven MT sliding. N = 11 events for hFLt, N = 9 events for kMBXt. G) Average MT-sliding rate calculated for bundles at different ATP concentrations. N = 6 events for each condition. Error bars are SD, values are reported in Table 4.
We next sought to determine how kinesin-5 minitetramers compared with full-length kinesin-5 during MT sliding. We therefore identified crosslinked bundles with aligned MTs and overlap lengths of at least two microns and monitored the positions of the mobile MT with respect to the full-length or mini-tetramer kinesin-5 motors during these sliding events across a range of conditions. As we were unable to reliably form bundles using kMBX motors in our assays, we focused on comparing the crosslinking and sliding mechanics of hFLt to kMBXt motors. Both motors were able to slide MTs apart efficiently (Figure 5C). We also observed that the mobile-free MT occasionally paused or exhibited reduced velocity for brief stretches when undergoing sliding by hFLt motors (Figure 5C left panel; Supplemental Figure S5, A–B and Movie S5). In contrast, MT sliding generated by the kMBXt motor tended to exhibit more consistently continuous MT-sliding motility with faster velocities throughout (Figure 5C; Supplemental Figure S5, C–D, Movie S6). It is not clear how the pausing in the case of hFLt motors relates to the movement of motor clusters along MTs. To examine this difference, we determined the frequency of observed pauses or velocity decreases across many MT sliding examples and found that MT bundles in the case of hFLt motor-condition exhibited pauses 0.21 ± 0.06 times per μm of distance traveled, while kMBXt motors paused at a lower rate of 0.08 ± 0.04 times per μm which is three-fold lower than the pausing exhibited by hFLt motors during MT sliding (Figure 5F). Finally, we calculated the MT sliding velocities across a range of ATP concentrations for both constructs (Figure 5G). We found that the MT sliding velocity was approximately twice the speed of motor velocity along single MTs, consistent with the previously reported finding that both pairs of motor domains at each of the bipolar end of kinesin-5 motors undergoing processive motility along their respective MT in the opposite direction, leading to a two-fold MT sliding motility compared with the motility generated along single MTs (Kapitein et al., 2005). Importantly, we observed that the average sliding velocity of kMBXt motor-driven MT sliding at 100 and 250 μM ATP was about 10% higher than that hFLt motors (Table 4). This suggests that hFLt motors may have more compliance than kMBXt motor leading to less than 10%, lower than maximal two-fold single MT-motility velocity during MT sliding. Together, these results indicate that, while the kinesin-5 mini-tetramers are not as efficient at initially crosslinking aligning two MTs as native full-length kinesin-5, they are capable at sliding MTs as with less three-fold less pausing and 10% higher velocities compared with those generated by full-length kinesin-5, which also assemble into clusters during MT sliding.
TABLE 4:
MT sliding velocities for Native and minitetramer kinesin-5 motors.
| ATP (μM) | hFLt | kMBXt |
|---|---|---|
| 10 | N/A | 23 ± 11 nm/s (n = 9) |
| 25 | N/A | 35 ± 15 nm/s (n = 6) |
| 50 | N/A | 51 ± 12 nm/s (n = 11) |
| 100 | 69 ± 11 nm/s (n = 13) | 82 ± 17 nm/s (n = 15) |
| 250 | 91 ± 14 nm/s (n = 17) | 103 ± 21 nm/s (n = 14) |
Movie S5.
Three examples of MT sliding by native kinesin‐5 (hFLt). The hFLt motors (green) slide a paired MT (red) along the anchored MT (magenta).
Movie S6.
Three examples of MT sliding by mini‐tetramer‐kinesin‐5 (kMBXt). The kMBXt motors (green) slide a paired MT (red) along the anchored MT (magenta).
Finally, we determined the role of multi-motor clustering for the full length and minitetramer kinesin-5 during the MT-sliding process (Supplemental movie S5–S6). We used normalized-fluorescence linescan analyses for the GFP or NG channel at multiple time-points during MT sliding events to evaluate the clustering of these motors during MT sliding events. This analysis revealed that both kMBXt and hFLt motors formed multimotor clusters while sliding MTs. However, hFLt motors tended to exhibit more mobile-cluster peaks suggesting movement of or within clusters, in contrast to the kMBXt motors which were more static during MT sliding events over similar time-periods (Figure 5, D and E). These observations are consistent with clustering a critical aspect of kinesin-5 binding along MTs during MT sliding motility. Differences in cluster kMBXt and hFLt motor mobilities during MT sliding suggest differences in the force-transmission processes of these two motor systems which are influenced by the distinct length of their bipolar minifilaments.
