Abstract
Background
Glioblastoma (GBM) is a highly aggressive and invasive brain tumor associated with high patient mortality. A large fraction of GBM tumors have been identified as epidermal growth factor receptor (EGFR) amplified and ~50% also are EGFRvIII mutant positive. In a previously reported multicenter phase II study, we have described the response of recurrent GBM (rGBM) patients to dacomitinib, an EGFR tyrosine kinase inhibitor (TKI). As a continuation of that report, we leverage the tumor cargo-encapsulating extracellular vesicles (EVs) and explore their genetic composition as carriers of tumor biomarker.
Methods
Serum samples were longitudinally collected from EGFR-amplified rGBM patients who clinically benefitted from dacomitinib therapy (responders) and those who did not (nonresponders), as well as from a healthy cohort of individuals. The serum EV transcriptome was evaluated to map the RNA biotype distribution and distinguish GBM disease.
Results
Using long RNA sequencing, we show enriched detection of over 10 000 coding RNAs from serum EVs. The EV transcriptome yielded a unique signature that facilitates differentiation of GBM patients from healthy donors. Further analysis revealed genetic enrichment that enables stratification of responders from nonresponders prior to dacomitinib treatment as well as following administration.
Conclusion
This study demonstrates that genetic composition analysis of serum EVs may aid in therapeutic stratification to identify patients with dacomitinib-responsive GBM.
Keywords: dacomitinib, EGFR, glioblastoma, liquid biopsy, RNA sequencing
Key Points.
Serum EV RNA differentiates healthy individuals from GBM patients.
Dacomitinib administration alters serum EV transcriptome.
DNMT3A, ZNF35, and LAMTOR2 expression bears potential to predict dacomitinib response in EGFR-amplified recurrent GBM patients.
Importance of the Study.
Therapies targeting tyrosine kinase inhibitors (TKIs) have failed to demonstrate efficacy for the majority of patients with GBM. However, for a small population of patients, EGFR inhibitors, such as dacomitinib, have been effective in targeting the disease. Identification of dacomitinib responders thus becomes beneficial to improve planning of patient treatment course. This study longitudinally quantifies the RNA signatures of serum extracellular vesicles from patients enrolled in NCT01112527 (treatment of recurrent GBM with dacomitinib) to develop a noninvasive stratification method to assess patient response. Enrichment analysis enables stratification of both dacomitinib response as well as patients from healthy donors. In particular, the identified select biomarkers, DNMT3A, ZNF35, and LAMTOR2, may enable discrimination of responders from nonresponders. While further validation through a larger cohort is warranted, this work provides the foundation for EV RNA-based selection of patients who may benefit from dacomitinib treatment.
Glioblastoma (GBM) is the most common primary brain tumor in adults with poor overall survival (OS). Current standard of care consists of maximal surgical debulking, followed by radiation therapy with concurrent and adjuvant temozolomide (TMZ) treatment.1 Patients experience an inevitable recurrence in 6–9 months posttherapy with a median postrecurrence survival of 3–6 months with no further effective treatment.2 In order to accurately diagnose the disease for management, the majority of patients undergo invasive brain surgery or tissue biopsy for molecular diagnosis to stratify options of therapy.3 Disease status is subsequently monitored via magnetic resonance imaging.
A subset of aggressive GBMs (30%–50%) harbor mutations, rearrangements, splicing alterations, and amplification of the epidermal growth factor receptor (EGFR) gene. The most frequent of these mutations is EGFRvIII, an in-frame deletion in the extracellular domain (ECD) of the EGFR gene which results in the constitutive activation of the intracellular tyrosine kinase domain. EGFRvIII expression leads to cellular proliferation, migration, and invasion of tumor cells.4,5 In the realm of cancer therapy drugs, tyrosine kinase inhibitors (TKIs) are known to drive inhibition of these signal transduction cascades and have demonstrated successful therapeutic responses in multiple types of cancer.6 Dacomitinib (PF-00299804) is one such drug that was approved by the US Food and Drug Administration (FDA) for the first-line treatment of patients with EGFR exon 19 deletion or exon 21 L858R mutated metastatic non-small cell lung cancer.7 Early TKI clinical trials in unselected patients with GBM reported limited efficacy8,9 and have attributed inefficiency to intertumoral heterogeneity, redundancy of intracellular signaling pathways, and off-target effects on ERB2 and ERB4.
In order to monitor therapeutic efficacy, liquid biopsy has increasingly become one of the most powerful tool sets, with several FDA-approved companion diagnostics to date. Liquid biopsies offer minimally analyses of molecules and vesicular structures shed by cancer cells as a means to diagnose and monitor cancers longitudinally and throughout therapy.10 All cells, including tumor cells, release small membrane-bound nanoparticles called extracellular vesicles (EVs) which contain proteins of parental cells and encapsulate native, cell-specific genetic cargo, including mRNA, miRNA, and other noncoding RNA.11 Recent advances in RNA sequencing combined with the improved understanding of EV RNA technologies have allowed the full mapping of the circulating EV RNA transcriptome in biofluids, providing insight into tumor dynamics over the course of the disease and therapy.12
We have previously evaluated the efficacy of dacomitinib in a multiarmed, multi-institutional clinical trial (NCT01112527).13 Dacomitinib is a second generation, irreversible, oral small-molecule EGFR TKI with superior pharmacokinetic properties that has demonstrated enhanced blood–brain barrier (BBB) penetration in patients with EGFR-amplified recurrent GBM (rGBM).14 In a preplanned, secondary analysis of the clinical trial, we have evaluated the potential of EV RNA-based liquid biopsy to determine tumor presence, assist patient stratification, and predict response to therapy in the setting of the dacomitinib treatment in patients with EGFR-amplified rGBM. We performed RNA sequencing analysis on long RNA extracted from patient serum samples pretreatment and 1-month postinitiation of treatment to determine EV RNA signatures. Here, using EV transcriptome sequencing, we (1) successfully distinguished patients with GBM from healthy controls, (2) identified RNA biomarkers for dacomitinib response prediction, and (3) show that dacomitinib responders can be prospectively identified from nonresponders, as defined by 6-month progression-free survival (PFS).
