Abstract
At this time an amino acid substitution at position 276 in the TEM-1 enzyme is associated with an additional substitution at position 69 in natural β-lactamase-inhibitor-resistant (IRT) β-lactamases. The effect of the Asn276→Asp substitution on resistance was assessed with the Asn276Asp variant, generated by site-directed mutagenesis. The mutant was resistant to β-lactamase inhibitors, but the MICs of amoxicillin combined with clavulanic acid or tazobactam were strikingly different for E. coli strains producing the Asn276Asp variant and those producing naturally occurring IRTs with single or double substitutions. The inhibitory effects of clavulanic acid and tazobactam were the same in IRTs with substitutions at position 69 (IRT-5 and IRT-6). The effect of clavulanic acid on the MICs of amoxicillin for the Asn276Asp variant was greater than that of tazobactam. In IRTs with double substitutions, at positions 69 plus 276 (IRT-4, IRT-7, and IRT-8) or 69 plus 275 (IRT-14), tazobactam was a more potent inhibitor than clavulanic acid. The effect of the Asn276→Asp substitution on the values of the kinetic constants and the concentration required to inhibit by 50% the hydrolysis of benzylpenicillin confirms that this single mutation is responsible for resistance to β-lactamase inhibitors. Molecular modeling of the Asn276Asp mutant shows that Asp276 can form two salt bonds with Arg244 close to the penicillin-binding cavity. The addition of the Asp276 mutation to that preexisting at position 69 confers a higher selective advantage to bacteria, as shown by the reduction in β-lactamase inhibitor efficiencies of the double variants. Therefore, the emergence of multiple mutations in TEM β-lactamases by virtue of the use of β-lactamase inhibitors increases selection pressure resulting in the convergent evolution of resistant strains.
The β-lactam antibiotics are the most frequently prescribed antimicrobial agents in clinical practice. The enzymes of the β-lactamase family of gram-negative bacteria play a significant role in the development of resistance to β-lactam antibiotics. The frequent occurrence of β-lactamase genes in readily transmissible plasmids and their possible integration into bacterial chromosomes is of concern in the management of antimicrobial therapy in the community and in hospital centers. The evolution of extended-spectrum resistance is mediated by mutant derivatives of the TEM and SHV β-lactamases (17).
Strains resistant to inhibitors of β-lactamases and containing TEM-derived β-lactamases have been described (2, 3, 5, 7, 12, 19, 39, 42). These β-lactamase-inhibitor-resistant (IRT) TEM β-lactamases are encoded by blaIRT genes that carry mutations affecting the kinetic properties of the enzymes by altering the binding of both the β-lactams and the β-lactamase inhibitors (24). The structure-function relationships of some of these mutations have been investigated with variants generated in vitro (3, 16, 21, 29, 38). The effect of the sequence variation at position 276 of the TEM-1 β-lactamase (Asn276→Asp) has been examined by this approach (38) to assess enzyme kinetics and the resistance pattern in qualitative terms. This substitution has never occurred alone in natural variants and is always accompanied by substitution of the methionine at position 69 (Met69) in β-lactamase.
We studied the quantitative effects of the IRT variants with single and double substitutions on resistance to antibiotic-inhibitor combinations. We generated an Asn276→Asp variant (hereafter designated Asn276Asp) by site-directed mutagenesis. We compared its phenotypic characteristics with those of various naturally occurring IRTs (IRT-5, IRT-6, IRT-7, and IRT-8 and the reference IRTs IRT-4 [7] and IRT-14 [10]) and the in vitro-generated variants M69L (16) and W165R (33).
(This work was presented in part at the 16th Interdisciplinary Meeting on Anti-Infectious Chemotherapy, Paris, France, 1996 [11].)
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage.
The characteristics of the bacterial strains, plasmids, and phage used in this study are given in Table 1. The phagemid pBluescript-II KS(−) (Stratagene Cloning Systems, La Jolla, Calif.), which encodes the TEM-1 β-lactamase, was used for site-directed mutagenesis. This phagemid was propagated in Escherichia coli RZ1032 to obtain the single-stranded template DNA for site-directed mutagenesis. Mutated DNA (pMBS276D) was introduced into the bacterial host E. coli XL-1 Blue, which was then tested for antibiotic susceptibility. The E. coli strains producing IRT-5 (P30), IRT-6 (P9), IRT-7 (P11), IRT-8 (P12), IRT-4 (7), and IRT-14 (10) and the control strain expressing the TEM-1 (R111) β-lactamase were used for MIC assays.
TABLE 1.