DISCUSSION
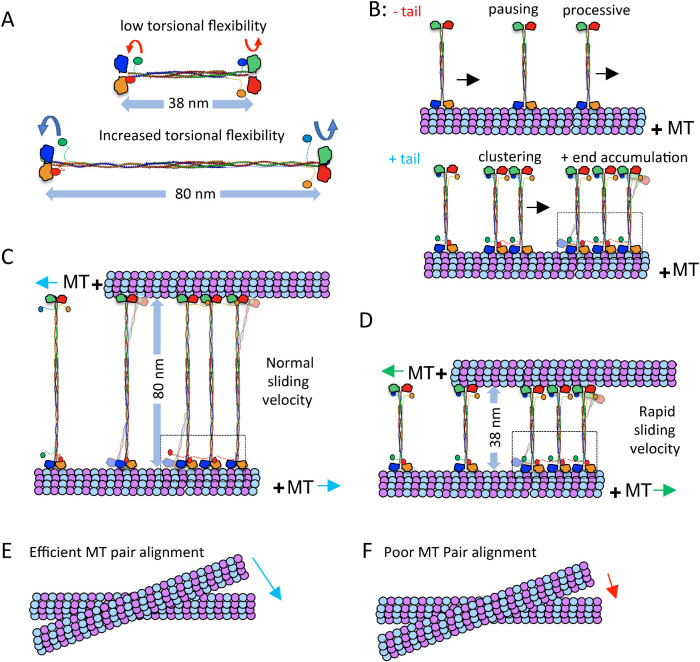
Here we compared the motility and structure of full-length and structure-inspired minitetramer kinesin-5 motors, revealing the roles of both the C-terminal tail and bipolar-tetrameric minifilament domains in governing the conserved kinesin-5 MT-sliding motility mechanism. We also demonstrate that the structural and biochemical features of these domains are highly conserved across all eukaryotic kinesin-5 motors. Our BASS-XL X-ray structure reveals that it possesses the unique capacity to form a force-bearing junction for two coiled coils and motors positioned at opposite ends of its central bipolar-tetrameric structure. Guided by this structure, we designed and studied Human and D. melanogaster kinesin-5 minitetramers and compared their behavior to full-length kinesin-5 (Bodrug et al., 2020). These kinesin-5 minitetramers recapitulate critical aspects of full-length kinesin-5 including multimotor clustering and MT sliding and shed new light on how features impact the kinesin-5 mechanism. These studies, in conjunction with our previous study, reveal the critical roles for the kinesin-5 tail domain and the length of the central-bipolar minifilament in MT crosslinking and sliding. By comparing these motors to full-length kinesin-5 with and without the tail domain, we show how these two adaptions dial their force transmission during MT sliding (Bodrug et al., 2020). These structural adaptations are critical for kinesin-5 function in bipolar-mitotic spindle assembly, organization, and elongation (Figure 6).
FIGURE 6:
Kinesin-5 MT sliding motility is tuned by the tail to motor mediated clustering and the length of the bipolar minifilament. A) Kinesin-5 mini-tetramers are 38 nm in length while native kinesin-5 motors are 60–80 nm in length. The decreased minifilament length leads to a decrease in torsional flexibility in the minitetramers, compared with the high-torsional flexibility of native kinesin-5. B) Top panel, kinesin-5 minitetramers, without the tail domain, show processive motility punctuated by pauses and short residence time at MT plus-ends. The bottom panel, kinesin-5 minitetramers, with the tail domain, show increased pausing, coupled with motor clustering mediated by cross motor-tail interactions between multiple minitetramers. C) MT sliding motility mediated by native kinesin-5 leads to 80-nm separation between paired MTs, which slide apart with normal sliding velocity, which is punctuated by pauses and is lower than twice the velocity of each motor ends along each MT. D) MT sliding motility by the kinesin-5 minitetramers leads to 38-nm separation between the paired MTs and a more efficient MT sliding motility that approaches closely to twice the velocity of each motor end. E) The native kinesin-5 motor MT pair alignment is efficient due to the torsional flexibility of the minifilament F) Kinesin-5 mini-tetramer MT pair alignment is poor due to decreased torsional flexibility of its shortened minifilament.
The kinesin-5 tail domain-binding regulates the motor domain mechanochemical cycle and promotes slower motility and increased processivity
We have previously demonstrated that the kinesin-5 tail domains down-regulate MT-activated ATP hydrolysis of the motor domain by stabilizing its MT-bound nucleotide-free state (Bodrug et al., 2020). This interaction leads to slower processive motility, longer run lengths, and frequent pauses along MTs (Figures 2 and 3). As with full length kinesin-5 (Bodrug et al., 2020), the presence of the tail domains in both D. melanogaster and human minitetramers leads to slow motility with frequent pauses in which motors are statically bound to MTs (Figure 2). Motor pauses represent strongly bound states with extended lifetimes in which we postulate that the leading-motor domain is in the no nucleotide state, while the trailing-motor domain is either in the ADP-Pi or ADP state. This is similar to the so-called ATP gate or stepping gate (Andreasson et al., 2012; Cross and McAinsh, 2014; Bodrug et al., 2020). This is also somewhat like motor motility in the presence of a mixture of ATP and AMP-PNP, in which comparable switches in motility are observed (Vugmeyster et al., 1998; Subramanian and Gelles, 2007). We suggest that docking of the kinesin-5 tail onto the motor domain serves as an externally imposed gate that is highly conserved and which enhances pausing and promotes assembly into clusters mediated via encounters between motile and paused motors along MTs. We further hypothesize that transinteractions between the tails of one kinesin-5 tetramer and the motor domains of other tetramers are likely to be the source of this multimotor clustering. Interestingly, multimotor clustering has also been described for the budding yeast, Saccharomyces cerevisiae, and orthologue Cin8 and has been proposed to mediate its minus-end to plus-end MT-motility directionality reversal (Shapira et al., 2017; Singh et al., 2018; Pandey et al., 2021a; Pandey et al., 2021b).
The kinesin-5 tail domain drives the formation of multi-motor clusters with different motile properties than single motors on the lattice and at plus-ends
Our studies reveal that by increasing the frequency of pauses, the likelihood of encounters between motile and paused motors also increases to promote the formation of multimotor clusters (Bodrug et al., 2020). For the subset of interactions that lead to clustering, we speculate that these motors are engaged with the same protofilament, while motor crossing events which do not result in clusters likely involve proteins moving on separate protofilaments. Once within a cluster, our analyses suggest multiple motors move together as assemblies, though the nature of the motor-tail interactions within these structures remains unclear and warrants future studies (Figure 4). Larger clusters lead to longer run lengths likely due to the increase in the availability of motor domains within the cluster, as evidenced by the observed increase in total run lengths in kMBXt and hMBXt motors compared with kMBX and hMBX motors. Similarly, the lifetime of motors at MT plus-ends increases with cluster size and kMBXt or hMBXt clusters was found to move at slower and less variable speeds than single motors (Figure 4). Accumulation of kinesin-5 at MT plus-ends has been described previously, although with different domain requirements than those described here (Chen and Hancock, 2015; Chen et al., 2019). We interpret this MT plus-end accumulation to be due to the extended lifetime of kinesin-5 strongly-bound motor states which are enhanced by the tail-to-motor interactions and the resulting motor clusters at MT ends (Vershinin et al., 2007; Furuta et al., 2013). Finally, our MT crosslinking and sliding analyses show that both full-length and mini-tetramer kinesin-5 form clusters during MT sliding.