Methods
Patient Cohort Design
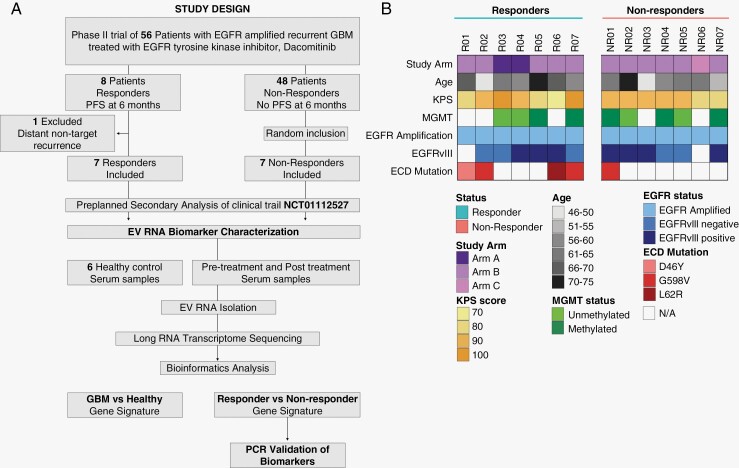
The study population (n = 14) consisted of a subset of patients enrolled in a multicenter, open-label, 3-arm nonrandomized clinical trial (NCT01112527). Patients in the trial were enrolled at Massachusetts General Hospital (MGH; Boston, MA), Brigham and Women’s Hospital (BWH; Boston, MA), Beth Israel Deaconess Medical Center (BIDMC; Boston, MA), Cleveland Clinic (Cleveland, OH), and Henry Ford Hospital (Detroit, MI). Enrollment criteria for all patients included a minimum age requirement of 18 years, histologically confirmed diagnosis of primary or secondary rGBM (World Health Organization 2016, grade IV), and the presence of EGFR gene amplification in the most recent tumor specimen as per the inclusion and exclusion criteria of the clinical trial.13 Arm A of the study included first-recurrence patients who were candidates for surgical resection and had not previously undergone anti-vascular endothelial growth factor (anti-VEGF) therapy (n = 2). Arm B included patients in a 2-stage phase II trial who had first-recurrence EGFR-amplified GBM and were anti-VEGF treatment naive (n = 11). Arm C consisted of first-recurrence patients with unlimited prior therapies following a bevacizumab-containing regimen (n = 1). Patients received 45 mg dacomitinib once daily orally with or without food continuously across a 28-day cycle or until disease progression or drug intolerance. Radiographic responses in the clinical trial were assessed using MacDonald criteria. For this longitudinal study of EV RNA-based liquid biopsy, a clinical response to dacomitinib therapy was defined as a PFS of at least 6 months. For further details on the clinical trial, see Chi et al. 13 Patient characteristics are depicted in Supplementary Table 1.
Ethics Statement
This trial was conducted in compliance with the Declaration of Helsinki and with the International Conference on Harmonization Good Clinical Practice Guidelines protocol. Approval by the Institutional Review Boards and/or Independent Ethics Committees at each of the participating investigational centers was attained. All patients provided written, informed consent prior to study participation.
Patient Serum Collection and Processing
Blood samples from patients were collected prior to therapy (pretreatment) and 1 month after the initiation of daily dacomitinib dosing (posttreatment). The blood was left to clot for 30 min at room temperature (RT) and serum was isolated, according to the manufacturer’s recommendations (BD Vacutainer, Franklin Lakes, NJ) within 2 h of collection. Serum was filtered by slow passage through a 0.8 μm syringe filter (Millipore, Billerica, MA), aliquoted into 1.8 mL cryotubes (Fisher Scientific, Waltham, MA), and stored at −80°C until use.
Healthy Control Characteristics
The control serum samples used in this study consisted of 6 pools of healthy (with no known cancer or glioma) human serum of approximately 3 mL volume per pool. Each serum pool consisted of 0.3 mL of serum from 10 different individuals of different age, sex, and racial backgrounds (n = 60 individuals total). These samples were obtained from BioIVT (formerly BioreclamationIVT, Westbury, NY). Donor characteristics can be found in Supplementary Table 2. The average age of serum donors was 37.6 years.
EV RNA Isolation
Extraction and analysis of the serum samples were carried out in 1 central laboratory by Exosome, Inc., a Bio-Techne Brand (Waltham, MA). Serum EV RNA was extracted from the samples using ExoLution RNA extraction technology (Exosome Diagnostics, Waltham, MA) on a median serum input of 2 mL per patient or pooled control sample.
NextGen Sequencing
Total RNASeq library construction and sequencing were performed at Exosome Diagnostics, Inc. (Waltham, MA) using a proprietary EV long RNA sequencing platform. Briefly, isolated RNA samples (n = 34) were first treated with DNase to remove any trace amounts of co-purified DNA present in the sample. Post-DNA digestion, synthetic RNA spike-in controls (ERCC Exfold Spike-In Mix, Life Technologies, Carlsbad, CA) were added to each sample, with Timepoint 1 samples receiving Spike-In Mix 1 and Timepoint 2 samples receiving Spike-In Mix 2. Both Spike-In Mix suspensions are manufacturer preformulated ERCC RNA blends of 92 unlabeled and polyadenylated transcripts ranging from 250 to 2000 nts in length. Synthetic RNA is derived and traceable from NIST-certified DNA plasmids and is typically used to control the quality of the starting material, the RNA yield, the platform employed, and the RNA Seq operation. The exoRNA/synthetic spike-in mix was then reverse transcribed using a combination of random hexamers and oligo-dT primers. Second strand synthesis and addition of adapters were performed using a polymerase chain reaction (PCR)-based approach. Libraries were then subjected to 2 rounds of cleanups using AMPureXP beads (Beckman Coulter, Indianapolis, IN). Post cleanup, rRNA derived fragments were enzymatically depleted from the libraries. Finally, ribodepleted libraries were amplified by PCR and purified with AMPureXP beads. Quantification was performed using Agilent Bioanalyzer 2100 High Sensitivity Assay and Qubit 1X dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA). Post normalization, libraries were combined into 4 pools and sequenced on Illumina NextSeq500 using 2 × 150 cycles read length chemistry. The 4 sequencing runs produced an output of 994 million read pairs with an average sequencing depth of 35.5 million read pairs per sample.
Bioinformatics
Two 150 paired-end reads were aligned to the human genome (hg38) with the Gencode v25 gene model using STAR (version 2.5.2a). Alignments to the transcriptome output from STAR were used as input to Salmon (version 0.8.1) with position bias and guanine–cytosine bias corrections to obtain read counts and transcripts per million at the transcript level. These transcript-level values were then aggregated to the gene level using the tximport package in Bioconductor. Differential expression (DEX) analysis was performed on raw read counts per gene using DESeq2 with the various covariates. The P-values were corrected using Benjamini–Hochberg. Genes were reported as differentially expressed if a statistical significance of P ≤ .05 was achieved.
