Abstract
Background
Epigenetic clocks constructed from DNA methylation patterns have emerged as excellent predictors of aging and aging-related health outcomes. Iron, a crucial element, is meticulously regulated within organisms, a phenomenon referred as iron homeostasis. Previous researches have demonstrated the sophisticated connection between aging and iron homeostasis. However, their causal relationship remains relatively unexplored.
Results
Through two-sample Mendelian randomization (MR) utilizing the random effect inverse variance weighted (IVW) method, each standard deviation (SD) increase in serum iron was associated with increased GrimAge acceleration (GrimAA, BetaIVW = 0.27, P = 8.54E−03 in 2014 datasets; BetaIVW = 0.31, P = 1.25E−02 in 2021 datasets), HannumAge acceleration (HannumAA, BetaIVW = 0.32, P = 4.50E−03 in 2014 datasets; BetaIVW = 0.32, P = 8.03E−03 in 2021 datasets) and Intrinsic epigenetic age acceleration (IEAA, BetaIVW = 0.34, P = 5.33E−04 in 2014 datasets; BetaIVW = 0.49, P = 9.94E−04 in 2021 datasets). Similar results were also observed in transferrin saturation. While transferrin manifested a negative association with epigenetic age accelerations (EAAs) sensitivity analyses. Besides, lack of solid evidence to support a causal relationship from EAAs to iron-related biomarkers.
Conclusions
The results of present investigation unveiled the causality of iron overload on acceleration of epigenetic clocks. Researches are warranted to illuminate the underlying mechanisms and formulate strategies for potential interventions.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-023-01575-w.
Keywords: Iron metabolism, Iron homeostasis, Senescence, Aging, Epigenetic age acceleration, Mendelian randomization
Introduction
Aging is a natural process of all organisms characterized by loss of physiological integrity, function decline and vulnerable to death in a time-dependent manner [1]. Iron is one of the most essential transition metals in human body [2]. The balance of iron metabolism, also known as iron homeostasis, is strictly regulated due to its crucial role in erythropoiesis, oxidative phosphorylation and redox reaction [3]. Evidences have connected altered iron homeostasis with biological aging. For example, epidemiological research reported that over 10% of both men and women aged 65 years or older were anemic in the US, in which iron deficiency made up approximately 20% of all anemia cases [4]. Chronic inflammation of the elder people might also contribute to the alteration of serum iron biomarkers, causing iron deficiency and impaired iron mobilization [3, 5]. On the other hand, cellular iron accumulation in older individuals was observed. Serum level of ferritin, which reflected the storage of iron, was reported to be increasing with age and negatively associated with telomere length [6, 7]. Iron overload in cell induced the accumulation of lipofuscin, which was considered one of the hallmarks of aging and could be cytotoxic [5, 8]. However, little was known about the change of total iron content of body with age, as well as the relationship between iron homeostasis and aging.
The lifespan of organism is programed by its genetic information and influenced by its environment [9]. Distinct from conservative DNA sequence, the epigenome regulated gene expression based on a range of chemical modifications that were reversible [10]. Epigenetic alteration was proposed as one of the twelve hallmarks of aging and participated in the pathogenesis of several age-related diseases [1]. At present, the best characterized mark of epigenome is DNA methylation, a methyl group added on the fifth carbon of a cytosine residue, which could also be influenced by environmental factors, including the passage of time [11]. Thus, the epigenetic clocks, based on the DNA methylation status of human and their chronological age and health-related outcomes, were built to discover the impact of both genetic and environmental factors on human aging [12]. Epigenetic age acceleration (EAA) was used to describe individuals with greater epigenetic-clock-estimated age than their true chronological age, indicating worse health outcome [13]. Although iron homeostasis was in connection with aging, no research regarding the relationship between epigenetic clocks or EAA and iron homeostasis has been conducted.
Utilizing outcomes from genome-wide association studies (GWAS), Mendelian randomization (MR) has been widely used in discovering causality between exposure factors and outcomes and has a better performance in controlling confounding and reverse causation [14]. To achieve MR analysis, single nucleotide polymorphisms (SNPs) are selected as instrumental variables (IVs) with three rules, IVs must be (1) associated with the exposure; (2) independent of all confounders of the exposure–outcome association; and (3) independent of the outcome [15]. McCartney et al. conducted a GWAS of four epigenetic clocks, and subsequent MR analysis identified several risk factors of EAAs [16]. Based on the GWAS statistics, Pan et al. have reported bidirectional causal relationships between EAAs and kidney function [17]. To our knowledge, no MR analysis has been conducted exploring the causal relationship between iron homeostasis and EAAs.
In this study, we conducted a two-sample MR analyses with summarized GWAS data mentioned above to investigate the causal relationship between iron homeostasis and EAAs. The results might provide new evidence for further research on intervening iron homeostasis of the elderly and its potential mechanics.
Results
Genetic instruments selection
Selection of qualified SNPs from summarized GWAS data was conducted, and the number of SNPs in every process is presented in Additional file 1: Figure S1. Notably, lack of qualified SNPs from pancreas iron content, thus 183 of SNPs that met the threshold of 5E−06 were elevated. Total F-statistics of SNPs were all larger than 10, indicating strong IVs.
Causal effects of plasmatic iron biomarkers on EAAs
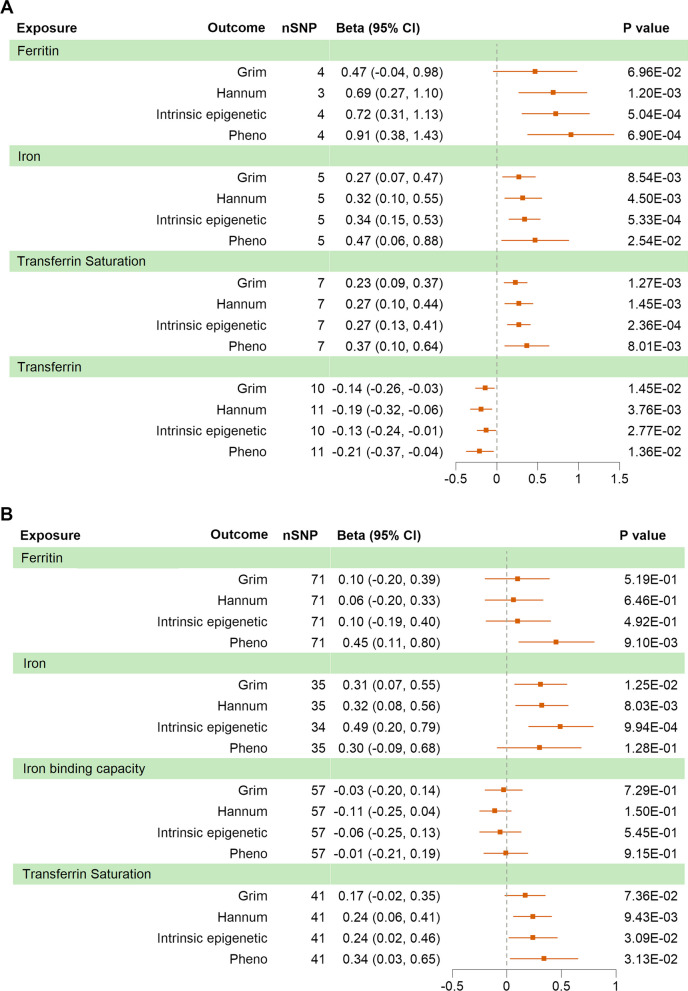