Taken together, we propose that kinesin-5 tail induced motor clustering is essential for amplifying and coalescing the motile forces of individual kinesin-5 tetramers during MT sliding. We postulate that kinesin-5 clusters and single motors may be fulfilling different roles during mitotic spindle assembly and elongation. For example, clusters of kinesin-5 may be present between parallel MTs, such as near the spindle poles, where they could assist in MT capture early in spindle assembly. Kinesin-5 clusters may be selectively retained in this region because they move slower than single motors. In contrast, faster-moving single motors may “escape” and travel towards MT plus-ends, localizing them in the region of antiparallel MT overlap in the spindle midzone where they may merge again to form multimotor clusters. Here, these motors could facilitate spindle pole separation as clustered motor assemblies. Finally, the kinesin-5 tail domain contains a conserved BimC box or CDK1 phosphorylation site (Threonine 926 in human kinesin-5), and the more distal regulatory regions of the tail domain (i.e., the KEN box, D box, and Nek6 phosphorylation site; Blangy et al., 1995; Rapley et al., 2008; Bertran et al., 2011; Drosopoulos et al., 2014). The phosphorylation of the kinesin-5 tail by Cdk1 kinase has been shown to increase the affinity of the motor for MTs in vitro (Cahu et al., 2008). We postulate that such phosphorylation may enhance the tail-motor interaction by adding a negative charge to the tail, potentially increasing its affinity for the motor domain, which is positively charged near the ATP binding site. The impact of phosphorylation on the kinesin-5 tail domain warrants further study.
The length of the Kinesin-5 bipolar minifilament directly regulates force transmission during MT-sliding motility
Finally, our studies elucidate the crucial role of the 60–80 nm Kinesin-5 bipolar central minifilament in the MT sliding mechanism. The kinesin-5 minitetramers are half the length of native kinesin-5 and are therefore likely both stiffer and have lower-torsional flexibility due to the shorter dimeric coiled-coils on either side of the BASS domain. This shorter minifilament may enhance the force coupling between the bipolar ends of kinesin-5 as they engage the two MTs they crosslink. However, the decreased length and torsional flexibility between each end of the kinesin-5 minitetramers appears to impede efficient MT alignment into MT bundles, leading to the observed defects in the initial crosslinking. Once aligned, however, we propose that the shorter minifilament leads to a higher maximal MT-sliding velocity due to the increased compliance and stiffness of the minitetramers.
Our studies also show the relationship between the kinesin-5 minifilament length and the tail-induced regulation in modulating MT crosslinking and sliding motility. Both full-length and minitetrameric kinesin-5 form clusters during MT-sliding motility, but the absence of the tail and shorter minifilament length nearly eliminated MT crosslinking and sliding. In contrast, tailless full-length kinesin-5 was still able to crosslink and slide MTs (Bodrug et al., 2020). These data also show that the tail-motor clustering likely enhances force transmission by kinesin-5 likely by focusing and synchronizing individual motors, and future studies of direct force measurement are warranted.
In addition to its role in sliding antiparallel MTs, kinesin-5 motors have been suggested to act as a brake to slow the rate of MT sliding by other motors, both during anaphase (Saunders et al., 2007; Rozelle et al., 2011) and in nonmitotic cells (Myers and Baas, 2007; Nadar et al., 2008; Falnikar et al., 2011; Lin et al., 2011; Nadar et al., 2012). Moreover, the ability of kinesin-5 motors to act as a brake between parallel MT or quickly sliding antiparallel MTs has been demonstrated in vitro (Shimamoto et al., 2015). The tail domain-docking onto the motor domain induces pausing and motor clustering and may contribute to the capacity of kinesin-5′s brake-like behavior by increasing the time these motors spend in a strongly bound state on the MT lattice and promoting efficient MT crosslinking and sliding. We speculate that a motor is less likely to detach when subjected to loads while it is in tail-induced clusters and paused states than while moving as individual tetramers, allowing them to withstand substantial loads when acting as a brake.
Taken together, our studies identify how the unique conserved structural adaptations of the kinesin-5 tail and tetramerization domains enable its role in organizing the mitotic spindle. The tail domain promotes motor pausing and clustering. Single motors and clusters behave differently, suggesting that they may fulfill different roles in spindle assembly. The relatively long length of the minifilament provides the molecule with the flexibility required to efficiently crosslink and slide MTs.
MATERIALS AND METHODS
Protein Production, X-ray crystallography, and Model building
The KLP61F minifilament extended region (residues 597–833) was expressed and purified in BL21 Escherichia coli as described in (Kashina et al., 1996; Scholey et al., 2014). Bacterial pellets were lysed using a microfluidizer in (300 mM KCl, 50 mM HEPES, 1 mM MgCl2, 3 mM β-mercaptoethanol with protease inhibitors). The bacterial lysate was clarified by centrifugation at 18k rpm for 30 min at 4°C. Ni-NTA affinity was used to purify BASS-XL, and passage over HiTrap Q HP cation exchange in low salt (70 mM KCl, 50 mM HEPES, 1 mM MgCl2) was used to remove contaminants where BASS-XL eluted in the flow through. A second Ni-NTA affinity step was used in conjunction with 10K Amicon Filters to concentrate the BASS-XL. The concentrated BASS-XL tetramer was applied on a HiLoad 16/600 Superdex 200 gel filtration column using an AKTA Purifier (GE Healthcare). Crystallization conditions were screened using a Mosquito Robot (TTP Labtech) by mixing 100 nL of protein with 100 nL precipitant conditions. Crystals were obtained and refined in 0.01 M FeCl3, 0.1M sodium citrate pH 5.6, 12% Jeffamine M-600 at 18°C and cryoprotected with 20% glycerol. Crystals were diffracted at the SSRL 11-1 beamline and showed highly anisotropic X-ray diffraction. Crystals adopt space group C2 with four molecules in the asymmetric unit. We used 4.4 Å as the high-resolution cutoff to avoid excessive loss of completeness. The diffraction data was truncated using boundaries determined via the Anisotropic server (Strong et al., 2006). The BASS-XL structure was determined using molecular replacement using the previously determined BASS model (Kashina et al., 1996; Scholey et al., 2014). Data from each monomer were combined using noncrystallographic symmetry and were averaged and refined using PHENIX with cycles of the model building using the coot program (Emsley et al., 2010; Liebschner et al., 2019). The individual-positional coordinates and anisotropic B-factor were refined with automatic weight optimization in the final stage. The final model includes the BASS core domain with extended parallel helices at the N-terminal end.
