Abstract
Nervous systems exhibit dramatic diversity in cell morphology and size. How neurons regulate their biosynthetic and secretory machinery to support such diversity is not well understood. Endoplasmic reticulum exit sites (ERESs) are essential for maintaining secretory flux, and are required for normal dendrite development, but how neurons of different size regulate secretory capacity remains unknown. In Caenorhabditis elegans, we find that the ERES number is strongly correlated with the size of a neuron’s dendritic arbor. The elaborately branched sensory neuron, PVD, has especially high ERES numbers. Asymmetric cell division provides PVD with a large initial cell size critical for rapid establishment of PVD’s high ERES number before neurite outgrowth, and these ERESs are maintained throughout development. Maintenance of ERES number requires the cell fate transcription factor MEC-3, C. elegans TOR (ceTOR/let-363), and nutrient availability, with mec-3 and ceTOR/let-363 mutant PVDs both displaying reductions in ERES number, soma size, and dendrite size. Notably, mec-3 mutant animals exhibit reduced expression of a ceTOR/let-363 reporter in PVD, and starvation reduces ERES number and somato-dendritic size in a manner genetically redundant with ceTOR/let-363 perturbation. Our data suggest that both asymmetric cell division and nutrient sensing pathways regulate secretory capacities to support elaborate dendritic arbors.
How neurons regulate secretory machinery to support vastly different cell sizes is not well understood.
We show that Endoplasmic Reticulum Exit Site (ERES) number scales with somato-dendritic size in C. elegans neurons, and that this scaling requires cell fate transcription factors, nutrients, and C. elegans TOR.
This work provides a foundation for further investigation into the molecular mechanisms regulating neuron cell size and secretory capacity.
INTRODUCTION
Neurons exhibit a large diversity in morphology and size. A single nervous system contains small neurons with simple morphology and very large neurons with extremely complex neurites. How neurons adjust their biosynthetic and secretory activity to match their morphology and size is not well understood. It is well known that cell types with secretory functions such as pancreatic acinar cells, have expanded rough Endoplasmic Reticulum (ER) and Golgi apparatus (Gorelick and Jamieson, 2012). Work in secretory cells and mammalian cell lines demonstrates that early secretory organelle size and number is associated with changes in secretory cargo load (Griffiths et al., 1985; Clermont et al., 1993; Farhan et al., 2008a). However, it is not currently understood how postmitotic neurons of different types regulate secretory capacity to accommodate divergent cell sizes in vivo.
Examination of endoplasmic reticulum exit sites (ERESs) may offer a valuable proxy for gauging secretory capacity in neurons in vivo. ERESs export correctly folded proteins from the ER, via active cargo selection and bulk flow through a system of vesicles or tubules, to subsequent secretory compartments such as the Golgi (Barlowe and Helenius, 2016; Peotter et al., 2019; Weigel et al., 2021). As the first step in the secretory pathway, ERESs are intimately linked to a cell’s overall secretory flux (Barlowe and Helenius, 2016). Indeed, ERES’ size and number have been shown to increase with elevated secretory load (Farhan et al., 2008a). Specialized cell types with high biosynthetic and secretory capacities can display dozens to hundreds of ERES, while simpler cells can display as few as a single ERES (Stephens, 2003; Yelinek et al., 2009; Warren, 2013; Saegusa et al., 2022). In Drosophila melanogaster neurons, genes involved in ERES function have been shown to play an important role in dendritic arbor development (Ye et al., 2007).
The worm, Caenorhabditis elegans, offers an attractive system for studying how neurons regulate secretory capacity to support diverse cell size. C. elegans hermaphrodites have 302 neurons of widely varying size and complexity, many of which can be genetically manipulated on the individual cell level. In addition, the transparent C. elegans allows for visualization of secretory organelle formation and dynamics within a single neuron throughout its development. The vast majority of C. elegans neurons a have simple cell morphology with one or two unbranched neurites (White et al., 1986). However, the multimodal sensory neuron PVD develops an expansive dendritic arbor with over 150 terminal branches that span nearly the entire length of the worm (Oren-Suissa et al., 2010; Smith et al., 2010). How PVD adapts the function or structure of its secretory pathway to accommodate the secretory flux required to establish such a large dendrite is not known.
In this study, we determined ERES number in a subset of C. elegans neurons of different sizes and morphological complexities. We report that neurons with larger dendrites had significantly higher ERES numbers in their somas, with PVD displaying the highest number of ERESs. Our data indicate that asymmetric cell division, resulting in a large initial soma size, establishes PVD’s high ERES number before neurite outgrowth. Subsequent maintenance of ERES number during PVD development is coupled with expansive somato-dentritic growth. Starvation, as well as disruption of the cell-fate transcription factor MEC-3 and the master metabolic regulator C. elegans TOR (ceTOR/let-363), all resulted in a coordinated reduction of ERES number, dendritic, and soma size. Moreover, mec-3 perturbation reduced expression of a ceTOR/let-363 transcriptional reporter in PVD, and the impact of starvation on ERES number is redundant with that of the ceTOR/let-363-deletion in PVD. These data suggest that cell-fate transcription factors in PVD upregulate both initial cell size and nutrient sensing pathways, which together establish the high biosynthetic and secretory capacity required to generate and maintain a large dendritic arbor.
RESULTS
Larger neurons have more ER-exit sites
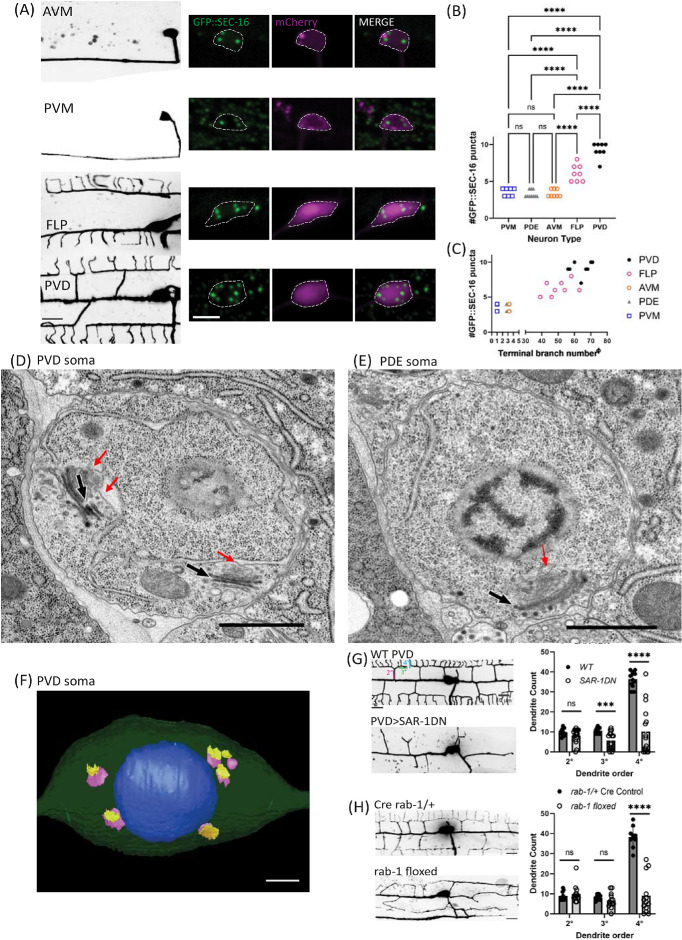
To understand the molecular mechanisms that generate the morphological diversity of neurons, we postulated that biosynthetic and membrane trafficking pathways are tuned to neuron size and complexity. To test this hypothesis, we visualized ERESs in neuronal types with different morphological complexity, using an endogenous, tissue-specific tag of ERES protein SEC-16 (GFP::FLPon::SEC-16). We observed that neurons with larger, more complex morphologies have significantly higher numbers of ERESs in their cell bodies or somas (Figure 1). Touch receptor neurons (TRNs) AVM and PVM, each with fewer than 5 terminal neurite branch points, typically have three to four distinct somatic ERESs. In contrast, significantly higher numbers of ERESs are found in the somas of multimodal sensory neurons FLP and PVD, each of which have highly branched dendrites with dozens of terminal neurite branch points (Figure 1, A–C). PDE, a smaller sensory neuron within the same lineage as PVD, has fewer ERESs than PVD. Comparing individual FLP and PVD neurons, we also found a trend that cells with higher branch numbers have more ERESs (Figure 1C). To validate the discrete punctate nature of the fluorescent ERES marker, we reconstructed PVD and PDE cell somas by serial section electron microscopy (EM). The EM images indicated six discrete Golgi stacks in PVD soma, each with one or two associated ERESs, corresponding well with results from fluorescence microscopy (Figure 1, D–F). We chose PVD as a system in which to further examine the relationship between ERES function, ERES number, and dendrite size, due to its large size, high ERES number, elaborately branched and quantifiable dendrites, and its well-characterized morphogenesis (Zou et al., 2016).