Bacterial strains and plasmids used or studied in this work investigation
| Strain, plasmid, or phage | Description, characteristics, or genotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| RZ1032 | Host for pBluescript; HfrKL16 PO/45 [lysA(61-62)] dut-1 ung-1 thi-1 relA1 Zbd-279::Tn10, supE44 | 25 |
| XL-1 Blue | Host for pBluescript with mutated β-lactamase; recA1 endA1 gyrA96 thi hsdR17 (rk−, mk+) supE44 relA1 λ−, lac mutant [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | 8 |
| K-12 J53/R111 | Amoxicillin-resistant strain producing TEM-1 | 27 |
| P9 | Clavulanate-resistant clinical strain producing IRT-6 | 12 and this work |
| P11, P12, P30 | Clavulanate-resistant clinical strains producing IRT-5 (P30), IRT-7 (P11), and IRT-8 (P12) | Cochin Hospital, Paris, France (this work) |
| Plasmids | ||
| pBluescript II KS(−) (pBSKS) | E. coli high-copy-number cloning vector; Aprori ColE1, ori M13 phage | GenBank accession no. X52329 |
| pMBS276D | High-copy-number recombinant plasmid encoding bla gene with Asp276 variation | This work |
| Phage R408 | ||
| M13 helper phage for packaging Bluescript M13 vectors | 35 |
Media and chemicals.
The bacteria used for site-directed mutagenesis were cultured in Luria-Bertani (Gibco BRL, Life Technologies, Paisley, Scotland) and 2× YT media (36). Brain heart infusion (Difco Laboratories, Detroit, Mich.) medium was used to culture bacteria for β-lactamase production. Mueller-Hinton agar (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France) was used for the MIC assay. The β-lactams used in this study for determination of the MICs and the values of the kinetic constants were those used in previous work (10).
Plasmid purification and bacterial transformation.
Plasmid DNA was isolated from E. coli by the alkaline lysis method (36). Competent E. coli cells (XL-1 Blue and RZ1032) were prepared and transformed with plasmid DNA by the calcium chloride method (15). Attempts to transfer the clinical plasmids encoding the blaIRT-5, blaIRT-6, blaIRT-7, and blaIRT-8 genes to E. coli were unsuccessful.
Single-stranded DNA preparation and site-directed mutagenesis of the TEM-1 β-lactamase.
We used the numbering of amino acid residues in the β-lactamase sequence proposed by Ambler et al. (1). Single-stranded plasmid DNA was prepared as described previously (36). Site-directed mutagenesis was carried out as described by Kunkel et al. (25) with an oligodeoxyribonucleotide with the intended substitution (underlined): 5′-ATG AAC GAG ATA GAC AGA T-3′. E. coli XL-1 Blue colonies containing the plasmid DNA with the mutant bla gene were selected for ampicillin and tetracycline resistance (Apr and Tcr, respectively).
Identification and characterization of mutants.
Plasmids were isolated from resistant clones and were tested for the presence of the intended mutation with a diagnostic Tsp509I enzyme as described previously (13). Plasmids testing positive were further sequenced (37) to confirm the presence of the expected mutation in the plasmid DNA (pMBS276D).
MIC assays and purification and IEF of β-lactamases.
Bacterial susceptibility to β-lactams was measured by the agar dilution test as described previously (10). Independent E. coli XL-1 Blue clones carrying pMBS276D or TEM-1 were grown overnight at 37°C in a large volume (3 to 5 liters) of brain heart infusion medium with continuous shaking. The preparation of cell-free lysates, enzyme purification (7, 40), reverse-phase high-performance liquid chromatography (34), analytical isoelectric focusing (IEF) (31), and β-lactamase detection (27) were carried out by published procedures. The enzymes used as controls in IEF were TEM-1 (R111) (27), TEM-2 (RP4) (32), IRT-1 (TEM-31), and IRT-2 (TEM-30) (40).
Kinetic parameter value determinations.
The values of Km, Ki and Vmax and the concentration required to inhibit by 50% the hydrolysis of benzylpenicillin (IC50) of the Asn276Asp β-lactamase were estimated at 37°C by computerized microacidimetry at pH 7.0 (26). The values of the kinetic parameters (Vmax and Km) were derived by weighted linear regression of these data. If the Vmax and Km values were very low they were simply not reported or Ki was measured by competitive inhibition with a reporter substrate, benzylpenicillin (1,000 μM), instead (9, 28). Purified β-lactamases were used for the kcat assay. The quantitative effect of inhibitors on the activities of the wild-type or mutant β-lactamases was assessed by determining the IC50. Five concentrations of inhibitors were used: from 10 to 300 μM for TEM-1 or 100 to 1,000 μM for the mutant (clavulanic acid), from 0.5 to 100 μM for TEM-1 or for the mutant (sulbactam), from 25 to 500 μM for TEM-1 or for the mutant (tazobactam), and from 2.5 to 10 μM for TEM-1 or for the mutant (brobactam). The β-lactamase was preincubated with the inhibitor for 10 min at 37°C in saline buffer at pH 7.0, and the assay was initiated by the addition of 1 mM benzylpenicillin. Inhibition data for clavulanic acid were plotted against the inhibitor: enzyme ratio to determine the partition ratio for inactivation from the extrapolated value for 100% inactivation. One unit of β-lactamase is the amount of enzyme that hydrolyzes 1 μmol of benzylpenicillin per min at 37°C under these experimental conditions.
Molecular modeling.
The crystal structure of the PC1 β-lactamase is available from the Protein Data Bank, Brookhaven National Laboratory, Brookhaven, Conn., under the entry 3BLM (20), as the structure of TEM-1 β-lactamase (1XPB) (18) and a closely related TEM-1 β-lactamase (1BTL) (23). Molecular modeling of the Asp276 TEM mutant was performed by using the Amber force field (41).