Engineering D. melanogaster and human minitetramer kinesin-5 motors
Human and D. melanogaster kinesin-5 minitetramers were designed using the BASS-XL X-ray structure as a template. For the KLP61F KMBX and KMBXt minitetramer constructs, the BASS XL was extended by 20 residues based on the heptad pattern observed in BASS-XL the structure (residues 597–833) and fused at its N-terminal end to the KLP61F motor and neck-linker domain (residues 1–369) and were either contained or lacked a C-terminal extension of the KLP61F tail domain (residues 910–1066) with a C-terminal tag. For the Eg5 mini-tetramer hMBX and hMBXt constructs the Homo sapiens Eg5 motor domain and neck linker (1–374) were fused to the N-terminal end of the D. melanogaster BASS-XL domain (597–799), either contained or lacked the Homo sapiens Eg5 C-terminal tail domain (913–1056), and a C-terminal 6x-His tag with mutations (C25V, C43S, C87A, C99A, N358C, C964S, and C1003S) to allow for the specific labeling of the motor at a single-reactive cysteine residue in the neck linker (N358C). The above KLP61F or Eg5 Minitetermers were expressed in using T7 expression in BL21 bacterial E. coli strains. Minitetramer transformed cells were grown at 37°C and then induced with 0.5 mM Isoproyl-thio-glucoside (IPTG) and then grown overnight at 19°C. Cells were pelleted and then lysed using lysis buffer (50 mM HEPES 250 mM KCl, pH 7.5, 1 mM MgCl2, 3 mM β-mercaptoethnaol) then clarified with centrifugation and then purified using Ni-IDA (Macherey Nagel, USA) then purified using lysis buffer with 100 mM Imidzaole. Minitetramer containing fractions were concentrated using amicon concentrators and then purified using Size exclusion chromatography using Superdex 200 increase (10/300) and fractions were collected using AKTA purifier (GE healthcare, USA). Mini-tetramer containing fractions were analyzed by SDS–PAGE and then concentrated and flash frozen using liquid nitrogen for further use. Tubulin was purified from Pork or Bovine brains (Castoldi and Popov, 2003). After purification, tubulin was cycled or labeled (with Alexa-546 or Alexa 647 [Thermo Fisher Scientific; Waltham MA], HiLyte 488 [AnaSpec; Fremont CA] or Biotin-LC-NHS [Thermo Fisher Scientific]) and then cycled before use. Unless otherwise stated, all chemicals and proteins were purchased from MilliporeSigma (Burlington, MA).
Motility assays for D. melanogaster Kinesin-5 (kMBX and kMBXt) minitetramers
Flow chambers were assembled from N 1.5 glass coverslips (0.16 mm thick; Ted Pella) that were cleaned with the Piranha protocol and functionalized with 2 mg/ml PEG-2000-silane containing 2 μg/ml biotin-PEG-3400-silane (Laysan Bio) suspended in 80% at pH 1 (Henty-Ridilla et al., 2016). After the flow chamber was assembled, 0.1 mg/ml NeutrAvidin (Thermo Fisher Scientific) was used to functionalize surfaces. Biotin and Alexa-Fluor-633-labeled porcine tubulin were generated in the laboratory as described (Al-Bassam, 2014) and were polymerized using the nonhydrolyzable GTP analog guanosine-5′-[(α,β)-methyleno] triphosphate (termed GMPCPP; Jena Biosciences) or using the MT-stabilizing drug, Paclitaxel (sigma). These MTs (100–200 μg/ml in BRB-80: 80 mM PIPES, 1 mM MgCl2, and 1 mM ETGA; pH 6.8, 1% glycerol, 0.5% pluronic-F127, 0.3 mg/ml casein, 3 mM BME, 4 mM ATP-MgCl2) were flowed into chambers and attached to glass via biotin-neutravidin linkage. Flow chambers were then extensively washed with imaging buffer (25 mM HEPES, 25–150 mM KCl, pH 7.5, 10 mM beta-mercatopethanol; 1% glycerol, 0.5% Pluronic-F127, 0.3 mg/ml casein, 3 mM BME, 4 mM ATP-MgCl2). Kinesin-5 MT-stimulated motility was reconstituted at 25°C by injecting 1–20 nM FL-Eg5-GFP combined with a photobleach-correction mixture into flow chambers (Telley et al., 2011). Movies were captured in TIRF mode using a Nikon Eclipse Ti microscope using 1.5 Na objective and an Andor IXon3 EM-CCD operating with three (488 nm, 560 nm, and 640 nm) emission filters using alternating filter wheel in 2 s increments operated using elements software (Nikon).