Random effect inverse variance weighted (IVW) analyses were carried out with iron-related traits from three meta-GWAS as exposures and EAAs as outcomes. Serum iron, ferritin and transferrin saturation were simultaneously obtained from two independent GWAS (namely 2014 datasets and 2021 datasets based on the year of publish). As plotted in Fig. 1A, B, genetically predicted serum iron was significantly associated with genetic predisposition to GrimAge acceleration (GrimAA, BetaIVW = 0.27 years per standard deviation (SD) increase in serum iron, P = 8.54E−03 in 2014 datasets; BetaIVW = 0.31, P = 1.25E−02 in 2021 datasets), HannumAge acceleration (HannumAA, BetaIVW = 0.32, P = 4.50E−03 in 2014 datasets; BetaIVW = 0.32, P = 8.03E−03 in 2021 datasets) and intrinsic epigenetic age acceleration (IEAA, BetaIVW = 0.34, P = 5.33E−04 in 2014 datasets; BetaIVW = 0.49, P = 9.94E−04 in 2021 datasets), but partially significant in PhenoAge acceleration (PhenoAA, BetaIVW = 0.47, P = 2.54E−02 in 2014 datasets; BetaIVW = 0.30, P = 1.28E−01 in 2021 datasets). Transferrin saturation of both datasets demonstrated significant association with HannumAA (BetaIVW = 0.27 years per SD increase in transferrin saturation, P = 1.45E−03 in 2014 datasets; BetaIVW = 0.24, P = 9.43E−03 in 2021 datasets), IEAA (BetaIVW = 0.27, P = 2.36E−04 in 2014 datasets; BetaIVW = 0.24, P = 3.09E−02 in 2021 datasets) and PhenoAA (BetaIVW = 0.37, P = 8.01E−03 in 2014 datasets; BetaIVW = 0.34, P = 3.13E−02 in 2021 datasets), but lacked significance with GrimAA in 2021 datasets (BetaIVW = 0.23, P = 1.27E−03 in 2014 datasets; BetaIVW = 0.17, P = 7.36E−02 in 2021 datasets). As for ferritin, significant results were observed in HannumAA (BetaIVW = 0.69 years per SD increase in ferritin, P = 1.20E−03), IEAA (BetaIVW = 0.72, P = 5.04E−04) and PhenoAA (BetaIVW = 0.91, P = 6.90E−04) from 2014 datasets, as well as PhenoAA (BetaIVW = 0.45, P = 9.10E−03) from 2021 datasets. The same negative direction of associations was obtained in genetically predicted transferrin from 2014 datasets and genetically predicted total iron binding capacity (TIBC) from 2021 datasets, the former was significant with GrimAA (BetaIVW = -0.14 years per SD increase in transferrin, P = 1.45E−02), HannumAA (BetaIVW = -0.19, P = 3.76E−03), IEAA (BetaIVW = −0.13, P = 2.77E−02) and PhenoAA (BetaIVW = − 0.21, P = 1.36E−02), while the latter was not.
Fig. 1.
MR analyses of 2014 (A) and 2021 (B) datasets with epigenetic aging accelerations. MR, Mendelian randomization; SNP, single nucleotide polymorphism
In sensitivity analyses, opposite directions were found in part of analyses of ferritin and TIBC from 2021 datasets. Notably, severe heterogeneity was detected in serum iron from 2014 datasets to PhenoAA (I2 = 66.5%). No pleiotropy was observed among all analyses. Results remained consistent after adjusting outliners in serum iron and transferrin saturation from 2021 datasets. Taken together, serum iron might be the potential cause of acceleration of GrimAge, HannumAge and intrinsic epigenetic age. Transferrin saturation was significantly associated with increasing of GrimAA, HannumAA, IEAA and PhenoAA. Ferritin was associated with elevated PhenoAA. All the above results were significant in two datasets and validated in sensitivity analyses. Serum transferrin demonstrated negative association with the four EAAs, but results of TIBC were only significant in IEAA and PhenoAA. Complete results are exhibited in Tables 1 and 2.
Table 1.
Sensitivity analyses of serum iron biomarkers in 2014 datasets with epigenetic aging accelerations
| Exposure | Outcome | Weighted median | MR-egger regression | Heterogeneitya | MR-PRESSO outlier detectb | Pleiotropyc | |||
|---|---|---|---|---|---|---|---|---|---|
| Beta (95% CI) | P value | Beta (95% CI) | P value | Beta (95% CI) | P Value | ||||
| Ferritin | Grim | 0.76 (0.23, 1.29) | 4.82E− 03 | 0.76 (− 0.33, 1.85) | 3.04E−01 | I2 = 39.6%; Cochrane Q = 5; P = 0.174 | No significant outliers | Intercept = -0.031; P = 0.603 | |
| Ferritin | Hannum | 0.82 (0.33, 1.31) | 1.04E−03 | 0.83 (− 0.25, 1.91) | 3.74E−01 | I2 = 0%; Cochrane Q = 2; P = 0.380 | No significant outliers | Intercept = -0.018; P = 0.812 | |
| Ferritin | IE | 0.75 (0.27, 1.24) | 2.29E−03 | 0.98 (0.14, 1.82) | 1.51E−01 | I2 = 1.6%; Cochrane Q = 3; P = 0.384 | No significant outliers | Intercept = -0.028; P = 0.554 | |
| Ferritin | Pheno | 0.80 (0.18, 1.41) | 1.18E−02 | 0.77 (− 0.43, 1.97) | 3.34E−01 | I2 = 6.7%; Cochrane Q = 3; P = 0.360 | No significant outliers | Intercept = 0.014; P = 0.820 | |
| Iron | Grim | 0.30 (0.06, 0.55) | 1.33E−02 | 0.49 (0.09, 0.90) | 9.77E−02 | I2 = 14.5%; Cochrane Q = 5; P = 0.322 | No significant outliers | Intercept = -0.045; P = 0.309 | |
| Iron | Hannum | 0.31 (0.08, 0.53) | 8.00E−03 | 0.54 (0.06, 1.01) | 1.15E−01 | I2 = 32.1%; Cochrane Q = 6; P = 0.208 | No significant outliers | Intercept = -0.043; P = 0.396 | |
| Iron | IE | 0.27 (0.02, 0.51) | 3.16E−02 | 0.65 (0.25, 1.06) | 5.09E−02 | I2 = 0%; Cochrane Q = 3; P = 0.480 | No significant outliers | Intercept = -0.064; P = 0.180 | |
| Iron | Pheno | 0.55 (0.24, 0.85) | 4.81E−04 | 0.60 (− 0.40, 1.6) | 3.24E−01 | I2 = 66.5%; Cochrane Q = 12; P = 0.018 | No significant outliers | Intercept = -0.027; P = 0.787 | |
| Transferrin saturation | Grim | 0.27 (0.10, 0.44) | 1.62E−03 | 0.28 (0.03, 0.52) | 7.80E−02 | I2 = 0%; Cochrane Q = 3; P = 0.856 | No significant outliers | Intercept = -0.011; P = 0.678 | |
| Transferrin saturation | Hannum | 0.28 (0.12, 0.44) | 7.46E−04 | 0.32 (0.01, 0.63) | 9.98E−02 | I2 = 30.3%; Cochrane Q = 9; P = 0.196 | No significant outliers | Intercept = -0.013; P = 0.716 | |
| Transferrin saturation | IE | 0.28 (0.11, 0.44) | 9.04E−04 | 0.41 (0.16, 0.66) | 2.36E−02 | I2 = 0%; Cochrane Q = 2; P = 0.870 | No significant outliers | Intercept = -0.035; P = 0.238 | |
| Transferrin saturation | Pheno | 0.36 (0.14, 0.58) | 1.49E−03 | 0.33 (− 0.19, 0.85) | 2.67E−01 | I2 = 56.8%; Cochrane Q = 14; P = 0.031 | No significant outliers | Intercept = 0.010; P = 0.869 | |
| Transferrin | Grim | − 0.05 (− 0.21, 0.12) | 5.68E−01 | − 0.18 (− 0.35, 0.01) | 8.77E−02 | I2 = 4%; Cochrane Q = 9; P = 0.403 | No significant outliers | Intercept = 0.009; P = 0.633 | |
| Transferrin | Hannum | − 0.12 (− 0.29, 0.05) | 1.79E−01 | − 0.15 (− 0.34, 0.05) | 1.82E−01 | I2 = 29.5%; Cochrane Q = 14; P = 0.164 | No significant outliers | Intercept = -0.012; P = 0.530 | |
| Transferrin | IE | − 0.03 (− 0.19, 0.13) | 6.71E−01 | − 0.17 (− 0.34, 0.02) | 9.29E−02 | I2 = 0.7%; Cochrane Q = 9; P = 0.432 | No significant outliers | Intercept = 0.011; P = 0.547 | |
| Transferrin | Pheno | − 0.17 (− 0.36, 0.03) | 9.24E−02 | − 0.19 (− 0.44, 0.07) | 1.83E−01 | I2 = 27.4%; Cochrane Q = 14; P = 0.184 | No significant outliers | Intercept = -0.005; P = 0.830 | |
aHeterogeneity in the random effect IVW methods was reported
bMR-PRESSO (NbDistribution = 10,000, P < 0.05)
cMR-Egger was used to detect Pleiotropy. There is no pleiotropy was observed among all analyses (P > 0.05)
CI, confidence interval, MR-PRESSO, Mendelian Randomization Pleiotropy RESidual Sum and Outlier, IE Intrinsic epigenetic
Table 2.