FIGURE 1:
Larger neurons have more ERESs in their somas. (A) Confocal-fluorescence microscopic images of neuron morphology on the left (scale bar = 10 µm), with closeups of soma and an endogenous ERES marker, GFP::SEC-16, on the right (scale bar = 5 µm). Background of PVM image was removed to more clearly show PVM morphology. (B) Quantification of number of GFP::SEC-16 puncta per soma in different cell types. Significance determined by one-way ANOVA, ****p < 0.0001, n ≥ 7 animals. (C) Quantification of number of GFP::SEC-16 puncta per soma in different cell types vs. number of terminal dendrites (see Methods). X-values have been nudged slightly to increase visibility of data points for PDE (x – 0.25) and AVM (x + 0.25). Pearson correlation between ERES number and terminal-dendrite number: r = 0.9282, R2 = 0.8615, p < 0.0001, n = 41 neurons with at least seven of each type. EM images of PVD (D), and PDE (E) soma. Red arrows indicate ERES. Black & white arrows indicate Golgi stacks. Scale bars = 1 µm. (F) 3D model of PVD soma constructed from EM images showing nucleus in blue, with cis (purple), and trans (yellow) cisternae of Golgi. Scale bar = 1 µm. (G) Confocal-fluorescence microscopic images of WT PVD and PVD with exogenously expressed SAR-1 dominant negative (SAR-1DN), with quantifications of secondary (2°), tertiary (3°), and quaternary (4°) dendrites in WT (n = 12 animals) and SAR-1-DN (n = 16 animals). Significance determined by multiple t tests, ***p < 0.001, ****p < 0.0001. (H) Confocal-fluorescence microscopic images of Cre rab-1/+ heterozygous control PVD (n = 10) and rab-1 floxed PVD (n = 13), with corresponding dendrite quantifications as in (I). Results were additionally repeated in at least two independent experiments.
ERES function is required for PVD dendrite size
To test the relationship between ERES function and dendrite size directly, we disrupted ERES function in PVD, whose elaborately branched dendrite allows for quantitative assessment of dendritic growth defects. PVD dendrite was dramatically reduced as a result of disrupting ERES function via two independent approaches (Figure 1). First, we expressed a dominant negative version of the ERES protein SAR-1 (SAR-1DN) specifically in PVD. SAR-1 is a small GTPase required for protein trafficking from ER to Golgi (Nakano and Muramatsu, 1989; Long et al., 2010). Overexpression of SAR-1DN severely disrupts dendrite growth in PVD (Figure 1G). Second, we made a tissue-specific PVD knockout of rab-1, which is required for ER to Golgi trafficking (Sannerud et al., 2006; Sato et al., 2006; Balklava et al., 2007). Similar to SAR-1-DN overexpression PVDs, rab-1-floxed PVDs exhibited severe branching defects (Figure 1H), suggesting that ERES function is critical for dendrite growth and branching in PVD, as in other systems (Ye et al., 2007).
Dendrite growth is not a prerequisite for increased secretory organelle abundance in PVD
Having established a strong association between ERES function, ERES number, and dendrite size, we hypothesized that the number of early secretory organelles might increase in response to rising secretory demand in expanding neurites. We therefore asked whether normal-dendrite outgrowth is required for high ERES number in PVD. To test this hypothesis, we disrupted several factors that are critical for PVD dendrite guidance (Liu and Shen, 2011; Dong et al., 2016; Zou et al., 2016). Perturbation of these guidance receptors and regulators including dma-1, hpo-30 and kpc-1, severely disrupts PVD dendrite morphology but does not alter the number of early secretory organelles (Supplemental Figure S1). Thus, feedback from dendrite outgrowth and precise arbor shape are not required for high secretory organelle abundance in PVD.
ERES number is coupled with dendrite size in PVD cell fate mutants
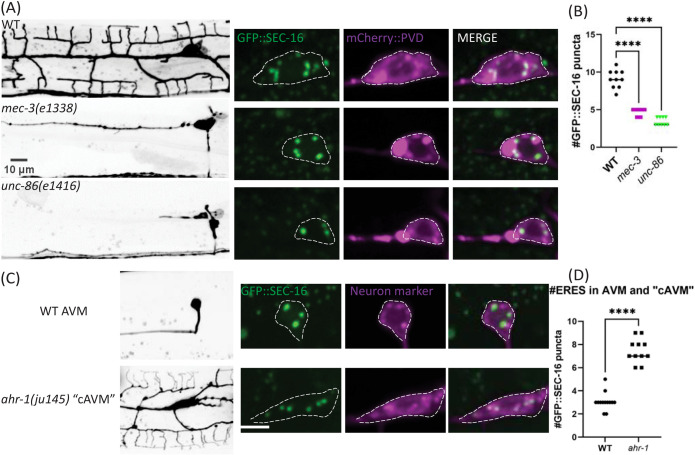
Given that feedback from an expanding dendritic arbor is not required for establishing secretory organelle number, we wondered whether PVD’s high ERES number is instead regulated by cell-fate transcription factors that drive PVD dendrite arborization. MEC-3 and UNC-86 are transcription factors that affect many aspects of PVD cell fate including dendrite growth. In mec-3(e1338) or unc-86 (e1416) mutants, PVD loses nearly all of its dendrites (Figure 2A; Tsalik and Hobert, 2003 Smith et al., 2010, 2013), and we find a corresponding reduction of ERES number in mutant PVD somas. We also tested whether cell-fate perturbations that increase dendritic size and complexity would result in elevated ERES numbers. Wild-type (WT) AVM has a simple neurite morphology. In transcription factor ahr-1(ju145) mutants, AVM becomes a cell with a large PVD-like dendritic arbor, cAVM (Figure 2C; Smith et al., 2013). Notably, cAVMs also have much higher ERES numbers than AVM (Figure 2D). These results indicate that transcription factors controlling dendrite size also specify ERES numbers, perhaps as part of cell-fate programming.
FIGURE 2:
Cell-fate mutants that alter neuron size perturb ERES abundance. (A and C) confocal-fluorescence microscopy of neuron morphology of WT and mutant worms on the left (scale bars = 10 µm, with closeups of soma and an endogenous ERES marker, GFP::SEC-16, on the right (scale bar = 5 µm). (B and D) Quantification of number of GFP::SEC-16 puncta per soma. Significance determined by one-way ANOVA (n ≥ 9 animals) in (B), and Student’s t test two-tailed in (D), ****p < 0.0001, n ≥ 11. Results were additionally repeated in at least two independent experiments.
ERES number is established early after PVD birth and maintained throughout PVD development
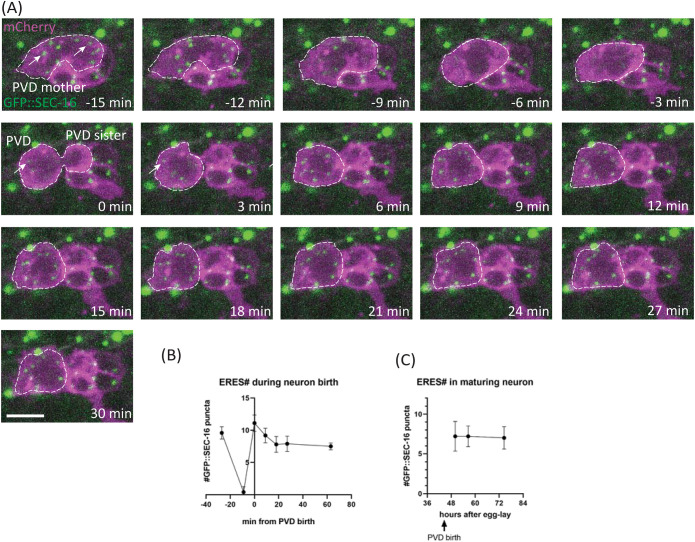
Given that secretory organelle number in PVD is dependent on cell-fate transcription factors but independent of dendrite growth, we predicted that ERES number is established early in PVD development. To test this prediction, we determined when ERES number is established, and how ERES number changes during neuron growth by conducting time-lapse recordings and timecourse experiments throughout PVD birth and development (Figure 3). These dual-color timelapse recordings of live animals allowed us to observe endogenous SEC-16 dynamics during PVD birth, and track ERES formation and numbers simultaneously with PVD morphology changes. We observed that high ERES number is established within the first 30 min of PVD birth, even before neurite outgrowth, and is subsequently maintained throughout PVD development (Figure 3, A–C). This rapid establishment of high ERES numbers is consistent with the idea that ERES number is determined via cell-fate programming rather than feedback from expanding neurites.
FIGURE 3:
ERES number is established rapidly (Phase one) after neuron birth and persists over time. (A) Time-lapse confocal-fluorescence microscopic images of PVD cell birth in WT L2 worms with Plin-32::mCherry::PH labeling neuron membranes and an endogenous fusion protein GFP::SEC-16 labeling ERESs (white arrows). Scale bar = 5 µm. (B) Quantification of ERES number from long-term time-lapse imaging during PVD birth, n ≥ 9 for each time point. (C) Quantification of ERES number after PVD birth, via confocal microscopy, as worm matures, n = 10 for each time point. Results were additionally repeated in at least two independent experiments.
In addition to pinpointing ERES emergence to a narrow time window shortly after PVD birth, our time-lapse analyses identified two distinct phases of ERES dynamics after PVD birth that might contribute to final ERES number in mature PVD: 1) ERES establishment and 2) maintenance. The establishment phase begins in the final stages of mitosis, when GFP::SEC-16 starts to coalesce rapidly into puncta that are clearly visible within 3 min of cytokinesis (Figure 3A). These puncta undergo a brief period of consolidation within the first several minutes of birth, establishing a high number of ERESs before neurite outgrowth (Figure 3, A and B). Subsequently, during the maintenance phase, ERES number remains at consistently high levels throughout PVD development (Figure 3C). Two central questions emerging from these observations are: 1) how high ERES numbers in PVD are established, and 2) how they are maintained throughout PVD development.