RESULTS
Isoelectric point.
The mutant β-lactamase encoded by pMBS276D was shown to have a pl of 5.2 by analytical IEF.
Susceptibility to β-lactams of E. coli producing Asn276Asp β-lactamase.
The MICs for E. coli Asn276Asp are presented in Table 2. For E. coli XL-1 Blue carrying pMBS276D, the MICs of amoxicillin were eightfold higher and those of ticarcillin and piperacillin fourfold higher than those for the wild-type in the presence of clavulanate. Tazobactam did not reduce the MICs of these β-lactams. Intriguingly, we found a striking difference in the MICs for the TEM-1 encoded by R111 and that encoded by pBSKS in the presence of tazobactam. Both plasmids were thought to encode the wild-type TEM-1 enzyme. The MICs of mecillinam, imipenem, and cephalosporins for the wild-type TEM-1 (pBSKS) and the Asn276Asp variant were similar.
TABLE 2.
β-Lactam MICs for E. coli strains
| Antibiotica | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| K-12 J53 TEM-1 (R111)b | XL-1 Bluec | XL-1 Blue TEM-1 (pBSKS)d | XL-1 Blue Asn276Asp (pMBS276D)e | |
| Amx | ≥8,192 | 4 | ≥8,192 | ≥8,192 |
| Amx + Clav (2 μg/ml) | 64 | 4 | 256 | 2,048 |
| Amx + Tazo (2 μg/ml) | 128 | 4 | 4,096 | ≥8,192 |
| Amx + Tazo (4 μg/ml) | 64 | 4 | 2,048 | 4,096 |
| Tic | ≥8,192 | 2 | ≥8,192 | ≥8,192 |
| Tic + Clav (2 μg/ml) | 128 | 2 | 256 | 1024 |
| Tic + Tazo (2 μg/ml) | 256 | 2 | ≥8,192 | ≥8,192 |
| Pip | 128 | 0.5 | 512 | 256 |
| Pip + Clav (2 μg/ml) | 2 | 0.5 | 2 | 8 |
| Pip + Tazo (2 μg/ml) | 16 | 0.5 | 128 | 128 |
| Mecillinam | 8 | 0.125 | 64 | 32 |
| Cephalothin | 16 | 4 | 32 | 16 |
| Cefoxitin | 4 | 2 | 2 | 2 |
| Cefotaxime | 0.03 | ≤0.015 | ≤0.015 | ≤0.015 |
| Imipenem | 0.125 | 0.125 | 0.125 | |
Amx, amoxicillin; Tic, ticarcillin; Pip, piperacillin; Clav, clavulanic acid; Tazo, tazobactam.
Strain producing TEM-1 (reference strain).
Susceptible recipient strain.
Strain producing TEM-1.
Strain producing TEM-1 with Asp276 substitution.
Effect of inhibitor-amoxicillin combination on IRTs characterized by one or two amino acid substitutions.
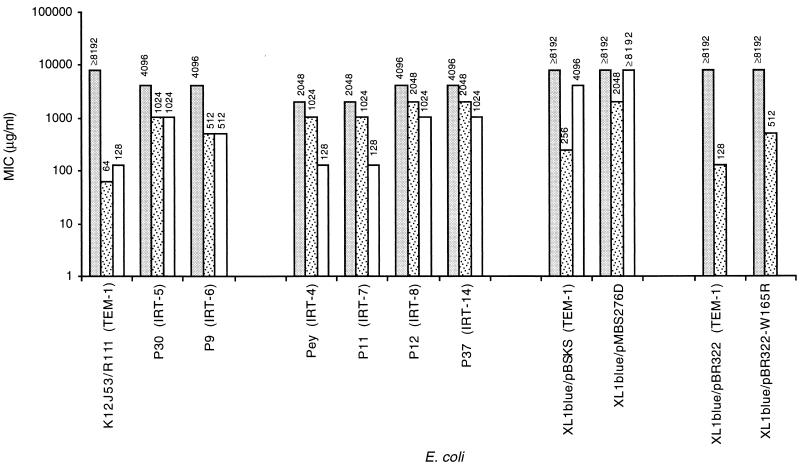
In the presence of β-lactamase inhibitors, the MICs of amoxicillin were fourfold lower for strains producing IRT-5 and eightfold lower for strains producing IRT-6. The effects of clavulanic acid and tazobactam on these single variants were the same (Fig. 1). The MICs of amoxicillin with clavulanic acid for the double variant IRT-7- and IRT-8-producing strains were reduced by half. In the presence of tazobactam, they were 16-fold lower for strains producing IRT-7 and 4-fold lower for strains producing IRT-8.
FIG. 1.