Sensitivity analyses of serum iron biomarkers in 2021 datasets with epigenetic aging accelerations
| Exposure | Outcome | Weighted median | MR-egger regression | Heterogeneitya | MR-PRESSO outlier detectb | Pleiotropyc | |||
|---|---|---|---|---|---|---|---|---|---|
| Beta (95% CI) | P Value | Beta (95% CI) | P value | Beta (95% CI) | P Value | ||||
| Ferritin | Grim | − 0.17 (− 0.57, 0.22) | 3.86E−01 | 0.33 (− 0.26, 0.92) | 2.77E−01 | I2 = 30.9%; Cochrane Q = 101; P = 0.008 | No significant outliers | Intercept = -0.010; P = 0.374 | |
| Ferritin | Hannum | − 0.08 (− 0.45, 0.30) | 6.85E−01 | 0.35 (− 0.17, 0.87) | 1.95E−01 | I2 = 17%; Cochrane Q = 84; P = 0.116 | No significant outliers | Intercept = -0.012; P = 0.217 | |
| Ferritin | IE | 0.07 (− 0.35, 0.49) | 7.53E−01 | 0.16 (− 0.43, 0.76) | 5.91E−01 | I2 = 30.7%; Cochrane Q = 101; P = 0.009 | -0.02 (-0.31, 0.27) | 8.88E−01 | Intercept = -0.003; P = 0.821 |
| Ferritin | Pheno | 0.24 (− 0.27, 0.75) | 3.58E−01 | 0.70 (0.02, 1.39) | 4.90E−02 | I2 = 17%; Cochrane Q = 84; P = 0.117 | No significant outliers | Intercept = -0.011; P = 0.418 | |
| Iron | Grim | 0.53 (0.18, 0.87) | 2.82E−03 | 0.45 (0.06, 0.84) | 3.01E−02 | I2 = 0%; Cochrane Q = 28; P = 0.743 | No significant outliers | Intercept = -0.011; P = 0.374 | |
| Iron | Hannum | 0.47 (0.12, 0.81) | 7.57E−03 | 0.55 (0.18, 0.93) | 6.71E−03 | I2 = 0%; Cochrane Q = 28; P = 0.75 | No significant outliers | Intercept = -0.019; P = 0.125 | |
| Iron | IE | 0.67 (0.29, 1.04) | 5.01E−04 | 0.75 (0.30, 1.21) | 2.79E−03 | I2 = 29.5%; Cochrane Q = 47; P = 0.056 | No significant outliers | Intercept = -0.021; P = 0.159 | |
| Iron | Pheno | 0.24 (− 0.21, 0.68) | 2.93E−01 | 0.45 (− 0.16, 1.07) | 1.60E−01 | I2 = 35.8%; Cochrane Q = 53; P = 0.02 | 0.36 (0.03, 0.69) | 3.96E−02 | Intercept = -0.012; P = 0.534 |
| Iron binding capacity | Grim | − 0.09 (− 0.32, 0.15) | 4.71E−01 | 0.05 (− 0.25, 0.34) | 7.66E−01 | I2 = 27.8%; Cochrane Q = 78; P = 0.03 | No significant outliers | Intercept = -0.007; P = 0.553 | |
| Iron binding capacity | Hannum | 0.01 (− 0.20, 0.22) | 9.18E−01 | − 0.11 (− 0.37, 0.14) | 3.94E−01 | I2 = 6.6%; Cochrane Q = 60; P = 0.333 | No significant outliers | Intercept = 0.001; P = 0.951 | |
| Iron binding capacity | IE | − 0.02 (− 0.28, 0.23) | 8.58E−01 | − 0.15 (− 0.49, 0.18) | 3.72E−01 | I2 = 42.3%; Cochrane Q = 97; P = 0.001 | No significant outliers | Intercept = 0.009; P = 0.501 | |
| Iron binding capacity | Pheno | − 0.17 (− 0.44, 0.10) | 2.19E−01 | − 0.06 (− 0.43, 0.30) | 7.29E−01 | I2 = 22.5%; Cochrane Q = 72; P = 0.071 | No significant outliers | Intercept = 0.005; P = 0.728 | |
| Transferrin saturation | Grim | 0.24 (− 0.01, 0.49) | 6.08E−02 | 0.18 (− 0.13, 0.49) | 2.65E−01 | I2 = 0%; Cochrane Q = 38; P = 0.547 | No significant outliers | Intercept = -0.001; P = 0.909 | |
| Transferrin saturation | Hannum | 0.26 (0.01, 0.52) | 4.38E−02 | 0.29 (− 0.02, 0.59) | 7.63E−02 | I2 = 0%; Cochrane Q = 37; P = 0.626 | No significant outliers | Intercept = -0.004; P = 0.700 | |
| Transferrin saturation | IE | 0.22 (− 0.04, 0.47) | 9.78E−02 | 0.35 (− 0.03, 0.73) | 7.80E−02 | I2 = 29.4%; Cochrane Q = 57; P = 0.042 | No significant outliers | Intercept = -0.009; P = 0.490 | |
| Transferrin saturation | Pheno | 0.38 (0.03, 0.73) | 3.13E−02 | 0.64 (0.11, 1.17) | 2.31E−02 | I2 = 45.1%; Cochrane Q = 73; P = 0.001 | 0.38 (0.11, 0.65) | 9.17E−03 | Intercept = -0.025; P = 0.183 |
aHeterogeneity in the random effect IVW methods was reported
bMR-PRESSO (NbDistribution = 10,000, P < 0.05)
cMR-Egger was used to detect pleiotropy. There is no pleiotropy was observed among all analyses (P > 0.05)
CI, confidence interval, MR-PRESSO, Mendelian Randomization Pleiotropy RESidual Sum and Outlier, IE Intrinsic epigenetic
Causal effects of organic iron content on EAAs
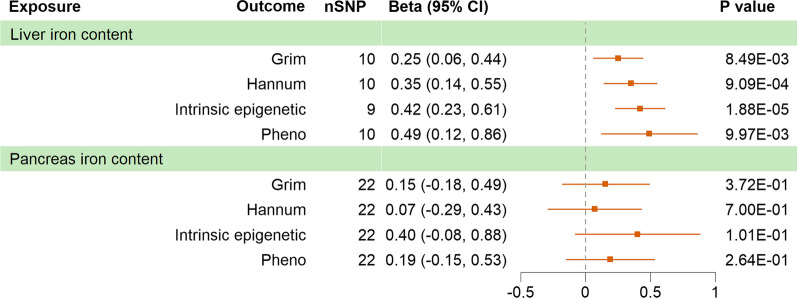
Genetically predicted liver iron content was associated with increased GrimAA (BetaIVW = 0.25 years per SD increase in liver iron content, P = 8.49E−03), HannumAA (BetaIVW = 0.35, P = 9.09E−04), IEAA (BetaIVW = 0.42, P = 1.88E−05) and PhenoAA (BetaIVW = 0.49, P = 9.97E−03), with no severe heterogeneity detected and no pleiotropy observed. One outliner (rs1799945) was observed by Mendelian Randomization Pleiotropy RESidual Sum and Outlier (MR-PRESSO), removing of which did not influence the result. However, negative results were reported in genetic predisposition to pancreas iron content, despite all analyses returned the same positive direction, as illustrated in Fig. 2 and Table 3. In summary, genetically predicted concentration of iron in liver was associated with acceleration of four epigenetic clocks.