Motility assays for human Kinesin-5 (hMBX and hMBXt) minitetramers
Coverslips (22 × 30 mm; Thermo Fisher Scientific) were cleaned by soaking in acetone for 10 min, sonicating in 50% methanol for 20 min, sonicating in 0.5M KOH for 20 min, and then rinsing in MilliQ water three times before drying using nitrogen gas. Cleaned coverslips were stored covered at room temperature. Immediately before silanization, coverslips were plasma treated on “high” (∼18W; 200mTorr) for 45 s after evacuating the chamber. Subsequently, they were soaked in PlusOne Repel-Silane ES (GE Healthcare; Chicago IL) for 20 min, transferred to 95% ethanol for 5 min, and then sonicated in fresh 95% ethanol for 10 min. Finally, they were dried again using nitrogen gas and stored covered at room temperature for up to 2 mo.
Taxol-stabilized MTs were prepared as follows: purified unlabeled, Alexa 647-labeled, and biotinylated tubulin were combined at 50 μM in BRB80 in a 50:2:1.5 ratio and supplemented with 2 mM GTP. After incubating at 37°C for 40 min, 40 μM Taxol (Cytoskeleton; Denver CO) was added, and MTs were incubated for an additional 30 min. Subsequently, MTs were pelleted by centrifugation (8000 rcf, 10 min, 25°C), and, after washing the pellet, resuspended in fresh BRB80 supplemented with 40 μM Taxol. The pelleting, washing, and resuspension were repeated twice sequentially.
Single-molecule motility assays were carried out as follows: Flow chambers with a volume of ∼20 μl were prepared from a silanized coverslip, a glass slide, and double-sided tape. NeutraAvidin was introduced at 20 μg/ml and incubated for 5 min, before blocking the remaining surface using 50 mg/ml Pluronic F-127 for 30 min. Stabilized MTs containing biotinylated tubulin were then flowed in at a concentration of ∼0.14 mg/ml and allowed to bind for 5 min, at which point any unbound MTs were removed by washing the chamber with BRB80 supplemented with 20 μM Taxol. Just before imaging, the assay buffer consisting of BRB80 supplemented with 20 μM Taxol, 0.5 mg/ml BSA, 10 mM DTT, 2 mM ATP, 0.5 mg/ml glucose oxidase, 7 mg/ml glucose, 0.2 mg/ml catalase, 0.2% wt/vol PEG, 40 mM potassium acetate, and ∼22 nM kinesin-5 (with or without the tail) was introduced. The flow chamber was then sealed at either end using vacuum grease and imaged immediately for no more than 30 min. Assays with AMP-PNP were performed identically, except that motors were diluted 36× more and the ATP was substituted with 2 mM AMP-PNP (Roche Diagnostics; Indianapolis IN).
TIRF microscopy imaging was performed using an Eclipse Ti-E inverted microscope (Nikon; Melville NY) equipped with diode lasers (100mW; 405 nm, 488 nm, 561 nm, and 640 nm; Coherent; Santa Clara CA), custom optics for TIRF, a 1.49 NA 100× objective, and an additional 1.5× lens to increase the magnification. Two-color image series were acquired by capturing images of the MTs (640 nm, 100 ms exposure, 1 mW) and the motors (561 nm, 200 ms exposure, 2 mW) in an alternating manner by alternating the laser illumination in synchrony with the rotating filter wheel using a custom-written LabView program, resulting in a frame rate of ∼1.2s–1. Images were captured on an iXon U897 EMCCD camera (Andor Technology; South Windsor CT).
Single-Molecule Analyses for D. melanogaster kinesin-5 (kMBX and kMBXt)
Motility, run length, and run-time analyses were carried out as follows: Image movie stacks were preprocessed with photobleach correction and image stabilization plugins using the program FIJI (Schindelin et al., 2012). For motility along individual MTs, individual kMBX and kMBXt motor-motility events were identified along anchored MTs based on kymographs generated for multiple channels. The FIJI kymograph TrackMate plugins (Schindelin et al., 2012) were used to measure particle motility rates and identify their run lengths as and run time. Large collections of motile events along fields of MTs were collected for kMBXt motors at 25, 50, 100, 125, and 150 mM KCl conditions and collected for kMBX motors at 50 and 100 mM KCl conditions (Table 2). Average MT parameters were determined by frequency binning the motility events in a range of conditions and then fitting these events using Gaussian distributions using the program Prism (Table 2). In general, all parameters fit single Gaussian distributions. Run lengths were fitted using exponential decay to identify the half-length for each motor condition. T-tests were performed to determine the significance of the differences observed.
The stoichiometry of kMBX and kMBXt motors per multimotor cluster were determined as previously described (Pandey et al., 2021a). Briefly, following correction for the uneven illumination of images and background subtraction, intensities of all NG-labeled kMBX or kMBXt in the first frame of a time-lapse sequence were measured using the TrackMate plugin of the ImageJ-Fiji software (Schindelin et al., 2012; Tinevez et al., 2017). Becasue kMBX and kMBXt are homotetrameric, each motor contains at least four NGs per tetrameric motor. The major peak of the intensity distribution histogram of these mNG-labeled minitetramers was fitted to a Gaussian distribution. The center of the Gaussian peak lay at ∼600 au, which corresponds to the average intensity of single kMBX or kMBXt motor containing one, two, three, or four fluorescent mNG, with each fluorescent mNG molecules contributing ∼240 au to the total intensity. Thus, the neon green-labeled motor population within this Gaussian peak likely represents single kMBX or kMBXt motors. By this method, we assigned intensity ranges for kMBX or kMBXt molecules fluorescence as < 960, 960 –1920, and >1920 for single minitetramer, pairs minitetramers, and higher order oligomers of minitetramers, respectively. All the fluorescence-intensity measurements to assign cluster size to a kMBX or kMBXt molecule were performed only in the first frame for each data set, thereby significantly reducing the possibility of photobleaching effects.