MICs for E. coli producing TEM-1 wild-type
enzymes, TEM-1 variant enzymes obtained by site-directed mutagenesis,
and natural IRT enzymes. The effects of clavulanate and tazobactam
combined with amoxicillin were determined. The following enzymes were
used: wild-type TEM-1 enzymes R111 (this study), pBSKS (this study),
and pBR322 (33); TEM-1 variants pMBS276D with Asn276Asp
(this study) and pBR322 with Trp165Arg (33); natural IRT
enzymes (this study) P30 producing IRT-5 (Met69Leu), P9 producing IRT-6
(Met69Val), P11 producing IRT-7 (Met69Val plus Asn276Asp), and P12
producing IRT-8 (Met69Ile plus Asn276Asp); and Pey producing IRT-4
(Met69Leu plus Asn276Asp) (7) and P37 producing IRT-14
(Met69Leu plus Arg275Gln) (10).
, amoxicillin;
 , amoxicillin plus clavulanate;
□, amoxicillin plus tazobactam.
, amoxicillin plus clavulanate;
□, amoxicillin plus tazobactam.
Effects of the Asn276→Asp substitution in TEM-1 β-lactamase on kinetic parameters.
The β-lactamase mutant Asn276Asp and the wild-type TEM-1 β-lactamase (Asn276) were produced in E. coli with a high-level expression vector. They were extracted from E. coli and purified. The specific activities were 5,300 U/mg for the mutant and 2,400 U/mg for TEM-1. The kinetic parameters for the Asn276Asp and TEM-1 β-lactamases were determined for 17 β-lactams and are summarized in Tables 3 and 4. The chief results were as follows described below.
TABLE 3.
Kinetic parameters for the hydrolysis of β-lactam substrates by the wild-type (pBSKS/TEM-1) and mutant (Asp276) β-lactamasesa
| Substrate | Wild-type (TEM-1)
|
Mutant
(Asn276Asp)
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1)b | Km (μM)c | kcat/Km (μM−1 · s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 · s−1) | |
| Benzylpenicillin | 1,160 | 28 | 41.2 | 1,980 | 105 | 18.9 |
| Amoxicillin | 1,080 | 32 | 33.8 | 1,620 | 276 | 5.9 |
| Ticarcillin | 139 | 13 | 10.7 | 269 | 113 | 2.4 |
| Piperacillin | 1,060 | 67 | 15.9 | 1,660 | 232 | 7.2 |
| Mezlocillin | 1,280 | 68 | 18.8 | 2,100 | 347 | 6.0 |
| Mecillinam | 822 | 1,730 | 0.48 | 436 | 793 | 0.55 |
| Imipenem | —d | NDe | ND | — | 198f | ND |
| Cephalothin | 312 | 341 | 0.9 | 273 | 389 | 0.7 |
| Cephaloridine | 1,630 | 1,020 | 1.6 | 1,830 | 397 | 4.6 |
| Ceftazidime | — | ND | ND | — | 261f | ND |
| Cefoperazone | 413 | 402 | 1.68 | 513 | 405 | 1.3 |
The kcat and Km of Asn276Asp for oxacillin, aztreonam, cefotaxime, ceftriaxone, cefoxitine, and cefuroxime could not be measured (there was no detectable hydrolysis or Vmax was <2% and Ki wash >500 μM).
The standard deviation s for the analysis were ±5%.
The standard deviation s were ±10% for Km values of <100 μM and ±20% for Km values of >100 μM.
—, no detectable hydrolysis.
ND, not determined.
Km was determined as Ki by the reporter substrate method. For more details, see Materials and Methods.
TABLE 4.
Ratio of kcat/Km of mutant enzymes with Asp276 and Leu69 substitutions and of IRT-4 and IRT-14 β-lactamases relative to that of the wild-type (pBSKS/TEM-1)a
| Substrate |
kcat/Km
ratio (%)
|
||||
|---|---|---|---|---|---|
| TEM-1 | Asn276Asp | M69Lb | IRT-4 Met69Leu, Asn276Asp)c | IRT-14 (Met69Leu, Arg275Gln)d | |
| Benzylpenicillin | 100 | 46 | 44 | 10.1 | 7.8 |
| Amoxicillin | 100 | 18 | 26 | 13.6 | 11.6 |
| Ticarcillin | 100 | 22 | 12 | 3.9 | 1.3 |
| Piperacillin | 100 | 45 | 52 | 5.5 | 6.5 |
| Mezlocillin | 100 | 32 | NDe | ND | ND |
| Mecillinam | 100 | 115 | ND | ND | ND |
| Cephalothin | 100 | 78 | 21 | 1.8 | 13 |
| Cephaloridine | 100 | 288 | 24 | 21.8 | 58.2 |
| Cefoperazone | 100 | 77 | 68 | ND | ND |
(i) Penicillins. The Asn276Asp enzyme hydrolyzed all penicillins tested except oxacillin. The kcat values of the mutant enzyme were 1.5- to 2-fold higher than those of TEM-1. The Km values of the mutant enzyme were four- to ninefold higher than those of the wild-type enzyme. Mecillinam was a poor substrate. Consequently, the kcat/Km values of the mutant enzyme for penicillins were reduced two- to sixfold.