Cell size at birth impacts ERES number in PVD
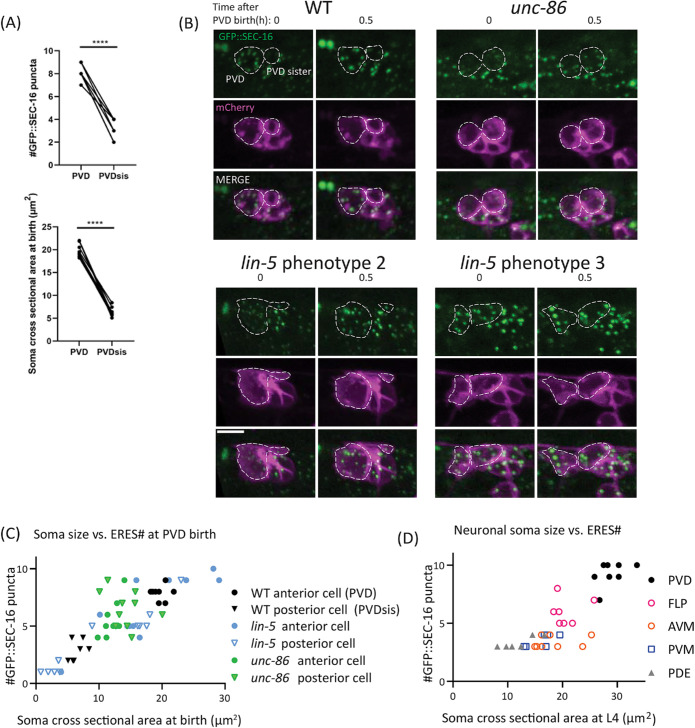
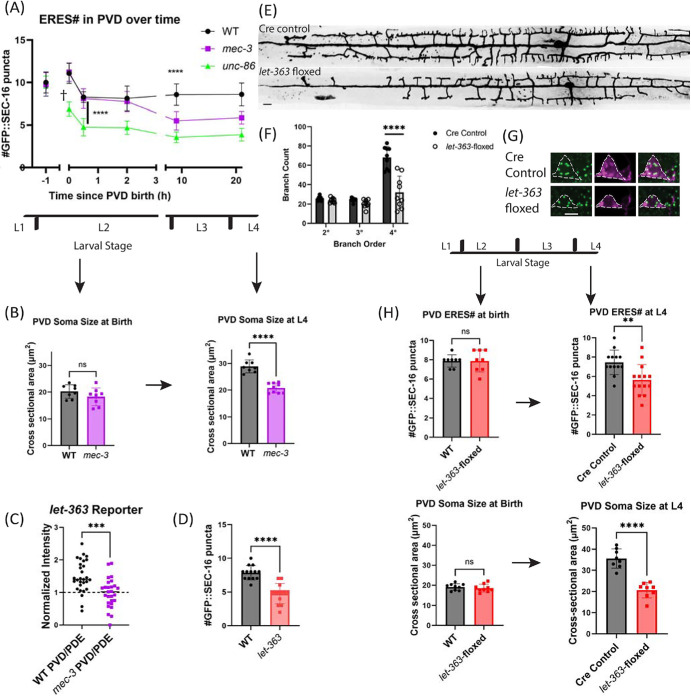
Because of the asymmetric cell division of PVD-mother observed in Figure 3A, we hypothesized that large cell-size at birth drives establishment of high ERES number. Consistent with previous findings, our time-lapse imaging showed that PVD is born significantly larger than its sister cell, PVD sis (Figure 3A, Figure 4, A and B upper left), which is eventually eliminated via apoptosis (Teuliere et al., 2018; unpublished data). We hypothesized that this asymmetric cell division is required for unequal distribution of cytoplasm, membranes, and ERES components, and may be responsible for seeding the high ERES number in PVD soon after birth. Consistent with this idea, the initial ERES number in PVD after consolidation is consistently higher than the ERES number in the smaller PVD sis (Figure 4A). To further test the impact of initial PVD size on ERES number, we examined initial ERES number under conditions in which asymmetric cell division was disrupted. In unc-86(e1416) mutants, PVD and PVD sis have similar cell size. We found that unc-86(e1416) mutant PVD and PVD sis showed similar ERES numbers, which fall in between the values of WT PVD and WT PVD sis (Figure 4B, top-right and C). These results are consistent with the notion that cells that are born larger form more ERESs. However, unc-86(e1416) mutants exhibit abnormalities in asymmetric cell division and PVD cell-fate pathways, both of which likely impact ERES number. It thus remained unclear whether the ERES phenotype in the unc-86(e1416) mutants is due to defects in asymmetric cell division or unrelated cell-fate pathways.
FIGURE 4:
Asymmetric-cell division is important for establishment phase of ERES number. (A) Quantification of ERES number shortly after birth (top), and soma cross-sectional area at birth (bottom) in PVD and PVD sister. Significance determined by Student’s t test two-tailed, ****p < 0.0001, n = 8 for each cell-type. (B) Representative images of WT and mutant worms at PVD birth and 0.5 h after birth. unc-86 mutants (top right) consistently disrupt asymmetry of PVD birth, displaying symmetric cell division of PVD mother. lin-5 mutants sometimes increase asymmetry of division (bottom left) and sometimes reduce asymmetry of division (bottom right). Scale bar = 5 µm. (C) Quantification of cross-sectional soma area at birth vs. ERES number 30 min after birth of PVD and PVD sister in WT and mutants. Pearson correlation between soma size and ERES number: r = 0.8404, R2 = 0.7062, p < 0.0001, n = 58 cells from 29 animals (WT: n = 8, lin-5: n = 11, unc-86: n = 10). (D) Quantification of cross-sectional soma area vs. ERES number across mature neuron types at larval stage L4. Pearson correlation between soma size and ERES number: r = 0.8449, R2 = 0.7138, p < 0.0001, n = 35 cells with at least six of each type. Results were additionally repeated in at least two independent experiments.
The impact of cell size on establishment of ERES number was directly tested using a lin-5 mutation. LIN-5/NUMA directly regulates asymmetric cell division and cell size by specifying spindle position during mitosis without directly involving transcription factors (Colombo et al., 2003; Gotta et al., 2003; Srinivasan et al., 2003). It has been shown that lin-5 mutations alter relative daughter cell size without changing anterior–posterior polarity (Jankele et al., 2021). To bypass the lethality of lin-5 null mutants, we generated a conditional lin-5 strain by tagging the endogenous lin-5 locus with a zf ubiquitination sequence (Armenti et al., 2014). We used the unc-86 promoter to express the ubiquitin ligase zif-1 to degrade zf-tagged LIN-5 in PVD mother shortly before PVD birth (Finney and Ruvkun, 1990). This manipulation displaces the spindle during PVD mother division, resulting in several categories of cell-division phenotypes that differ from animal to animal: 1) Failed cell division; 2) exaggerated asymmetry of daughter cells where the size difference between the daughter cells is larger than normal (Figure 4B lower left); 3) reduced asymmetry of daughter cells, where the daughter cells display a less pronounced size difference (Figure 4B lower right), and 4) WT-like asymmetry of daughter cells. We took advantage of these different phenotypes to compare initial ERES numbers in daughter cells of differing soma size at birth. We found a strong correlation between initial soma size and initial ERES number (Figure 4C), suggesting that asymmetric-cell division and large cell size at birth drives the establishment phase of high ERES numbers in PVD. Notably, we also observed a strong correlation between soma size and ERES number in mature neurons of different types (Figure 4D). Together with Figure 1C, this indicates that neurons of increasing dendrite size have larger somas and higher ERES numbers.
PVD cell-fate transcription factor MEC-3 is required for ERES number maintenance but not establishment
Having elucidated a mechanism for the establishment phase of high ERES number, we investigated mechanisms underlying the maintenance phase of ERES number during PVD development. Because MEC-3 acts downstream of UNC-86 (Chalfie and Au, 1989; Xue et al., 1992, 1993), and the mature PVD neuron in the mec-3(e1338)-mutant has a less severe phenotype than unc-86(e1416) (Figure 2B), we hypothesized that the mec-3(e1338) mutation would preserve asymmetric-cell division, but disrupt the later maintenance of ERES number. Consistent with this hypothesis, in mec-3(e1338) mutants, asymmetric cell division proceeds as normal, and we observe that the initial ERES number is similar to that of WT (Figure 5, A and B, left). However, by the L4 stage, mec-3(e1338) mutants showed dramatically reduced number of ERESs (Figure 2, A and B; Figure 5A), indicating that ERES number is actively maintained in a MEC-3-dependent manner as PVD develops. Indeed, time course analysis of mec-3(e1338) mutants show that ERES numbers in mec-3(e1338) are established at WT levels shortly after PVD birth, but gradually decline as the primary dendrite grows and the worm develops (Figure 5, A and B). Given the connection between soma size and initial ERES number observed in Figure 4, we wanted to test if mec-3(e1338) PVD soma size differs from WT. We observed that, like ERES number, initial soma size in WT and mec-3(e1338) does not significantly differ (Figure 5B, left). However, by L4, mec-3(e1338) PVD has a significantly smaller soma size than WT PVD (Figure 5B, right), further supporting an association between ERES number and soma size. While WT soma size increases as PVD develops, mec-3(e1338) soma size lacks that increase (Figure 5B). Notably, in unc-86(e1416) animals, where both asymmetric-cell division and cell-fate pathways are disrupted in PVD, we see a faster, more severe reduction in ERES number over time than in mec-3(e1338) mutants (Figure 5A). A reasonable interpretation of this observation is that both ERES establishment via asymmetric-cell division and ERES maintenance via cell-fate pathways are required for ERES number in the mature PVD neuron.