Manual Curation of Top 25 Gene Panel
Analysis of gene expression in normal brain tissue and GBM tissue was first performed using The Cancer Genome Atlas (TCGA) online database. Of the 285 genes that were enriched in the serum EV RNA from our current study, 65 of those genes were also found (via TCGA) to be significantly expressed in GBM tissue (fold change >3). Each 1 of the 65 genes was thoroughly researched in the literature using these filters: glioma, cancer hallmarks. If at least 1 study was found to describe the role of the specific gene in the context of glioma and at least on the hallmarks of cancer, the gene was included in the final list of 25 genes. Of the 40 genes not included, the genes were either not found to be involved with the hallmarks of cancer and/or with gliomas.
We then further explored the prognostic significance of the upregulated and down-regulated expression of each gene in GBM (> and <, respectively, of median expression) using the TCGA cohort. Kaplan–Meier analysis and survival disadvantage was assessed for each gene and significantly up- and down-regulated genes were chosen for further analysis.
TCGA Data Analyses
Analysis of gene expression in normal brain tissue and GBM tissue was performed using TCGA online database. Kaplan–Meier curves for GBM samples, stratified into a high and low expression of each gene of interest relative to the median expression of the group, were generated using the REpository of Molecular BRAin Neoplasia DaTa (REMBRANDT), accessed through betastasis.com with the Affymetrix Human Exon 1.0 ST platform.
Statistical Analysis
Statistical analysis was performed using unpaired two-tailed Student’s t-test in GraphPad Prism 8 software and P ≤ .05 was considered statistically significant. Confidence intervals were calculated using exact binomial distributions. The results are presented as the mean ± SD. (***P ≤ .001; ****P ≤ .0001).
Results
Clinical Trial Overview and Recovery and Analysis of Coding RNAs from Serum EVs
A cohort of adult recurrent EGFR-amplified GBM patients (n = 14) undergoing treatment with dacomitinib were included in the study. Of the original 56 patients with EGFR-amplified rGBM who participated in the clinical trial, 8 patients responded to therapy (PFS of at least 6 months; range of 7–8 months), while 48 patients did not respond to therapy (PFS of less than 6 months; range of 1–3 months). Seven patients met the PFS 6-month endpoint and were included in the responder group of this study. An 8th patient, despite achieving complete response of the target lesion, developed a nontarget recurrent lesion at the end of cycle 6 and thus did not meet the PFS6 endpoint (patient 16 in the clinical report13) and was excluded from the study. Of the 48 nonresponders, 7 patients were selected based on serum sample quality (no hemolysis) and a minimum of 2 mL serum and included in the nonresponder group.
(Figure 1A) Both responder and nonresponder population parameters were comparable at baseline; including age, EGFR amplification status, EGFRvIII positivity, and O[6]-methylguanine-DNA methyltransferase (MGMT) promoter methylation status (Supplementary Table 1). Additionally, 4 out of 7 responders had EGFR ECD mutations (G598V, L62R, and 2 patients with D46Y) whereas only 1 out of 7 nonresponders had an EGFR ECD mutation (G598V). The responders had significantly longer PFS (14 vs 2 months, P = .0003) and median OS from the time of dacomitinib therapy initiation (21.3 vs 10.3 months, P = .003), while the median OS from the time of diagnosis (34 vs 29.9 months, P = ns) was similar to nonresponders (Supplementary Table 1). The baseline demographic, histologic, and molecular characteristics of the GBM patient cohort is depicted in Figure 1B. The healthy donor cohort (n = 6) was designed to age match the patient cohort.
Figure 1.
Study and cohort design of GBM patients. EV RNA Biomarker analysis study design of the preplanned secondary analysis of the clinical trial NCT01112527 (A). Baseline patient demographic, histologic, and molecular characteristics of the GBM patient cohort (B). Healthy donor parameters can be found in Supplementary Table 2. EGFR, epidermal growth factor receptor; PFS, progression-free survival; EV, extracellular vesicle; GBM, glioblastoma.
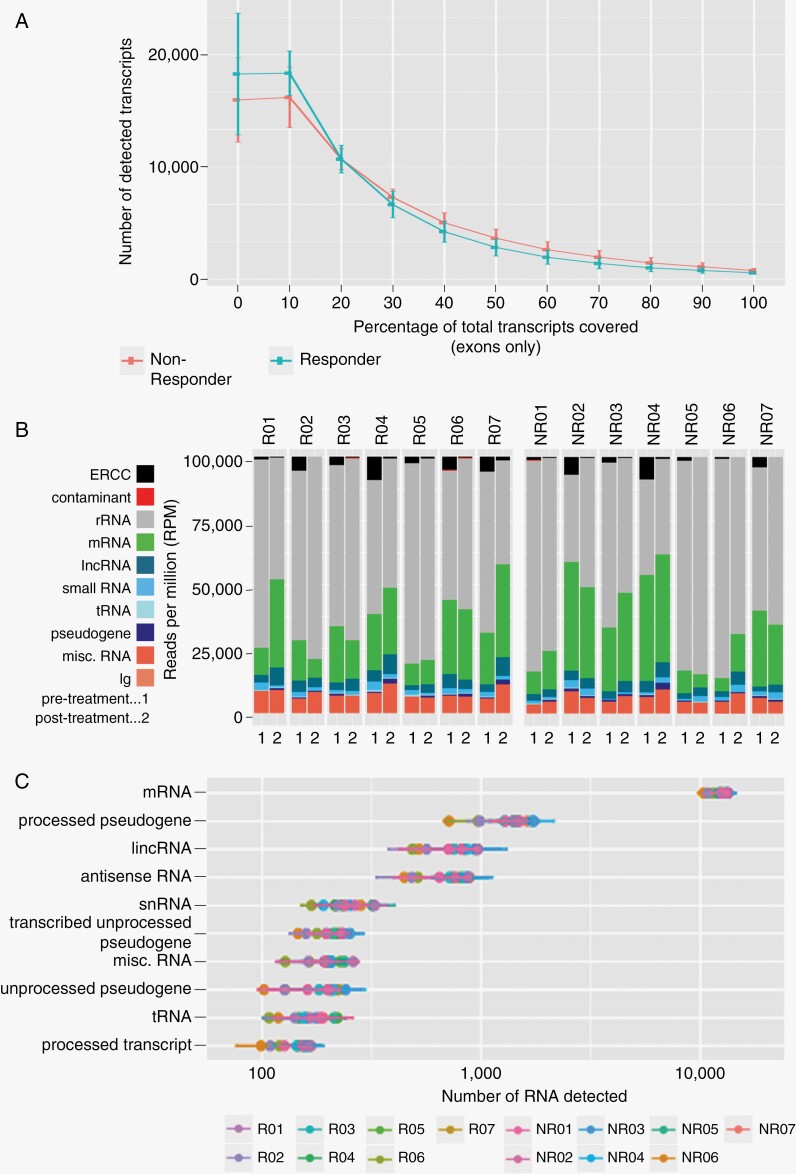