Single-molecule analyses of human kinesin-5 (hMBX and hMBXt) mini-tetramers
Image series were drift corrected using the MT images with the Image Stabilizer plug-in for ImageJ. The output coefficients were then applied to the motor images using the Image Stabilizer Log Applier plug-in. These transformation coefficients were saved and used for later steps in the analysis. For images presented in the text, the MT channel was bleach corrected using the Histogram Matching algorithm in ImageJ. Single-molecule tracking was performed on the original, unprocessed images using the TrackMate (Schindelin et al., 2012; Tinevez et al., 2017) plug-in for ImageJ. Briefly, subpixel localization was performed using a Laplacian of Gaussian filter with an estimated spot diameter of 0.4 μm to detect motors. Spots detected in sequential frames were then linked using the Linear Assignment Problem tracker using a maximum displacement of 0.6 μm, allowing for a two-frame absence of spots, and not permitting track merging or splitting. No filtering of the detected spots or tracks was done in ImageJ.
Quantitative analysis of motor-motility activities was performed as follows: MTs were manually traced out in ImageJ, and the x, y coordinates of these regions of interest (ROIs) were saved and imported into MATLAB. 20 Detected trajectories > 3 frames were then projected along the axis of their respective MT for run length and velocity analysis. Trajectories with a total run length < 150 nm were not included in the analysis to filter out statically bound motors (e.g., those adsorbed on the coverslip near a MT). All trajectories within 600 nm of another MT, or 600 nm of either end of the MT, were ignored in the analysis of pausing/motility. The local α-value analysis was performed on all trajectories with > 10 frames using drift-corrected (but not projected) x, y localizations. These were linearly interpolated to account for the slightly varying frame rate. α-values were calculated in sequential 8-frame windows using delays of two, three, and four frames. Change points were detected in the resulting α-values using the findchangepts function in MATLAB (The MathWorks; Natick MA) with a minimum improvement in the residual error of 0.3 and a minimum distance between consecutive change points of three frames. The means between detected change points were then evaluated and sections with a mean α-value > 1.1 were deemed processive. The localization in the original trajectory was then assigned as processive or paused based on the classification of the interpolated frame that was nearest to them in time. Subsequent analysis of intensities, velocities, and interpause run lengths was performed using these classifications. The fitting was done using maximum likelihood estimation (MLE). For the velocity histograms, normal distributions were fitted using the central 96% of the data. For pause-time estimates, distributions were fit using the lower 95% of the data because of the presence of a small number of inactive motors that are classified to have long “pauses”. For analysis of trajectories in the presence of AMP-PNP, no minimum-run length was used, and the α-value histogram was fit to the upper 95% of the data to ignore the skew.
MT plus-end motor localization intensity analyses were performed as follows: The duration of time motors/clusters were present at the plus-ends of MTs was quantified using ImageJ. Briefly, all unobstructed MT plus-ends (identified based on the direction of motor motility) were marked using point ROIs, which were added to the ROI Manager. These were then converted to circular ROIs with a diameter of 7 pixels centered on the point using a custom-written script. The intensity in these ROIs was traced over time using Multi Measure, ensuring that the recorded measurements included SD, min and max-gray value, the center of mass, integrated density, and mean-gray value. This process was also repeated for nine background spots spread throughout the field of view. The output from each movie was saved, and all data were imported into MATLAB for further analysis. A moving average was applied to the raw integrated density signal (IDS). In every ROI, a cluster/motor was deemed present in each frame of the movie if this filtered IDS in the ROI exceeded that of background ROIs by three SDs. Only cluster lifetimes > 9 frames were considered based on the approximate time taken for a motor to walk through the ROI. The lifetimes of any clusters present in the first or last frame of the movie were marked as censored for the subsequent lifetime analysis. Lifetime analysis was performed by fitting the empirical cumulative distribution function (CDF) to a mixture of two Weibull CDFs using MLE, taking censoring into account.
Reconstitution of full-length and minitetramer Kinesin-5 MT crosslinking and sliding
MTs for surface immobilization were generated via mixture of HiLyte 647 tubulin (TL670M), biotinylated tubulin (T333P), and unmodified tubulin (T240) at a ratio of 1:1:20 along with 1 mM GMPCPP. MTs were polymerized at 37°C for 1 h before clarification and stabilization in 30 uM Taxol following published protocols (Shimamoto et al., 2015). “Free” MTs for gliding were generated via a mixture of rhodamine tubulin (TL590M) and unmodified tubulin at a ratio of 1:20 along with 1 mM GMPCPP, and were polymerized, clarified, and stabilized in 30 uM Taxol following similar protocols.
The flow-chamber design and assay preparation were modified from a previously described protocol (Shimamoto et al., 2015). Antiparallel MT bundles were constructed using passivated glass coverslips coated with SVA-PEG at a ratio of 50 PEG:1 biotin-PEG. All reagents were prepared with 1X BRB80 buffer. Following each reagent flow-in and incubation, a flush with approximately three chamber volumes of BRB80 was performed. Reagents were introduced stepwise with the following order and incubation times: 1) 0.5 mg/ml neutravidin, 2 min; 2) 0.5 mg/ml alpha casein surface block, 3 min; 3) HiLyte-647 biotinylated MTs with 0.2 mg/ml alpha casein with no additional incubation and immediate flush; 4) kinesin-5 construct at a desired final concentration as reported with 0.2 mg/ml alpha casein, 2 min; 5) Rhodamine 561 MTs with 0.2 mg/ml alpha casein, 5 min, and the corresponding chamber flush included 1 mM TCEP bond breaker solution in BRB80; 6) imaging buffer (25 mM HEPES, 25–150 mM KCl, pH 7.5, 10 mM beta-mercatopethanol; 1% glycerol, 0.5% Pluronic-F127, 0.3 mg/ml casein, 3 mM BME, 4 mM ATP-MgCl2) with Oxygen Scavenging System (4.5 mg/ml glucose, 350 U/ml glucose oxidase, 34 U/ml catalase, 1 mM DTT) was then used to flush any unattached MTs. The chamber was then sealed with clear nail polish before experiments.