FIGURE 5:
MEC-3 and LET-363 maintain ERESs during neurite outgrowth in PVD. (A) Time course of ERES number in WT and cell-fate mutants. Points at t = 0 (PVD birth) were counted from time-lapse images to provide accurate timing information. Significance determined by one-way ANOVA, ****p < 0.0001, n ≥ 8 animals for each genotype at each time point. (B) Soma cross-sectional area of WT and mec-3 PVD at birth (left) and larval stage 4 (right). Significance determined by Student’s t test two-tailed ****p < 0.0001, n ≥ 8 animals. (C) Plet-363::GFP ceTOR transcriptional reporter intensity in PVD normalized to PDE intensity in WT and mec-3 animals at larval stage three. Significance determined by unpaired Student’s t test two-tailed ***p < 0.001, n = 29 (WT) & 26 (mec-3) animals. Data is from two independent experiments. (D) Quantification of number of GFP::SEC-16 puncta in WT vs. ceTOR/let-363(ok3018) constitutive mutant animal PVD somas. Significance determined by Student’s t test two-tailed ****p < 0.0001, n = 13 for each genotype. (E) Confocal-fluorescence microscopic images of PVD morphology in control and mutant worms (scale bar = 10 µm), with (F) quantification of branches. Significance determined by multiple t tests ****p < 0.0001. (G) Confocal-fluorescence microscopic images of PVD soma and endogenous ERES marker, GFP::SEC-16 in Pnhr-81::cre control worms (top) and conditionally floxed let-363 worms (bottom). Scale bar = 5 µm. (H) Quantification of number of GFP::SEC-16 puncta (top) and soma cross-sectional area (bottom) per PVD soma in control and mutant L2 (left) and L4 (right) worms. Significance determined by Student’s t test two-tailed, **p < 0.01, ****p < 0.0001, n ≥ 8 animals. Larval stages from youngest to oldest: L1, L2, L3, and L4. YA = Young Adult. Results were additionally repeated in at least one (C and H, left) or two independent experiments.
ceTOR/LET-363 expression is enhanced in PVD by MEC-3
Given the overall lack of somato-dendritic growth in mec-3(e1338) mutant PVD, we wondered whether MEC-3 might upregulate expression of master growth regulators. In the vertebrate, the master metabolic regulator mammalian target of rapamycin (mTOR) has been associated with overgrowth of neuronal somas in specific contexts (Kwon et al., 2001, 2003). TOR signaling has also been associated with cell size in a variety of cell types (Gonzalez and Rallis, 2017). We hypothesized that MEC-3 promotes expression or activity of C. elegans TOR (ceTOR) in PVD to promote increased biosynthetic and secretory capacity, and thus, ERES number in PVD. To test this hypothesis, we first examined a transcriptional reporter of let-363 developed by Long et al. (2002). To control for expression level differences between animals, we normalized ceTOR/let-363 reporter levels in PVD within each animal to reporter levels in PDE, which does not express MEC-3 (Way and Chalfie, 1989). We found that normalized reporter levels in WT PVD were significantly higher than in mec-3(e1338) (Figure 5C). These results suggest that MEC-3 enhances ceTOR/LET-363 expression in PVD.
ceTOR/LET-363 is required for maintenance of ERES number and somato-dendritic growth in PVD
Given TOR’s well-known role in promoting biosynthesis (Laplante and Sabatini, 2009; Nandagopal and Roux, 2015; Keith Blackwell et al., 2019), we wondered whether upregulation of ceTOR/LET-363 by MEC-3 in PVD might be responsible for PVD’s high ERES number and/or expansive somato-dendritic growth. First, we asked if let-363 activity is required for PVD’s high ERES number. In the constitutive let-363(ok3018) mutant, which exhibits a general growth arrest of C. elegans, we observe a reduction in ERES number in PVD soma (Figure 5D). To ask if LET-363 is required cell autonomously for ERES number, we built a conditional let-363 allele using the Cre-lox system to disrupt LET-363 specifically in PVD lineage cells (Figure 5, E–G). When let-363 was deleted, ERES number was normal at PVD birth, but significantly reduced by the time PVD reaches maturity at the L4 larval stage (Figure 5H). Similarly, the conditional let-363 mutant PVD also displayed normal soma size at PVD birth but reduced soma size by L4. Additionally, this allele showed a significant reduction in the size of PVD’s dendritic arbor compared with Cre controls (Figure 5E). The striking similarity between the conditional ceTOR/let-363 phenotypes and the mec-3(e1338) phenotypes argues that ceTOR/LET-363 is part of the MEC-3 dependent mechanisms necessary for PVD maturation.
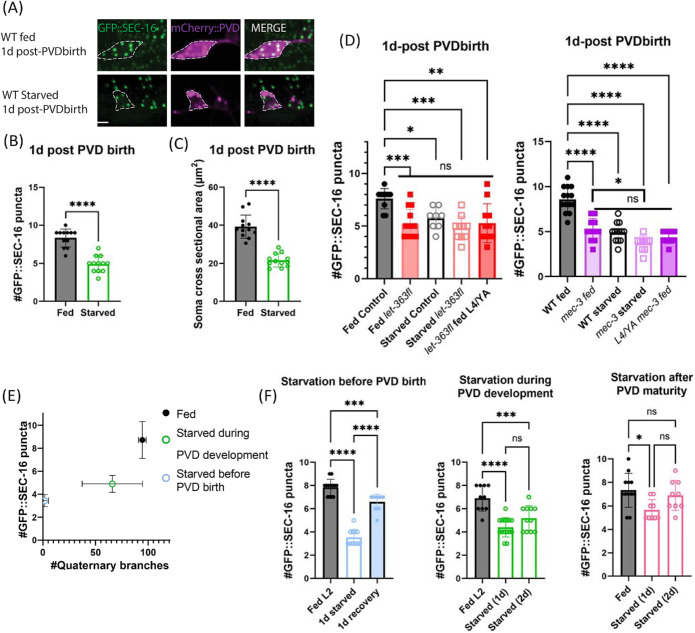
Starvation reduces ERES number and growth during critical period of development
Because TOR activity typically requires nutrient signals, and nutrient deprivation has been shown to disrupt ERESs (Zacharogianni et al., 2011), we hypothesized that TOR’s positive impact on ERES number in PVD requires the availability of nutrients (Baugh and Hu, 2020; González and Hall, 2017; Keith Blackwell et al., 2019). To test the effect of nutrient restriction on PVD ERES number, we removed worms from food just after PVD birth. The following day, starved worms displayed significantly fewer PVD ERESs than fed worms (Figure 6, A and B). In addition, PVD soma were significantly smaller in starved vs. fed worms (Figure 6C). To test whether the TOR-mediated and nutrient-mediated mechanisms on ERESs act in the same genetic pathway, we starved WT and let-363-floxed worms. We observed that there was no significant additional reduction of PVD ERESs in the let-363 worms upon starvation (Figure 6D, left). Similarly, there was no significant difference between WT-starved worms and let-363-starved worms (Figure 6D, left). These results are consistent with the notion that the impact of starvation on PVD is mediated at least in part through inactivation of ceTOR/let-363. Given our evidence of MEC-3-driven upregulation of LET-363 in PVD, we hypothesized that part of mec-3(e1338) mutant’s impact on ERES number in PVD is due to reduced LET-363 activity in these mutants. This hypothesis predicts that inactivation of TOR by starvation of mec-3(e1338) mutant animals would only have a small, if any, additional impact on ERES reduction compared with fed mec-3(e1338) mutant animals. We tested whether mec-3(e1338) and nutrient deprivation have independent effects on ERES number by starving mec-3(e1338) and WT worms. As expected, starved mec-3(e1338) worms only exhibit a small additional ERES reduction, as compared with WT starved worms and mec-3(e1338) fed worms (Figure 6D, right). This is consistent with the idea that ERES number reductions following mec-3(e1338) and starvation manipulations are partially but perhaps not fully redundant. Together, these findings suggest that MEC-3 drives a transcriptional program that includes increased ceTOR/let-363 expression, which helps to maintain PVD’s high ERES numbers in the presence of nutrients.
FIGURE 6:
Stage-specific nutrient deprivation reduces PVD ERES number and somatodendritic size. (A) Confocal-fluorescence microscopy closeups of PVD soma and endogenous ERES marker, GFP::SEC-16, 1 d after PVD birth in fed or 1-d starved WT animals (scale bar = 5 µm). PVDs in fed and starved animals were quantified for (B) number of PVD GFP::SEC-16 puncta (n = 12 per condition), and (C) soma size (n = 12 per condition) measured as described in Methods. Significance determined by Student’s t test two-tailed, ****p < 0.0001. (D) Number of GFP::SEC-16 puncta in PVD soma of fed vs. starved animals in Cre-control vs let-363-floxed (left, n ≥ 8 animals for each condition) and WT vs. mec-3 animals (right, n ≥ 10 animals for each condition). Animals were counted 1 d after PVD birth except where indicated. Fed animals at this time point were in late L3 or early L4 stages. (E) Quantification of PVD quaternary dendrite number vs. ERES number in either animals that were removed from food 7 h before PVD birth and starved for 1 d (pink, n = 9), animals that were removed from food just after PVD birth and starved for 1 d (green, n = 9), or animals that were fed for the duration of the experiment until 1 d after PVD birth at the L4 larval stage (black, n = 7). Points shown are mean values for each condition with standard deviations indicated by error bars. Pearson correlation between quaternary dendrite number and ERES number across conditions: r = 0.7122, R2 = 0.5072, p (two-tailed) < 0.0001. Color coding corresponds to diagram of starvation timelines in Supplemental Figure S3. (F) Number of GFP::SEC-16 puncta in WT animals removed from nutrients at different stages of PVD development, and starved for either 1 d and recovered (left, n ≥ 11), or 2 d (middle and right, n ≥ 9). Significance for (D) and (E) determined by one-way ANOVA, *p < 0.05,**p < 0.01,***p < 0.001, ****p < 0.0001. Results were additionally repeated in at least two independent experiments.