For the comparative study, 2 mL of serum was collected prior to therapy (T0) and at 1 month following dacomitinib treatment (T1). Pooled serum from healthy donors was used as a control group. EVs were isolated from serum and the RNA was subsequently extracted using ExolutionPlus technology. Following depletion of the majority of rRNAs during library preparation, we identified an average of 35.5 million read pairs of long RNAs per sample, with 20%–70% alignment of the reads to the human genome (hg38; Supplementary Figure 1A). Within the mapped genes of the GBM patients at both T0 and T1 (1 and 2, respectively), we elucidated a diverse landscape of RNA species including rRNA, mRNA, small RNA, tRNA, pseudogenes, lncRNA, and antisense transcripts. Despite rRNA depletion, this population was found as the most abundant species. Downstream analysis has excluded the detected rRNAs from the study to focus on long RNAs, specifically mRNAs. We confirmed, via coverage analysis, a similar number of detected transcripts across the cohorts within the study (Figure 2A). Additionally, although many of the transcripts demonstrated low transcript length coverage, a notable number of the transcripts (>6500) were detected with a minimum of 50% coverage, allowing for adequate summary of the long RNAs in each of the cohorts. The EV RNA profiles show a degree of inter- and intra-patient heterogeneity with a relative abundance of protein-coding mRNAs ranging between 10% and 50% (Figure 2B). Detailed analysis of the RNA landscape identified over 10 000 protein-coding genes, several hundreds of long intergenic noncoding RNAs (lncRNAs), as well as reliable detection of antisense RNAs across all patient samples (Figure 2C). A high correlation was observed between global (DESeq2) normalization and GAPDH normalization. We opted for GAPDH based normalization for all our subsequent analysis, for the ease of translating our findings to a PCR-based platform (Supplementary Figure 1B). By enabling the detection of thousands of mRNAs and long noncoding RNAs with high coverage from biofluid-derived EVs, our novel RNASeq workflow maximizes the repertoire of potential RNA-based biomarkers of GBM.
Figure 2.
Serum EV RNA quantification. Transcript length coverage (exonic) in serum EV-RNA of both responder and nonresponder patients (A). Biotype distribution, as quantified by reads per million (RPM), of transcriptome mapped EV RNA reads (B) of all patients pre- and post-treatment (1: pretreatment, 2: 1 month of administration). RNA detection of the top 10 most abundant RNA biotypes in serum EV-RNA for all patients (C).
EV RNA Signature Can Distinguish a Patient with GBM from Healthy Control
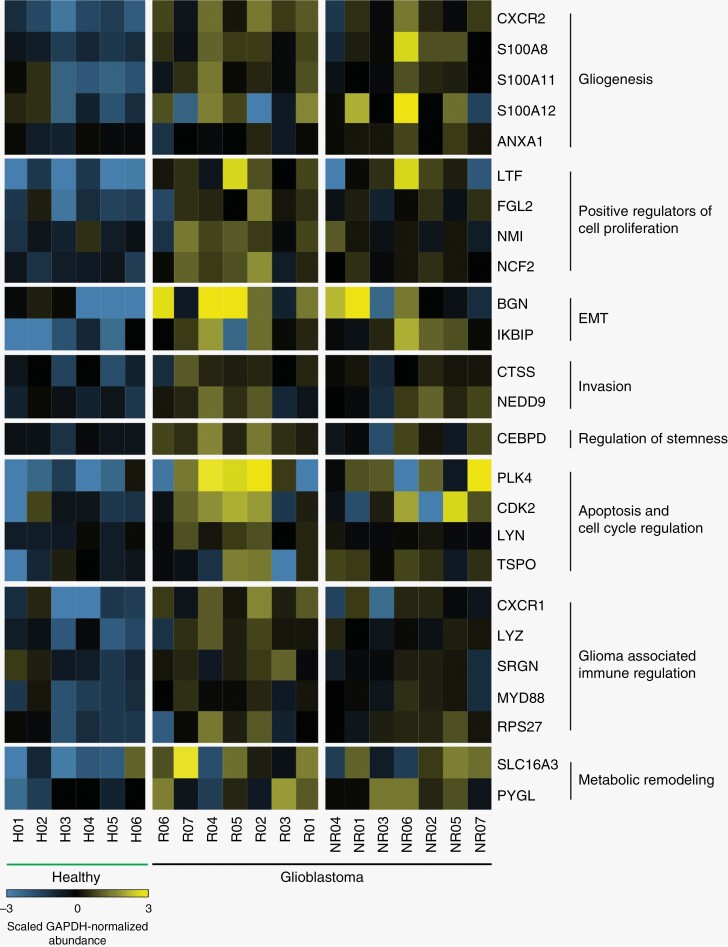
Serum EV RNA transcriptome profiles of pretreatment GBM patients (n = 14) and pooled healthy controls (n = 6) revealed 451 differentially expressed genes. Of these, 285 mRNAs were enriched and 166 mRNA were depleted in EVs derived from the plasma of GBM patients compared with healthy controls (Supplementary Figure 2). These genes were compared with the gene expression data derived from TCGA. Within the 285 genes that were enriched in EV RNA of GBM patients compared with healthy controls, 65 genes were highly expressed in GBM tissue as compared with normal brain tissue (fold change >3). However, of the 166 genes that were depleted in EV RNA of GBM patients compared with healthy controls, only 7 genes were highly expressed in GBM tissue when compared with normal brain tissue (fold change >3). Through evaluation of these 65 genes enriched in the tumor (TCGA) and plasma (present study) of GBM patients, we manually curated a list of the 25 genes that retain tumorigenic functionality and are included within the hallmarks of the GBM genetic signature (Table 1; see Methods). We further explored the prognostic significance of the upregulated and down-regulated expression of each gene in GBM (> and <, respectively, of median expression) using the TCGA cohort. Kaplan–Meier analysis of the upregulated expression of LTF, S100A8, and SLC16A3 in GBM patients conferred a survival disadvantage (Supplementary Figure 3). The 25 gene signature is representative of critical pathways in 8 hallmarks of cancer including gliogenesis (CXCR2; S100A8, S100A11, S100A12, ANXA1), positive regulators of cell proliferation (LTF, FGL2, NMI, NCF2), epithelial–mesenchymal transition (BGN, IKBIP), invasion (CTSS, NEDD9), regulation of stemness (CEBPD), apoptosis and cell cycle regulation (PLK4, CDK2, LYN, TSPO), glioma-associated immune regulation (CXCR1, LYZ, SRGN, MYD88, RPS27), metabolic remodeling (SLC16A3, PYGL; Figure 3).