Fig. 2.
MR analyses of organic iron content to epigenetic aging accelerations. MR, Mendelian randomization; SNP, single nucleotide polymorphism
Table 3.
Sensitivity analyses of organic iron content with epigenetic aging accelerations
| Exposure | Outcome | Weighted median | MR-egger regression | Heterogeneitya | MR-PRESSO outlier detectb | Pleiotropyc | |||
|---|---|---|---|---|---|---|---|---|---|
| Beta (95% CI) | P value | Beta (95% CI) | P value | Beta (95% CI) | P Value | ||||
| Liver iron content | Grim | 0.28 (0.05, 0.50) | 1.63E−02 | 0.27 (− 0.03, 0.56) | 1.12E−01 | I2 = 0%; Cochrane Q = 8; P = 0.538 | No significant outliers | Intercept = -0.003; P = 0.892 | |
| Liver iron content | Hannum | 0.35 (0.13, 0.57) | 2.01E−03 | 0.42 (0.08, 0.76) | 3.97E−02 | I2 = 21.1%; Cochrane Q = 11; P = 0.249 | No significant outliers | Intercept = -0.014; P = 0.598 | |
| Liver iron content | IE | 0.39 (0.16, 0.62) | 8.75E−04 | 0.32 (0.02, 0.62) | 7.22E−02 | I2 = 0%; Cochrane Q = 7; P = 0.520 | No significant outliers | Intercept = 0.019; P = 0.421 | |
| Liver iron content | Pheno | 0.39 (0.09, 0.69) | 1.19E−02 | 0.21 (− 0.36, 0.77) | 4.95E−01 | I2 = 58.5%; Cochrane Q = 22; P = 0.010 | 0.43 (0.12, 0.74) | 2.54E−02 | Intercept = 0.052; P = 0.241 |
| Pancreas iron content | Grim | 0.11 (− 0.35, 0.57) | 6.35E−01 | 0.57 (− 0.29, 1.43) | 2.10E−01 | I2 = 4.2%; Cochrane Q = 22; P = 0.404 | No significant outliers | Intercept = -0.029; P = 0.316 | |
| Pancreas iron content | Hannum | 0.08 (− 0.42, 0.58) | 7.46E−01 | 0.50 (− 0.41, 1.41) | 2.96E−01 | I2 = 9.9%; Cochrane Q = 23; P = 0.328 | No significant outliers | Intercept = -0.030; P = 0.328 | |
| Pancreas iron content | IE | 0.35 (− 0.30, 1.00) | 2.87E−01 | 0.45 (− 0.84, 1.75) | 5.00E−01 | I2 = 16.4%; Cochrane Q = 25; P = 0.242 | No significant outliers | Intercept = -0.004; P = 0.931 | |
| Pancreas iron content | Pheno | 0.18 (− 0.29, 0.66) | 4.48E−01 | 0.35 (− 0.51, 1.21) | 4.35E−01 | I2 = 0%; Cochrane Q = 17; P = 0.715 | No significant outliers | Intercept = -0.011; P = 0.701 | |
aHeterogeneity in the random effect IVW methods was reported
bMR-PRESSO (NbDistribution = 10,000, P < 0.05)
cMR-Egger was used to detect pleiotropy. There is no pleiotropy observed among all analyses (P > 0.05)
CI, confidence interval, MR-PRESSO, Mendelian Randomization Pleiotropy RESidual Sum and Outlier, IE Intrinsic epigenetic
Causal effects of EAAs on iron-related traits
To analyze the possible reverse causality, IVW method followed by sensitivity analyses were also performed with EAAs as exposure. The IVW results of EAAs on 2014 datasets, 2021 datasets and organic iron content are pictured in Additional file 1: Figures S2, S3 and S4, with sensitivity analyses on Additional file 1: Tables S1, S2 and S3, respectively. Briefly, none of the genetically predicted EAAs was associated with any of the seven iron-related traits. The sensitivity analyses did not reveal significantly association either. No pleiotropy was observed in every MR-PRESSO test, suggesting reliable results. Collectively, lack of enough evidence to support the causality of EAA on iron-related traits.
Discussion
In present research, we explored the causal relationship between iron-related biomarkers and epigenetic clocks. In the iron-related traits, genetically predicted serum iron was associated with increasing GrimAA, HannumAA and IEAA. Transferrin carries and transports most of the serum iron to organs and tissues by combining with transferrin receptor of cytomembrane. Serum transferrin was negatively associated with EAAs, despite lack of validation in another GWAS. TIBC is used to describe the maximal capacity of iron transportation in circulation [18]. Transferrin saturation refers to the proportion of transferrin binding with iron, which is normally around 20–40%, derived as serum iron divided by TIBC [3, 19]. Transferrin saturation was associated with all increased EAAs in present study, while TIBC was only significantly associated with IEAA and PhenoAA. Ferritin is responsible for the storage of iron and holds the largest amount of non-functional iron [5]. Serum ferritin from both datasets was associated with PhenoAA, which was supported by results from previous observational study [6]. Liver is rich in iron and plays important role in iron homeostasis. Ferroportin (FPN) is the only cellular exporter of iron, which is regulated by hepcidin synthetized and secreted by liver [20]. Liver is also responsible for recycling iron from aged erythrocytes with Kupffer cells, which export iron to transferrin [3]. In our results, increased liver iron content is associated with all four types of EAAs, which was in agreement with previous results of ferritin levels and ferritin iron saturation in the liver of rats increased with age [21]. However, pancreas iron content displayed no association with EAAs. Although it has been proposed that inflammaging, the aging-driven systemic inflammation, induced the increase of ferritin and was responsible for the iron deficiency and many other diseases [5]. The results did not support the causal relationship of EAA on neither plasmatic nor organic iron content, suggesting other mechanisms involved. Taken together, higher iron concentration in transport and storage were both associated with increased epigenetic age, while lack of concrete evidence to support the causal inference conversely.