Given the nutrient requirement for PVD’s high ERES number and the correlation between dendrite size and ERES number in Figure 1, we predicted that dendrite size would scale with ERES number at different levels of nutrient availability. To test this prediction, we quantified ERES number and total quaternary dendrite number in animals with variable nutrient availability during PVD outgrowth (Supplemental Figure S3). In the first condition, to maximally reduce available nutrients for PVD growth, animals were removed from food before PVD birth. In the second condition, to partially reduce available nutrients during PVD growth, animals were removed from food shortly after PVD birth during dendrite outgrowth (as in Figure 6, A–D). In the third condition, to test for the requirement of nutrients on ERES number in mature PVD, animals were removed from food at the L4 stage, after PVD reached maturity. In the fourth and control condition, animals were left on food throughout the experiment. Consistent with our prediction, ERES number and quaternary dendrite number tend to scale with one another in response to the amount of nutrients available during PVD development (Figure 6E). Notably, the reduction of ERES number due to starvation was eliminated in mature starved animals (Figure 6F), raising the possibility that ERES number stabilizes with age.
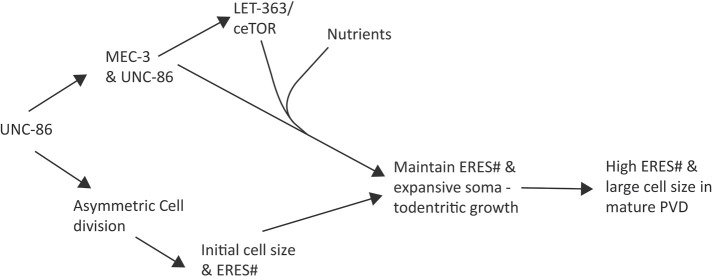
In summary, we show that ERES dynamics are organized into two phases, both of which contribute to high ERES number in PVD. During the establishment phase, UNC-86 and LIN-5 drive asymmetric-cell division to generate a large PVD soma at the expense of a small and dying sister cell. Our data suggest that the large soma leads to high ERES number in PVD due to cytoplasmic scaling. During the maintenance phase, as neurites emerge, MEC-3 drives ceTOR/LET-363, which, in the presence of nutrients, maintains high ERES number and promotes growth of the somato-dendritic compartment. Our results suggest a model in which transcription factors, master metabolic regulators, and nutrient availability coordinate to specify developmental parameters including soma size, ERES number, and dendrite size, which together determine neuronal cell fate (Figure 7).
FIGURE 7:
Proposed model for PVD-neuron size control. A schematic summary of the molecular pathways that regulate somato-dendritic size and ERES number in PVD.
DISCUSSION
In this study, we show that ERES number and somato-dendritic size are coordinately regulated, and identify mechanisms by which large, complex neurons establish and maintain elevated ERES numbers. ERESs export the majority of correctly folded cargo from the ER, and their structure and function is thus critical for maintaining secretory capacity to enable cellular growth and health, particularly in neurons (Barlowe and Helenius, 2016; Peotter et al., 2019; Tang, 2021). We observed a strong correlation between ERES number and dendrite size across cell types and cell-fate mutants, with PVD, C. elegans’ largest neuron, displaying the highest number of ERESs. ERESs, thus may be regarded as reliable indicators of neuronal biosynthetic and transport activity. We discovered that high ERES number in PVD is established rapidly after neuron birth, before neurite outgrowth, and is actively maintained throughout PVD development, using both time-lapse imaging and genetic perturbation experiments. Establishment of high ERES number is driven by asymmetric cell division and the resultant large cell size at birth. In turn, maintenance of ERES number is regulated by the transcription factor MEC-3, nutrient availability, and C. elegans TOR (ceTOR/let-363), a major nutrient sensor and regulator of cell growth. Our data support the notion that transcription factors and master growth regulators control ERES number, soma size, and dendrite size in a coordinated manner to drive neuronal cell-fate determination. In this context, asymmetric-cell division can be viewed as the initiating event of neuronal cell-fate determination, due to uneven distribution of key resources to daughter cells.
We demonstrate in PVD that ERES number is determined by cell fate pathways rather than feedback from growing neurites. Previous study has shown that extrinsic cues such as target-derived neurotrophins regulate survival and neuronal morphogenesis, demonstrating a feedback regulation mechanism (Purves et al., 1988; Fawcett and Keynes, 1990). We hypothesized that increasing secretory demand from a developing dendrite might upregulate ERES number. The PVD dendritic arbors are not only unusually large among worm neurons but also precisely target the space between skin and muscles (Zou et al., 2016). In the dendrite-guidance factor and receptor mutants, the dendritic arbor is drastically smaller and fails to grow into the skin–muscle junctions (Liu and Shen, 2011; Dong et al., 2016; Zou et al., 2016, 2018). However, we found these mutants did not show abnormalities in the number of early secretory organelles in PVD. These results argue strongly that the size, shape, and precise targeting of PVD dendrites are dispensable for the determination of somatic ERES number. Consistent with this notion, we demonstrate that GFP::SEC-16 puncta emerge rapidly during PVD birth, and establish high ERES numbers before neurite outgrowth. These findings suggest that secretory organelle number, and likely secretory capacity, is encoded into a neuron’s identity, acting as part of the cell-intrinsic programming that helps to drive a cell’s fate. Thus, while local neurite outgrowth is highly regulated by extrinsic growth factors and cues, overall secretory capacity, the “engine” underlying neurite growth, may be largely determined by cell-fate programs driving global nutrient sensing pathways (see below).
Whether high ERES numbers in C. elegans neurons correspond specifically to increased dendritic complexity (i.e., degree of branching), or more generally to increased cell-surface area or volume is an open question. Given the technical challenges of using fluorescence microscopy to measure cell membrane areas or volumes of entire extended neurons with complex morphologies, we used terminal branch number as a proxy for cell-surface area. While the assumptions made by this approximation are reasonable for the cell-types examined, the same assumptions may not hold for other cell-types. For example, the CAN neuron has a very simple morphology, but its processes have large diameters that result in very large estimations of CAN total cell-volume (Froehlich et al., 2021). Anecdotally, we have observed a large cell body in the region where CAN soma is located, that has high ERES numbers (unpublished data). We surmise that high ERES number results from the elevated biosynthesis required for large neuron surface areas in general, rather than being specifically related to high levels of neuronal branching. However, we cannot rule out the possibility that the morphological specifications and specialized proteins required for elaborate-dendritic branching specifically impact secretory rate and/or ERES number.
We report that cell-fate programming, governed by transcription factors UNC-86 and MEC-3, drives PVD ERES number in two phases: 1) establishment and 2) maintenance. In the establishment phase, our direct-imaging experiments show that PVD inherits a large soma due to asymmetric-cell division. We used different approaches to manipulate the size of PVD soma at birth and found that there is a strong correlation between initial cell-size and ERES number. In this phase, ERESs emerge rapidly as mitosis concludes, and the establishment of ERES numbers requires both the cell-fate transcription factor UNC-86 and regulators of cell-division plane including LIN-5/NUMA. Asymmetric-cell division has been previously shown to differentially distribute cell-fate transcription factors and organelles (Li et al., 2016; Vertii et al., 2018). It is tempting to speculate that uneven distribution of ERES components, such as SEC-16 and ER membranes, could provide the necessary seeds to establish persistent differences in secretory capacities. Given that PVD sister undergoes apoptosis soon after birth, there may be a preferential sorting of progrowth factors away from PVD sister to PVD, such that PVD sister is, in a sense, sacrificed to boost PVD’s growth potential. In the maintenance phase, MEC-3 and sufficient nutrients are needed to actively maintain ERES number. In mec-3(e1338) mutants, as well as starvation conditions, the number of ERESs decreases during this phase. The differences in ERES number phenotypes between unc-86(e1416) and mec-3(e1338) are consistent with the differential expression of these two transcription factors. Previous study shows that UNC-86 is expressed early, before PVD birth, and later activates MEC-3, which forms a heterodimer with UNC-86 to activate lineage-specific transcriptional programs (Chalfie and Au, 1989; Finney and Ruvkun, 1990; Xue et al., 1992, 1993; Duggan et al., 1998). These expression patterns are consistent with our findings that UNC-86 regulates the establishment of ERES number, while MEC-3 is only required for the maintenance phase. In a possible third phase, ERES numbers become stable because late-starvation, in contrast to early-starvation, does not lead to a reduction of ERESs.
We show ceTOR/LET363 acts downstream of cell-fate transcription factors during the maintenance phase, integrating nutrient-availability information to maintain ERES number and promote somato-dendritic growth of PVD. This work raises the possibility that cell-fate transcription factors could modulate ceTOR expression levels, which, in the presence of sufficient nutrients, set biosynthetic, and secretory capacities required for a particular cell-fate. Such a function would indicate a new-cell autonomous role for TOR as a regulator of physiological neuron-cell size. TOR’s function and signaling pathways in growth and nutrient sensing has been intensively studied in various contexts, but its role in nonpathological growth of neurons in vivo is not well understood. Kwon et al. (2003) found that abnormal soma expansion in granule cells of PTEN-deficient mice was dependent on TOR. Choi et al. (2008) showed that rapamycin-mediated inhibition of TOR in cultured-embryonic rat hippocampal neurons impaired axon growth but not dendrite growth. We report a cell-specific requirement for ceTOR/let-363 in the normal physiological growth of PVD. ceTOR/let-363′s role in PVD appears to coordinate soma size, ERES number, and dendrite size, suggesting it may be a cell-intrinsic master regulator for biosynthesis, secretion, and cell size in PVD. This impact on soma size differs from the finding by Kwon et al. (2003) that TOR disruption did not impact WT soma size in murine granule cells. Several factors may account for the apparent discrepancy in these results. Granule cells are among the smallest cells in vertebrate brains and their physiological growth may therefore be less sensitive to TOR (D’Angelo, 2016). More broadly, TOR-dependent growth may vary between neuron types and species. Further study is required to test whether differential-TOR activity is a general feature of neuron size and fate regulation.