MT bundles were imaged using three-channel TIRF microscopy using the following exposure times and laser lines from a Nikon LUNA four-channel laser launch module: HiLyte-647 MTs: 640 nm laser (60% power, 200 ms exposure); GFP-tagged kinesin-5: 488 nm laser (30% power, 100 ms exposure); and rhodamine MTs: 561 nm laser (30% power, 100 ms exposure). Imaging was performed on a Nikon Ti-E inverted microscope with a CFI Apo 100X/1.49NA oil immersion TIRF objective. Images were acquired using a Photometric Prime 95B camera controlled with Nikon NIS Elements software. Analysis of fluorescent data and generation of intensity linescan data sets were performed using FIJI (ImageJ) tools (Schindelin et al., 2012).
Supplementary Material
Acknowledgments
The authors thank Jonathan Scholey (Molecular Cellular Biology, UC-Davis) for the inspiration on the project. J.A.B. thanks Richard McKenney (Molecular Cellular biology, UC-Davis) for the critical reading of this manuscript. BASS-XL diffraction data were collected at the Stanford Radiation Laboratory (SSRL) using 11-1 beamline. We thank Peter Dunten, Ana Gonzalez, and Tzanko Doukov (SSRL) for their help with data collection. The Authors thank MD Ashaduzzaman (Molecular Cellular Biology, UC-Davis) for Mass photometry measurements. This research was supported in part by the Israel Science Foundation grant (ISF-386/18) awarded to L.G.; the National Science Foundation (NSF-1615991) and the United States - Israel Binational Science Foundation grant (BSF-2015851), awarded to L.G. and J.A.B., respectively; A United States - Israel Binational Science Foundation grant (BSF-2019008), awarded to L.G. and S.R.; and the National Institutes of Health (NIH- GM11283), awarded to J.A.B. S.F is supported by the Rensselaer Polytechnic Institute School of Science Startup funds. A.G.H is supported by the Canadian Institutes of Health Research (CIHR) grant PJT-159490 and the Natural Sciences and Engineering Research Council of Canada (NSERC) grant RGPIN-2020-04608. Crystallographic data and structure coordinates are deposited at the RCSB (PDB ID: 7S5U).
Abbreviations used:
- BASS
bipolar assembly
- BASS-XL
extended bipolar assembly domain
- Eg5
human kinesin-5
- FL
full-length kinesin-5 without the tail
- FLt
full-length kinesin-5 with the tail
- GFP
green fluorescent protein
- IPTG
Isoproyl-thio-glucoside
- MBX
kinesin-5 minitetramer without a tail
- MBXt
kinesin-5 minitetramer with tail
- MT
microtubule
- NG
neon green
- TIRF
total internal reflection fluorescence
- TMR
Tetra-methyl Rhodamine.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-07-0287) on August 23, 2023.
REFERENCES
- Acar S, Carlson DB, Budamagunta MS, Yarov-Yarovoy V, Correia JJ, Ninonuevo MR, Jia W, Tao L, Leary JA, Voss JC, et al. (2013). The bipolar assembly domain of the mitotic motor kinesin-5. Nat Commun 4, 1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J (2014). Reconstituting Dynamic Microtubule Polymerization Regulation by TOG Domain Proteins. Methods Enzymol 540, 131–148. [DOI] [PubMed] [Google Scholar]
- Andreasson JOL, Clancy BE, Behnke-Parks W, Rosenfeld SS, Block SM (2012). A Universal Pathway for Kinesin Stepping. Biophys J 102, 369a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcizet D, Meier B, Sackmann E, Radler JO, Heinrich D (2008). Temporal Analysis of Active and Passive Transport in Living Cells. Phys Rev Lett 101, 248103. [DOI] [PubMed] [Google Scholar]
- Beausang JF, Goldman YE, Nelson PC (2011). Changepoint Analysis for Single-Molecule Polarized Total Internal Reflection Fluorescence Microscopy Experiments. Method Enzymol 487, 431–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertran MT, Sdelci S, Regue L, Avruch J, Caelles C, Roig J (2011). Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J 30, 2634–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d’Herin P, Harper M, Kress M, Nigg EA (1995). Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83, 1159–1169. [DOI] [PubMed] [Google Scholar]
- Bodrug T, Wilson-Kubalek EM, Nithianantham S, Thompson AF, Alfieri A, Gaska I, Major J, Debs G, Inagaki S, Gutierrez P, et al. (2020). The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding. Elife 9, e51131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahu J, Olichon A, Hentrich C, Schek H, Drinjakovic J, Zhang CJ, Doherty-Kirby A, Lajoie G, Surrey T (2008). Phosphorylation by Cdk1 Increases the Binding of Eg5 to Microtubules In Vitro and in Xenopus Egg Extract Spindles. Plos One 3, e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M, Popov AV (2003). Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif 32, 83–88. [DOI] [PubMed] [Google Scholar]
- Chen GY, Mickolajczyk KJ, Hancock WO (2016). The Kinesin-5 Chemomechanical Cycle Is Dominated by a Two-heads-bound State. J Biol Chem 291, 20283–20294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Cleary JM, Asenjo AB, Chen Y, Mascaro JA, Arginteanu DFJ, Sosa H, Hancock WO (2019). Kinesin-5 Promotes Microtubule Nucleation and Assembly by Stabilizing a Lattice-Competent Conformation of Tubulin. Curr Biol 29, 2259–2269.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Hancock WO (2015). Kinesin-5 is a microtubule polymerase. Nat Commun 6, 8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh M, Reissner M, Bugiel M, Lipka E, Herrmann A, Roy B, Muller S, Schaffer E (2018). Phragmoplast Orienting Kinesin 2 Is a Weak Motor Switching between Processive and Diffusive Modes. Biophys J 115, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross RA, McAinsh A (2014). Prime movers: the mechanochemistry of mitotic kinesins. Nat Rev Mol Cell Bio 15, 257–271. [DOI] [PubMed] [Google Scholar]
- Drosopoulos K, Tang C, Chao WCH, Linardopoulos S (2014). APC/C is an essential regulator of centrosome clustering. Nat Commun 5, 3686. [DOI] [PubMed] [Google Scholar]