To utilize iron in physiological process while avoid its toxicity to the organism, the concentration of iron is strictly modulated in different types of cells, which is typically named as iron homeostasis [3]. Although iron deficiency occurred commonly in the elderly, accumulation of iron in certain organs and tissues was observed, including brain, liver, spleen, kidney, and skeletal muscles, indicating malfunction of iron homeostasis [5, 22]. Iron deposition in senescent cells was observed, which was found associated with increased risk of age-related diseases, including malignancies, neurological disorders and cardiovascular diseases [23].
Several mechanisms have been found to be involved in iron accumulation in senescent cells and its cytotoxic effect. In cells, labile iron pool (LIP) functions as transfer station of imported free iron to stored, and utilized iron. The level of ferritin in senescent cells was elevated, as a result of impaired ferritinophagy, the process of ferritin degradation in lysosome, causing iron sequestration [24]. On the other hand, increased LIP induces the production of reactive oxygen species (ROS), which leads to oxidative stress, lipid peroxidation, and DNA damage, and finally causes damage to cell and promotes cell death [25]. Lipofuscin is also induced by labile iron, the accumulation of which could jeopardize lysosome and promote apoptosis [26]. Ferroptosis, promoted by increased free iron, is involved in malignancies and neurodegeneration [27]. In mitochondria, iron is used to produce heme or iron–sulfur clusters [28]. Overload of iron in mitochondria is related to oxidative stress and malfunction of mitochondria [23].
At the individual level, longevity of organisms could be extended by regulating iron absorption and metabolism. Inhibition of iron absorption has been observed to prolong lifespan in Drosophila and C. elegans [29, 30]. Reduction of mitochondrial iron in C. elegans could also extend the lifespan [31]. C. elegans, Drosophila, and mice fed with iron-chelating agents demonstrated increased the average lifespan [32]. Deregulation of iron-related genes, including inositol phosphosphingolipid phospholipase C (ISC1), MET18 in yeast, the homologue of methyl-methanesulfonate sensitivity protein 19 (MMS19) in human, frataxin (FXN), have been reported to be associated with shorten lifespan of in vivo models [33–35]. The mammalian target of rapamycin (mTOR) is an important regulator of cell growth and proliferation. Iron activates mTOR, which could be reversed by iron chelators. The activation of mTOR results in iron accumulation via hepcidin, which could be reversed by rapamycin, which was reported to extend the lifespan of mice [36, 37].
In previous study, DNA methylation has been found in connection with iron metabolism. For example, in vitro experiment showed DNA methylation of iron sensing genes modulated the expression of HAMP, who encodes hepcidin [38]. Tibetans with iron overload had a higher ratio of methylation in cytosine-guanine dinucleotide (CpG) compared with normal controls [39]. Iron deficiency was associated with altered DNA methylation in hippocampus of neonate [40]. Serum ferritin of maternal early pregnancy was inversely associated with three CpGs in cord blood [41]. Hemochromatosis (HFE) mutation, which caused hereditary hemochromatosis featured as iron deposition in cells, resulted in attenuated DNA methyltransferase activity and decreased brain methylation in mice [42].
This study is the first MR analysis on iron homeostasis and aging. Epigenetic clock and telomere length have been considered as the most plausible candidates of biological age predictors [12]. In MR study, leukocyte telomere length was commonly used as traits for aging, which could also be influenced by environmental factors [43]. Although telomere length has been widely validated in different kinds of health outcomes, its predictive power was low [12]. Currently, no MR analysis focusing on iron homeostasis and telomere length has been reported. EAA provided more abundant information of impact by both genetic and environmental factors, making it an excellent trait for aging and aging-related outcomes. To make sure external validity of the results, two independent GWAS for serum iron biomarkers were employed, as well as one GWAS for organic iron content. Moreover, only GWAS based on individuals of European ancestry were applied in present study to avoid potential bias due to population stratification.
This study also has several limitations. First, not all four EAAs demonstrated unanimous results in some MR analyses. This is probably due to heterogeneity of EAAs, as they were trained based on different tissues and clinical outcomes. To make results concrete, sensitivity analyses were performed and conclusion would be made only when all analyses exhibited the same direction and no pleiotropy was reported. Second, lack of evidence to support iron content in liver or pancreas to represent iron content of whole body. However, no other GWAS represented body iron content was available. Positive result of liver iron content partially reflected the storage of body, as a compensation to serum ferritin. Third, lack of enough genome-wide significant (P < 5E−08) SNPs when applying pancreas iron content as exposure, which might affect the strength of IVs. No F-statistic of pancreas iron content IVs was less than 20; thus, bias due to weak instrument was unlikely.
Present study demonstrated the potential influence of impaired iron homeostasis, particularly iron overload status, on DNA methylation alteration of human, which represented biological aging and higher mortality risk.
Conclusions
In a nutshell, our results demonstrated the potential causal relationship of iron overload to accelerate epigenetic clocks. Further researches are required to elucidate the mechanisms and additionally intervene health outcomes via iron homeostasis.
Methods
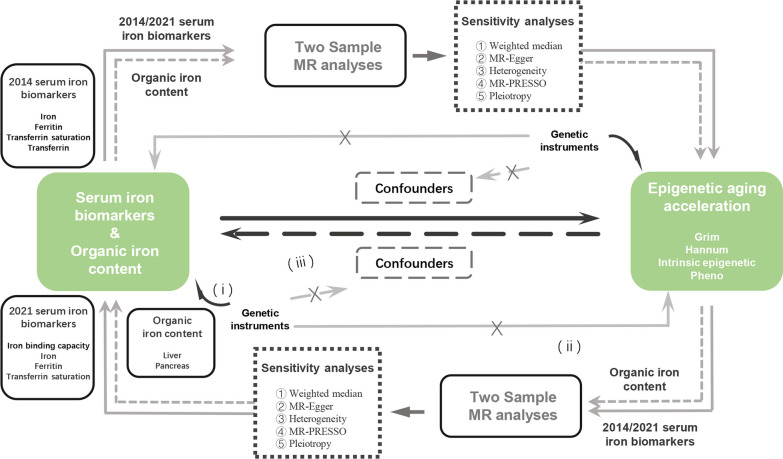
Overview of present study is shown in Fig. 3. The causal relationship of iron homeostasis to EAAs and the reverse causality was explored, with totally eighty of two-sample MR analyses were performed. Sensitivity analyses of each analysis were also carried out.
Fig. 3.
Workflow of present study and basic assumptions of MR analysis. Genetic instruments must be (i) associated with the exposure; (ii) independent of confounders; (iii) independent of the outcome. MR, Mendelian randomization, MR-PRESSO, Mendelian Randomization Pleiotropy RESidual Sum and Outlier
GWAS for iron-related traits
A total of five serum iron biomarkers and two organic iron contents were selected to represent the status of iron metabolism. Benyamin et al. reported a meta-GWAS (n = 48,972) for iron homeostasis with four traits, serum level of iron, level of ferritin, level of transferrin and its saturation based on 19 cohorts [44]. Another similar meta-GWAS was constructed by Bell et al., in which individuals of three studies in Ice land, UK and Denmark were enrolled and serum iron (n = 163,551), ferritin (n = 246,139), transferrin saturation (n = 131,471) and TIBC (n = 135,430) were introduced [19]. Liu et al. reported GWAS for iron content in liver (n = 11,069) and pancreas (n = 5,525) by using high-throughput MRI combining deep learning with imaging and clinical data from UK biobank [45].