(ii) Cephalosporins. The kcat and Km values of the enzyme were unaffected except for those for cephaloridine, for which the kcat/Km value of the mutant was three-fold higher than that of the wild type. Aztreonam, expanded-spectrum cephalosporins, and cefoxitin were not substrates for these β-lactamases. The catalytic efficiencies (kcat/Km) of the Asn276Asp β-lactamase for penicillins were less than 50% of those of the TEM-1 enzyme (Table 4). The catalytic efficiencies of the Asn276Asp β-lactamase for mecillinam and cephalosporins were comparable to or higher than those of TEM-1. The IC50s for TEM-1 and the mutant β-lactamases are given in Table 5. Clavulanic acid was 100 times more active than sulbactam against the mutant enzyme, 2.5 times more active than tazobactam, and 4 times less active than brobactam. The IC50s for the mutant were sevenfold higher with clavulanic acid, twofold higher with tazobactam, and similar to those for TEM-1 with sulbactam and brobactam. The inactivation of the mutant was progressive, with partition ratio of 950 for inactivation by clavulanic acid, compared to a ratio of 120 for the TEM-1 β-lactamase.
TABLE 5.
IC50s for hydrolysis of benzylpenicillin
| Inhibitor | IC50
(μM)a
|
|
|---|---|---|
| TEM-1 | Asn276Asp | |
| Clavulanic acid | 0.02 | 0.13 |
| Sulbactam | 10 | 13.4 |
| Tazobactam | 0.15 | 0.33 |
| Brobactam | 0.02 | 0.03 |
The standard deviation for the analysis was ±5%.
Molecular modeling.
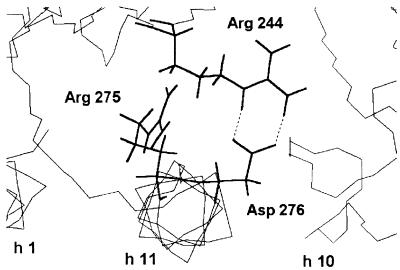
Figure 2, based on the crystal structure of TEM-1 β-lactamase (18, 23), shows the stereo view of the environment of Asp276 and interactions with Arg244 of the Asn276Asp mutant, in which two salt bonds can be formed between those amino acids.
FIG. 2.
Molecular modeling of the Asn276Asp mutant of TEM-1: stereo view of the environment of Asp276 and interactions with Arg244. Hydrogen bonds are indicated by dashed lines. The backbones of α-helices h1, h2, and h10 are shown (at the bottom) as thin lines.
DISCUSSION
Previous work has stressed the importance of the Asn276→Asp change in β-lactamase for conferring resistance to clavulanic acid (38). This change also decreases the substrate affinity and catalytic efficiency of β-lactamase. However, this substitution has never been found alone in strains with naturally occurring IRT and is always accompanied by another substitution, at position 69, which contributes on its own to the IRT phenotype. Thus, site-directed mutagenesis at position 276 was used to investigate the role of this position in inhibitor-resistant phenotypes. The previous study evaluated only the qualitative effects of antibiotic-inhibitor combinations.
In this study we investigated the effects of the Asn276→Asp substitution, generated by site-directed mutagenesis of the blaTEM-1 gene (the variant was Asn276Asp), on other functional characteristics of TEM-1 β-lactamase and, in particular, on the MICs and IC50s. We also compared the properties of this enzyme with those of naturally occurring variants to assess the contribution of the amino acid change to the resistance phenotype.
The observed differences in susceptibility to clavulanate and tazobactam of E. coli bearing pBSKS and E. coli bearing R111 (Table 1) are probably due to the higher copy number of pBSKS, as observed previously for E. coli resistant to β-lactamase inhibitors (30). However, the Asn276Asp variant was more resistant than the wild type TEM-1 to clavulanate and tazobactam (Table 2). Interestingly, this enhanced resistance to β-lactamase inhibitors has already been observed by determination of the MICs for the Asn276→Gly variant of OHIO-1 (4). This substitution did not affect the level of resistance to penicillins and cephalosporins according to the observed MICs (Table 2). Clavulanate and tazobactam had different effects on naturally occurring IRT enzymes, depending on whether the strains had a single substitution (at position 69) or double substitutions (at both position 69 and position 275 or 276) in their β-lactamases (Fig. 1). Indeed, clavulanic acid was more potent against strains producing IRT enzymes with single substitutions than against those with double substitutions. Tazobactam was more potent than clavulanic acid against strains producing IRT enzymes with double substitutions. The double substitution (positions 69 and 276) was also present in IRT-10 along with a third substitution at position 165 (19), which was itself responsible for an IRT phenotype (W165R) (33) (Fig. 1). These observations might be in agreement with the mechanisms, proposed by Imtiaz et al. (22), that differentiate inhibition by clavulanic acid from that by tazobactam.
Some changes in the functional properties of the enzyme caused by the Asp276 mutation may not be apparent, because determination of the MIC takes into account the whole response of bacteria. However, kinetic results were consistent with the MICs for the mutant enzyme. Higher Km and kcat values were recorded in these studies. This demonstrated the close relationship between enzyme properties and resistance to β-lactams. We also found that the Asn276Asp mutant enzyme had lower catalytic efficiencies (kcat/Km) than TEM-1 due to the higher Km values for penicillins (Table 3), suggesting that the substrates may interact less efficiently with the mutant enzyme. Our kinetic data were consistent with those previously reported for the same mutation in a different plasmid construct and host bacterium (38). Single substitutions at positions 69 (mutant M69L) (16) and 276 (mutant Asn276Asp) independently caused similar reductions in the catalytic efficiency of TEM-1. There was a greater reduction in catalytic efficiency when mutations at both positions occurred together, as in mutant IRT-4 (7). There was a similar reduction in catalytic efficiency with the doubly substituted IRT-14 mutant, demonstrating the importance of the mutation at position 275 in conferring the IRT phenotype (10) (Table 4).