Several potential downstream mechanisms could explain the reduction of ERES number in ceTOR/let-363 mutants and starved animals. TOR plays well-established roles in promoting protein translation and lipid biosynthesis, while inhibiting autophagy (Laplante and Sabatini, 2009; Nandagopal and Roux, 2015; Keith Blackwell et al., 2019; Deleyto-Seldas and Efeyan, 2021). It is possible that a reduction in protein translation and the resultant reduction of secretory flux subsequently reduces ERES number (Farhan et al., 2008b). In addition, a lack of TOR-mediated lipid synthesis, or a disinhibition of ER-phagy (Hamasaki et al., 2005; Zhao et al., 2015) may restrict the size of the soma and ER membrane, constraining the distance between ERESs, and thus increase probability of ERES fusion to reduce ERES number (Farhan et al., 2008a; Tillmann et al., 2015; Speckner et al., 2021). Another possibility is that lack of TOR activity disinhibits autophagy of ERESs directly, which may help to reduce secretion during starvation. Zacharogianni et al. (2011) show that amino-acid starvation in cell lines results in a TORC1 complex-independent dispersal of ERESs. Together with our data suggesting a ceTOR-dependent ERES reduction, these results support the notion that secretory organelles respond to starvation via a number of different pathways depending on variables such as the particular types of nutrient perturbations or organisms under study.
The rapid emergence of GFP::SEC-16 during cell birth is reminiscent of phase-transition dynamics characteristic of liquid–liquid phase separation (LLPS). Indeed, our observations of ERES establishment in PVD are consistent with several characteristics of LLPS, and suggest that regulation of biophysical properties of ERES components may play a role in determining ERES number. For instance, the dependence of initial ERES number on PVD cell size is consistent with the inherent size scaling behavior of LLPS condensates (Brangwynne, 2013). The tools developed here enable additional studies to further assess the emerging role of LLPS in the establishment and maintenance of ERESs in multicellular organisms (Bevis et al., 2002; Farhan et al., 2008a; Heinzer et al., 2008; Tillmann et al., 2015; Alberti et al., 2019; Peotter et al., 2019; Gallo et al., 2020; Speckner et al., 2021).
Taken together, our data suggest a model for neuronal-cell size control. Cell-fate transcription factors in PVD drive asymmetric-cell division to establish a large-initial cell size, which, in turn, determines high ERES number, perhaps through LLPS size-scaling. ERES number is subsequently maintained via a transcription factor-mediated modification of TOR levels, which regulates the nutrient-dependent biosynthetic and secretory capacity required for PVD’s neuronal fate. Our data also provide an additional perspective for future investigation into why neurons are particularly sensitive to mutations in early-secretory pathway proteins (Tang, 2021). The upregulation of early-secretory pathway structures shown here suggests that larger neurons may be particularly reliant on high, efficient secretory output. Large postmitotic neurons require a high-minimum secretory output to establish and maintain the expansive morphologies critical to their function. Thus, relatively small defects in secretory capacity may disproportionately impact form and function of large neurons. Thus, investigation into how large neurons regulate secretory structure and function will offer valuable insights into the heightened vulnerability of nervous systems to early-secretory pathway defects.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
C. elegans Strains
All strains were grown on nematode growth medium (NGM) plates seeded with OP50 Escherichia coli at 20°C unless otherwise stated for experimental conditions. N2 Bristol was used as WT strain. Mutant alleles and transgenes used in this study are listed in Supplemental Table S1. lin-5 mutants were kindly provided by Dr. Xing Liang.
Plasmids
Plasmid constructs for C. elegans expression were generated in pSM delta vector as a backbone, which was kindly provided by Dr. Andrew Fire. KE51(Pmec-17::mScarlet) & Plin-32::FLP, and pCER218 (ser2prom3:: sar-1(T35N)) constructs were kindly provided by Dr. Kelsie Eichel and Dr. Claire Richardson. DNA constructs were assembled into plasmid constructs using Gibson cloning methods. pCER218 (Pser2prom3:: sar-1(T35N), called sar-1DN in text) was made by amplifying the worm genomic sar-1, cloning it into the pSM delta vector and introducing a T35N mutation in a highly conserved residue in the GTPase domain (Kuge et al., 1994) to make sar-1(T35N) via site-directed mutagenesis.
C. elegans transformation
Transgenic extrachromosomal arrays were generated using a standard microinjection protocol (Mello and Fire, 1995). In short, plasmid constructs and coinjection marker were mixed and injected to distal arm of the gonad of young adult animals. F1 animals were then screened for transgenic lines. The details of transgenic arrays can be found on Supplemental Table S1.
CRISPR Genome editing
Clustered regularly interspaced short palindromic repeats (CRISPR)knock-ins were generated by direct gonadal injection of Cas9 protein (Integrated DNA Technologies [IDT]) duplexed with tracrRNA and targeting crRNA (IDT) along with respective repair templates (Paix et al., 2017). For long insertions (>150bp), knock-in templates (GFP-FLPon-SEC-16A.2) were PCR amplified from pSK–FLPon–GFP vectors (McDonald et al., 2020) with ultramer oligonucleotides (IDT) containing 100 bp of flanking-genome homology. PCR products were purified by ethanol precipitation or PCR cleanup and used for injection at 300 ng/μl concentrations. For short insertions (loxp sites), synthetic single-stranded DNA oligos (IDT Ultramers) were used as templates and injected at 600 nM concentrations. F1 animals were screened by PCR genotyping. Heterozygous F1 animals were homozygoused and the insertions were verified by Sanger sequencing. The following guide RNA sequences were used for genome edits: GFP-FLPon-SEC-16A.2: TGTTGCCAATAGAAGCTCAT; let-363-loxp: tttttctatattcgaaccaa & tgatattttgaaatatgcaa; rab-1-loxp: gaggaatcgatgcgagaatg & gtgaaagagtgccgaagagg.
C. elegans synchronization, staging, and nutrient deprivation
Gravid adults were washed off from eight to ten 6-cm plates with M9 buffer. Animals were then bleached in hypochlorite solution to obtain embryos. Embryos were washed with M9 buffer two to three times and then resuspended in M9 and allowed to hatch overnight into L1-arrested animals. L1-arrest animals were kept for maximum of 24 h before plating onto OP50-seeded NGM plates.
For the nutrient-deprivation protocol, freshly seeded, OP50 NGM plates were treated with ultraviolet (UV) light to kill OP50 bacteria (using a CL-1000S UV Crosslinker with 254 nm bulbs at the maximum energy setting), preventing additional bacterial growth to allow for more precisely timed starvation. Arrested L1s were plated onto UV-treated OP50 NGM plates, with varying amounts of OP50 in order to vary developmental stage of starvation. Animals were then monitored over the next 2 d, and at the appropriate stage (either 7 h before PVD-birth, 1–2 h following PVD-birth, or after PVD reached maturity) animals were either allowed to continue growing on starved plates or removed from OP50 onto either nutrient-free NGM plates (for starvation), or OP50-seeded NGM plates (fed controls). Data from fed and starved animals was collected at time points indicated in Supplemental Figure S3.
For time-course experiments, animals were synchronized as described above and examined under a Zeiss Axioplan fluorescence scope with 63X Plan-Apochromat 63 ×/1.4 NA objective for counting of ERES at indicated time points.
To image early PVD morphogenesis events (L2 animals), L1-arrest animals were grown on OP50-seeded NGM plates for 25–28 h depending on the strain at ∼20°C to reach L2 stage. After L2 stage had been reached, animals were monitored every 30 min under Zeiss Axioplan fluorescene scope with 63X Plan-Apochromat 63 ×/1.4 NA objective. Animals were mounted on slides for imaging when the majority of the animals reached the desired developmental stage (PVD precursor stage). To image mature PVDs and PDEs and TRNs, L4 animals, identified based on vulval development, were mounted on slides.
Microscopy
For still confocal images, hermaphrodite worms at L2 or L4 stage worms were immobilized on 2–3% agarose pads using 10 mM levamisole in M9 buffer. Images were obtained on a spinning disk system (3i) with a CSU-W1 spinning disk (Yokogawa), 405-nm, 488-nm, and 561-nm solid-state lasers, a C-Apochromat 63 ×/1.2 NA water-immersion objective, and a Prime95B camera (Photometrics). Z-stacks with 0.25–0.5 μm sections spanning the imaged neurons were taken. For live imaging of developing worms, L2 stage hermaphrodite worms were immobilized on 7.5% agarose pads using 2.5 mM levamisole in M9 buffer. Slides were sealed with VALAP (Vaseline, lanolin, paraffin, 1:1:1) to prevent agarose pad from desiccation. Live imaging was performed with the same system. Z-stacks with 0.4 μm sections were obtained every 3 min for maximum duration of 6 h.
Image Analysis
ERES were counted manually from unmodified Z-stacked images, using the neuron plasma membrane marker mCherry::PH(PLCγ), or cytoplasmic mScarlet to identify cells of interest (COI), and counting the number of endogenous GFP::SEC-16A.2 puncta in COI somas. Two puncta that appeared to be in contact but had clearly separate centers of highest intensity were defined as doublets and counted as two ERESs. Counting criteria were the same for counts made in Z-stack images taken on the 3i spinning disk confocal and counts made on animals directly under the Zeiss Axioplan microscope under the same magnification. For fluorescence intensity quantifications of nuclear-localized Plet-363::GFP, total fluorescence intensity values were obtained at Regions of Interest (ROIs) of SUM-projected Z-stacks of PVD and PDE nuclei using ImageJ.