GWAS for epigenetic clocks
To measure biological aging, GWAS (n = 34,710) of four types of EAAs reported by McCartney et al. were exploited, which integrated data from 29 studies [16]. To predict chronological age based on the status of DNA methylation, Hannum et al. and Horvath et al. established epigenetic clocks, respectively, now both known as the first generation of epigenetic clocks. The former was built using CpG markers of whole blood cells, which were processed with the Illumina Infinium HumanMethylation450 BeadChip, and the latter applying 51 types of human tissues and cell types analyzed by Illumina Infinium HumanMethylation27 or 450 BeadChip [46, 47]. IEAA, derived from the Horvath clock, was developed to alleviate the influence of varied blood components [16]. To better link epigenetic alterations with age-related outcomes, PhenoAge and GrimAge were developed, which were usually called the second generation of epigenetic clocks. PhenoAge was trained and validated with morbidity and mortality risk (phenotypic age), with DNA methylation sequencing of whole blood from published databases, which exceeded the first generation in predicting mortality [48]. GrimAge was trained on mortality, using DNA methylation data of blood samples from the Framingham heart study (FHS) Offspring Cohort, outperforming the other three in predicting years of time to death [49]. All GWAS included in present study are listed in Table 4.
Table 4.
Description of GWAS statistic included in present study
| Authors | PMID | Traits | Measurement | Data adjustment | Sample size | Sample source | Population | Sex |
|---|---|---|---|---|---|---|---|---|
| Benyamin et al. (the 2014 datasets) | 25352340 | Serum ferritin | Mean, SD | Age, principal component scores and other study specific covariates, for each sex separately, adjustments for each cohort were available in the supplementary of original article | 23,986 (Discovery) + 24,986 (Replication) | Genetics of Iron Status Consortium, the Discovery cohorts were composed of 11 cohorts in 9 participating centers, and 8 additional cohorts for the Replication cohorts | European ancestry | Males and Females |
| 25352340 | Serum iron | |||||||
| 25352340 | Serum transferrin | |||||||
| 25352340 | Transferrin Saturation | |||||||
| Bell et al. (the 2021 datasets) | 33536631 | Serum ferritin | Rank-based inverse normal transformed SD | Adjusted for age using a generalized additive model. Additionally, for UK cohort the biomarkers were adjusted for menopausal status, ABO blood group, BMI, smoking levels, alcohol levels and iron supplementation status | 246,139 | Ice land (deCODE genetics), UK (INTERVAL study) and Denmark (Danish Blood Donor Study) | European ancestry | Males and Females |
| 33536631 | Serum iron | 163,551 | ||||||
| 33536631 | Iron binding capacity | 135,430 | ||||||
| 33536631 | Transferrin saturation | 131,471 | ||||||
| Liu et al. | 34128465 | Liver iron content | Mean, SD | Age at imaging visit, age squared, sex, imaging center, scan date, scan time, and genotyping batch | 11,069 | UK Biobank | European ancestry | Males and females |
| 34128465 | Pancreas iron content | Mean, SD | 5,525 | |||||
| McCartney et al. | 34187551 | GrimAA | Mean, SD | Adjustments for each cohort were available in the supplementary of original article | 34,710 | 29 studies from UK (n = 9), USA (n = 8), Netherlands (n = 3), Finland (n = 2), and Australia (n = 1), Denmark (n = 1), Estonia (n = 1), Germany (n = 1), Italy (n = 1), Sweden (n = 1) and Switzerland (n = 1) | European ancestry | Males and females |
| 34187551 | HannumAA | 34,710 | ||||||
| 34187551 | Horvath (IEAA) | 34,710 | ||||||
| 34187551 | PhenoAA | 34,710 |
SD, standard deviation; GrimAA, GrimAge acceleration; HannumAA, HannumAge acceleration; IEAA, intrinsic epigenetic age acceleration; PhenoAA; PhenoAge acceleration
Genetic instruments selection criteria
SNPs associated with exposure at a genome-wide significant (P < 5E−08) level were applied in identifying IVs. Restriction was loosened to a threshold of 5E−06 if IVs were less than three in IVW analysis. SNPs in linkage disequilibrium (LD, r2 > 0.01, clump window < 10,000 kb) were discriminated and abandoned to keep used SNPs independent based on 1000 Genomes LD reference panel in European ancestry. SNPs with potential weak instrument bias (F-statistic < 10) were removed. SNPs significantly associated with outcome (P < 5E−08) were excluded to avoid violation of MR principles. Elimination of palindromic SNPs was performed using R package “TwoSampleMR” [50]. All the IVs of every MR analysis are listed in Additional file 2: Tables S4–S83.
MR analyses
To verify the true causality between iron homeostasis and aging, IVW method was applied in this research, as it provided stable causal inference regardless of heterogeneity [51]. Weighted median, MR-Egger regression, heterogeneity test, Cochrane’s Q test and MR-PRESSO were utilized to assess the robustness of IVW. Weighted median model is able to generate consistent estimates, in which more than half of the analytical weights are derived from valid IVs [51]. MR-Egger regression allows pleiotropy in more than half of IVs, while the statistical power is influenced [52]. MR-PRESSO corrects bias due to horizontal pleiotropic outliers [53]. To estimate heterogeneity among SNPs for exposures, Cochrane’s Q test was performed. The conclusion of causal inference was drawn if the same direction results of IVW and all sensitivity analyses were presented, besides no horizontal pleiotropic effect in the intercept test of MR-Egger regression. Finally, to evaluate the strength of IVs for exposures, total F-statistics were calculated [54].
Statistical analyses
All MR analyses were performed in R 4.2.2 (https://www.R-project.org/). R package “TwoSampleMR” (https://github.com/MRCIEU/TwoSampleMR) [50] “MRPRESSO” (https://github.com/rondolab/MR-PRESSO) [53] were used. A two-sided significance level was set as P value < 0.05 for all statistical testing.
Supplementary Information
Additional file 1: Figure S1. Procedure of IVs selection. Figure S2. MR analyses of epigenetic aging accelerations with 2014 datasets serum iron biomarkers. Figure S3. MR analyses of epigenetic aging accelerations with 2021 datasets serum iron biomarkers. Figure S4. MR analyses of epigenetic aging accelerations with organic iron content. Table S1. Sensitivity analyses of epigenetic aging accelerations with serum iron biomarkers in 2014 datasets. Table S2. Sensitivity analyses of epigenetic aging accelerations with serum iron biomarkers in 2021 datasets. Table S3. Sensitivity analyses of epigenetic aging accelerations with organic iron content.
Additional file 2: Tables S4–S83. Instrumental SNPs for serum iron biomarkers, organic iron content and epigenetic aging accelerations in the univariable Mendelian randomization analysis.
Acknowledgements
Our sincere thanks to Benyamin et al. [44], Bell et al. [19], Liu et al. [45] and McCartney et al. [16] for publicly publishing their summary statistics and developers of the R software and all the related algorithms and websites. Participants included in this study are also appreciated.