Table 1.
Distinct Differentially Enriched Genes in Serum from GBM Patients Compared with Healthy Controls
| Gene | Symbol | Plasma (Present Study) | Tissue (TCGA) | OS (TCGA) | ||||
|---|---|---|---|---|---|---|---|---|
| log2 (FC) | P | log2 (FC) | GBM | Healthy | High vs Low | P | ||
| Biglycan | BGN | 3.94 | 1.37E-02 | 4.99 | 12.40 | 10.10 | No | .81 |
| Lactotransferrin | LTF | 3.64 | 2.19E-05 | 74.40 | 10.50 | 4.24 | Yes | .008 |
| Polo Like Kinase 4 | PLK4 | 3.50 | 9.70E-03 | 4.48 | 7.34 | 5.18 | No | .91 |
| S100 Calcium Binding Protein A12 | S100A12 | 2.91 | 3.41E-02 | 3.76 | 2.63 | 0.72 | No | .28 |
| S100 Calcium Binding Protein A8 | S100A8 | 2.77 | 2.56E-05 | 4.57 | 7.68 | 5.49 | Yes | .01 |
| C-X-C Motif Chemokine Receptor 2 | CXCR2 | 2.69 | 7.31E-09 | 10.10 | 4.99 | 1.65 | NA | |
| IKBKB Interacting Protein | IKBIP | 2.63 | 1.69E-04 | 7.85 | 8.89 | 5.91 | No | .07 |
| Solute Carrier Family 16 Member 3 | SLC16A3 | 2.31 | 4.30E-02 | 4.05 | 10.10 | 8.10 | Yes | .01 |
| S100 Calcium Binding Protein A11 | S100A11 | 2.16 | 8.64E-05 | 8.93 | 11.40 | 8.26 | No | .08 |
| Cyclin Dependent Kinase 2 | CDK2 | 2.10 | 4.51E-02 | 14.10 | 9.54 | 5.73 | No | .08 |
| C-X-C Motif Chemokine Receptor 1 | CXCR1 | 2.08 | 2.09E-02 | 3.96 | 3.21 | 1.22 | NA | |
| Translocator Protein | TSPO | 2.03 | 2.20E-02 | 4.10 | 10.30 | 8.28 | No | .21 |
| Lysozyme | LYZ | 1.96 | 2.90E-10 | 8.80 | 9.60 | 6.46 | No | .81 |
| Glycogen Phosphorylase L | PYGL | 1.95 | 5.34E-03 | 9.10 | 10.50 | 7.31 | No | .20 |
| Fibrinogen Like 2 | FGL2 | 1.55 | 1.38E-03 | 3.16 | 9.26 | 7.60 | No | .69 |
| Neural Precursor Cell expressed, Developmentally Down-Regulated 9 | NEDD9 | 1.44 | 2.25E-03 | 3.10 | 10.10 | 8.43 | No | .19 |
| MYD99 Innate Immune Signal Transduction Adaptor | MYD88 | 1.39 | 1.71E-02 | 6.69 | 10.10 | 7.33 | No | .89 |
| CCAAT Enhancer Binding Protein Delta | CEBPD | 1.37 | 1.14E-02 | 4.05 | 10.50 | 8.44 | No | .46 |
| Ribosomal Protein S27 | RPS27 | 1.34 | 1.00E-02 | 16.70 | 8.09 | 4.03 | No | .08 |
| Serglycin | SRGN | 1.23 | 2.35E-02 | 3.20 | 11.20 | 9.47 | No | .01 |
| Cathepsin S | CTSS | 1.10 | 5.62E-03 | 5.67 | 10.50 | 7.98 | No | .25 |
| N-Myc and STAT Interactor | NMI | 1.08 | 1.67E-02 | 5.90 | 8.38 | 5.82 | No | .06 |
| Neutrophil Cytosolic Factor 2 | NCF2 | 1.06 | 5.75E-03 | 4.79 | 8.38 | 6.11 | No | .06 |
| Annexin A1 | ANXA1 | 1.04 | 3.25E-02 | 20.40 | 12.60 | 8.28 | No | .13 |
| LYN Proto-Oncogene, Src Family Tyrosine | LYN | 0.80 | 2.20E-02 | 3.33 | 9.95 | 8.22 | No | .56 |
Analysis was based on a cutoff of a 3-fold increase in relative expression in GBM tumor tissue compared with healthy brain tissue.
Abbreviations: TCGA, the cancer genome atlas; FC, fold change; NA, not available.
Figure 3.
Differential gene expression profiles of healthy and GBM patient serum EV transcriptomes. The top 25 genes enriched in serum EV RNA of patients with GBM (n = 14; responders, 7; nonresponders, 7) as compared with healthy controls (n = 6) were selected for analysis. Total reads were normalized to GAPDH expression via log2. Top 285 enriched mRNAs were compared with the gene expression profiles of GBM and healthy tissue in The Cancer Genome Atlas (TCGA). Twenty-five of the 65 mRNAs enriched in serum EVs and GBM tissue (TCGA) were identified and their relevance to GBM summarized.