In the evolutionary process of IRTs, the amino acid substitution of the methionine at position 69 with leucine, valine, or isoleucine appears to be crucial in the first stages of the emergence of IRTs (12). The addition of the Asp276 mutation to the preexisting mutation at position 69 should confer a higher selective advantage to the bacterium. Similar selection processes probably operate for the mutation at position 275 (Arg275→Gln) (a similar reduction in catalytic efficiency for IRT-14; Table 4), for which no site-directed mutagenesis or a functional assay has so far been done. The Trp165→Arg substitution, which accounts for resistance to inhibitors (29, 33), would further contribute to such a selection process. This substitution occurs with the two other substitutions (positions 69 and 276) in the naturally occurring IRT-10 (19). Thus, the strain producing the IRT-10 β-lactamase may have undergone stronger selection pressure than the others.
The possible mechanism of the resistance generated by the substitutions at positions 275 and 276 is discussed below. These amino acid positions are not located close to the conserved boxes in class A enzymes. In TEM-1, they are at the beginning of helix h11 (23). In the crystal structure of PC1 β-lactamase (20), as the structures of TEM-1 β-lactamase (18) and a closely related TEM-1 β-lactamase (23) show, Arg244 is located on β sheet s-4 and its guanidinium side chain is close to the penicillin-binding cavity. In PC1, Asp276 forms a salt bond with Arg244, whereas in TEM-1, Asn276 forms hydrogen bonds with the guanidinium of Arg244. Molecular modeling of the Asp276 TEM mutant suggests that the Asp276 residue of the mutant forms a salt bond, as shown in Fig. 2, very similar to that observed with the PC1 enzyme. Nevertheless, the PC1 β-lactamase is susceptible to clavulanic acid. In fact, the interactions of clavulanic acid with the PC1 and TEM β-lactamases are not the same. With the PC1 enzyme, clavulanic acid forms a stable acyl enzyme, a cis-enamine susceptible to rearrangement into a decarboxylated trans-enamine (14). In the case of TEM enzymes, as shown with the TEM-2 enzyme, the acyl enzyme is not stable and acylation of Ser70 is followed by cross-linking of the enzyme with serine 130, as observed by electrospray ionization mass spectroscopy (6). Thus, in IRTs, it is credible that the formation of the acyl enzyme is still possible. This is shown in kinetic experiments with a transient inhibition, but the acyl enzyme almost totally reactivates because the level of cross-linking seems very low.
Therefore, IRT enzymes with double substitutions are the result of convergent evolution, because each substitution itself causes the overall resistance phenotype. There are differences in the behaviors of IRT enzymes with single and double substitutions toward different inhibitors. This highlights the importance of considering such enzyme sequence variations in strategies to design new inhibitor molecules.
ACKNOWLEDGMENTS
We thank SmithKline Beecham for providing clavulanic acid, Lederle for providing tazobactam, and Pfizer for providing sulbactam. We are grateful to C. Deloménie for the gift of the RZ1032 and XL-1 Blue strains and for helpful discussions about the site-directed mutagenesis method. We thank L. Gilly for performing purification of the β-lactamases.
This work was supported in part by a BQR from Conseil Scientifique UFR Médecine Cochin and a grant from the Institut National de la Santé et de la Recherche Médicale (INSERM). During this study, M.M.C. was supported first by the European Human Capital and Mobility Program and later by PRAXIS XXI research fellowships provided by Junta Nacional de Investigação Cientifica e Tecnológica, Lisbon, Portugal.