Author contributions
ZHW and YL contributed to preliminary literature search and study design. Data retrieve was conducted by ZHW, MRZ, YFX and TFL. Statistical analysis was carried out by YL, SXZ and YBY. ZHW, SLC and WHL contributed to interpretation of results and original draft of manuscript. YL illustrated figures. YHL, WCY and YY contributed to the writing revise of manuscript. All authors have read and suggested revisions of manuscript, and approved submission of the final version.
Funding
This study was supported by the Sichuan Science and Technology Program (2023YFQ0002 to YH L) in study design, the Sichuan Science and Technology Program (2023YFG0127 to Y Y) in data collection and results interpretation, and the Sichuan Provincial Foundation of Science and Technology (2023NSFSC1867 to SX Z) in results interpretation.
Availability of data and materials
No original data were generated in present study. The datasets supporting the conclusions of this article are available in the Edinburgh DataShare (https://datashare.ed.ac.uk/handle/10283/3645) for GWAS of EAAs [16], GWAS Catalog (https://www.ebi.ac.uk/gwas/publications/25352340) for 2014 datasets [44], deCODE genetics (https://www.decode.com/summarydata/) for 2021 datasets [19], NHGRI-EBI GWAS Catalog for GWAS for organic iron content [45] (accession numbers GCST90016666-GCST90016676, http://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/GCST90016001-GCST90017000/GCST90016676/).
Declarations
Ethics approval and consent to participate
All principles in the Declaration of Helsinki were strictly followed in this study. All cited summarized GWAS statistics from published studies and publicly available database were approved by a relevant review board.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhihao Wang and Yi Liu are joint first authors.
References
- 1.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. Hallmarks of aging: an expanding universe. Cell. 2023;186(2):243–278. doi: 10.1016/j.cell.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Weber KA, Achenbach LA, Coates JD. Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat Rev Microbiol. 2006;4(10):752–764. doi: 10.1038/nrmicro1490. [DOI] [PubMed] [Google Scholar]
- 3.Coffey R, Ganz T. Iron homeostasis: an anthropocentric perspective. J Biol Chem. 2017;292(31):12727–12734. doi: 10.1074/jbc.R117.781823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guralnik JM, Eisenstaedt RS, Ferrucci L, Klein HG, Woodman RC. Prevalence of anemia in persons 65 years and older in the United States: evidence for a high rate of unexplained anemia. Blood. 2004;104(8):2263–2268. doi: 10.1182/blood-2004-05-1812. [DOI] [PubMed] [Google Scholar]
- 5.Zeidan RS, Han SM, Leeuwenburgh C, Xiao R. Iron homeostasis and organismal aging. Ageing Res Rev. 2021;72:101510. doi: 10.1016/j.arr.2021.101510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casale G, Bonora C, Migliavacca A, Zurita IE, de Nicola P. Serum ferritin and ageing. Age Ageing. 1981;10(2):119–122. doi: 10.1093/ageing/10.2.119. [DOI] [PubMed] [Google Scholar]
- 7.Liu B, Sun Y, Xu G, Snetselaar LG, Ludewig G, Wallace RB, Bao W. Association between body iron status and leukocyte telomere length, a biomarker of biological aging, in a nationally representative sample of US adults. J Acad Nutr Dietetics. 2019;119(4):617–625. doi: 10.1016/j.jand.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Höhn A, Jung T, Grimm S, Grune T. Lipofuscin-bound iron is a major intracellular source of oxidants: role in senescent cells. Free Rad Biol Med. 2010;48(8):1100–1108. doi: 10.1016/j.freeradbiomed.2010.01.030. [DOI] [PubMed] [Google Scholar]
- 9.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genetics. 2018;19(6):371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 10.Kane AE, Sinclair DA. Epigenetic changes during aging and their reprogramming potential. Crit Rev Biochem Mol Biol. 2019;54(1):61–83. doi: 10.1080/10409238.2019.1570075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seale K, Horvath S, Teschendorff A, Eynon N, Voisin S. Making sense of the ageing methylome. Nat Rev Genet. 2022;23(10):585–605. doi: 10.1038/s41576-022-00477-6. [DOI] [PubMed] [Google Scholar]
- 12.Jylhävä J, Pedersen NL, Hägg S. Biological age predictors. EBioMedicine. 2017;21:29–36. doi: 10.1016/j.ebiom.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson J, Russ TC, Clarke TK, Howard DM, Hillary RF, Evans KL, Walker RM, Bermingham ML, Morris SW, Campbell A, et al. A meta-analysis of genome-wide association studies of epigenetic age acceleration. PLoS Genet. 2019;15(11):e1008104. doi: 10.1371/journal.pgen.1008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yarmolinsky J, Wade KH, Richmond RC, Langdon RJ, Bull CJ, Tilling KM, Relton CL, Lewis SJ, Davey Smith G, Martin RM. Causal inference in cancer epidemiology: what is the role of mendelian randomization? Cancer Epidemiol Biomarkers Prev. 2018;27(9):995–1010. doi: 10.1158/1055-9965.Epi-17-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labrecque J, Swanson SA. Understanding the assumptions underlying instrumental variable analyses: a brief review of falsification strategies and related tools. Curr Epidemiol Rep. 2018;5(3):214–220. doi: 10.1007/s40471-018-0152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCartney DL, Min JL, Richmond RC, Lu AT, Sobczyk MK, Davies G, Broer L, Guo X, Jeong A, Jung J, et al. Genome-wide association studies identify 137 genetic loci for DNA methylation biomarkers of aging. Genome Biol. 2021;22(1):194. doi: 10.1186/s13059-021-02398-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y, Sun X, Huang Z, Zhang R, Li C, Anderson AH, Lash JP, Kelly TN. Effects of epigenetic age acceleration on kidney function: a Mendelian randomization study. Clin Epigenetics. 2023;15(1):61. doi: 10.1186/s13148-023-01476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasvosve I, Delanghe J. Total iron binding capacity and transferrin concentration in the assessment of iron status. Clin Chem Lab Med. 2002;40(10):1014–1018. doi: 10.1515/cclm.2002.176. [DOI] [PubMed] [Google Scholar]
- 19.Bell S, Rigas AS, Magnusson MK, Ferkingstad E, Allara E, Bjornsdottir G, Ramond A, Sørensen E, Halldorsson GH, Paul DS, et al. A genome-wide meta-analysis yields 46 new loci associating with biomarkers of iron homeostasis. Commun Biol. 2021;4(1):156. doi: 10.1038/s42003-020-01575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiao B, Sugianto P, Fung E, Del-Castillo-Rueda A, Moran-Jimenez MJ, Ganz T, Nemeth E. Hepcidin-induced endocytosis of ferroportin is dependent on ferroportin ubiquitination. Cell Metab. 2012;15(6):918–924. doi: 10.1016/j.cmet.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulvik BE, Berenshtein E, Konijn AM, Grinberg L, Vinokur V, Eliashar R, Chevion MM. Aging is an organ-specific process: changes in homeostasis of iron and redox proteins in the rat. Age (Dordr) 2012;34(3):693–704. doi: 10.1007/s11357-011-9268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busti F, Campostrini N, Martinelli N, Girelli D. Iron deficiency in the elderly population, revisited in the hepcidin era. Front Pharmacol. 2014;5:83. doi: 10.3389/fphar.2014.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian Y, Tian Y, Yuan Z, Zeng Y, Wang S, Fan X, Yang D, Yang M. Iron metabolism in aging and age-related diseases. Int J Mol Sci. 2022 doi: 10.3390/ijms23073612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masaldan S, Clatworthy SAS, Gamell C, Meggyesy PM, Rigopoulos AT, Haupt S, Haupt Y, Denoyer D, Adlard PA, Bush AI, et al. Iron accumulation in senescent cells is coupled with impaired ferritinophagy and inhibition of ferroptosis. Redox Biol. 2018;14:100–115. doi: 10.1016/j.redox.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dixon SJ, Stockwell BR. The role of iron and reactive oxygen species in cell death. Nat Chem Biol. 2014;10(1):9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 26.Brunk UT, Terman A. Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Rad Biol Med. 2002;33(5):611–619. doi: 10.1016/s0891-5849(02)00959-0. [DOI] [PubMed] [Google Scholar]