- Duselder A, Fridman V, Thiede C, Wiesbaum A, Goldstein A, Klopfenstein DR, Zaitseva O, Janson ME, Gheber L, Schmidt CF (2015). Deletion of the Tail Domain of the Kinesin-5 Cin8 Affects Its Directionality. J Biol Chem 290, 16841–16850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnikar A, Tole S, Baas PW (2011). Kinesin-5, a mitotic microtubule-associated motor protein, modulates neuronal migration. Mol Biol Cell 22, 1561–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K, Furuta A, Toyoshima YY, Amino M, Oiwa K, Kojima H (2013). Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proc Natl Acad Sci USA 110, 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner AE, Santen L, Rieger H, Shaebani MR (2016). Run-and-pause dynamics of cytoskeletal motor proteins. Sci Rep-Uk 6, 37162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Rankova A, Eskin JA, Kenny K, Goode BL (2016). Accelerated actin filament polymerization from microtubule plus ends. Science 352, 1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt ER, Gheber L, Kingsbury T, Hoyt MA (2006). Homotetrameric form of Cin8p, a Saccharomyces cerevisiae kinesin-5 motor, is essential for its in vivo function. J Biol Chem 281, 26004–26013. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF (2005). The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114–118. [DOI] [PubMed] [Google Scholar]
- Kashina AS, Scholey JM, Leszyk JD, Saxton WM (1996). An essential bipolar mitotic motor. Nature 384, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Liu M, Son YJ, Himes BT, Snow DM, Yu WQ, Baas PW (2011). Inhibition of Kinesin-5, a Microtubule-Based Motor Protein, As a Strategy for Enhancing Regeneration of Adult Axons. Traffic 12, 269–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh T, Gluszek AA, Welburn JPI (2018). Microtubule end tethering of a processive kinesin-8 motor Kif18b is required for spindle positioning. J Cell Biol 217, 2403–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KA, Baas PW (2007). Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J Cell Biol 178, 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadar VC, Ketschek A, Myers KA, Gallo G, Baas PW (2008). Kinesin-5 Is Essential for Growth-Cone Turning. Curr Biol 18, 1972–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadar VC, Lin S, Baas PW (2012). Microtubule Redistribution in Growth Cones Elicited by Focal Inactivation of Kinesin-5. J Neurosci 32, 5783–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey H, Reithmann E, Goldstein-Levitin A, Al-Bassam J, Frey E, Gheber L (2021a). Drag-induced directionality switching of kinesin-5 Cin8 revealed by cluster-motility analysis. Sci Adv 7, eabc1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey H, Singh SK, Sadan M, Popov M, Singh M, Davidov G, Inagaki S, Al-Bassam J, Zarivach R, Rosenfeld SS, Gheber L. (2021b). Flexible microtubule anchoring modulates the bi-directional motility of the kinesin-5 Cin8. Cell Mol Life Sci 78, 6051–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapley J, Nicolas M, Groen A, Regue L, Bertran MT, Caelles C, Avruch J, Roig J (2008). The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J Cell Sci 121, 3912–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozelle DK, Hansen SD, Kaplan KB (2011). Chromosome passenger complexes control anaphase duration and spindle elongation via a kinesin-5 brake. J Cell Biol 193, 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthardt N, Lamb DC, Brauchle C (2011). Single-particle Tracking as a Quantitative Microscopy-based Approach to Unravel Cell Entry Mechanisms of Viruses and Pharmaceutical Nanoparticles. Mol Ther 19, 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Powers J, Strome S, Saxton WM (2007). Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol 17, R453–R454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JE, Nithianantham S, Scholey JM, Al-Bassam J (2014). Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers. Elife 3, e02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira O, Goldstein A, Al-Bassam J, Gheber L (2017). A potential physiological role for bi-directional motility and motor clustering of mitotic kinesin-5 Cin8 in yeast mitosis. J Cell Sci 130, 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto Y, Forth S, Kapoor TM (2015). Measuring Pushing and Braking Forces Generated by Ensembles of Kinesin-5 Crosslinking Two Microtubules. Dev Cell 34, 669–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Pandey H, Al-Bassam J, Gheber L (2018). Bidirectional motility of kinesin-5 motor proteins: structural determinants, cumulative functions and physiological roles. Cell Mol Life Sci 75, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang SS, Phillips M, Cascio D, Eisenberg D (2006). Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA 103, 8060–8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Gelles J (2007). Two distinct modes of processive kinesin movement in mixtures of ATP and AMP-PNP. J Gen Physiol 130, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley IA, Bieling P, Surrey T (2011). Reconstitution and Quantification of Dynamic Microtubule End Tracking In Vitro Using TIRF Microscopy. Methods Mol Biol 777, 127–145. [DOI] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW (2017). TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90. [DOI] [PubMed] [Google Scholar]
- Vershinin M, Carter BC, Razafsky DS, King SJ, Gross SP (2007). Multiple-motor based transport and its regulation by Tau. Proc Natl Acad Sci USA 104, 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vugmeyster Y, Berliner E, Gelles J (1998). Release of isolated single kinesin molecules from microtubules. Biochemistry 37, 747–757. [DOI] [PubMed] [Google Scholar]
- Weinger JS, Qiu M, Yang G, Kapoor TM (2011). A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr Biol 21, 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac AL, Goldman YE, Holzbaur ELF, Ostap EM (2013). Local Cytoskeletal and Organelle Interactions Impact Molecular-Motor-Driven Early Endosomal Trafficking. Curr Biol 23, 1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.