REFERENCES
- 1.Ambler R P, Coulson A F W, Frère J-M, Ghuysen J-M, Joris B, Forsman M, Levesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991;276:269–272. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belaaouaj A, Lapoumeroulie C, Caniça M M, Vedel G, Névot P, Krishnamoorthy R, Paul G. Nucleotide sequences of the genes coding for the TEM-like β-lactamases IRT-1 and IRT-2 (formerly called TRI-1 and TRI-2) FEMS Microbiol Lett. 1994;120:75–80. doi: 10.1111/j.1574-6968.1994.tb07010.x. [DOI] [PubMed] [Google Scholar]
- 3.Blazquez J, Baquero M-R, Canton R, Alos I, Baquero F. Characterization of a new TEM-type β-lactamase resistant to clavulanate, sulbactam, and tazobactam in a clinical isolate of Escherichia coli. Antimicrob Agents Chemother. 1993;37:2059–2063. doi: 10.1128/aac.37.10.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonomo R A, Dawes C G, Knox J R, Shlaes D M. β-Lactamase mutations far from the active site influence inhibitor binding. Biochim Biophys Acta. 1995;1247:121–125. doi: 10.1016/0167-4838(94)00188-m. [DOI] [PubMed] [Google Scholar]
- 5.Bret L, Chanal C, Sirot D, Labia R, Sirot J. Characterization of an inhibitor-resistant enzyme IRT-2 derived from TEM-2 β-lactamase produced by Proteus mirabilisstrains. J Antimicrob Chemother. 1996;38:183–191. doi: 10.1093/jac/38.2.183. [DOI] [PubMed] [Google Scholar]
- 6.Brown R P A, Aplin R T, Schofield C J. Inhibition of TEM-2 β-lactamase from Escherichia coliby clavulanic acid: observation of intermediates by electrospray ionization mass spectrometry. Biochemistry. 1996;35:12421–12432. doi: 10.1021/bi961044g. [DOI] [PubMed] [Google Scholar]
- 7.Brun T, Péduzzi J, Caniça M M, Paul G, Névot P, Barthélémy M, Labia R. Characterization and amino acid sequence of IRT-4, a novel TEM-type enzyme with a decreased susceptibility to β-lactamase inhibitors. FEMS Microbiol Lett. 1994;120:111–118. doi: 10.1111/j.1574-6968.1994.tb07016.x. [DOI] [PubMed] [Google Scholar]
- 8.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia colistrain with beta-galactosidase selection. Bio Techniques. 1987;5:376–378. [Google Scholar]
- 9.Bush K, Sykes R B. Methodology for the study of β-lactamases. Antimicrob Agents Chemother. 1986;30:6–10. doi: 10.1128/aac.30.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caniça M M, Barthélémy M, Gilly L, Labia R, Krishnamoorthy R, Paul G. Properties of IRT-14 (TEM-45), a newly characterized mutant of TEM-type β-lactamases. Antimicrob Agents Chemother. 1997;41:374–378. doi: 10.1128/aac.41.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caniça M M, Deloménie C, Labia R, Krishnamoorthy R, Paul G. Program and abstracts of the 16th Interdisciplinary Meeting on Anti-Infectious Chemotherapy. 1996. Characterization of the mutant produced by site-directed mutagenesis generating the substitution Asn-276→Asp in the critical proximity of Arg-244 in TEM-1 β-lactamase, abstr. 24/C3; p. 100. [Google Scholar]
- 12.Caniça M M M, Lu C Y, Krishnamoorthy R, Paul G C. Molecular diversity and evolution of blaTEM genes encoding β-lactamases resistant to clavulanic acid in clinical E. coli. J Mol Evol. 1997;44:57–65. doi: 10.1007/pl00006121. [DOI] [PubMed] [Google Scholar]
- 13.Caroff N, Caniça M M M, Gilly L, Krishnamoorthy R, Paul G. Program and abstracts of the 15th Interdisciplinary Meeting on Anti-Infectious Chemotherapy. 1995. Detection of a ponctual mutation in the blaTEM gene using an artificially created restriction site, abstr. 360/P24; p. 228. [Google Scholar]
- 14.Chen C C H, Herzberg O. Inhibition of β-lactamase by clavulanate. Trapped intermediates in cryocrystallographic studies. J Mol Biol. 1992;224:1103–1113. doi: 10.1016/0022-2836(92)90472-v. [DOI] [PubMed] [Google Scholar]
- 15.Cohen S N, Chang A C Y, Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coliby R-factor DNA. Proc Natl Acad Sci USA. 1972;69:2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaire M, Labia R, Samama J-P, Masson J-M. Site-directed mutagenesis at the active site of Escherichia coliTEM-1 β-lactamase. Suicide inhibitor-resistant mutants reveal the role of arginine 244 and methionine 69 in catalysis. J Biol Chem. 1992;267:20600–20606. [PubMed] [Google Scholar]
- 17.Du Bois S K, Marriott M S, Amyes S G B. TEM- and SHV-derived extended-spectrum β-lactamases: relationship between selection, structure and function. J Antimicrob Chemother. 1995;35:7–22. doi: 10.1093/jac/35.1.7. [DOI] [PubMed] [Google Scholar]
- 18.Fonzé E, Charlier P, Toth Y, Vermeire M, Raquet X, Dubus A, Frère J-M. TEM-1 β-lactamase structure solved by molecular replacement and refined structure of the S235A mutant. Acta Crystallogr. 1995;51:682–694. doi: 10.1107/S0907444994014496. [DOI] [PubMed] [Google Scholar]