- 27.Tang D, Chen X, Kang R, Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31(2):107–125. doi: 10.1038/s41422-020-00441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braymer JJ, Lill R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J Biol Chem. 2017;292(31):12754–12763. doi: 10.1074/jbc.R117.787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massie HR, Aiello VR, Williams TR. Inhibition of iron absorption prolongs the life span of Drosophila. Mech Ageing Dev. 1993;67(3):227–237. doi: 10.1016/0047-6374(93)90001-8. [DOI] [PubMed] [Google Scholar]
- 30.Schiavi A, Maglioni S, Palikaras K, Shaik A, Strappazzon F, Brinkmann V, Torgovnick A, Castelein N, De Henau S, Braeckman BP, et al. Iron-starvation-induced mitophagy mediates lifespan extension upon mitochondrial stress in C. elegans. Curr Biol CB. 2015;25(14):1810–1822. doi: 10.1016/j.cub.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 31.Ren Y, Yang S, Tan G, Ye W, Liu D, Qian X, Ding Z, Zhong Y, Zhang J, Jiang D, et al. Reduction of mitoferrin results in abnormal development and extended lifespan in Caenorhabditis elegans. PLoS ONE. 2012;7(1):e29666. doi: 10.1371/journal.pone.0029666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen LR, Parnell LD, Ordovas JM, Lai CQ. Curcumin and aging. BioFactors (Oxford, England) 2013;39(1):133–140. doi: 10.1002/biof.1086. [DOI] [PubMed] [Google Scholar]
- 33.Almeida T, Marques M, Mojzita D, Amorim MA, Silva RD, Almeida B, Rodrigues P, Ludovico P, Hohmann S, Moradas-Ferreira P, et al. Isc1p plays a key role in hydrogen peroxide resistance and chronological lifespan through modulation of iron levels and apoptosis. Mol Biol Cell. 2008;19(3):865–876. doi: 10.1091/mbc.e07-06-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen YQ, Liu XG, Zhao W, Cui H, Ruan J, Yuan Y, Tu Z. MET18 deficiency increases the sensitivity of yeast to oxidative stress and shortens replicative lifespan by inhibiting catalase activity. BioMed Res Int. 2017;2017:7587395. doi: 10.1155/2017/7587395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Britti E, Delaspre F, Feldman A, Osborne M, Greif H, Tamarit J, Ros J. Frataxin-deficient neurons and mice models of Friedreich ataxia are improved by TAT-MTScs-FXN treatment. J Cell Mol Med. 2018;22(2):834–848. doi: 10.1111/jcmm.13365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangan D. Iron: an underrated factor in aging. Aging. 2021;13(19):23407–23415. doi: 10.18632/aging.203612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharp PA, Clarkson R, Hussain A, Weeks RJ, Morison IM. DNA methylation of hepatic iron sensing genes and the regulation of hepcidin expression. PLoS ONE. 2018;13(5):e0197863. doi: 10.1371/journal.pone.0197863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Q, Ge Z, Fu S, Wan S, Shi J, Wu Y, Zhang Y. DNA methylation plays an important role in iron-overloaded Tibetans. Genes Genetic Syst. 2022;97(2):55–66. doi: 10.1266/ggs.21-00006. [DOI] [PubMed] [Google Scholar]
- 40.Schachtschneider KM, Liu Y, Rund LA, Madsen O, Johnson RW, Groenen MA, Schook LB. Impact of neonatal iron deficiency on hippocampal DNA methylation and gene transcription in a porcine biomedical model of cognitive development. BMC Genomics. 2016;17(1):856. doi: 10.1186/s12864-016-3216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taeubert MJ, de Prado-Bert P, Geurtsen ML, Mancano G, Vermeulen MJ, Reiss IKM, Caramaschi D, Sunyer J, Sharp GC, Julvez J, et al. Maternal iron status in early pregnancy and DNA methylation in offspring: an epigenome-wide meta-analysis. Clin Epigenetics. 2022;14(1):59. doi: 10.1186/s13148-022-01276-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye Q, Trivedi M, Zhang Y, Böhlke M, Alsulimani H, Chang J, Maher T, Deth R, Kim J. Brain iron loading impairs DNA methylation and alters GABAergic function in mice. FASEB J. 2019;33(2):2460–2471. doi: 10.1096/fj.201801116RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang D, Lin S, He J, Wang Q, Zhan Y. Association between COVID-19 and telomere length: a bidirectional Mendelian randomization study. J Med Virol. 2022;94(11):5345–5353. doi: 10.1002/jmv.28008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benyamin B, Esko T, Ried JS, Radhakrishnan A, Vermeulen SH, Traglia M, Gögele M, Anderson D, Broer L, Podmore C, et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat Commun. 2014;5:4926. doi: 10.1038/ncomms5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Basty N, Whitcher B, Bell JD, Sorokin EP, van Bruggen N, Thomas EL, Cule M. Genetic architecture of 11 organ traits derived from abdominal MRI using deep learning. Elife. 2021 doi: 10.7554/eLife.65554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10(4):573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. 2019;11(2):303–327. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genetic Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanderson E, Spiller W, Bowden J. Testing and correcting for weak and pleiotropic instruments in two-sample multivariable Mendelian randomization. Stat Med. 2021;40(25):5434–5452. doi: 10.1002/sim.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Procedure of IVs selection. Figure S2. MR analyses of epigenetic aging accelerations with 2014 datasets serum iron biomarkers. Figure S3. MR analyses of epigenetic aging accelerations with 2021 datasets serum iron biomarkers. Figure S4. MR analyses of epigenetic aging accelerations with organic iron content. Table S1. Sensitivity analyses of epigenetic aging accelerations with serum iron biomarkers in 2014 datasets. Table S2. Sensitivity analyses of epigenetic aging accelerations with serum iron biomarkers in 2021 datasets. Table S3. Sensitivity analyses of epigenetic aging accelerations with organic iron content.
Additional file 2: Tables S4–S83. Instrumental SNPs for serum iron biomarkers, organic iron content and epigenetic aging accelerations in the univariable Mendelian randomization analysis.
Data Availability Statement
No original data were generated in present study. The datasets supporting the conclusions of this article are available in the Edinburgh DataShare (https://datashare.ed.ac.uk/handle/10283/3645) for GWAS of EAAs [16], GWAS Catalog (https://www.ebi.ac.uk/gwas/publications/25352340) for 2014 datasets [44], deCODE genetics (https://www.decode.com/summarydata/) for 2021 datasets [19], NHGRI-EBI GWAS Catalog for GWAS for organic iron content [45] (accession numbers GCST90016666-GCST90016676, http://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/GCST90016001-GCST90017000/GCST90016676/).