Electron microscopy (EM)
WT N2 worms were prepared for conventional EM by high pressure freezing/freeze-substitution. Worms in M9 containing 20% BSA and E. coli were frozen in 100-μm well-specimen carriers (Type A) opposite a hexadecane-coated flat carrier (Type B) using a BalTec HPM 01 high-pressure freezer (BalTec, Lichtenstein). Freeze-substitution in 1%-OsO4, 0.1% of uranyl acetate, 1% of methanol in acetone, containing 3% of water (Walther and Ziegler, 2002; Buser and Walther, 2008) was carried out with a Leica AFS2 unit. Following substitution, samples were rinsed in acetone, infiltrated, and then polymerized in Eponate 12 resin (Ted Pella, Inc, Redding, CA). Serial 50 nm sections were cut with a Leica UCT ultramicrotome using a Diatome diamond knife, picked up on Pioloform coated slot grids and stained with uranyl acetate and Sato’s lead (Sato, 1968). Sections were imaged with an FEI Tecnai T12 TEM at 120 kV using a Gatan 4k x 4 k camera. TrakEM2 in Fiji was used to align serial sections (Cardona et al., 2012; Saalfeld et al., 2012; Schindelin et al., 2012). Modeling of serial sections was performed with IMOD (Kremer et al., 1996). Criteria for identification of ERESs in Electron Micrographs: ERESs can be unambiguously identified by serial EM reconstruction of the PVD cell body. ERESs show the characteristic-tubulovesicular structures located where ER lumen is expanded and ER membrane is free of ribosomes, adjacent to the Golgi stacks.
Neuron Soma Measurements
Z-stack still images taken on the 3i spinning disk microscope, as described above, were projected into a single plane using ImageJ software. ROIs were drawn manually around the projected soma, using the neuron plasma membrane marker mCherry::PH(PLCγ), or cytoplasmic mScarlet to identify the COI, which develop with highly stereotyped polarities and positions. In Figure 3 and Figure 4 time-lapse imaging experiments, plasma membrane localized mCherry was used to identify the cytokinesis-cleavage furrow resulting in PVD’s birth, and ROIs were drawn around PVD and PVD sister as described above, in the time frame showing the conclusion of cytokinesis (determined by the point at which the two sisters become and remain clearly separated by membrane mCherry). The areas of these ROI’s were measured in μm2 as an estimation of the maximal soma cross-section of each COI in the xy plane.
Quantification of dendritic arbor
For Figure 1C, all terminal branches in AVM, PVM, PDE, and FLP were counted in images of L4 animals. For all PVD dendrite quantifications, images were straightened with the ImageJ software (US National Institutes of Health) using PVD primary dendrite as a reference. For WT PVD in Figure 1C, quaternary branches were counted within a rectangular ROI extending 150 μm anterior of PVD soma and 30 μm posterior of PVD soma. For SAR-1-DN and control L4 animals (Figure 1I), a 100-μm long rectangular ROI was placed starting at the center of PVD soma extending 100 μm anteriorly, containing the entire width of the animal, while for rab-1(wy1375) (Figure 1H), SAR-1DN, and control young-adult animals, a 100-μm long rectangular ROI was placed with PVD soma at the center, and the number of secondary, tertiary, and quaternary branches within ROIs were counted. For let-363-floxed and control L4 animals, all branches anterior of PVD soma were counted. Any protrusion from the primary dendrite was classified as a secondary branch, and any correctly oriented protrusion from the tertiary dendrite was classified as a quaternary branch.
Quantification and statistical analysis
Quantifications were blinded in any cases where mutant phenotypes were not easily observable. All statistical analysis was done using GraphPad Prism 9 software. All statistical parameters can be found in the corresponding figure legends. No statistical methods were used to predetermine the sample size.
Materials and data availability
All reagents and data are available upon request.
Supplementary Material
Acknowledgments
We thank A.A. for conversations and advice; C.W. and F.M. for strain FR297 ( Long et al., 2002); J.L. for assistance with long-term time-lapse imaging; L.T. for guidance receptor mutant morphology images; C.R. and members of the Shen Lab for conversation and reagents; V.P. for excellent technical support. This work was supported by the Howard Hughes Medical Institute and the National Institute of Neurological Disorders and Stroke (1R01NS082208) and the Phil & Penny Knight Initiative for Brain Resilience at the Wu Tsai Neurosciences Institute, Stanford University.
Abbreviations used:
- C. elegans
Caenorhabditis elegans
- cAVM
converted AVM
- ceTOR
Caenorhabditis elegans target of rapamycin
- COI
cell of interest
- CRISPR
clustered regularly interspaced short palindromic repeats
- DN
dominant negative
- EM
electron microscopy
- ER
endoplasmic reticulum
- ERES
endoplasmic reticulum exit site
- GFP
green fluorescent protein
- GTP
guanosine triphosphate
- L4
larval stage 4
- LLPS
liquid-liquid phase separation
- mTOR
mammalian target of rapamycin
- NGM
nematode growth medium
- PVD sis
PVD sister cell
- ROI
Region of Interest
- TORC1
target of rapamycin complex 1
- TRN
touch receptor neuron
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-03-0090) on August 9, 2023.
REFERENCES
- Alberti S, Gladfelter A, Mittag T (2019). Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 176, 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenti ST, Lohmer LL, Sherwood DR, Nance J (2014). Repurposing an endogenous degradation system for rapid and targeted depletion of C. elegans proteins. Development 141, 4640–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balklava Z, Pant S, Fares H, Grant BD (2007). Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol 9, 1066–1073. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Helenius A (2016). Cargo Capture and Bulk Flow in the Early Secretory Pathway. Annu Rev Cell Dev Biol 32, 197–222. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hu PJ (2020). Starvation Responses Throughout the Caenorhabditiselegans Life Cycle. Genetics 216, 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevis BJ, Hammond AT, Reinke CA, Glick BS (2002). De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat Cell Biol 2002 410 4, 750–756. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP (2013). Phase transitions and size scaling of membrane-less organelles. J Cell Biol 203, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser C, Walther P (2008). Freeze-substitution: The addition of water to polar solvents enhances the retention of structure and acts at temperatures around -60°C. J Microsc 230, 268–277. [DOI] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, Douglas RJ (2012). TrakEM2 Software for Neural Circuit Reconstruction. PLoS One 7, e38011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Au M (1989). Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science 243, 1027–1033. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Di Nardo A, Kramvis I, Meikle L, Kwiatkowski DJ, Sahin M, He X (2008). Tuberous sclerosis complex proteins control axon formation. Genes Dev 22, 2485–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont Y, Xia L, Rambourg A, Turner JD, Hermo L (1993). Structure of the Golgi apparatus in stimulated and nonstimulated acinar cells of mammary glands of the rat. Anat Rec 237, 308–317. [DOI] [PubMed] [Google Scholar]
- Colombo K, Grill SW, Kimple RJ, Willard FS, Siderovski DP, Gönczy P (2003). Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science (80- ) 300, 1957–1961. [DOI] [PubMed] [Google Scholar]
- D’Angelo E (2016). Granule cells and parallel fibers. In: Essentials of cerebellum and cerebellar disorders, ed. Gruol DL, Koibuchi N, Manto M, Molinari M, Schmahmann JD, Shen Y, Switzerland: Springer International, 177–182. [Google Scholar]
- Deleyto-Seldas N, Efeyan A (2021). The mTOR–Autophagy Axis and the Control of Metabolism. Front Cell Dev Biol 9, 1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Chiu H, Park YJ, Zou W, Zou Y, Özkan E, Chang C, Shen K (2016). Precise regulation of the guidance receptor DMA-1 by KPC-1/Furin instructs dendritic branching decisions. Elife 5. doi: 10.7554/ELIFE.11008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan A, Ma C, Chalfie M (1998). Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development 125, 4107–4119. [DOI] [PubMed] [Google Scholar]
- Farhan H, Weiss M, Tani K, Kaufman RJ, Hauri HP (2008a). Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J 27, 2043–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H, Weiss M, Tani K, Kaufman RJ, Hauri HP (2008b). Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J 27, 2043–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JW, Keynes RJ (1990). Peripheral nerve regeneration. Annu Rev Neurosci 13, 43–60. [DOI] [PubMed] [Google Scholar]
- Finney M, Ruvkun G (1990). The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63, 895–905. [DOI] [PubMed] [Google Scholar]
- Froehlich JJ, Rajewsky N, Ewald CY (2021). Estimation of C. elegans cell- and tissue volumes. microPublication Biol 2021. doi: 10.17912/MICROPUB.BIOLOGY.000345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo R, Rai A, Pelkmans L (2020). DYRK3-Controlled Phase Separation Organizes the Early Secretory Pathway. bioRxiv 2020.02.10.941757. [Google Scholar]
- González A, Hall MN (2017). Nutrient sensing and TOR signaling in yeast and mammals. EMBO J 36, 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez S, Rallis C (2017). The TOR signaling pathway in spatial and temporal control of cell size and growth. Front Cell Dev Biol 5, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick FS, Jamieson JD (2012). Structure–function Relationships in the Pancreatic Acinar Cell. Physiol Gastrointest Tract, Two Vol Set 1341–1360. doi: 10.1016/B978-0-12-382026-6.00049-X [DOI] [Google Scholar]
- Gotta M, Dong Y, Peterson YK, Lanier SM, Ahringer J (2003). Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr Biol 13, 1029–1037. [DOI] [PubMed] [Google Scholar]