- 19.Henquell C, Chanal C, Sirot D, Labia R, Sirot J. Molecular characterization of nine different types of mutants among 107 inhibitor-resistant TEM β-lactamases from clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1995;39:427–430. doi: 10.1128/aac.39.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herzberg O. Refined crystal structure of β-lactamase from Staphylococcus aureusPC1 at 2.0 Å resolution. J Mol Biol. 1991;217:701–719. doi: 10.1016/0022-2836(91)90527-d. [DOI] [PubMed] [Google Scholar]
- 21.Imtiaz U, Billings E, Knox J R, Manavathu E K, Lerner S A, Mobashery S. Inactivation of class A β-lactamases by clavulanic acid: the role of arginine-244 in a proposed nonconcerted sequence of events. J Am Chem Soc. 1993;115:4435–4442. [Google Scholar]
- 22.Imtiaz U, Billings E M, Knox J R, Mobashery S. A structure-based analysis of the inhibition of class A β-lactamases by sulbactam. Biochemistry. 1994;33:5728–5738. doi: 10.1021/bi00185a009. [DOI] [PubMed] [Google Scholar]
- 23.Jelsch C, Mourey L, Masson J-M, Samama J-P. Crystal structure of Escherichia coliTEM-1 β-lactamase at 1.8 Å resolution. Proteins Struct Funct Genet. 1993;16:364–383. doi: 10.1002/prot.340160406. [DOI] [PubMed] [Google Scholar]
- 24.Knox J R. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob Agents Chemother. 1995;39:2593–2601. doi: 10.1128/aac.39.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 26.Labia R, Andrillon J, Le Goffic F. Computerized microacidimetric determination of β-lactamase Michaelis-Menten constants. FEBS Lett. 1973;33:42–44. doi: 10.1016/0014-5793(73)80154-1. [DOI] [PubMed] [Google Scholar]
- 27.Labia R, Barthélémy M, Fabre C, Guionie M, Péduzzi J. Kinetic studies of three R-factor mediated β-lactamases. In: Hamilton-Miller J M T, Smith J T, editors. Beta-lactamases. London, United Kingdom: Academic Press; 1979. pp. 429–442. [Google Scholar]
- 28.Labia R, Lelievre V, Peduzzi J. Inhibition kinetics of three R-factor-mediated β-lactamases by a new β-lactam sulfone (CP 45899) Biochim Biophys Acta. 1980;611:351–357. doi: 10.1016/0005-2744(80)90071-6. [DOI] [PubMed] [Google Scholar]
- 29.Lenfant F, Petit A, Labia R, Maveyraud L, Samana J-P, Masson J-M. Site-directed mutagenesis of β-lactamase TEM-1. Investigating the potential role of specific residues on the activity of Pseudomonas-specific enzymes. Eur J Biochem. 1993;217:939–946. doi: 10.1111/j.1432-1033.1993.tb18324.x. [DOI] [PubMed] [Google Scholar]
- 30.Martinez J L, Vicente M F, Delgado-Iribarren A, Perez-Diaz J C, Baquero F. Small plasmids are involved in amoxicillin-clavulanate resistance in Escherichia coli. Antimicrob Agents Chemother. 1989;33:595. doi: 10.1128/aac.33.4.595-a. . (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthew M, Harris A M, Marshall M J, Ross G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 32.Matthew M, Hedges R W. Analytical isoelectric focusing of R factor-determined β-lactamases: correlation with plasmid compatibility. J Bacteriol. 1976;125:713–718. doi: 10.1128/jb.125.2.713-718.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petit A, Jullian E, Labia R. Substitution du tryptophane-165 par une arginine dans la beta-lactamase TEM-1. C R Acad Sci Paris. 1991;312:993–997. [Google Scholar]
- 34.Reynaud A, Péduzzi J, Barthélémy M, Labia R. Cefotaxime-hydrolysing activity of the β-lactamase of Klebsiella oxytocaD488 could be related to a threonine residue at position 140. FEMS Microbiol Lett. 1991;81:185–192. doi: 10.1016/0378-1097(91)90301-p. [DOI] [PubMed] [Google Scholar]
- 35.Russel M, Kidd S, Kelley M R. An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene. 1986;45:333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saves I, Burlet-Schiltz O, Swarén P, Lefèvre F, Masson J-M, Promé J-C, Samama J-P. The asparagine to aspartic acid substitution at position 276 of TEM-35 and TEM-36 is involved in the β-lactamase resistance to clavulanic acid. J Biol Chem. 1995;270:18240–18245. doi: 10.1074/jbc.270.31.18240. [DOI] [PubMed] [Google Scholar]
- 39.Stapleton P, Wu P-J, King A, Shannon K, French G, Phillips I. Incidence and mechanisms of resistance to the combination of amoxicillin and clavulanic acid in Escherichia coli. Antimicrob Agents Chemother. 1995;39:2478–2483. doi: 10.1128/aac.39.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vedel G, Belaaouaj A, Gilly L, Labia R, Philippon A, Névot P, Paul G. Clinical isolates of Escherichia coliproducing TRI β-lactamases: novel TEM-enzymes conferring resistance to β-lactamase inhibitors. J Antimicrob Chemother. 1992;30:449–462. doi: 10.1093/jac/30.4.449. [DOI] [PubMed] [Google Scholar]
- 41.Weiner S J, Kollman P A, Case D A, Singh U C, Ghio C, Alagona G, Profeta S J, Weiner P. A new force field for molecular mechanical simulation of nucleic acids and proteins. J Am Chem Soc. 1984;106:764–784. [Google Scholar]
- 42.Zhou X Y, Bordon F, Sirot D, Kitzis M-D, Gutmann L. Emergence of clinical isolates of Escherichia coliproducing TEM-1 derivatives or an OXA-1 β-lactamase conferring resistance to β-lactamase inhibitors. Antimicrob Agents Chemother. 1994;38:1085–1089. doi: 10.1128/aac.38.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]