Serum EV RNA Signature Can Identify Responders to Dacomitinib Therapeutics
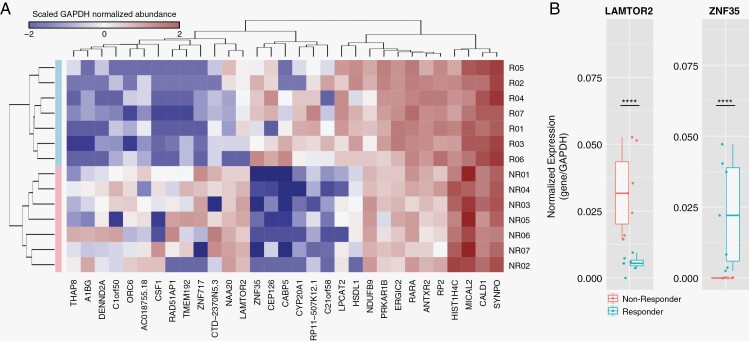
We next analyzed the serum EV RNA transcriptome profiles of GBM patient pretreatment (n = 14), to evaluate the possibility of distinguishing dacomitinib responders from nonresponders. Interestingly, we identified 8 genes (ZNF35, CEP126, CABP5, CYP20A1, RP11-507K12.1, C21orf58, LPCAT2, HSDL1) that are significantly enriched in responders and 13 genes (THAP8, A1BG, DENND2A, C1orf50, ORC6, AC018755.18, CSF1, RAD51AP1, TMEM192, ZNF717, CTD-2370N5.3, NAA20, LAMTOR2) that are significantly enriched in nonresponders (Figure 4A). The same genes are identified regardless of the choice of either GAPDH normalization (current study) or global (DESeq2) normalization13; we opted for GAPDH normalization in order to simplify follow up of these signatures by methods such as digital PCR. Additionally, we have also identified a set of 10 genes (NDUFB9, PRKAR1B, ERGIC2, RARA, RP2, HIST1H4C, MICAL2, CALD1, SYNPO) that are stably expressed in both responders and nonresponders, which can be used as normalizers. Specifically, the EV mRNAs ZNF35 and LAMTOR2 distinguish responders from nonresponders with a high degree of statistical significance (P-adjusted = 2.6E-8 and 2.4E-6, respectively; Figure 4B).
Figure 4.
Notable gene expression in serum EV RNA prior to dacomitinib administration. Phylogenetic plotting of genetic expression in responders (R, n = 7) and nonresponders (NR, n = 7) at T0 (A). Normalized gene expression of LAMTOR2 and ZNF35 (B) at T0 in responders and nonresponders. Statistical comparison of responder gene expression and non-responder gene expression was performed with P value adjustment (LAMTOR2: P = 2.5E-6, ZNF35: P = 2.6E-8).
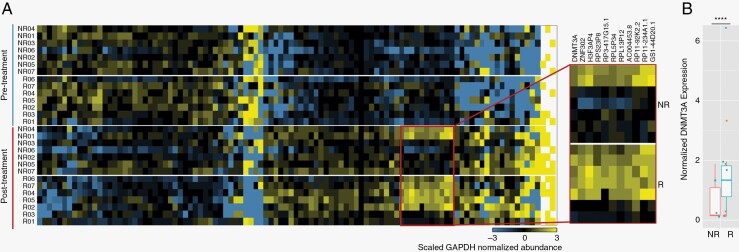
We observed a distinct gene expression profile in the serum of all patient’s posttreatment as compared with pretreatment (Figure 5A). Interestingly, in both responders and nonresponders, administration of dacomitinib induced similar genetic expression shifts across all patients, regardless of response status. A larger shift in fold change was found in the responder patient cohort (responder: 3.83, nonresponder: 2.65; Supplementary Figure 4). Via analysis of sera EV profiles following 1 month of treatment, we identified a small group of genes (DNMT3A, ZNF302, HF3AP4, RPS23P8, RP3-417G15.1, RPL5P34, RPL13P12, AC004453.8, RP11-92K2.2, RP11-234A1.1, GS1-44D20.1) that are enriched in the EVs of responders, as compared with nonresponders, in the posttreatment sera which may be potential markers of treatment response. DNMT3A (DNA methyltransferase 3 alpha) is significantly enriched (P-adjusted = 1.8E-4) in the posttreatment serum of dacomitinib responders (Figure 5B).
Figure 5.
EV transcriptome following dacomitinib administration. Gene expression profiles in serum EVs of EGFR-amplified GBM patients posttreatment (T1; A). Normalized gene expression of DNMT3A of responders and nonresponders at T1 (B). Statistical comparison was performed with P value adjustment (P = 1.8E-4).
Discussion
Accurate molecular diagnosis of brain tumors at first-time diagnosis and at recurrence is largely dependent on the feasibility of obtaining tissue that involves invasive brain surgery. Emerging evidence over the past decade has delineated the intrinsic nature of intratumoral heterogeneity and the evolutionary landscape of GBM over the course of disease and therapy.15–18 Moreover, most of the clinical trials stratify patients for therapeutic intervention at recurrence based on the genomic status of the primary tumor at initial diagnosis as opposed to the molecular status of the recurrent tumor. This can be attributed to the complex nature of tissue biopsy and the increased morbidity associated with repeated brain biopsies over the disease course. The need for accurate molecular diagnosis has, in turn, driven the demand for alternative methodologies including minimally invasive liquid biopsy-based strategies to provide a comprehensive molecular profile of the tumor, over time, and therapy.
The role of EVs has been described in health and diseases. Specifically, in the context of GBM, EVs are implicated in tumor proliferation, angiogenesis, metabolic reprogramming, evasion of immune surveillance, and acquired drug resistance.19 As such, characterization of the full repertoire of EV cargo is imperative to understanding the biological functions of the cell of origin and identifying potential biomarkers for diagnostic and prognostic significance. However, the majority of recent exploratory sequencing studies in EVs have focused primarily on characterizing small RNA fractions, reporting a relatively small proportion and poor transcript coverage of long RNA.12,20–22 This has led to a preconception that EVs mostly harbor miRNAs, tRNAs, or short fragments of mRNA and lncRNA.23,24 Here, we employed a novel platform to characterize long RNAs (including transcripts longer than 100 nt) derived from EVs and demonstrated detection of a range of RNA species, including rRNA, mRNA, small RNA, tRNA, pseudogenes, lncRNA, and antisense transcripts. Furthermore, we incorporated rRNA depletion to minimize rRNA detection. Our methodology yielded a broader range of mRNA coverage as compared with previous small RNA sequencing studies,25,26 resulting in detection of a myriad of mRNAs that had previously remained undetectable in EVs. In sharp contrast with previous reports, our study reports detection of over 6500 transcripts, with high coverage, from biofluid-derived EVs.12,20–22
Whole transcriptome analysis of serum EV RNA reveals significant DEX between the genes of GBM patients as compared with healthy controls. Our unique 25 mRNA gene signature enriched in the serum EVs of GBM patients is representative of critical pathways in several hallmarks of cancer, including gliogenesis, positive regulators of cell proliferation, epithelial–mesenchymal transition (EMT), invasion, regulation of stemness, apoptosis, cell cycle regulation, glioma-associated immune regulation, and metabolic remodeling. In the context of gliogenesis, CXCR2, S100A8, S100A11, S100A12, ANXA1 mRNAs are enriched in the serum EVs of GBM patients as compared with those derived from healthy controls. One possible limitation of this finding is the age range of the healthy controls, which includes individual younger compared with the patient populations.