- Griffiths G, Pfeiffer S, Simons K, Matlin K (1985). Exit of newly synthesized membrane proteins from the trans cisterna of the Golgi complex to the plasma membrane. J Cell Biol 101, 949–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Noda T, Baba M, Ohsumi Y (2005). Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6, 56–65. [DOI] [PubMed] [Google Scholar]
- Heinzer S, Wörz S, Kalla C, Rohr K, Weiss M (2008). A model for the self-organization of exit sites in the endoplasmic reticulum. J Cell Sci 121, 55–64. [DOI] [PubMed] [Google Scholar]
- Jankele R, Jelier R, Gönczy P (2021). Physically asymmetric division of the c. Elegans zygote ensures invariably successful embryogenesis. Elife 10, 1–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith Blackwell T, Sewell AK, Wu Z, Han M (2019). TOR Signaling in Caenorhabditis elegans Development, Metabolism, and Aging. Genetics 213, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996). Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116, 71–76. [DOI] [PubMed] [Google Scholar]
- Kuge O, Dascher C, Orci L, Rowe T, Amherdt M, Plutner H, Ravazzola M, Tanigawa G, Rothman JE, Balch WE (1994). Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J Cell Biol 125, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Zhu X, Zhang J, Baker SJ (2003). mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc Natl Acad Sci USA 100, 12923–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Zhu X, Zhang J, Knoop LL, Tharp R, Smeyne RJ, Eberhart CG, Burger PC, Baker SJ (2001). Pten regulates neuronal soma size: A mouse model of Lhermitte-Duclos disease. Nat Genet 29, 404–411. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM (2009). An Emerging Role of mTOR in Lipid Biosynthesis. Curr Biol 19, R1046–R1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L-B, Lei H, Arey RN, Li P, Liu J, Murphy CT, Xu XZS, Shen K (2016). The Neuronal Kinesin UNC-104/KIF1A Is a Key Regulator of Synaptic Aging and Insulin Signaling-Regulated Memory. Curr Biol 26, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu OW, Shen K (2011). The transmembrane LRR protein DMA-1 promotes dendrite branching and growth in C. elegans. Nat Neurosci 2011 151 15, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KR, Yamamoto Y, Baker AL, Watkins SC, Coyne CB, Conway JF, Aridor M (2010). Sar1 assembly regulates membrane constriction and ER export. J Cell Biol 190, 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Spycher C, Han ZS, Rose AM, Müller F, Avruch J (2002). TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr Biol 12, 1448–1461. [DOI] [PubMed] [Google Scholar]
- McDonald NA, Fetter RD, Shen K (2020). Assembly of synaptic active zones requires phase separation of scaffold molecules. Nat 2020 5887838 588, 454–458. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A (1995). Chapter 19 DNA Transformation. Methods Cell Biol 48, 451–482. [PubMed] [Google Scholar]
- Nakano A, Muramatsu M (1989). A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol 109, 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandagopal N, Roux PP (2015). Regulation of global and specific mRNA translation by the mTOR signaling pathway. Translation 3, e983402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren-Suissa M, Hall DH, Treinin M, Shemer G, Podbilewicz B (2010). The fusogen EFF-1 controls sculpting of mechanosensory dendrites. Science 328, 1285–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Seydoux G (2017). Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods 121–122, 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peotter J, Kasberg W, Pustova I, Audhya A (2019). COPII-mediated trafficking at the ER/ERGIC interface. Traffic doi: 10.1111/tra.12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves D, Snider WD, Voyvodic JT (1988). Trophic regulation of nerve cell morphology and innervation in the autonomic nervous system. Nat 1988 3366195 336, 123–128. [DOI] [PubMed] [Google Scholar]
- Saalfeld S, Fetter R, Cardona A, Tomancak P (2012). Elastic volume reconstruction from series of ultra-thin microscopy sections. Nat Methods 2012 97 9, 717–720. [DOI] [PubMed] [Google Scholar]
- Saegusa K, Matsunaga K, Maeda M, Saito K, Izumi T, Sato K (2022). Cargo receptor Surf4 regulates endoplasmic reticulum export of proinsulin in pancreatic β-cells. Commun Biol 2022 51 5, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R, Marie M, Nizak C, Dale HA, Pernet-Gallay K, Perez F, Goud B, Saraste J (2006). Rab1 defines a novel pathway connecting the pre-Golgi intermediate compartment with the cell periphery. Mol Biol Cell 17, 1514–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sato M, Audhya A, Oegema K, Schweinsberg P, Grant BD (2006). Dynamic regulation of caveolin-1 trafficking in the germ line and embryo of Caenorhabditis elegans. Mol Biol Cell 17, 3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T (1968). A modified method for lead staining of thin sections. J Electron Microsc (Tokyo) 17, 158–159. [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 2012 97 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, O’Brien T, Chatzigeorgiou M, Clay Spencer W, Feingold-Link E, Husson SJ, Hori S, Mitani S, Gottschalk A, Schafer WR, Miller DM (2013). Sensory neuron fates are distinguished by a transcriptional switch that regulates dendrite branch stabilization. Neuron 79, 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Watson JD, Spencer WC, O’Brien T, Cha B, Albeg A, Treinin M, Miller DM (2010). Time-lapse imaging and cell-specific expression profiling reveal dynamic branching and molecular determinants of a multi-dendritic nociceptor in C. elegans. Dev Biol 345, 18–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speckner K, Stadler L, Weiss M (2021). Unscrambling exit site patterns on the endoplasmic reticulum as a quenched demixing process. Biophys J 120, 2532–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan DG, Fisk RM, Xu H, Van den Heuvel S (2003). A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev 17, 1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DJ (2003). De novo formation, fusion and fission of mammalian COPII-coated endoplasmic reticulum exit sites. EMBO Rep 4, 210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL (2021). Defects in early secretory pathway transport machinery components and neurodevelopmental disorders. Rev Neurosci 32, 851–869. [DOI] [PubMed] [Google Scholar]
- Teuliere J, Kovacevic I, Bao Z, Garriga G (2018). The Caenorhabditis elegans gene ham-1 regulates daughter cell size asymmetry primarily in divisions that produce a small anterior daughter cell. doi: 10.1371/journal.pone.0195855 [DOI] [PMC free article] [PubMed]
- Tillmann KD, Reiterer V, Baschieri F, Hoffmann J, Millarte V, Hauser MA, Mazza A, Atias N, Legler DF, Sharan R, et al. (2015). Regulation of Sec16 levels and dynamics links proliferation and secretion. J Cell Sci 128, 670–682. [DOI] [PubMed] [Google Scholar]
- Tsalik EL, Hobert O (2003). Functional mapping of neurons that control locomotory behavior in Caenorhabditis elegans. J Neurobiol 56, 178–197. [DOI] [PubMed] [Google Scholar]
- Vertii A, Kaufman PD, Hehnly H, Doxsey S (2018). New dimensions of asymmetric division in vertebrates. Cytoskeleton 75, 87–102. [DOI] [PubMed] [Google Scholar]
- Walther P, Ziegler A (2002). Freeze substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. J Microsc 208, 3–10. [DOI] [PubMed] [Google Scholar]
- Warren G (2013). Transport through the Golgi in Trypanosoma brucei. Histochem Cell Biol 140, 235–238. [DOI] [PubMed] [Google Scholar]
- Way JC, Chalfie M (1989). The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev 3, 1823–1833. [DOI] [PubMed] [Google Scholar]
- Weigel AV, Chang CL, Shtengel G, Xu CS, Hoffman DP, Freeman M, Iyer N, Aaron J, Khuon S, Bogovic J, et al. (2021). ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell 184, 2412–2429.e16. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S (1986). The Structure of the Nervous System of the Nematode Caenorhabdites Elegans. Phil Trans R Soc London Ser B, Biol Scien 314. http://www.wormatlas.org/ver1/MoW_built0.92/toc.html [DOI] [PubMed] [Google Scholar]
- Xue D, Finney M, Ruvkun G, Chalfie M (1992). Regulation of the mec-3 gene by the C.elegans homeoproteins UNC-86 and MEC-3. EMBO J 11, 4969–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Tu Y, Chalfie M (1993). Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science 261, 1324–1328. [DOI] [PubMed] [Google Scholar]
- Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN (2007). Growing dendrites and axons differ in their reliance on the secretory pathway. Cell 130, 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelinek JT, He CY, Warren G (2009). Ultrastructural Study of Golgi Duplication in Trypanosoma brucei. Traffic 10, 300–306. [DOI] [PubMed] [Google Scholar]
- Zacharogianni M, Kondylis V, Tang Y, Farhan H, Xanthakis D, Fuchs F, Boutros M, Rabouille C (2011). ERK7 is a negative regulator of protein secretion in response to amino-acid starvation by modulating Sec16 membrane association. EMBO J 30, 3684–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang H, Liu W, Duan X, Shang W, Xia D, Tong C (2015). Sec22 regulates endoplasmic reticulum morphology but not autophagy and is required for eye development in Drosophila. J Biol Chem 290, 7943–7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Dong X, Broederdorf TR, Shen A, Kramer DA, Shi R, Liang X, Miller DM, Xiang YK, Yasuda R, et al. (2018). A Dendritic Guidance Receptor Complex Brings Together Distinct Actin Regulators to Drive Efficient F-Actin Assembly and Branching. Dev Cell 45, 362–375.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Shen A, Dong X, Tugizova M, Xiang YK, Shen K (2016). A multi-protein receptor-ligand complex underlies combinatorial dendrite guidance choices in C. elegans. Elife 5. doi: 10.7554/ELIFE.18345 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.