Although dacomitinib was not effective for the majority of patients with EGFR-amplified GBM, a small subset of patients (n = 8, 14%) had clinical benefit by remaining progression-free for at least 6 months, with 5 patients remaining progression free for at least 1 year. This fraction is significantly lower than other EGFR amplified and/or mutated cancers (lung,27 HNSCC28). Similarly, a majority of the clinical trials focusing on therapeutic options for GBM have had minimal success owing to multiple factors: inter- and intra-tumoral heterogeneity, evolution of therapy-resistant clonal subpopulations, reacquisition of GBM stem cell stemness, the impenetrable nature of the BBB and intrinsic drug efflux mechanisms, metabolic adaptations, and enhanced repair of drug-induced DNA damage.29 Thus, predicting the subset of patients who could respond to a particular therapy is critical for stratifying targeted therapies to the potential patients who would benefit. In this context, we evaluated the feasibility of a serum EV RNA signature that can identify the potential responders to dacomitinib therapy. Analysis of serum EVs at baseline identified 8 genes (ZNF35, CEP126, CABP5, CYP20A1, RP11-507K12.1, C21orf58, LPCAT2, HSDL1) that are enriched in responders and 13 genes (THAP8, A1BG, DENND2A, C1orf50, ORC6, AC018755.18, CSF1, RAD51AP1, TMEM192, ZNF717, CTD-2370N5.3, NAA20, LAMTOR2) enriched in nonresponders, as previously reported.13 Notably, ZNF35 is significantly enriched in the serum EVs obtained from responders. ZNF35 is a part of the zinc-finger protein family and ontologically involved in DNA transcription transactivation. Further study is required to discern its mechanistic involvement in GBM response to dacomitinib. We have additionally found that LAMTOR2 (Late Endosomal/Lysosomal Adaptor, MAPK and MTOR Activator 2), a key activator of MAPK (mitogen-activated protein kinase) and mTOR (mammalian target of rapamycin) signaling, was enriched in serum EVs of nonresponders. Upregulation of LAMTOR2 is likely indicative of increased MAPK and mTOR signaling, both of which have been previously implicated as important resistance mechanisms in the context of EGFR-TKIs.30,31 Moreover, multiple co-activation of the EGFR receptor tyrosine kinase generates redundant downstream signaling, further limiting single TKI efficacy.32
Our findings also suggest dynamic alteration of the EV transcriptome due to dacomitinib administration. We detected a significant increase in DNMT3A RNA levels in posttreatment serum EVs. DNMTs are key methylators that establish DNA methylation patterns for gene expression regulation. Additionally, DNMT3A has been described to be significantly overexpressed in GBM compared with normal brain tissue.33 Alterations in DNMT3A are implicated in epigenetic dysregulation that enhances growth, proliferation, and survival in multiple cancers.34,35 Higher levels of DNMT3A expression in responders posttreatment could be indicative of methylation changes in response to dacomitinib therapy.
These sequencing results suggest that assessment of DNMT3A, ZNF35, and LAMTOR2 levels in serum EVs will provide insight into patient response both prior to treatment and after 1 month of dacomitinib administration. Further validations on serum EVs, however, will be needed to verify biomarker expression. Similarly, tumor tissue was not available from the select patients whose plasma was sequenced either because tumor resection was not clinically indicated/possible or because not enough tissue was obtained. Finally, although limited by patient cohort size, this study has established a series of biomarkers that may provide the foundation for future discovery of drug responsiveness in cancer.
Conclusion
We report here a novel approach for serum EV-derived long RNA sequencing and quantification. While further characterization of candidate genes is warranted, these results still bear promising utility for the identification of potential GBM biomarkers for disease detection, patient stratification, and prediction of disease response to dacomitinib. In the context of clinical validation, dacomitinib, a candidate drug that failed to achieve endpoint efficacy measures in initial EGFR-amplified GBM patients, may warrant a second look in a new prospective clinical trial following baseline serum EV RNA-based selection of GBM patients who may benefit from dacomitinib treatment.
Supplementary Material
Acknowledgments
We would like to thank the patients and their families for their participation in this study.
Contributor Information
Anudeep Yekula, Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Tiffaney Hsia, Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Robert R Kitchen, Exosome Diagnostics, Inc., a Bio-Techne Brand, Waltham, Massachusetts, USA.
Sudipto K Chakrabortty, Exosome Diagnostics, Inc., a Bio-Techne Brand, Waltham, Massachusetts, USA.
Wei Yu, Exosome Diagnostics, Inc., a Bio-Techne Brand, Waltham, Massachusetts, USA.
Syeda M Batool, Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Brian Lewis, Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Antoni J Szeglowski, Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Ralph Weissleder, Center for Systems Biology, Massachusetts General Hospital, Boston, Massachusetts, USA.
Hakho Lee, Center for Systems Biology, Massachusetts General Hospital, Boston, Massachusetts, USA.
Andrew S Chi, Department of Neurology, Massachusetts General Hospital, Boston, Massachusetts, USA.
Tracy Batchelor, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts, USA.
Bob S Carter, Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Xandra O Breakefield, Department of Neurology, Massachusetts General Hospital, Boston, Massachusetts, USA.
Johan Skog, Exosome Diagnostics, Inc., a Bio-Techne Brand, Waltham, Massachusetts, USA.
Leonora Balaj, Department of Neurosurgery, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Funding
This work is supported by grants [U01 CA230697, B.S.C., L.B.], [R01 CA239078 and CA237500, B.S.C., L.B.], [P01 CA069246, B.S.C., X.O.B.], and [R35 CA232103, X.O.B.]. The funding sources had no role in the writing of the manuscript or decision to submit the manuscript for publication. The authors have not been paid to write this article by any entity. The corresponding author has full access to the manuscript and assumes final responsibility for the decision to submit for publication.
Conflict of interest statement
J.S., R.K., S.C., and W.Y. are employees of Exosome Diagnostics, a Bio-Techne Brand. A.S.C. is an employee of Bright Peak Therapeutics. None of the authors declare any conflicts of interests.
This paper follows a previous clinical publication [DOI: 10.1200/PO.19.00295] and provides detailed analysis on serum EV transcriptomic biomarkers to predict response to dacomitinib in EGFR-amplified recurrent glioblastoma.
Authorship statement
Conception and design: X.O.B., J.S., L.B. Project administration and supervision: X.O.B., J.S., L.B. Collection and assembly of data: A.Y., T.H., R.R.K., S.K.C., W.Y., S.M.B., A.J.S., B.L., T.B., B.S.C., A.S.C., X.O.B., J.S., L.B. Data analysis and interpretation: A.Y., T.H., R.R.K., S.K.C., W.Y., S.M.B., R.W., H.L., T.B., B.S.C., A.S.C., X.O.B., J.S., L.B. Manuscript writing: all authors. Final approval of manuscript: all authors.
Data availability
Source data for figures are provided within extended data figures. Code is available upon reasonable request.
References
- 1. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Gruber ML, Buster WP.. Temozolomide in combination with irinotecan for treatment of recurrent malignant glioma. Am J Clin Oncol. 2004;27(1):33–38. [DOI] [PubMed] [Google Scholar]
- 3. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Talasila KM, Soentgerath A, Euskirchen P, et al. EGFR wild-type amplification and activation promote invasion and development of glioblastoma independent of angiogenesis. Acta Neuropathol. 2013;125(5):683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heimberger AB, Hlatky R, Suki D, et al. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res. 2005;11(4):1462–1466. [DOI] [PubMed] [Google Scholar]
- 6. Arora A, Scholar EM.. Role of tyrosine kinase inhibitors in cancer therapy. J Pharmacol Exp Ther. 2005;315(3):971–979. [DOI] [PubMed] [Google Scholar]
- 7. Shirley MD. First global approval. Drugs. 2018;78(18):1947–1953. [DOI] [PubMed] [Google Scholar]
- 8. Brandes AA, Franceschi E, Tosoni A, Hegi ME, Stupp R.. Epidermal growth factor receptor inhibitors in neuro-oncology: hopes and disappointments. Clin Cancer Res. 2008;14(4):957–960. [DOI] [PubMed] [Google Scholar]
- 9. Reardon DA, Wen PY, Mellinghoff IK.. Targeted molecular therapies against epidermal growth factor receptor: past experiences and challenges. Neuro Oncol. 2014;16(suppl 8): viii7–viii13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Palmirotta R, Lovero D, Cafforio P, et al. Liquid biopsy of cancer: a multimodal diagnostic tool in clinical oncology. Ther Adv Med Oncol. 2018;10:175883591879463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Niel G, D’Angelo G, Raposo G.. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–228. [DOI] [PubMed] [Google Scholar]
- 12. Huang X, Yuan T, Tschannen M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14(1):319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chi AS, Cahill DP, Reardon DA, et al. Exploring predictors of response to dacomitinib in EGFR-amplified recurrent glioblastoma. JCO Precis Oncol. 2020;4:593–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zahonero C, Aguilera P, Ramírez-Castillejo C, et al. Preclinical test of dacomitinib, an irreversible EGFR inhibitor, confirms its effectiveness for glioblastoma. Mol Cancer Ther. 2015;14(7):1548–1558. [DOI] [PubMed] [Google Scholar]
- 15. Kim H, Zheng S, Amini SS, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25(3):316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim J, Lee IH, Cho HJ, et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell. 2015;28(3):318–328. [DOI] [PubMed] [Google Scholar]
- 17. Wang J, Cazzato E, Ladewig E, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee JK, Wang J, Sa JK, et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet. 2017;49(4):594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yekula A, Yekula A, Muralidharan K, et al. Extracellular vesicles in glioblastoma tumor microenvironment. Front Immunol. 2019;10:3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nolte-’t Hoen ENM, Buermans HPJ, Waasdorp M, et al. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40(18):9272–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yuan T, Huang X, Woodcock M, et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep. 2016;6(1):19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Quek C, Bellingham SA, Jung CH, et al. Defining the purity of exosomes required for diagnostic profiling of small RNA suitable for biomarker discovery. RNA Biol. 2017;14(2):245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chakrabortty SK, Kitchen RR, Coticchia CM, et al. Abstract LB-226: exosomal liquid biopsy reveals mRNA and lincRNA biomarkers in early stage breast cancer patient plasma. Cancer Res. 2018;78(13_Supplement):LB-226. [Google Scholar]
- 24. Amorim MG, Valieris R, Drummond RD, et al. A total transcriptome profiling method for plasma-derived extracellular vesicles: applications for liquid biopsies. Sci Rep. 2017;7(1):14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wei Z, Batagov AO, Schinelli S, et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun. 2017;8(1):1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Srinivasan S, Yeri A, Cheah PS, et al. Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell. 2019;177(2):446–462.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lavacchi D, Mazzoni F, Giaccone G.. Clinical evaluation of dacomitinib for the treatment of metastatic non-small cell lung cancer (NSCLC): current perspectives. Drug Des Devel Ther. 2019;13:3187–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abdul Razak AR, Soulières D, Laurie SA, et al. A phase II trial of dacomitinib, an oral pan-human EGF receptor (HER) inhibitor, as first-line treatment in recurrent and/or metastatic squamous-cell carcinoma of the head and neck†. Ann Oncol. 2013;24(3):761–769. [DOI] [PubMed] [Google Scholar]
- 29. Noch EK, Ramakrishna R, Magge R.. Challenges in the treatment of glioblastoma: multisystem mechanisms of therapeutic resistance. World Neurosurg. 2018;116(116):505–517. [DOI] [PubMed] [Google Scholar]
- 30. Ma P, Fu Y, Chen M, et al. Adaptive and acquired resistance to EGFR inhibitors converge on the MAPK pathway. Theranostics. 2016;6(8):1232–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fang W, Huang Y, Gu W, et al. PI3K-AKT-mTOR pathway alterations in advanced NSCLC patients after progression on EGFR-TKI and clinical response to EGFR-TKI plus everolimus combination therapy. Transl Lung Cancer Res. 2020;9(4):1258–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318(5848):287–290. [DOI] [PubMed] [Google Scholar]
- 33. Etcheverry A, Aubry M, de Tayrac M, et al. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics. 2010;11(1):701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lachance G, Uniacke J, Audas TE, et al. DNMT3a epigenetic program regulates the HIF-2α oxygen-sensing pathway and the cellular response to hypoxia. Proc Natl Acad Sci U S A. 2014;111(21):7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang W, Xu J.. DNA methyltransferases and their roles in tumorigenesis. Biomarker Res. 2017;5(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for figures are provided within extended data figures. Code is available upon reasonable request.